Abstract
Calgranulin B is known to be involved in tumor development, but the underlying molecular mechanism is not clear. To gain insight into possible roles of calgranulin B, we screened for calgranulin B-interacting molecules in the SNU-484 gastric cancer and the SNU-81 colon cancer cells. Calgranulin B-interacting partners were identified by yeast two-hybrid and functional information was obtained by computational analysis. Most of the calgranulin B-interacting partners were involved in metabolic and cellular processes, and found to have molecular function of binding and catalytic activities. Interestingly, 46 molecules in the network of the calgranulin B-interacting proteins are known to be associated with cancer and FKBP2 was found to interact with calgranulin B in both SNU-484 and SNU-81 cells. Polyubiquitin-C encoded by UBC, which exhibited an interaction with calgranulin B, has been associated with various molecules of the extracellular space and plasma membrane identified in our screening, including Na-K-Cl cotransporter 1 and dystonin in SNU-484 cells, and ATPase subunit beta-1 in SNU-81 cells. Our data provide novel insight into the roles of calgranulin B of gastrointestinal cancer cells, and offer new clues suggesting calgranulin B acts as an effector molecule through which the cell can communicate with the tumor microenvironment via polyubiquitin-C.
Introduction
Calprotectin is heterotetrameric calgranulin A and B complex that were noncovalently bonded without a peptide bridge between two subunits. The amounts of calprotectin in blood or extracellular body fluids are reportedly increased under many pathological conditions, such as rheumatoid arthritis, inflammatory bowel diseases, viral infection, microbial infection, tumors, and many inflammatory conditions [1].
Various functions of calprotectin have been reported, such as stimulation of fibroblast growth and beta 2-integrin-mediated neutrophil adhesion, neutrophil chemoattraction, and macrophage deactivation [2–5]. Calprotectin is also believed to function in altering the cytoskeleton and cell shape, transducing signals, and modulating intracellular calcium.
One of the two units of calprotectin, calgranulin B, is a small calcium-binding protein that is mainly found in granulocytes, monocytes, and activated keratinocytes [6–10]. It has also emerged as a marker for non-inflammatory pathological conditions, such as tumor development. Calgranulin B is reportedly overexpressed in various tumor types, including ovarian cancer, head and neck tumors, pulmonary carcinoma, and prostate cancer [11]. In addition, it is secreted by intestinal monocytes and epithelial cells, and elevated levels of calgranulin B have been detected in stool samples from colorectal cancer patients. We previously reported that calgranulin B is a candidate fecal marker for the diagnosis of colorectal cancer [12], and more recently showed that combining the fecal occult blood test (the established means of colorectal cancer screening) with calgranulin B screening can increase the sensitivity of colorectal cancer detection [13]. However, the intracellular molecular mechanism underlying the involvement of calgranulin B in tumor development is unknown.
Here, we set out to investigate the role of calgranulin B in gastrointestinal cancer by identifying calgranulin B-interacting partners in cancer cell lines.
Materials and methods
Human cell lines
SNU-81 colorectal carcinoma cells, SNU-484 gastric carcinoma cells, and HEK293 human embryonic kidney cells were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea).
Yeast two hybrid (Y2H)
The full-length cDNA of human calgranulin B was PCR amplified and cloned into the pGBKT7 vector (containing the GAL4 DNA-binding domain). The pGBKT7-calgranulin B construct did not show any autonomous transcriptional activation or cytotoxicity following transformation into the yeast strain, Y2H Gold.
SNU-484, SNU-81, and HEK293 cells were used to construct cDNA libraries in the pGADT7-Rec vector (containing a GAL4 activation domain) using Matchmaker Library Construction and Screening kits (Clontech, Santa Clara, CA, USA). Each library was then transformed into the Y187 yeast strain, and Y2H screening was performed using the Matchmaker Two-Hybrid system (Clontech). Positive clones were selected based on their ability to grow on synthetic dropout (SD) medium/-LTH/X-α-Gal (TDO). Their cDNA inserts were PCR amplified, sequenced, and subjected to BLAST alignment. Interaction between the bait and identified prey clones was verified by co-transforming the purified prey plasmid plus the bait pGBKT7-calgranulin B construct into Y2H Gold cells, and then selecting clones on SD/-LTHA/ X-α-Gal medium. Co-transformation of pGBKT7-p53 plus pGADT7-SV40 was used as a positive control, while co-transformation of pGBKT7-p53 with empty pGADT7 vector was used as a negative control.
Gene ontology and top disease information analysis
Computational analysis was applied to all of the molecules identified as interacting with calgranulin B in the three cell lines. Gene ontology (GO) analysis of the relevant biological processes, cellular components, and molecular functions was performed using the Protein Analysis Through Evolutionary Relationships program (PANTHER, www.pantherdb.org), which refers to a curated database of protein families, functions and pathways [14,15]. GO terms assigned into identified molecules were classified according to their function.
The most highly represented diseases and disorders information was obtained from Ingenuity Pathway Analysis (IPA, www.quiagen.com/ingenuity), which determines functions of identified molecules from literature-based information.
Protein class and pathway analysis
Furthermore, the protein class levels and pathways of all identified molecules were assessed using the PANTHER pathway program, which refers to a database containing: 165 expertly curated metabolic and signaling pathways; 20,851 proteins directly associated with these pathways, along with an evidence code; and 3569 distinct literature references (http://pid.nci.nih.gov/2009/090414/full/pid.2009.1.shtml).
Molecular interaction and network analysis
IPA, which determines the interactions and functions of identified gene products based on the information available in the literature, was used to map the identified molecules with respect to their interconnections. IPA scans input molecules to identify networks using the Ingenuity Pathways Knowledge Base (IPKB) for interaction between identified molecules. Molecular interactions are shown by grey lines, the identified calgranulin B-interacting molecules are highlighted in purple, and molecules that have been associated with cancer are indicated by dotted blue lines.
STRING analysis (https://www.string-db.org) was used to find direct and indirect interactions between all identified molecules interacting with calgranulin B in the extracellular space and plasma membrane. STRING is a database which provides direct and indirect associations from computational prediction, and interactions from other databases for interaction analysis [16,17]. STRING score is calculated from the combination of all predictions and network analysis was set at a medium confidence. Eight different colored lines were used to represent the types of evidence for associations, as follows: green, neighborhood evidence; red, gene fusion; blue, co-occurrence; black, co-expression; purple, experimental; light blue, database; yellow, text mining; sky blue, protein homology.
