Abstract
Objectives
To identify SNPs associated with switching from an ACE-inhibitor to an angiotensin receptor blocker (ARB).
Methods
Two cohorts of patients starting ACE-inhibitors were identified within the Rotterdam Study in the Netherlands and the GoDARTS study in Scotland. Cases were intolerant subjects who switched from an ACE-inhibitor to an ARB, controls were subjects who used ACE-inhibitors continuously for at least 2 years and did not switch. GWAS using an additive model was run in these sets and results were meta-analysed using GWAMA.
Results
972 cases out of 5 161 ACE-inhibitor starters were identified. 8 SNPs within 4 genes reached the GWAS significance level (P<5×10-8) in the meta-analysis (RBFOX3, GABRG2, SH2B1 and MBOAT1). The strongest associated SNP was located in an intron of RBFOX3, which contains a RNA binding protein (rs2061538: MAF=0.16, OR=1.52[95%CI: 1.32-1.76], p=6.2x10-9).
Conclusions
These results indicate that genetic variation in abovementioned genes may increase the risk of ACE-inhibitors induced adverse reactions.
Keywords: ACE inhibitors, ACE-inhibitor intolerance, adverse drug reaction, cough, angioedema, Genome Wide Association Study
Introduction
Angiotensin converting enzyme inhibitors (ACE-inhibitors) are one of the most frequently prescribed groups of medications for the management of high blood pressure, heart failure and renal disease [1]. While ACE-inhibitors are generally prescribed for lifetime treatment, a cohort study showed that 32.4% of patients halted their medication likely due to adverse drug reactions (ADRs) within a median 336 days follow up time [2]. The most common ACE-inhibitor induced ADR is a persistent, dry cough and the most severe one is life threatening angioedema of lips, tongue and upper airway [3]. There is evidence suggesting genetic predisposition to these ADRs; ACE-inhibitor induced cough occurs with higher incidence in East Asian patients (23%) compared with Caucasians (5–11%) [4, 5]. The ACE-inhibitor induced angioedema rate is higher in black patients than in white patients and angioedema patients often have affected relatives [6, 7].
The mechanism of ACE-inhibitor induced cough and angioedema is not completely understood. ACE-inhibitors inhibit Angiotensin I Converting Enzyme (ACE) that cleaves several target proteins including angiotensin I and pro-inflammatory kinins. The blood pressure modification takes place through angiotensin I [8]. Accumulation of these inflammatory kinins is hypothesized to be the main reason of ACE-inhibitor induced angioedema and cough [9, 10]. For two decades, multiple candidate genes studies have tested the associations between ACE-inhibitor induced cough and genetic variation in ACE and bradykinin pathways, of which the insertion-deletion (I/D) variation in the ACE gene has been investigated most frequently [11–14]. A meta-analysis of 12 such studies, did not find a statistically significant association for the ACE I/D polymorphism [15]. Studies on ACE-inhibitor induced angioedema have also been conducted with the same approach; 3 of them found a statistically significant association between ACE-inhibitor induced angioedema and single nucleotide polymorphisms (SNPs) in the XPNPEP2 gene [16–18]. One study showed that the bradykinin receptor2 (B2) -9/+9 polymorphism is associated with both ACE-inhibitor induced cough and angioedema [19]. However generally, most of the candidate gene approach studies have been difficult to replicate and their results should be interpreted with caution [20]. The only genome wide association study (GWAS) on 175 ACE-inhibitor induced angioedema cases and 489 controls that also used ACE-inhibitors, found no genome-wide association, which might be due to the small sample size [21]. For ACE-inhibitor induced cough, the only GWAS with 1 595 cases and 5 485 controls identified genome-wide significant associations in KCNIP4 gene at chromosome 4 (rs145489027, p=1.0x10-8) which was replicated in 2 independent populations [22].
Based on the probable similar mechanism of ACE-inhibitor induced ADRs (cough and angioedema), this study aims to use a GWAS approach to identify SNPs associated with intolerance of ACE-inhibitors defined as switching of an ACE-inhibitor to an angiotensin receptor blocker (ARB) as a marker for ADRs [23].
Methods
Study population
This study was performed in 2 separate European populations:
-
A)
The Rotterdam study in the Netherlands has been described in detail previously [24, 25]. In summary, it is an ongoing cohort, composed of three different sub-cohorts (RS1, RS2, and RS3), started in 1990 in Ommoord a suburb of Rotterdam that has included 14 926 subjects aged 45 years or older (72.0 % of 20 744 eligible invited people). The Rotterdam Study has been approved by the medical ethics committee according to the Wet Bevolkingsonderzoek: ERGO (Population Study Act: Rotterdam Study), executed by the Ministry of Health, Welfare and Sports of the Netherlands. All participants gave informed consent to participate in the study and to obtain information from treating physicians and pharmacies, separately.
-
B)
The (Go-DARTS study) which is a genetic sub-study of The Diabetes Audit and Research Tayside, Scotland (DARTS) that has been described and validated in previous publications [26]. In summary, this project was based on linking clinical records by a patient-specific identifier, allowing the creation and maintenance of sophisticated regional health informatics systems. The DARTS project electronically followed all residents in Tayside, since January 1996 (n=391 274 including 7 596 individuals with diabetes) through linking the clinical datasets with a high degree of reliability and accuracy. Collection and analysis of data in DARTS and Go-DARTS was approved by the East of Scotland Research and Ethics Committee, in compliance with the declaration of Helsinki.
