Abstract
This article presents a unified mathematical framework for inference in graphical models, building on the observation that graphical models are algebraic varieties. From this geometric viewpoint, observations generated from a model are coordinates of a point in the variety, and the sum-product algorithm is an efficient tool for evaluating specific coordinates. Here, we address the question of how the solutions to various inference problems depend on the model parameters. The proposed answer is expressed in terms of tropical algebraic geometry. The Newton polytope of a statistical model plays a key role. Our results are applied to the hidden Markov model and the general Markov model on a binary tree.
This article presents a unified mathematical framework for probabilistic inference with statistical models, such as graphical models. Our approach is summarized by the following theses:
Statistical models are algebraic varieties.
Every algebraic variety can be tropicalized.
Tropicalized statistical models are fundamental for parametric inference.
1. Algebraic Statistics, Tropical Geometry, and Inference
By a statistical model, we mean a family of joint probability distributions for a collection of discrete random variables Y = {Y1,...,Yn}. Thesis i states that many families of interest can be characterized by polynomials in the joint probabilities pσ1...σn = Prob(Y1 = σ1,...,Yn = σn). Although the variety defined by these polynomials contains points that are not in the model (for example, points with negative coordinates), the emerging field of algebraic statistics (1, 2) offers practical and useful algorithms for studying statistical models.
Tropicalization means replacing the arithmetic operations (+, ×) by the operations (min, +). This process captures the essence of what happens when the joint probabilities pσ1...σn are replaced by their logarithms. The tropicalization of an algebraic variety is a piecewise-linear set that has many features familiar from algebraic geometry (3, 4). In particular, the tropicalization of a statistical model is a piecewise-linear set in the space with logarithmic coordinates -log(pσ1...σn).
Thesis iii states that tropical algebraic geometry of statistical models is of fundamental interest in analyzing the behavior of inference algorithms under the variation of model parameters. By inference, we mean the evaluation of one or more coordinates of a single point on the algebraic variety, in either (+, ×) or (min, +) arithmetic. This evaluation corresponds to a form of inference that is used for graphical models in statistical learning theory (5), but it differs from other (more classical) notions of inference in mathematical statistics. By parametric inference, we mean the analysis of the dependence of inference on parameters.
To give a more concrete discussion of parametric inference, it is useful to focus on directed graphical models. A directed graphical model (or Bayesian network) is a finite directed acyclic graph G with two kinds of vertices (observed variables Y = {Y1,...,Yn} and hidden variables X = {X1,..., Xm}), where each edge is labeled by a transition matrix whose entries are linear forms in some parameters. The rules of discrete probability express the observed probabilities pσ1...σn as polynomials of a degree ≤ E in the parameters, where E is the number of edges of G. The polynomials parameterize the graphical model as an algebraic variety.
The following are two types of inference questions from statistical learning theory for graphical models.
-
The calculation of marginal probabilities:
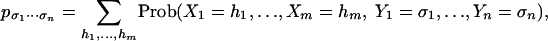
and
-
The calculation of maximum a posteriori (MAP) log probabilities:
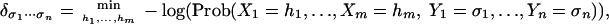
where the hi range over all of the possible assignments for the hidden random variables Xi. Together, these two primitives can be used effectively to solve a range of other statistical learning inference problems, including the calculation of conditional probabilities and other quantities of interest. The key to these statistical learning inference questions for graphical models is the sum-product algorithm (6), which is also known as the generalized distributive law (7). This polynomial-time algorithm (in the case that the graph has constant clique size) is used, both in ordinary arithmetic (+, ×) and in tropical arithmetic (min, +), to solve problems 1 and 2 efficiently. For more background on the sum-product algorithm, and for connections to message passing and the junction tree algorithm, see ref. 5.
Although the sum-product algorithm provides efficient solutions to the basic inference problems 1 and 2, it only applies to one coordinate
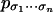 of one distribution at a time. We are interested in the parametric versions of the inference problems. They can be phrased as follows:
of one distribution at a time. We are interested in the parametric versions of the inference problems. They can be phrased as follows: Find all parameter values for a model that result in the same values for all
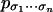 .
.-
Given observations Y = σ and hidden data X = h, identify all parameter values such that h is the most likely explanation for the observations σ.
As we will show, the following modeling problems are related fundamentally to problems 3 and 4:
Which (parameter-independent) relations on the probabilities
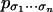 does the model imply?
does the model imply?Describe the tropicalization of the variety that corresponds to a graphical model.
Problem 5 asks for the ideal of polynomial invariants of a statistical model (1). Invariants have been investigated in phylogenetics (8, 9), where they can help to identify good trees for aligned DNA sequences.
The primary goal of our study is to give a practical answer to problem 4 for graphical models. Our main algorithmic result is an efficient procedure for parametric inference that can be viewed as a polytopal analog of the sum-product algorithm. The efficiency is based on the complexity estimates for Newton polytopes that we derive in section 4. The resulting polytope propagation algorithm is applied to problems in biological sequence analysis in ref. 10.
The mathematics developed in sections 3 and 4 is of independent interest. It also furnishes tools for parametric inference (problems 3 and 4) and parametric modeling (problems 5 and 6), which are applicable to a wide range of statistical problems. We demonstrate this point of view by analyzing the hidden Markov model (HMM) and the general Markov model on a binary tree in sections 2 and 5, respectively.