Results
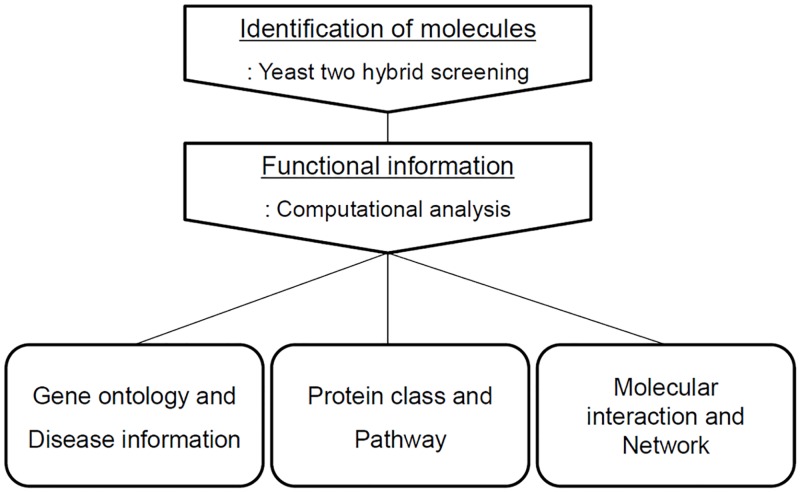
Calgranulin B-interacting molecules were successfully identified and various computational analyses draw functional information, which could provide the biological role of calgranulin B in the context of a complex molecular interaction network. The whole scheme of the experimental procedures was represented in Fig 1.
Fig 1. Overview of the experimental procedure.
A scheme of the experimental workflow used for identification and functional analysis of calgranulin B interacting molecules was illustrated.
Identification of calgranulin B-interacting partners using a yeast two-hybrid system
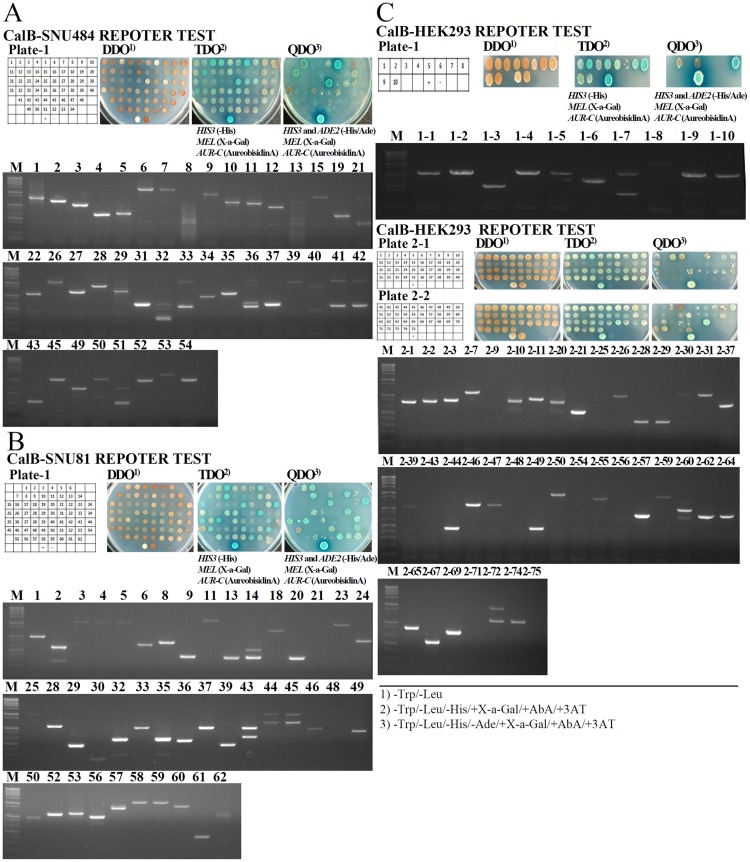
cDNA libraries were derived from the SNU-484 human gastric cancer cell line, the SNU-81 human colon cancer cell line, and the HEK293 normal human kidney cell line, and calgranulin B-interacting partners were screened using the bait plasmid, pGBKT7-calgranulin B. Interactions of bait and prey proteins were examined by assessing growth on two selective media with different levels of restrictiveness: TDO (-His/X-a-Gal/AbA/) and QDO (-His/-Ade/X-a-Gal/AbA) (Fig 2). Screening of 1.55x107 and 1.87x107 primary transformants obtained from SNU-484 and SNU-81 cells, respectively, yielded 25 and 27 positive clones, respectively (Fig 2A and 2B, upper). These clones were isolated, sequenced and aligned using the NCBI BLAST alignment search tool. We found that 14 and 13 of the clones, respectively, were in-frame; these were selected as potential candidate calgranulin B-interacting partners in gastric and colon cancers, respectively (Fig 2A and 2B, lower). We also performed two screenings in HEK293 cells, and obtained 1.76x107 and 1.41x107 primary transformants that yielded 10 and 34 positive clones, respectively. Four and 15 of the positive clones were found to be in-frame (Fig 2C), and were subjected to further analysis to reveal the biological meaning and relationship with diseases in the context of an interaction with calgranulin B. All of the selected candidate calgranulin B-interacting genes identified in the three cell lines by Y2H assay system are listed in Table 1.
Fig 2. Yeast two-hybrid screening of calgranulin B-interacting partners.
Positive interactions were identified through growth in selective media with different levels of restrictiveness. Positive clones were isolated and sequenced from the SNU-484 human gastric cancer cell line (A), the SNU-81 human colon cancer cell line (B), and the HEK293 human kidney cell line (C).
Table 1. List of identified significant genes interacting with calgranulin B.