Phenotype
For both study populations similar phenotype definitions were applied for cases and control selection:
Cases: Patients who switched to an ARB during ACEI treatment.
Controls: Patients, who started ACE-inhibitors, and continued treatment for at least 2 years. They did not discontinue or switch their ACE-inhibitors during the follow up.
For defining continuation, discontinuation or switching, a maximum of 6 months gap between 2 prescription periods was considered. These definitions were validated in our previous study as the best marker of ACE-inhibitor induced ADRs within the prescription databases [23].
Genotyping
Within the Rotterdam study a total of 12 453 subjects were genotyped with Illumina 500(+duo) and Illumina 610 quad and 11 496 subjects passed genotyping quality control. Exclusion criteria for SNPs were a call rate <98%, Hardy-Weinberg p-value <1 × 10-6, minor allele frequency <0.01%, excess autosomal heterozygosity >0.336, sex mismatch and outlying identity-by-state clustering estimates. Data was imputed with the 1000-Genomes reference panel (phase 1, version 3) using MACH version 1.0.15/1.0.16.
Within the Go-DARTS study, subjects were genotyped on the Affymetrix 6.0 (Affymetrix, Santa Clara, CA, USA) or Illumina HumanOmniExpress (Illumina, San Diego, CA, USA) platforms. Both platforms were imputed using IMPUTE2 and the 1000 Genomes reference panel [27]. Subjects were excluded if they fulfilled any of the following criteria: SNPs call rate < 95%, sample call rate < 95%, outliers identified by IBS clustering analysis and gender discordant individuals. SNPs deviating from Hardy–Weinberg equation (P<1 × 10-6) or with an Info Score <0.4 were excluded.
Data analyses
The primary single SNP tests of association were performed using logistic regression assuming an additive genetic model, adjusting for age and gender. PLINK v1.07 was used for the Dutch cohort [28] and SNPTEST-v2.5-beta was used for the Scottish cohort [29]. The fixed effect meta-analyses were done at both sites using the inverse variance weighting, in the Netherlands using METAL and Scotland using GWAMA [30, 31]. The final SNP list in the Netherlands analysis was filtered based on the index of heterogeneity (I2 <60) and the number of cohorts that covered a SNP (more than two cohorts) [32]. The final values presented in this study are from the analyses in Scotland because GWAMA provides the odds ratios and does not require further calculations; however the consistency of the results at both sites was considered for the most significantly associated SNPs. Data of SNPs around the most significant gene were visualized using LocusZoom [33]. All other analyses were performed using SAS v9.3 (SAS Institute, Cary, NC, USA). R packages were used to plot the graphs. Metafor R package used for forest plot [34] and qqman package for Manhattan and QQ plot [35].