| Cell line | ID | Description | Frame | Reporter expression | |||
|---|---|---|---|---|---|---|---|
| MEL | AUR-C | HIS3 | ADE2 | ||||
| SNU-484 | 3 | A-kinase-interacting protein 1 isoform b [Homo sapiens] | +1 | +++ | +++ | +++ | +++ |
| 4 | PREDICTED: thioredoxin, mitochondrial isoform X1 [Homo sapiens] | +1 | +++ | +++ | +++ | - | |
| 7 | pyruvate dehydrogenase protein X component, mitochondrial isoform 2 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 9 | 60S ribosomal protein L18 isoform 1 [Homo sapiens] | +1 | ++ | ++ | ++ | + | |
| 11 | ribosomal protein L11, isoform CRA_a [Homo sapiens] | +1 | + | ++ | ++ | - | |
| 12 | A-kinase-interacting protein 1 isoform b [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 28 | ran-binding protein 9 [Homo sapiens] | +1 | +++ | +++ | +++ | ++ | |
| 29 | NAG13 [Homo sapiens] | +1 | + | +++ | +++ | ++ | |
| 31 | afadin isoform X17 [Homo sapiens] | +1 | ++ | +++ | ++ | +++ | |
| 34 | peptidyl-prolyl cis-trans isomerase FKBP2 precursor [Homo sapiens] | +1 | ++ | +++ | +++ | ++ | |
| 35 | procollagen C-endopeptidase enhancer 1 precursor [Homo sapiens] | +1 | +++ | +++ | +++ | + | |
| 37 | ribosomal L1 domain-containing protein 1 [Homo sapiens] | +1 | + | +++ | +++ | ++ | |
| 39 | dystonin, isoform CRA_f [Homo sapiens] | +1 | + | ++ | ++ | - | |
| 50 | COMM domain-containing protein 1 [Homo sapiens] | +1 | ++ | ++ | ++ | - | |
| SNU-81 | 1 | succinate dehydrogenase [ubiquinone] iron-sulfur subunit, mitochondrial precursor [Homo sapiens] | +1 | +++ | +++ | +++ | +++ |
| 5 | probable tRNA pseudouridine synthase 1 [Homo sapiens] | +1 | + | ++ | ++ | - | |
| 6 | Homo sapiens chromosome 19, alternate assembly CHM1_1.1 | +1 | +++ | +++ | +++ | - | |
| 18 | NEDD4-like E3 ubiquitin-protein ligase WWP2 isoform WWP2-N [Homo sapiens] | +1 | ++ | ++ | ++ | - | |
| 24 | 60S ribosomal protein L39 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 46 | OCIA domain containing 2, isoform CRA_b [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 49 | peptidyl-prolyl cis-trans isomerase FKBP2 precursor [Homo sapiens] | +1 | ++ | ++ | ++ | ++ | |
| 50 | OCIA domain-containing protein 2 isoform 2 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 52 | integrin beta-1-binding protein 1 isoform 1 [Homo sapiens] | +1 | ++ | + | + | - | |
| 57 | PREDICTED: lysophosphatidic acid phosphatase type 6 isoform X1 [Homo sapiens] | +1 | ++ | ++ | ++ | ++ | |
| 58 | fructose-bisphosphate aldolase C [Homo sapiens] | +1 | ++ | ++ | ++ | + | |
| 60 | sodium/potassium-transporting ATPase subunit beta-1 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 62 | signal peptidase complex subunit 2 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| HEK293 | 1–2 | KIAA1109 protein | +1 | ++ | +++ | +++ | ++ |
| 1–4 | KIAA1109 protein | +1 | ++ | +++ | +++ | ++ | |
| 1–6 | splicing factor, arginine/serine-rich 2, interacting protein, isoform CRA_b | +1 | +++ | +++ | +++ | - | |
| 1–8 | zinc finger protein 737 isoform X3 | +1 | +++ | +++ | +++ | +++ | |
| 2–1 | rho GDP-dissociation inhibitor 1 isoform a [Homo sapiens] | +1 | +++ | +++ | +++ | ++ | |
| 2–3 | DNA replication licensing factor MCM7 isoform 2 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 2–4 | DNA replication licensing factor MCM7 isoform 2 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 2–7 | AT-rich interactive domain-containing protein 2 isoform X1 | +1 | ++ | +++ | +++ | +++ | |
| 2–11 | differentially expressed in FDCP 8 homolog isoform 2 [Homo sapiens] | +1 | ++ | +++ | +++ | ++ | |
| 2–28 | DHX30 protein [Homo sapiens] | +1 | +++ | +++ | +++ | + | |
| 2–31 | reticulon-4 receptor precursor [Homo sapiens] | +1 | ++ | +++ | +++ | + | |
| 2–37 | 60S ribosomal protein L12 [Homo sapiens] | +1 | + | ++ | ++ | - | |
| 2–50 | NOMO3 protein, partial [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 2–55 | ran-binding protein 10 isoform X1 [Homo sapiens] | +1 | +++ | +++ | +++ | +++ | |
| 2–57 | peroxiredoxin-6 [Homo sapiens] | +1 | ++ | ++ | ++ | + | |
| 2–59 | CEV14 [Homo sapiens] | +1 | ++ | +++ | +++ | ++ | |
| 2–60 | PREDICTED: COP9 signalosome complex subunit 1 isoform X13 [Homo sapiens] | +1 | ++ | ++ | ++ | - | |
| 2–62 | 60S ribosomal protein L12 [Homo sapiens] | +1 | ++ | ++ | ++ | ++ | |
| 2–65 | probable hydrolase PNKD isoform 3 precursor [Homo sapiens] | +1 | ++ | ++ | ++ | + | |
Gene ontology and top diseases information
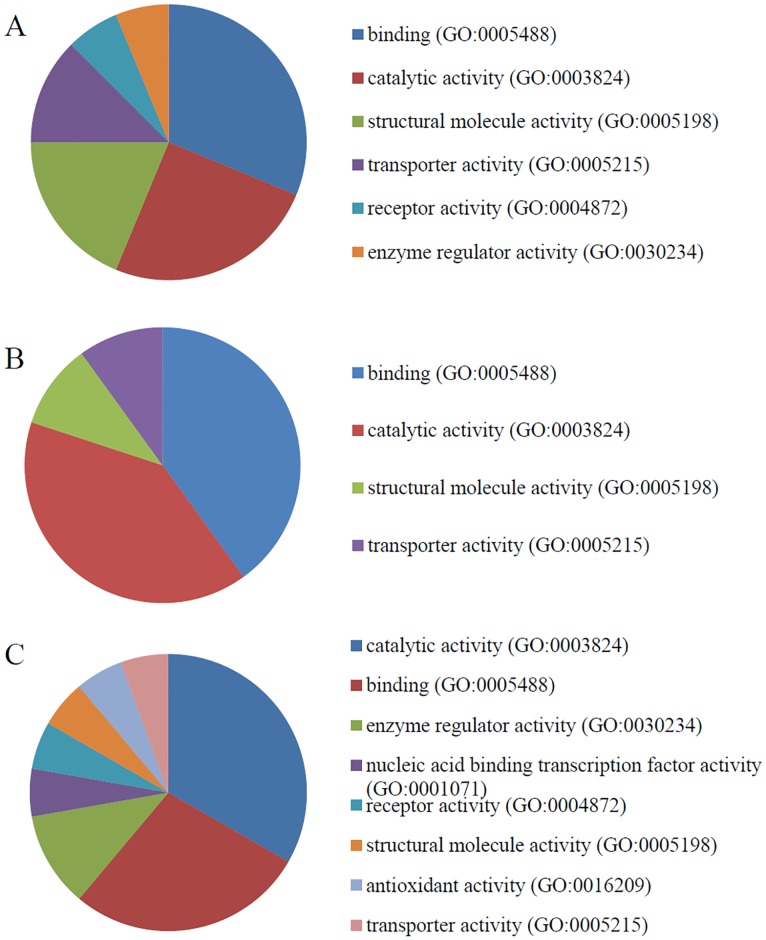
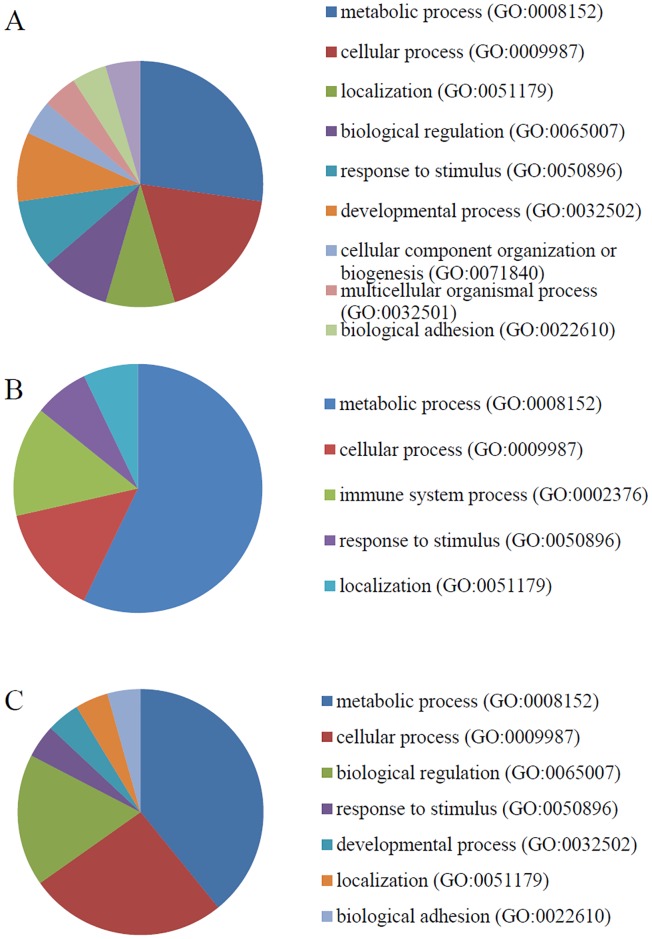
PANTHER was used to identify the GO terms associated with the molecular function, biological activity, and cellular component(s) associated with each candidate calgranulin B-interacting protein. The results of this analysis (Figs 3–5) revealed that calgranulin B appears to be associated with slightly different molecular functions, biological processes and cellular components in SNU-484, SNU-81, and HEK293 cells. However, in all three lines, binding (GO:0005488) and catalytic activity (GO:0003824) were the most highly represented molecular functions, with representations of 31.3% and 33.3% in SNU-484 and HEK293 cells, respectively and 40% for both cases in SNU-81 cells. The next most common functions were structural molecule activity (GO:0005198) in SNU-484 cells, enzyme regulation (GO:0030234) in HEK293 cells, and structural molecule activity (GO:0005198) and transportation (GO:00052158) in SNU-81 cells (Fig 3).