Results
A total of 710 cases of ACE-inhibitor intolerant patients and 3 599 tolerant controls in the Genetics of Diabetes Audit and Research in Tayside Scotland (Go-DARTS) population and 262 cases and 590 controls in the population of the Rotterdam study were analysed separately and subsequently meta-analysed. 2004 patients from Go-DARTS population were genotyped using the Illumina chip (GD1) and the rest (2305 patients) were genotyped using the Affymetrix chip (GD2). Three sub populations within Rotterdam study, RS1, RS2 and RS3 had 630, 170 and 52 patients respectively). In both cohorts the mean age of included patients was not statistically significantly different between cases and controls. The proportion of females was significantly higher within cases compared to controls in both cohorts (Table 1).
Table 1.
General characteristics of the included ACE-inhibitors starters
| GoDARTS | Rotterdam study | ||||||
|---|---|---|---|---|---|---|---|
| Case (n=710) | Control (n=3599) | P-value | Case (n=262) | Control (n=590) | P-value | ||
| gender | Male | 51.4% | 59.8% | <0.001 | 33.59% | 53.2% | <0.001 |
| Female | 48.6% | 40.2% | 66.41% | 46.8% | |||
| Mean age years [SD] | 62.77 [9.98] | 62.45 [10.84] | 0.4631 | 64.47 [6.79] | 65.15 [7.69] | 0.2177 | |
SD: standard deviation
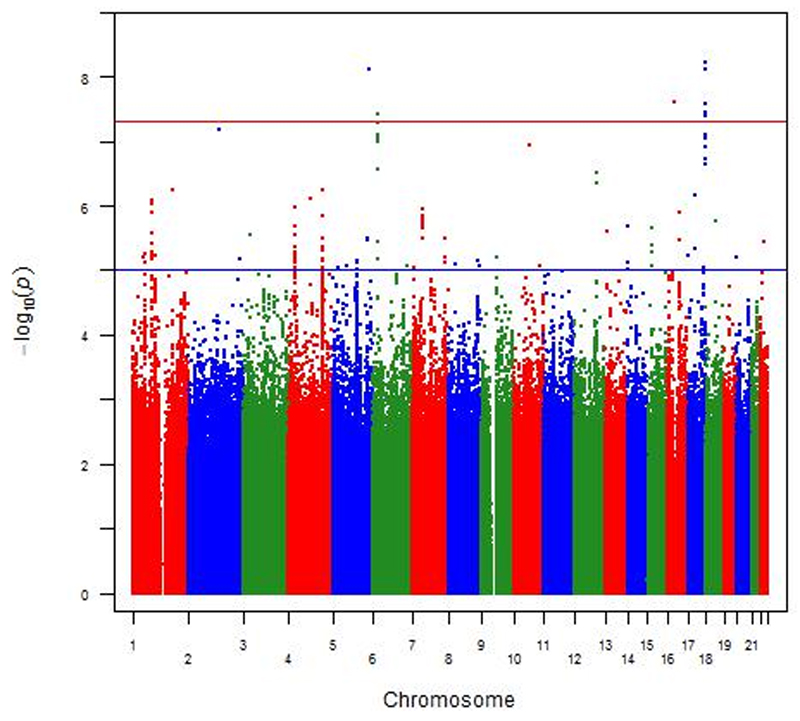
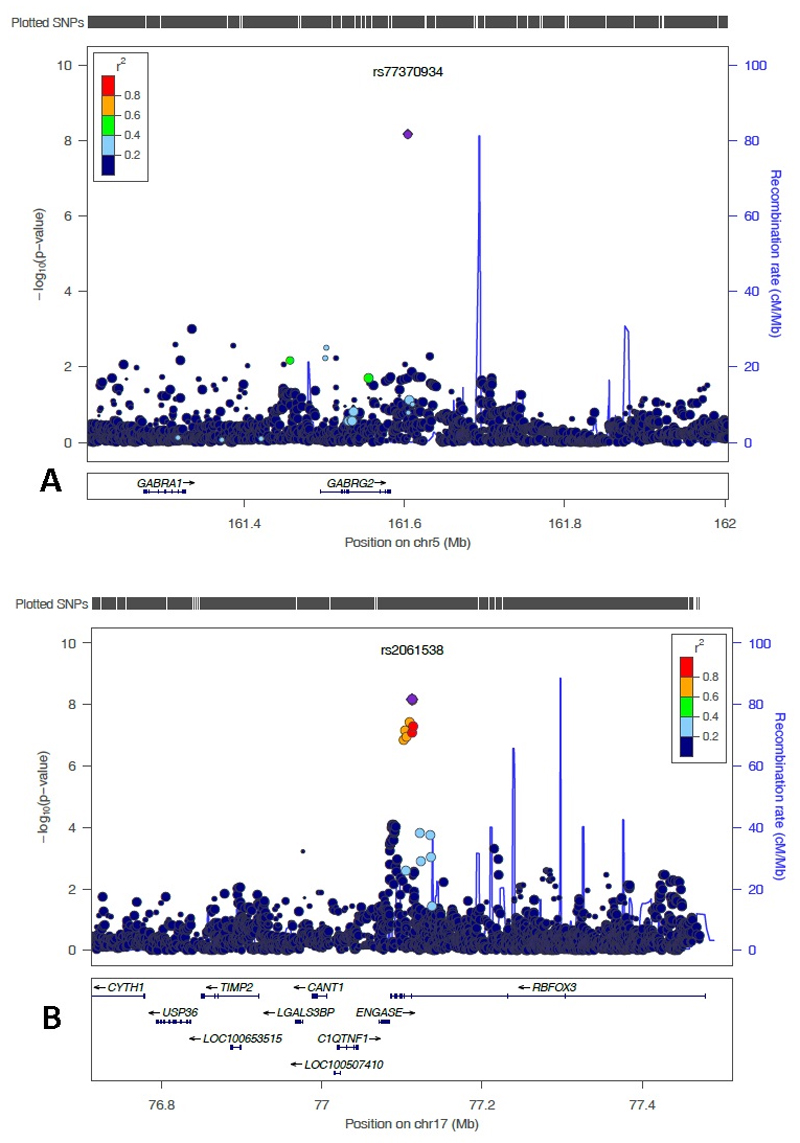
In the meta-analysis of both cohorts using multivariable regression analyses adjusting for gender and age, 8 SNPs located on chromosome 5 (one SNP), 6 (one SNP), 16 (one SNP) and 17 (five SNPs) reached genome-wide significance level (P-value less than 5x10-08) (Figure 1 and 2). Table 2 shows the details of the most statistically significantly associated SNPs. From these SNPs, two were only available in the Go-DARTS population (rs192613545 and the insertion/deletion polymorphism on chromosome 17 position 77112502). A List of the most significantly associated SNPs which reached P-value of less than 10-05 in meta-analysis, is available in the supplement in Table 1. The most significantly associated SNP (rs2061538) was located within the gene RBFOX3 (RNA Binding Protein, Fox-1 Homolog (C. Elegans) 3). There were several other strongly associated SNPs in high linkage disequilibrium (LD) with this SNP in that region (Figure 3A). The second most statistically significant SNP (rs77370934) was located within the gene GABRG2 (Gamma-Aminobutyric Acid Receptor Subunit Gamma-2), however, there were no other SNPs with a high level of LD in that locus (Figure 3B).
Figure 1.
Manhattan plot of genotyped SNPs associated with ace-inhibitor intolerance using an additive model adjusted for age and gender. The red line indicates the genome-wide significance threshold of alpha=5x10-8.
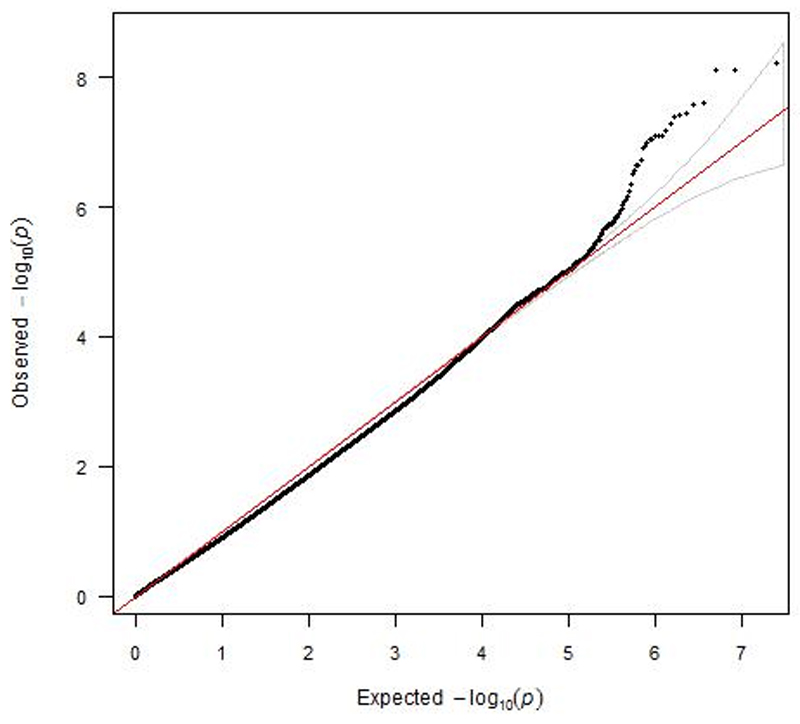
Figure 2.
A QQ plot for SNP associations from a meta-analysis of GWAS of ACE-inhibitor intolerance using an additive model adjusted for age and gender. (Lambda=0.88)
Table 2.
Most significantly associated SNPs
| SNP | Chr | Position | MA | MAF | OR | 95% CI | P-value | Gene |
|---|---|---|---|---|---|---|---|---|
| rs2061538 | 17 | 77112562 | G | 0.16 | 1.52 | 1.3-1.7 | 6.2x10-09 | RBFOX3 |
| rs77370934 | 5 | 161604254 | G | 0.03 | 3.16 | 2.1-4.6 | 7.7x10-09 | GABRG2 |
| rs56209714 | 17 | 77113268 | G | 0.14 | 1.54 | 1.3-1.7 | 7.9x10-09 | RBFOX3 |
| rs192613545 | 16 | 28863901 | T | 0.07 | 2.33 | 1.7-3.1 | 2.5x10-08 | SH2B1 |
| chr17:77112502:I | 17 | 77112502 | C | 0.14 | 1.62 | 1.3-1.9 | 2.7x10-08 | |
| rs62063838 | 17 | 77114028 | C | 0.17 | 1.47 | 1.2-1.6 | 3.7x10-08 | RBFOX3 |
| rs10946364 | 6 | 20177222 | T | 0.39 | 1.34 | 1.2-1.4 | 3.8x10-08 | MBOAT1 |
| rs56044629 | 17 | 77109653 | G | 0.14 | 1.51 | 1.3-1.7 | 4.2x10-08 | RBFOX3 |
SNP: single nucleotide polymorphism, Chr: chromosome, MA: minor allele, MAF: minor allele frequency, OR: odds ratio, CI: confidence interval,
A List of the most significantly associated SNPs which reached P-value of less than 10-05 in meta-analysis, is available as the supplementary material.
Figure 3.
LocusZoom plot of most strongly associated SNPs from the meta-analysis located in A) the region of most significantly associated genes
A) The RBFOX3 (chromosome 17 centred around SNP rs2061538 (shown in purple). Linkage disequilibrium (based on r2 values) with respect to rs2061538 are based on the CEU reference population.
B) The GABRG2 (chromosome 5 centred around SNP rs77370934 (shown in purple). Linkage disequilibrium (based on r2 values) with respect to rs77370934 are based on the CEU reference population.
There were also genome wide statistically significant SNPs within the MBOAT1 gene (Membrane Bound O-Acyltransferase Domain Containing 1) and SH2B1 gene (SH2B Adaptor Protein 1).