Fig 3. Molecular function gene ontology (GO) terms overrepresented among the calgranulin B-interacting partners.
The distributions of the molecular functions represented by the calgranulin B-interacting molecules identified in SNU-484 (A), SNU-81 (B), and HEK293 (C) cells are shown.
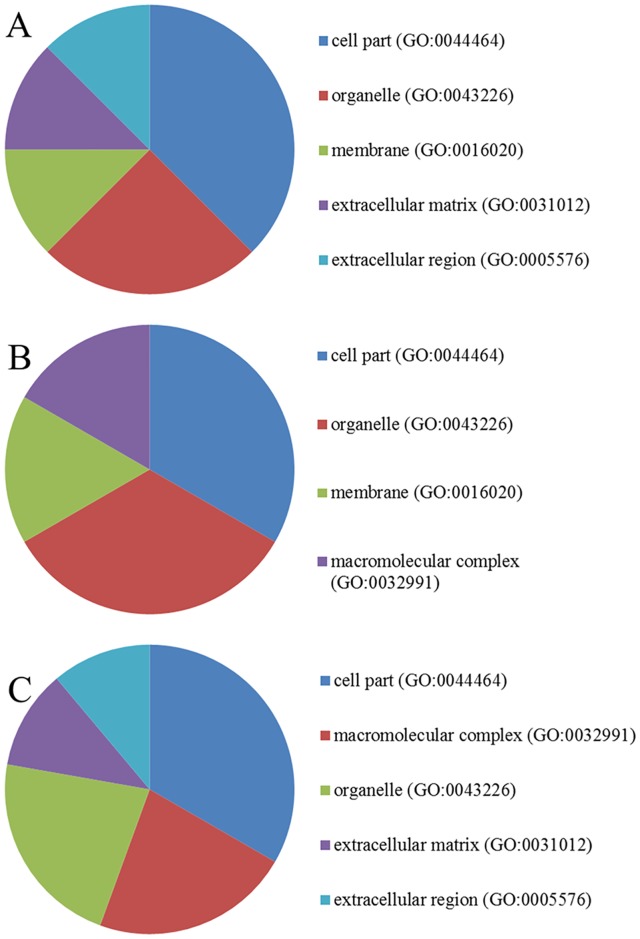
Fig 5. Cellular components associated with the calgranulin B-interacting molecules.
The calgranulin B-interacting molecules identified in SNU-484 (A), SNU-81 (B), and HEK293 (C) cells were categorized according to cellular component information.
The calgranulin B-interacting molecules were also classified according to their related biological activities/processes (i.e., recognized series of events or molecular functions; www.geneontology.org). In all three cell lines, the most highly represented biological process was metabolic process (GO:0008152), followed by cellular processes (GO:0009987) in SNU-484 and HEK293 cells and immune processes (GO:0002376) in SNU-81 cells (Fig 4).
Fig 4. Biological processes overrepresented among the calgranulin B-interacting partners.
The distributions of the biological processes represented by the calgranulin B-interacting molecules identified in SNU-484 (A), SNU-81 (B), and HEK293 (C) cells are shown.
The cellular component portion of the GO analysis was used to classify the candidate proteins by their known locations at the levels of subcellular structure and macromolecular complexes (www.geneontology.org). Our results revealed that the calgranulin B-interacting molecules were largely localized in the cell part (GO:0044464) and organelle (GO:0043226) components of SNU-484 and SNU-81 cells, while they were distributed to the cell part (GO: 0044464), macromolecular complex (GO: 0032991) and organelle (GO: 0043226) components of HEK293 cells (Fig 5).
Ingenuity Pathway Analysis (IPA) was used to identify the most highly represented diseases and disorders among the calgranulin B-interacting proteins of each cell line. Cancer was the most highly represented disease/disorder in SNU-81 and HEK293 cells, whereas cardiovascular disease was the most highly represented disease/disorder in SNU-484 cells. The full results of this analysis are presented in Table 2.
Table 2. List of diseases and disorders associated with molecules identified in three cell lines.
| Cell line | Name | p-value | # molecules |
|---|---|---|---|
| SNU-484 | Cardiovascular Disease | 6.45E-04–6.45E-04 | 1 |
| Connective Tissue Disorders | 6.45E-04–6.45E-04 | 1 | |
| Dermatological Diseases and Conditions | 6.45E-04–6.45E-04 | 1 | |
| Developmental Disorder | 6.45E-04–6.45E-04 | 2 | |
| Hematological Disease | 1.35E-02–6.45E-04 | 2 | |
| SNU-81 | Cancer | 4.91E-02–5.91E-04 | 5 |
| Connective Tissue Disorders | 1.41E-02–5.91E-04 | 3 | |
| Developmental Disorder | 1.41E-02–5.91E-04 | 2 | |
| Gastrointestinal Disease | 3.49E-03–5.91E-04 | 2 | |
| Hereditary Disorder | 1.41E-02–5.91E-04 | 2 | |
| HEK 293 | Cancer | 4.13E-02–8.60E-04 | 3 |
| Developmental Disorder | 3.47E-02–8.60E-04 | 2 | |
| Hereditary Disorder | 4.13E-02–8.60E-04 | 2 | |
| Infectious Diseases | 8.60E-02–8.60E-04 | 1 | |
| Neurological Disease | 3.55E-02–8.60E-04 | 4 |
Protein classes and pathways related to the identified calgranulin B-interacting partners
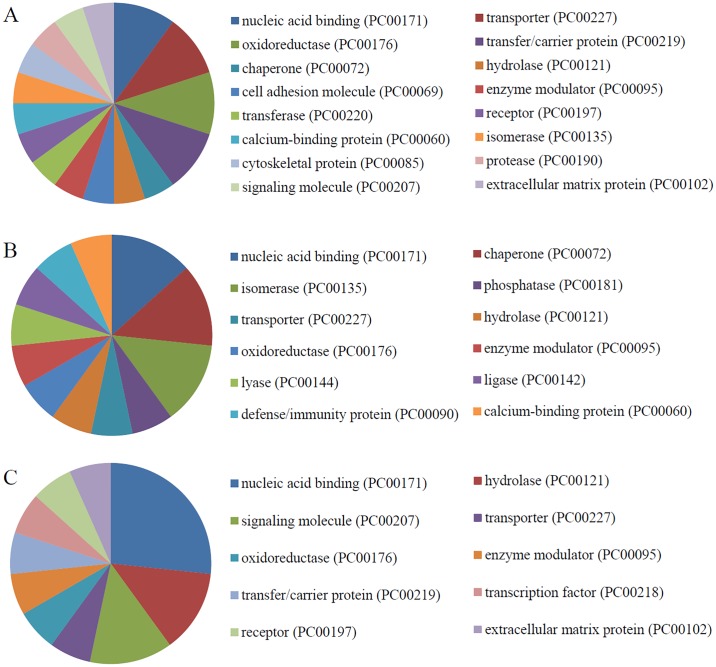
The protein class distribution in each cell line was evaluated by PANTHER analysis (Fig 6). Our results revealed that nucleic acid binding (PC00171) was the most highly represented protein class in all three cell lines, followed by: transporter (PC00227), oxidoreductase (PC00176), and transfer/carrier (PC00219) functions in SNU-484 cells (Fig 6A); chaperone (PC00072) and isomerase (PC00135) functions in SNU-81 cells (Fig 6B); and hydrolase (PC00121) and signaling (PC00207) functions in HEK293 cells (Fig 6C).
Fig 6. PANTHER analysis of protein classes overrepresented among the calgranulin B-interacting partners.
Protein class distributions of the calgranulin B-interacting molecules identified in SNU-484 (A), SNU-81 (B), and HEK293 (C) cells are shown.
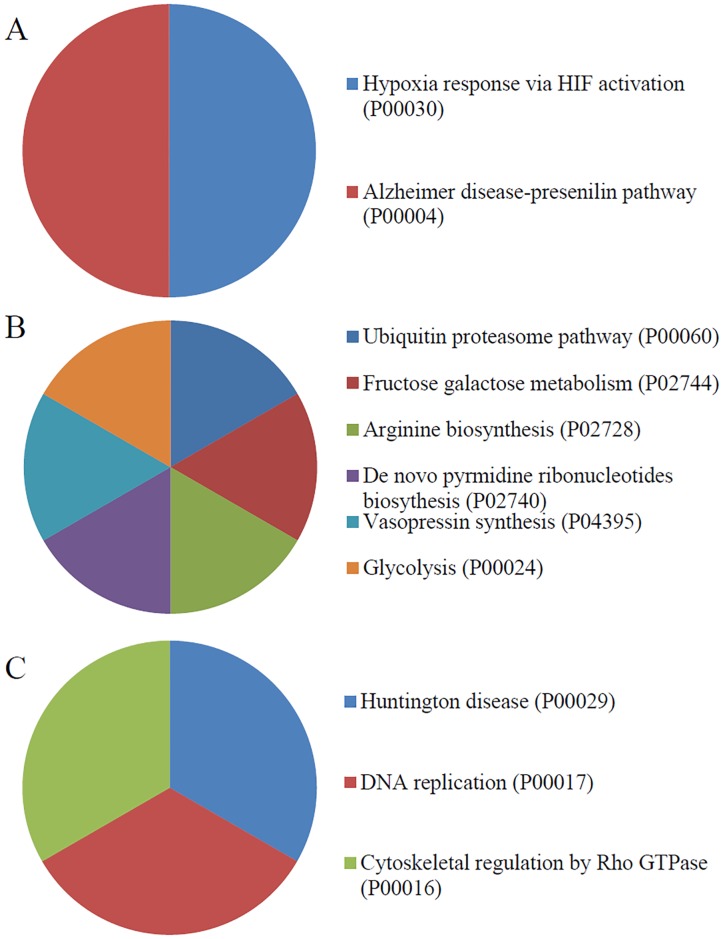
PANTHER pathway analysis was used to identify pathways that were highly represented among the calgranulin B-interacting partners identified in SNU-484, SNU-81, and HEK293 cells. Interestingly, the results differed among the three cell lines. In SNU-484 cells, the calgranulin B-interacting partners were often involved in the Alzheimer disease-presenilin pathway (P00004), the hypoxia response via HIF activation (P00030) (Fig 7A). In SNU-81 cells, the overrepresented pathways included the ubiquitin proteasome pathway (P00060), the fructose-galactose metabolism pathway (P02744), and the arginine biosynthesis pathway (P02728), the de novo pyrimidine ribonucleotides biosynthesis (P02740), Vasopressin synthesis (P04395), and Glycolysis (P00024) (Fig 7B). In HEK293 cells, the overrepresented pathways included those related to Huntington disease (P00029), DNA replication (P00017), and Cytoskeletal regulation by Rho GTPase (P00016) (Fig 7C).
Fig 7. PANTHER analysis of pathways overrepresented among the calgranulin B-interacting partners.
The pathway distributions of the calgranulin B-interacting molecules identified in SNU-484 (A), SNU-81 (B), and HEK293 (C) cells are shown.
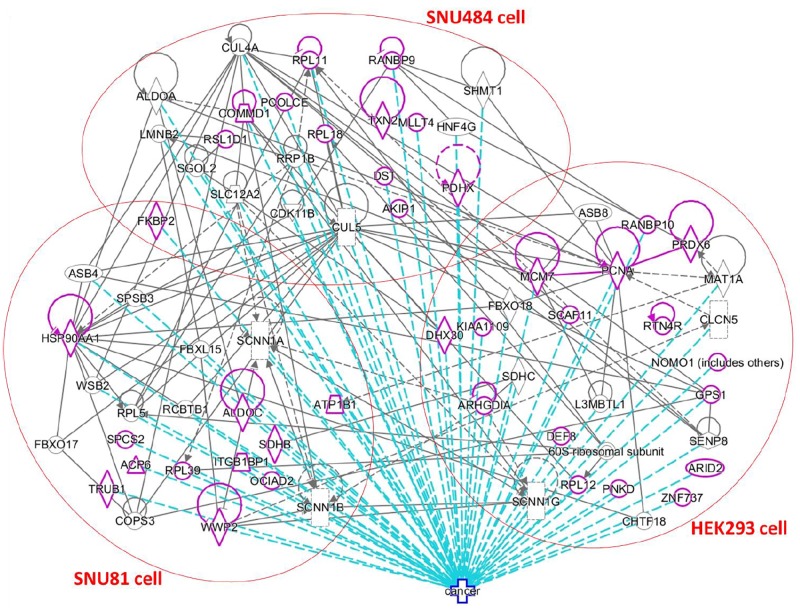
Molecular network analysis and cancer-related calgranulin B-interacting proteins
To obtain a more comprehensive view of the molecular network related to the calgranulin B-interacting molecules identified herein, we used IPA to analyze their direct and indirect molecular interactions. The results are shown in Fig 8, where molecular interactions are indicated with grey lines, the identified calgranulin B-interacting molecules are highlighted in purple, the relevant cell lines are indicated in red circles, and the 46 calgranulin B-interacting molecules that have been associated with cancer are indicated with dotted blue lines. All candidate molecules interacting with calgranulin B were unique in three cell lines except FKBP2 that was found in two cancer cell lines, SNU-484 and SNU-81. However molecular network analysis revealed they had direct and/or indirect interaction across 3 cell lines and most of them were involved in cancer disease development.