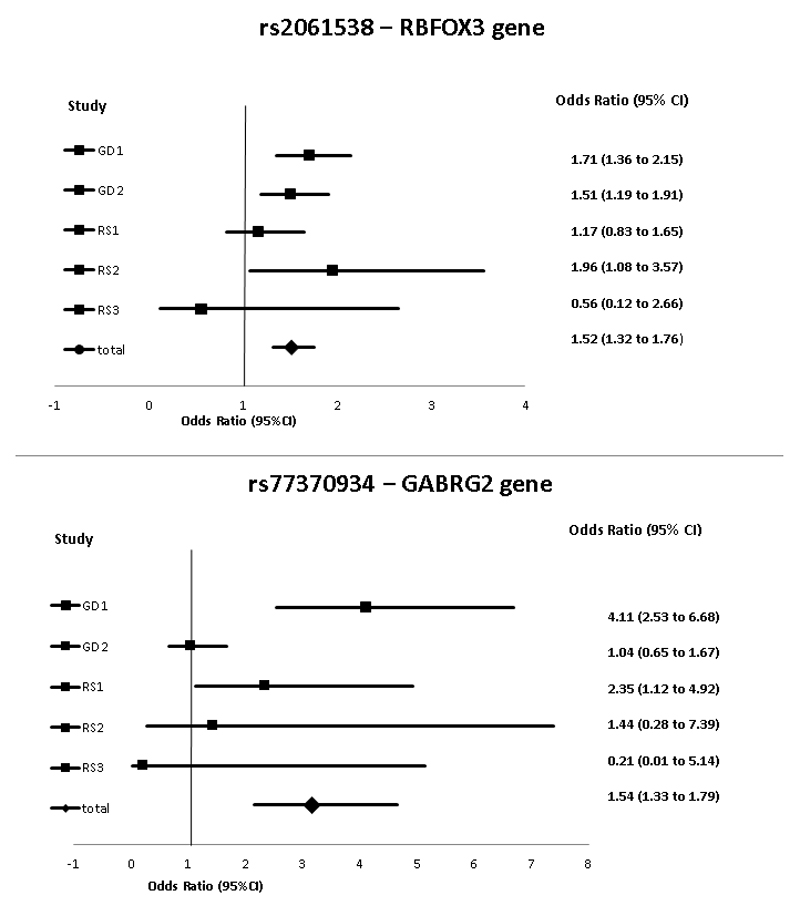
Figure 4 presents the odds ratio and the 95% confidence interval (95% CI) for the two most statistically significantly associated SNPs for the different sub studies of the Rotterdam study and the Go-DARTS population. Except for the RS3 which is the smallest subpopulation, the effect directions were concordant between the populations.
Figure 4.
The forest plot from the meta-analyses of most strongly associated SNPs
GD: GoDARTS, RS: Rotterdam study, CI: confidence interval
A high level of consistency was observed for the meta-analyses results from both sites using the GWAMA and METAL, particularly for the most significantly associated SNPs.
Discussion
Our study describes a large GWAS study investigating SNP variants associated with switching of an ACE-inhibitor to an ARB as a marker for ACE inhibitor induced ADRs. All phenotype data for this study were derived from clinical settings that incorporate either the prescription data system (GoDARTS) or the pharmacy drug dispensing database (Rotterdam study). We found statistically significant associations with SNPs located within the genes RBFOX3, GABRG2, SH2B1 and MBOAT1. These are novel candidate genes which may play a role in the adverse drug reactions to ACE-inhibitors.
The SNPs showing the strongest association with the phenotype are located on chromosome 17 within the gene RBFOX3. This is a member of the RBFOX family that in mammals consists of three members: RBFOX1, RBFOX2 and RBFOX3. RBFOX3 is expressed specifically in neuronal cells. This protein contains an RNA recognition motif that binds specifically to an RNA element, UGCAUG and regulates alternative pre-mRNA splicing. Alternative splicing of pre-mRNA is an important mechanism for post-transcriptional regulation of gene expression and has increasingly been appreciated as a major mechanism to generate diversity of gene products in higher eukaryotes [36, 37].
The other most strongly associated SNP was located on chromosome 5 within the gene GABRG2 which encodes a gamma-aminobutyric acid (GABA) receptor. GABA is the major inhibitory neurotransmitter in the mammalian nervous system, where it acts at GABA-A receptors. GABA-A receptors are pentameric, consisting of proteins from several subunit classes: alpha, beta, gamma, delta and rho [38]. There are several studies proving the effects of GABA receptor agonists in decreasing the sensitivity to cough both in animal models and in humans. This makes them a possible target for cough treatment [39]. Dicpinigaitis et al showed that Baclofen (as a GABA receptor agonist) can supress cough induced by ACE-inhibitors [40]. They also proved in a prospective clinical trial that baclofen can inhibit capsaicin-induced cough [41].
SH2B1 (sarcoma (Src) homology 2 (SH2) B adaptor protein 1) is a member of a family of scaffold proteins implicated in signalling downstream of a variety of receptor tyrosine kinases and cytokine receptors [42]. Variations in this gene have been reported to be associated with obesity [43]; however its role in the abnormal glucose homeostasis has not been proved [44]. The significant association of this gene with the intolerance of ACE-inhibitors needs to be further investigated because there was no previous report of this gene contributing in cough or angioedema.
MBOAT1 (membrane bound O-acyltransferase domain containing 1) belongs to the superfamily of MBOAT that transfer organic compounds, usually fatty acids onto hydroxyl groups of membrane-embedded targets [45]. This trans-membrane protein has been reported to be involved in developmental processes [46].
The main hypothesized mechanism of ACE-inhibitor induced ADRs (mainly cough and angioedema) is stimulation of sensory nerve resulting from the accumulation of inflammatory mediators that are normally cleaved by the ACE [3]. This hypothesis has served as the basis for candidate gene studies that have focused on variation in inflammatory pathways, however findings of those candidate gene studies were replicated inconsistently and the meta-analyses of loci that had enough studies, did not find the significant effect for the insertion/deletion polymorphism within ACE gene [15]. Hypothesis free GWA studies may lead to finding novel loci to be associated with ADRs of ACE-inhibitors. The only available large GWAS on ACE-inhibitor induced cough found an association with Kv Channel Interacting Protein 4 (KCNIP4) which is predominantly expressed in nervous systems [22]. However the only available GWAS on the ACE-inhibitor induced angioedema with 175 ACE-inhibitor induced angioedema cases and 489 controls could not find any significant association on a genome wide level which could be due to the relatively small sample size and lack of the power [21]. Our results suggest that an important source of variation may be directly related to the sensory nerves themselves, because both GABRG2 and RBFOX3 genes are playing a role in the central and peripheral nervous systems as well. These findings are in line with the previous GWAS on ACE-inhibitor induced cough [22].