Fig 8. Network analysis and cancer associations of the calgranulin B-interacting molecules.
In the generated network, molecular interactions are shown by grey lines, the identified calgranulin B-interacting molecules are highlighted in purple, and molecules that have been associated with cancer are indicated by dotted blue lines.
Calgranulin B-interacting proteins associated with the extracellular space and plasma membrane
Among the identified calgranulin B-interacting proteins, those known to be localized to the extracellular space and plasma membrane were further characterized by network analysis to reveal interactive relationship with calgranulin B. As shown in Fig 9, calgranulin B (S100A9) was found to directly interact with polyubiquitin-C (encoded by UBC), which is known to directly interact with various other calgranulin B-interacting partners, including dystonin (encoded by DST) in SNU-484 cells, ATPase subunit beta-1 (ATP1B1, encoded by ATP1B1) in SNU-81 cells, and nodal modulator 1 (encoded by NOMO1) in HEK293 cells.
Fig 9. Communication of calgranulin B with extracellular environment via polyubiquitin-C(UBC) in SNU-484, SNU-81, and HEK293 cells.
Direct interactions with calgranulin B (S100A9) were predicted using STRING analysis, and are indicated by different colored lines: green, red, blue, black, purple, light blue, yellow, sky blue based on the types of evidence for associations.
Discussion
Yeast two-hybrid screening analysis is a molecular biology technique that enables researchers to uncover protein-protein and protein-DNA interactions by testing for physical interactions between relevant fragments. Y2H screening has been steadily used to identify new interacting partners for a protein of interest, or they may confirm binary protein-protein interactions, which play crucial roles in almost all biological processes [18]. Here, we used Y2H analysis to identify candidate calgranulin B-interacting molecules (Fig 2), and subjected these proteins to PANTHER analysis of: their predicted activities, which emphasized binding and catalytic activity (Figs 3 and 6); their cellular involvements, which emphasized metabolic and cellular processes (Fig 4); and their cellular localizations (Fig 5). Molecular function, biological activity and cellular components of identified molecules showed slight differences among three cell lines but most terms were common in all three cell lines tested. To determine further biological characteristics of calgranulin B-interacting proteins, previously reported calgranulin B-interacting molecules on the BioGRID database (http://thebiogrid.org) were analyzed by PANTHER and found to have correlation with the present data. For example, the most highly represented molecular function; cellular component, biological process, and protein class of the previously reported 92 molecules are same with our present study as binding, cell part, metabolic process, and nucleic acid binding, respectively (data not shown).
However, signaling pathways involved calgranulin B interacting partners were totally different in each cell line (Fig 7) providing an involvement of unique signaling cascades. Ubiquitin proteasome pathway represented in SNU-81 colon cell is very interesting considering internalization pathway of calgranulin B in colon cancer cells. Ubiquitination, one of the post-translational modifications of proteins can lead to targeting for degradation by the proteasome machinery as well as can serve as a sorting signal for regulation of internalization such as activation of receptor endocytosis [19,20]. In addition, ubiquitination of integral plasma membrane proteins triggers their rapid internalization into endocytic pathway [21]. Recently we found that calgranulin B released from immune cells can be internalized specifically into colon cancer cells and suppressed cell proliferation [22] but the molecular mechanism of internalization pathway was not clear. Ubiquitin proteasome pathway proposed in this study could provide a possible mechanism of calgranulin B internalization in colon cancer cells.
Among the calgranulin B-interacting molecules known to be involved in cancer (Fig 8, purple), FKBP2 (FK506-binding protein 2) was identified from both SNU-484 and SNU-81 cells. FKBP2, which is a member of the immunophilin protein family, contributes to immunoregulation and is thought to function as an ER chaperone and a component of membrane cytoskeletal scaffolding [23]. The FKBP2 gene has been associated with pathological osteoporosis [24], and its protein product was suggested to be associated with the development of type 2 diabetes [25]. Northern blotting revealed that FKBP2 is expressed in tissues that are predisposed to hyperplasia in multiple endocrine neoplasia type 1 (MEN1) patients; however, mutation analysis of MEN1 kindreds and sporadic tumors excluded FKBP2 as a candidate gene for MEN1 [26]. No previous report has suggested any molecular cooperation between FKBP2 and calgranulin B. Here, we show for the first time that calgranulin B may have FKBP2-mediated function in gastrointestinal cancer cells.
No previous study has exhibited a direct link between polyubiquitin-C (encoded by UBC) and cancer. However, many studies have shown that polyubiquitin-C interacts with various cancer-associated effector molecules, including cdk1 [27], E2F1 [28], epidermal growth factor receptor (EGFR) [29], HDAC3 [27], HIF1A [30], Mdm2 [31], NOTCH1 [32], and p53 [29]. Our present results with previous proteome analysis reveal that polyubiquitin-C interacts with calgranulin B [33–35], suggesting that polyubiquitin-C may act as an important mediator that connects calgranulin B to various proteins in the extracellular space and plasma membrane, such as Na-K-Cl cotransporter 1 (NKCC1, encoded by SLC12A2) and dystonin (encoded by DST) in SNU-484 cells, and ATPase subunit beta-1 (ATP1B1, encoded by ATP1B1) in SNU-81 cells (Fig 9). These interactions with polyubiquitin-C have been reported in the previous studies [34,36–39], which suggest that calgranulin B may communicate with the tumor microenvironment via polyubiquitin-C.
NKCC1 is a membrane-transport protein that governs the active bi-directional transport of sodium, potassium, and chloride [40]. NKCC1 expression has been shown to predict poor prognosis in lung adenocarcinoma [41] and affect the G2/M checkpoint in esophageal squamous cell carcinoma [42]. In glioma cells, inhibition of NKCC1 was found to reduce invasion [43,44] and augment temozolomide-induced apoptosis [45]. Blockade of NKCC1 function also diminished the proliferation of poorly differentiated human gastric cancer cells by affecting G0/G1 phase [42]. It is not yet known whether the interaction between calgranulin B and NKCC1 blocks or augments the function of NKCC1, but studies showing that calgranulin B and calprotectin exert pro-apoptotic effects [46–48] may suggest that calgranulin B inhibits the activity of NKCC1 in gastric cancer cells.
Dystonin, which is a neural isoform of bullous pemphigoid antigen 1, has an N-terminal actin-binding domain and is essential for maintaining the cytoskeletal integrity of neurons [49]. No previous study has linked dystonin to gastric cancer. Further work is needed to clarify the potential for calgranulin B to communicate with the extracellular environment via dystonin.
Among the identified proteins, ATPase subunit beta-1 is unique in that it may interact with calgranulin B to facilitate colon cancer progression (Fig 8) and/or communicate directly with polyubiquitin-C (Fig 9). ATPase is a well-known membrane protein that is responsible for maintaining the electrochemical gradients of Na and K ions across the plasma membrane. It can also be a target of TGF-beta1 during TGF-beta1-induced epithelial-to-mesenchymal transition (EMT) [50], and participate in the drug sensitization of cancer cells [51]. In contrast to the interaction between calgranulin B and NKCC1 we do not expect to see a calgranulin B-mediated change in ATPase activity. However, our overall data demonstrate that ATPase subunit beta-1 is a most promising candidate for future efforts to study the potential function of calgranulin B in colon cancer cells.
In sum, we herein screened for calgranulin B-interacting partners and found that they included both cytosolic and plasma membrane proteins. The molecular linkages identified herein suggest that calgranulin B may have multiple anti-tumor functions acting against the progression of gastrointestinal cancer. Our novel findings suggest that further functional studies are warranted to investigate the signaling downstream of the encounters between calgranulin B and its interacting partners.
Acknowledgments
We thank Dr. Seho Cha for analysis, processing, and interpretation of the data and helpful discussion in revising the article.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by grants from the National Cancer Center, Korea (NCC-1410870-3) and the Soonchunhyang University Research Fund.
References
- 1.Nakatani Y, Yamazaki M, Chazin WJ, Yui S. Regulation of S100A8/A9 (calprotectin) binding to tumor cells by zinc ion and its implication for apoptosis-inducing activity. Mediators Inflamm. 2005: 280–292. 10.1155/MI.2005.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguiar-Passeti T, Postol E, Sorg C, Mariano M. Epithelioid cells from foreign-body granuloma selectively express the calcium-binding protein MRP-14, a novel down-regulatory molecule of macrophage activation. J Leukoc Biol. 1997;62: 852–858. [DOI] [PubMed] [Google Scholar]
- 3.Ryckman C, McColl SR, Vandal K, de Medicis R, Lussier A, Poubelle PE, et al. Role of S100A8 and S100A9 in neutrophil recruitment in response to monosodium urate monohydrate crystals in the air-pouch model of acute gouty arthritis. Arthritis Rheum. 2003;48: 2310–2320. 10.1002/art.11079 [DOI] [PubMed] [Google Scholar]
- 4.Shibata F, Miyama K, Shinoda F, Mizumoto J, Takano K, Nakagawa H. Fibroblast growth-stimulating activity of S100A9 (MRP-14). Eur J Biochem. 2004;271: 2137–2143. 10.1111/j.1432-1033.2004.04129.x [DOI] [PubMed] [Google Scholar]
- 5.Newton RA, Hogg N. The human S100 protein MRP-14 is a novel activator of the beta 2 integrin Mac-1 on neutrophils. J Immunol. 1998;160: 1427–1435. [PubMed] [Google Scholar]
- 6.Roth J, Teigelkamp S, Wilke M, Grun L, Tummler B, Sorg C. Complex pattern of the myelo-monocytic differentiation antigens MRP8 and MRP14 during chronic airway inflammation. Immunobiology. 1992;186: 304–314. 10.1016/S0171-2985(11)80259-7 [DOI] [PubMed] [Google Scholar]
- 7.Lagasse E, Clerc RG. Cloning and expression of two human genes encoding calcium-binding proteins that are regulated during myeloid differentiation. Mol Cell Biol. 1988;8: 2402–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz E, Muhlebach MS, Tessier PA, Alexis NE, Duncan Hite R, Seeds MC, et al. Different expression ratio of S100A8/A9 and S100A12 in acute and chronic lung diseases. Respir Med. 2008;102: 567–573. 10.1016/j.rmed.2007.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J Leukoc Biol. 2007;81: 28–37. 10.1189/jlb.0306170 [DOI] [PubMed] [Google Scholar]
- 10.Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. 2003;170: 3233–3242. [DOI] [PubMed] [Google Scholar]
- 11.Kapoor S. Beyond interstitial lung disease: the rapidly evolving role of Calgranulin B as a biomarker of systemic malignancies. Inflammation. 2009;32: 70-009-9104-8. [DOI] [PubMed] [Google Scholar]
- 12.Yoo BC, Shin YK, Lim SB, Hong SH, Jeong SY, Park JG. Evaluation of calgranulin B in stools from the patients with colorectal cancer. Dis Colon Rectum. 2008;51: 1703–1709. 10.1007/s10350-008-9381-6 [DOI] [PubMed] [Google Scholar]
- 13.Kim BC, Joo J, Chang HJ, Yeo HY, Yoo BC, Park B, et al. A predictive model combining fecal calgranulin B and fecal occult blood tests can improve the diagnosis of colorectal cancer. PLoS One. 2014;9: e106182 10.1371/journal.pone.0106182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8: 1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35: D247–52. 10.1093/nar/gkl869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43: D447–52. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28: 3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagemans J, Lavigne R. Identification of protein-protein interactions by standard gal4p-based yeast two-hybrid screening. Methods Mol Biol. 2015;1278: 409–431. 10.1007/978-1-4939-2425-7_27 [DOI] [PubMed] [Google Scholar]
- 19.Bulut GB, Sulahian R, Ma Y, Chi NW, Huang LJ. Ubiquitination regulates the internalization, endolysosomal sorting, and signaling of the erythropoietin receptor. J Biol Chem. 2011;286: 6449–6457. 10.1074/jbc.M110.186890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piper RC, Dikic I, Lukacs GL. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb Perspect Biol. 2014;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 1997;11: 1215–1226. [DOI] [PubMed] [Google Scholar]
- 22.Kim K, Kim KH, Roh K, Yoo BC, Ku JL, Shin YK, et al. Antitumor effects of calgranulin B internalized in human colon cancer cells. Oncotarget. 2016;7: 20368–20380. 10.18632/oncotarget.7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin YJ, Albers MW, Lane WS, Bierer BE, Schreiber SL, Burakoff SJ. Molecular cloning of a membrane-associated human FK506- and rapamycin-binding protein, FKBP-13. Proc Natl Acad Sci U S A. 1991;88: 6677–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borsy A, Podani J, Steger V, Balla B, Horvath A, Kosa JP, et al. Identifying novel genes involved in both deer physiological and human pathological osteoporosis. Mol Genet Genomics. 2009;281: 301–313. 10.1007/s00438-008-0413-7 [DOI] [PubMed] [Google Scholar]