This study is a large GWAS on the intolerance of ACE-inhibitors within a population of European ancestry. However the direct relevance of our findings with ACE-inhibitor induced ADRs is not clear yet and needs to be further investigated, these findings, if replicated in other populations, can improve our understanding of the biological mechanism of ACE-inhibitor induced ADRs. Furthermore, it will help to identify those patients at high-risk to develop ACE-inhibitor induced ADRs including angioedema, which is a life threatening event. We recently showed that approximately 50% of ACE-inhibitor users continue ACE-inhibitors after the first episode of angioedema [47]; Identification of those patients at high risk could help physicians guide their treatment choice. ACE-inhibitor induced cough is not as life threatening as angioedema but it can be misdiagnosed and mistreated which significantly decreases the compliance of patients and might finally result in unsuccessful drug therapy [48, 49]. Therefore in the context of precision medicine, the ultimate application of these findings within the clinic would be the prediction of susceptible patients and treating them with an alternative medication with comparable effect such as ARBs [50].
An important limitation of this study is defining phenotype based on the electronic medical records which can potentially lead to misclassification of cases and controls. However in a validation study, the proxy marker for cases showed a positive predictive value of 68.3% for probable ACE-inhibitor induced ADRs [23]. This study also cannot detect associations for rare SNPs (MAF< 0.01%). The study results are restricted to the European ancestor populations.
In conclusion, this study used a GWAS to identify SNP variants associated with ACE-inhibitor intolerance as a marker of ADRs. We identified SNPs in the genes RBFOX3, GABRG2, SH2B1 and MBOAT1 as potential candidates for ACE inhibitor induced ADRs. Due to the fact that this is a hypothesis generating study, the functional role of significantly associated genes was not investigated; therefore future studies are needed to replicate our findings in addition to the epigenetic and molecular studies are needed to explore the functional roles of variations within genes reported in this study specifically the GABRG2 gene for which several clinical studies also showed its role in susceptibility to cough [39–41]. The standard clinical criteria have been described for ACE-inhibitor induced angioedema [51] and to make it possible to combine results it would be good if new genetic association studies would use this standard phenotype in the future.
Supplementary Material
Acknowledgments
This research was conducted as a part of the Personalisation of treatment In Cardiovascular disease through next generation sequencing in Adverse Drug Reactions (PREDICTION-ADR) consortium. The PREDICTION-ADR project is supported by the European Union FP7 Grant no. 602108. F.W.A. is supported by the UCL Hospitals NIHR Biomedical Research Centre and by a Dekker scholarship (Junior Staff Member 2014T001) from the Dutch Heart Foundation.
For the GoDARTS study: We are grateful to all the participants in this study, the general practitioners, the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The study complies with the Declaration of Helsinki. We acknowledge the support of the Health Informatics Centre, University of Dundee for managing and supplying the anonymised data and NHS Tayside, the original data owner. The Wellcome Trust United Kingdom Type 2 Diabetes Case Control Collection (GoDARTS) was funded by The Wellcome Trust (072960/Z/03/Z, 084726/Z/08/Z, 084727/Z/08/Z, 085475/Z/08/Z, 085475/B/08/Z) and as part of the EU IMI-SUMMIT program.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Khalil ME, Basher AW, Brown EJ, Jr, Alhaddad IA. A remarkable medical story: benefits of angiotensin-converting enzyme inhibitors in cardiac patients. J Am Coll Cardiol. 2001;37:1757–1764. doi: 10.1016/s0735-1097(01)01229-3. [DOI] [PubMed] [Google Scholar]
- [2].Morimoto T, Gandhi TK, Fiskio JM, Seger AC, So JW, Cook EF, et al. An evaluation of risk factors for adverse drug events associated with angiotensin-converting enzyme inhibitors. J Eval Clin Pract. 2004;10:499–509. doi: 10.1111/j.1365-2753.2003.00484.x. [DOI] [PubMed] [Google Scholar]
- [3].Israili ZH, Hall WD. Cough and angioneurotic edema associated with angiotensin-converting enzyme inhibitor therapy. A review of the literature and pathophysiology. Ann Intern Med. 1992;117:234–242. doi: 10.7326/0003-4819-117-3-234. [DOI] [PubMed] [Google Scholar]
- [4].Chan WK, Chan TY, Luk WK, Leung VK, Li TH, Critchley JA. A high incidence of cough in Chinese subjects treated with angiotensin converting enzyme inhibitors. Eur J Clin Pharmacol. 1993;44:299–300. doi: 10.1007/BF00271377. [DOI] [PubMed] [Google Scholar]