- 25.Lu H, Yang Y, Allister EM, Wijesekara N, Wheeler MB. The identification of potential factors associated with the development of type 2 diabetes: a quantitative proteomics approach. Mol Cell Proteomics. 2008;7: 1434–1451. 10.1074/mcp.M700478-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimmond S, Weber G, Larsson C, Walters M, Teh B, Shepherd J, et al. Exclusion of the 13-kDa rapamycin binding protein gene (FKBP2) as a candidate gene for multiple endocrine neoplasia type 1. Hum Genet. 1995;95: 455–458. [DOI] [PubMed] [Google Scholar]
- 27.Tan F, Lu L, Cai Y, Wang J, Xie Y, Wang L, et al. Proteomic analysis of ubiquitinated proteins in normal hepatocyte cell line Chang liver cells. Proteomics. 2008;8: 2885–2896. 10.1002/pmic.200700887 [DOI] [PubMed] [Google Scholar]
- 28.Zhou F, Zhang L, Wang A, Song B, Gong K, Zhang L, et al. The association of GSK3 beta with E2F1 facilitates nerve growth factor-induced neural cell differentiation. J Biol Chem. 2008;283: 14506–14515. 10.1074/jbc.M706136200 [DOI] [PubMed] [Google Scholar]
- 29.Sehat B, Andersson S, Girnita L, Larsson O. Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis. Cancer Res. 2008;68: 5669–5677. 10.1158/0008-5472.CAN-07-6364 [DOI] [PubMed] [Google Scholar]
- 30.Andre H, Pereira TS. Identification of an alternative mechanism of degradation of the hypoxia-inducible factor-1alpha. J Biol Chem. 2008;283: 29375–29384. 10.1074/jbc.M805919200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanchuk SM, Mondal S, Rutka JT. p14ARF interacts with DAXX: effects on HDM2 and p53. Cell Cycle. 2008;7: 1836–1850. 10.4161/cc.7.12.6025 [DOI] [PubMed] [Google Scholar]
- 32.Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3: e2735 10.1371/journal.pone.0002735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, et al. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci U S A. 2007;104: 12011–12016. 10.1073/pnas.0704849104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danielsen JM, Sylvestersen KB, Bekker-Jensen S, Szklarczyk D, Poulsen JW, Horn H, et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol Cell Proteomics. 2011;10: M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Large-scale analysis of the human ubiquitin-related proteome. Proteomics. 2005;5: 4145–4151. 10.1002/pmic.200401280 [DOI] [PubMed] [Google Scholar]
- 36.Lee KA, Hammerle LP, Andrews PS, Stokes MP, Mustelin T, Silva JC, et al. Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J Biol Chem. 2011;286: 41530–41538. 10.1074/jbc.M111.248856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimura SH, Iwasaka S, Schwarz W, Takeyasu K. Fast degradation of the auxiliary subunit of Na+/K+-ATPase in the plasma membrane of HeLa cells. J Cell Sci. 2008;121: 2159–2168. 10.1242/jcs.022905 [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Yang J, Li S, Zhang J, Zheng J, Hou W, et al. N-myc downstream-regulated gene 2, a novel estrogen-targeted gene, is involved in the regulation of Na+/K+-ATPase. J Biol Chem. 2011;286: 32289–32299. 10.1074/jbc.M111.247825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stes E, Laga M, Walton A, Samyn N, Timmerman E, De Smet I, et al. A COFRADIC protocol to study protein ubiquitination. J Proteome Res. 2014;13: 3107–3113. 10.1021/pr4012443 [DOI] [PubMed] [Google Scholar]
- 40.Haas M. The Na-K-Cl cotransporters. Am J Physiol. 1994;267: C869–85. [DOI] [PubMed] [Google Scholar]
- 41.Sun PL, Jin Y, Park SY, Kim H, Park E, Jheon S, et al. Expression of Na+-K+-2Cl- cotransporter isoform 1 (NKCC1) predicts poor prognosis in lung adenocarcinoma and EGFR-mutated adenocarcinoma patients. QJM. 2015. [DOI] [PubMed] [Google Scholar]
- 42.Shiozaki A, Nako Y, Ichikawa D, Konishi H, Komatsu S, Kubota T, et al. Role of the Na (+)/K (+)/2Cl(-) cotransporter NKCC1 in cell cycle progression in human esophageal squamous cell carcinoma. World J Gastroenterol. 2014;20: 6844–6859. 10.3748/wjg.v20.i22.6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haas BR, Sontheimer H. Inhibition of the Sodium-Potassium-Chloride Cotransporter Isoform-1 reduces glioma invasion. Cancer Res. 2010;70: 5597–5606. 10.1158/0008-5472.CAN-09-4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garzon-Muvdi T, Schiapparelli P, ap Rhys C, Guerrero-Cazares H, Smith C, Kim DH, et al. Regulation of brain tumor dispersal by NKCC1 through a novel role in focal adhesion regulation. PLoS Biol. 2012;10: e1001320 10.1371/journal.pbio.1001320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Algharabil J, Kintner DB, Wang Q, Begum G, Clark PA, Yang SS, et al. Inhibition of Na(+)-K(+)-2Cl(-) cotransporter isoform 1 accelerates temozolomide-mediated apoptosis in glioblastoma cancer cells. Cell Physiol Biochem. 2012;30: 33–48. 10.1159/000339047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li C, Chen H, Ding F, Zhang Y, Luo A, Wang M, et al. A novel p53 target gene, S100A9, induces p53-dependent cellular apoptosis and mediates the p53 apoptosis pathway. Biochem J. 2009;422: 363–372. 10.1042/BJ20090465 [DOI] [PubMed] [Google Scholar]
- 47.Ghavami S, Eshragi M, Ande SR, Chazin WJ, Klonisch T, Halayko AJ, et al. S100A8/A9 induces autophagy and apoptosis via ROS-mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res. 2010;20: 314–331. 10.1038/cr.2009.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin F, Song Y, Li Z, Zhao L, Zhang Y, Geng L. S100A8/A9 induces apoptosis and inhibits metastasis of CasKi human cervical cancer cells. Pathol Oncol Res. 2010;16: 353–360. 10.1007/s12253-009-9225-2 [DOI] [PubMed] [Google Scholar]
- 49.Dalpe G, Leclerc N, Vallee A, Messer A, Mathieu M, De Repentigny Y, et al. Dystonin is essential for maintaining neuronal cytoskeleton organization. Mol Cell Neurosci. 1998;10: 243–257. [DOI] [PubMed] [Google Scholar]
- 50.Rajasekaran SA, Huynh TP, Wolle DG, Espineda CE, Inge LJ, Skay A, et al. Na,K-ATPase subunits as markers for epithelial-mesenchymal transition in cancer and fibrosis. Mol Cancer Ther. 2010;9: 1515–1524. 10.1158/1535-7163.MCT-09-0832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tummala R, Wolle D, Barwe SP, Sampson VB, Rajasekaran AK, Pendyala L. Expression of Na,K-ATPase-beta(1) subunit increases uptake and sensitizes carcinoma cells to oxaliplatin. Cancer Chemother Pharmacol. 2009;64: 1187–1194. 10.1007/s00280-009-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.