- [5].Woo KS, Nicholls MG. High prevalence of persistent cough with angiotensin converting enzyme inhibitors in Chinese. Br J Clin Pharmacol. 1995;40:141–144. [PMC free article] [PubMed] [Google Scholar]
- [6].Mahoney EJ, Devaiah AK. Angioedema and angiotensin-converting enzyme inhibitors: are demographics a risk? Otolaryngol Head Neck Surg. 2008;139:105–108. doi: 10.1016/j.otohns.2008.03.029. [DOI] [PubMed] [Google Scholar]
- [7].Weber MA, Messerli FH. Angiotensin-converting enzyme inhibitors and angioedema: estimating the risk. Hypertension. 2008;51:1465–1467. doi: 10.1161/HYPERTENSIONAHA.108.111393. [DOI] [PubMed] [Google Scholar]
- [8].Bernstein KE, Ong FS, Blackwell WL, Shah KH, Giani JF, Gonzalez-Villalobos RA, et al. A modern understanding of the traditional and nontraditional biological functions of angiotensin-converting enzyme. Pharmacol Rev. 2012;65:1–46. doi: 10.1124/pr.112.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fox AJ, Lalloo UG, Belvisi MG, Bernareggi M, Chung KF, Barnes PJ. Bradykinin-evoked sensitization of airway sensory nerves: a mechanism for ACE-inhibitor cough. Nat Med. 1996;2:814–817. doi: 10.1038/nm0796-814. [DOI] [PubMed] [Google Scholar]
- [10].Molinaro G, Cugno M, Perez M, Lepage Y, Gervais N, Agostoni A, Adam A. Angiotensin-converting enzyme inhibitor-associated angioedema is characterized by a slower degradation of des-arginine(9)-bradykinin. J Pharmacol Exp Ther. 2002;303:232–237. doi: 10.1124/jpet.102.038067. [DOI] [PubMed] [Google Scholar]
- [11].Furuya K, Yamagachi E, Hirabayashi T, Itoh A, Hizawa N, Ohnuma N, Kawakami Y. Angiotensin-I-converting enzyme gene polymorphism and susceptibility to cough. Lancet. 1994;343:354. doi: 10.1016/s0140-6736(94)91190-8. [4] [DOI] [PubMed] [Google Scholar]
- [12].Grilo A, Saez-Rosas MP, Santos-Morano J, Sanchez E, Moreno-Rey C, Real LM, et al. Identification of genetic factors associated with susceptibility to angiotensin-converting enzyme inhibitors-induced cough. Pharmacogenet Genomics. 2011;21:10–17. doi: 10.1097/FPC.0b013e328341041c. [DOI] [PubMed] [Google Scholar]
- [13].Mas S, Gasso P, Alvarez S, Ortiz J, Sotoca JM, Francino A, et al. Pharmacogenetic predictors of angiotensin-converting enzyme inhibitor-induced cough: the role of ACE, ABO, and BDKRB2 genes. Pharmacogenetics and Genomics. 2011;21:531–538. doi: 10.1097/FPC.0b013e328348c6db. [DOI] [PubMed] [Google Scholar]
- [14].Mukae S, Itoh S, Aoki S, Iwata T, Nishio K, Sato R, Katagiri T. Association of polymorphisms of the renin-angiotensin system and bradykinin B2 receptor with ACE-inhibitor-related cough. J Hum Hypertens. 2002;16:857–863. doi: 10.1038/sj.jhh.1001486. [DOI] [PubMed] [Google Scholar]
- [15].Mahmoudpour SH, Leusink M, Putten L, Terreehorst I, Asselbergs FW, de Boer A, Maitland-van der Zee AH. Pharmacogenetics of ACE inhibitor-induced angioedema and cough: a systematic review and meta-analysis. Pharmacogenomics. 2013;14:249–260. doi: 10.2217/pgs.12.206. [DOI] [PubMed] [Google Scholar]
- [16].Woodard-Grice AV, Lucisano AC, Byrd JB, Stone ER, Simmons WH, Brown NJ. Sex-dependent and race-dependent association of XPNPEP2 C-2399A polymorphism with angiotensin-converting enzyme inhibitor-associated angioedema. Pharmacogenet Genomics. 2010;20:532–536. doi: 10.1097/FPC.0b013e32833d3acb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Duan QL, Nikpoor B, Dube M-, Molinaro G, Meijer IA, Dion P, et al. A variant in XPNPEP2 is associated with angioedema induced by angiotensin I-converting enzyme inhibitors. Am J Hum Genet. 2005;77:617–626. doi: 10.1086/496899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cilia La Corte AL, Carter AM, Rice GI, Duan QL, Rouleau GA, Adam A, et al. A functional XPNPEP2 promoter haplotype leads to reduced plasma aminopeptidase P and increased risk of ACE inhibitor-induced angioedema. Hum Mutat. 2011;32:1326–1331. doi: 10.1002/humu.21579. [DOI] [PubMed] [Google Scholar]
- [19].Moholisa RR, Rayner BR, Patricia Owen E, Schwager SL, Stark JS, Badri M, et al. Association of B2 Receptor Polymorphisms and ACE Activity With ACE Inhibitor-Induced Angioedema in Black and Mixed-Race South Africans. J Clin Hypertens (Greenwich) 2013;15:413–419. doi: 10.1111/jch.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- [21].Pare G, Kubo M, Byrd JB, McCarty CA, Woodard-Grice A, Teo KK, et al. Genetic variants associated with angiotensin-converting enzyme inhibitor-associated angioedema. Pharmacogenet Genomics. 2013;23:470–478. doi: 10.1097/FPC.0b013e328363c137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mosley JD, Shaffer CM, Van Driest SL, Weeke PE, Wells QS, Karnes JH, et al. A genome-wide association study identifies variants in KCNIP4 associated with ACE inhibitor-induced cough. Pharmacogenomics J. 2015 doi: 10.1038/tpj.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mahmoudpour SH, Asselbergs FW, de Keyser CE, Souverein PC, Hofman A, Stricker BH, et al. Change in prescription pattern as a potential marker for adverse drug reactions of angiotensin converting enzyme inhibitors. Int J Clin Pharm. 2015;37:1095–1103. doi: 10.1007/s11096-015-0159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA. Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol. 1991;7:403–422. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- [25].Hofman A, Brusselle GG, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, et al. The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol. 2015;30:661–708. doi: 10.1007/s10654-015-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Morris AD, Boyle DI, MacAlpine R, Emslie-Smith A, Jung RT, Newton RW, MacDonald TM. The diabetes audit and research in Tayside Scotland (DARTS) study: electronic record linkage to create a diabetes register. DARTS/MEMO Collaboration. BMJ. 1997;315:524–528. doi: 10.1136/bmj.315.7107.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- [28].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- [30].Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-288. 288-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36 [Google Scholar]
- [35].Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014 [Google Scholar]
- [36].Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 2011;39:3064–3078. doi: 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sieghart W, Sperk G. Subunit composition, distribution and function of GABA(A) receptor subtypes. Curr Top Med Chem. 2002;2:795–816. doi: 10.2174/1568026023393507. [DOI] [PubMed] [Google Scholar]
- [39].Chung KF. NMDA and GABA receptors as potential targets in cough hypersensitivity syndrome. Curr Opin Pharmacol. 2015;22:29–36. doi: 10.1016/j.coph.2015.03.002. [DOI] [PubMed] [Google Scholar]
- [40].Dicpinigaitis PV. Use of baclofen to suppress cough induced by angiotensin-converting enzyme inhibitors. Ann Pharmacother. 1996;30:1242–1245. doi: 10.1177/106002809603001106. [DOI] [PubMed] [Google Scholar]
- [41].Dicpinigaitis PV, Dobkin JB, Rauf K, Aldrich TK. Inhibition of capsaicin-induced cough by the gamma-aminobutyric acid agonist baclofen. J Clin Pharmacol. 1998;38:364–367. doi: 10.1002/j.1552-4604.1998.tb04436.x. [DOI] [PubMed] [Google Scholar]
- [42].Maures TJ, Kurzer JH, Carter-Su C. SH2B1 (SH2-B) and JAK2: a multifunctional adaptor protein and kinase made for each other. Trends Endocrinol Metab. 2007;18:38–45. doi: 10.1016/j.tem.2006.11.007. [DOI] [PubMed] [Google Scholar]
- [43].Pearce LR, Joe R, Doche ME, Su HW, Keogh JM, Henning E, et al. Functional characterization of obesity-associated variants involving the alpha and beta isoforms of human SH2B1. Endocrinology. 2014;155:3219–3226. doi: 10.1210/en.2014-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Prudente S, Copetti M, Morini E, Mendonca C, Andreozzi F, Chandalia M, et al. The SH2B1 obesity locus and abnormal glucose homeostasis: lack of evidence for association from a meta-analysis in individuals of European ancestry. Nutr Metab Cardiovasc Dis. 2013;23:1043–1049. doi: 10.1016/j.numecd.2013.05.001. [DOI] [PubMed] [Google Scholar]
- [45].Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25:111–112. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- [46].Dauwerse JG, de Vries BB, Wouters CH, Bakker E, Rappold G, Mortier GR, et al. A t(4;6)(q12;p23) translocation disrupts a membrane-associated O-acetyl transferase gene (MBOAT1) in a patient with a novel brachydactyly-syndactyly syndrome. Eur J Hum Genet. 2007;15:743–751. doi: 10.1038/sj.ejhg.5201833. [DOI] [PubMed] [Google Scholar]
- [47].Mahmoudpour SH, Asselbergs FW, Terreehorst I, Souverein PC, de Boer A, Maitland-van der Zee AH. Continuation of angiotensin converting enzyme inhibitor therapy, in spite of occurrence of angioedema. Int J Cardiol. 2015;201:644–645. doi: 10.1016/j.ijcard.2015.08.185. [DOI] [PubMed] [Google Scholar]
- [48].Vegter S, de Jong-van den Berg LT. Misdiagnosis and mistreatment of a common side-effect--angiotensin-converting enzyme inhibitor-induced cough. Br J Clin Pharmacol. 2010;69:200–203. doi: 10.1111/j.1365-2125.2009.03571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Vegter S, de Boer P, van Dijk KW, Visser S, de Jong-van den Berg LT. The effects of antitussive treatment of ACE inhibitor-induced cough on therapy compliance: a prescription sequence symmetry analysis. Drug Saf. 2013;36:435–439. doi: 10.1007/s40264-013-0024-z. [DOI] [PubMed] [Google Scholar]
- [50].Savarese G, Costanzo P, Cleland JG, Vassallo E, Ruggiero D, Rosano G, Perrone-Filardi P. A meta-analysis reporting effects of angiotensin-converting enzyme inhibitors and angiotensin receptor blockers in patients without heart failure. J Am Coll Cardiol. 2013;61:131–142. doi: 10.1016/j.jacc.2012.10.011. [DOI] [PubMed] [Google Scholar]
- [51].Wadelius M, Marshall SE, Islander G, Nordang L, Karawajczyk M, Yue QY, et al. Phenotype standardization of angioedema in the head and neck region caused by agents acting on the angiotensin system. Clin Pharmacol Ther. 2014;96:477–481. doi: 10.1038/clpt.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.