2. Algebraic Representation of HMMs
A graphical model is an algebraic variety that is presented as the image of a highly structured polynomial map f: Rd → Rm. Here, Rd is the space in which coordinates are the model parameters s1,..., sd, and Rm is the space in which coordinates 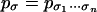 are the joint probabilities for the observed random variables. In applications, the integer m is much larger than the integer d; in fact, it is so large that one can only look at one coordinate pσ at a time. Each coordinate fσ = fσ(s1,..., sd) of the map f is a polynomial function in s1,..., sd. The efficient evaluation of these functions relies on the sum-product algorithm. Here, we study the (parametric) inference and modeling problems in the familiar context of the HMM.
are the joint probabilities for the observed random variables. In applications, the integer m is much larger than the integer d; in fact, it is so large that one can only look at one coordinate pσ at a time. Each coordinate fσ = fσ(s1,..., sd) of the map f is a polynomial function in s1,..., sd. The efficient evaluation of these functions relies on the sum-product algorithm. Here, we study the (parametric) inference and modeling problems in the familiar context of the HMM.
A discrete HMM has n observed states Y1,...,Yn taking on l possible values and n hidden states X1,..., Xn taking on k possible values. The HMM can be characterized by the following conditional independence statements for i = 1,..., n:
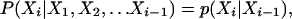 |
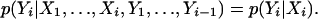 |
We consider the homogeneous model with uniform initial distribution, where all transitions Xi → Xi+1 are given by the same k × k matrix S = (sij) and all transitions Xi → Yi are given by the same k × l matrix T = (tij). Throughout our discussion, we disregard for simplicity the usual probabilistic hypothesis that S and T are nonnegative and that all row sums are 1.
Proposition 1. The HMM is the image of a map f: Rd → Rln, where d = k(k + 1) and each coordinate of f is a bihomogeneous polynomial of degree n - 1 in S and degree n in T.
Problem 3 is to compute the fibers of the map f. In statistics, this computation is called parameter identification. We use the term coordinate polynomials for the polynomials fσ that are coordinates of the map f.
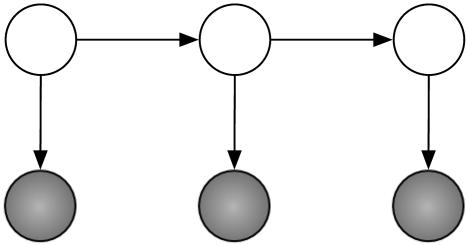
Our running example in this section is the case n = 3 with binary random variables (k = l = 2). The graph of this model is given in Fig. 1. The shaded nodes are the observed random variables.
Fig. 1.
The HMM of length 3.
Here, the parameter space is R8 with the coordinates s00, s01, s10, s11, t00, t01, t10, and t11, and it maps to R8 with the coordinates p000, p001, p010, p011, p100, p101, p110, and p111. The map f: R8 → R8 is given by the following:
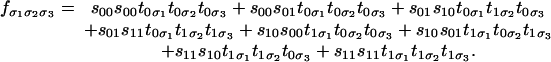 |
The HMM (i.e., the image of f) is the zero set of the following quartic polynomial:
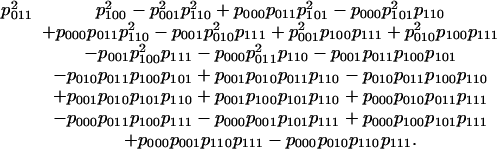 |
This polynomial was found by a Gröbner basis computation. See the discussion on implicitization in section 3 of ref. 11.
In general, the polynomial functions on Rln that vanish on the image of f are called the invariants of the model. They form a prime ideal If. In our example, If is generated by the quartic polynomial given above. Problem 5 is to compute generators of the ideal If. When ln and d are small, this can be done by using Gröbner bases, and in some cases it is possible to characterize If based on the structure of the model (see, for example, Conjecture 13), but in general, problem 5 is hard and the ideal If may remain unknown.
Here, tropical geometry comes in. The tropicalization of our map f is the map g: Rd → Rln defined by replacing products by sums and sums by minima in the formula for f. In our example (n = 3, k = l = 2), the tropicalization is the piecewise-linear map g: R8 → R8, (U, V) ↦ with the following:
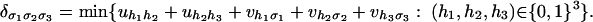 |
1 |
This minimum is attained by the most likely hidden data (ĥ1 ĥ2 ĥ3), given the observations (σ1, σ2, σ3) and given the parameters 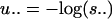 and
and 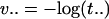 . The sequence (ĥ1, ĥ2, ĥ3) is known as the Viterbi sequence in the HMM literature (12). It solves problem 2 in section 1.
. The sequence (ĥ1, ĥ2, ĥ3) is known as the Viterbi sequence in the HMM literature (12). It solves problem 2 in section 1.
The key observation, which we discuss in more detail in section 4, is that the set of parameters (U, V) that selects the Viterbi sequence (ĥ1, ĥ2, ĥ3) is the normal cone at a vertex of the Newton polytope of the polynomial fσ1σ2σ3. This polytope is four-dimensional, it has eight vertices, and its normal fan represents the solution to problem 4 in section 1 when σ = σ1σ2σ3 is fixed.
We can also consider an extension of problem 4 in which σ = σ1σ2σ3 ranges over all possible observations. The solution is given by the Newton polytope of the map f. In our example, it is a five-dimensional polytope with 398 vertices, 1,136 edges, 1,150 two-faces, 478 three-faces, and 68 facets, namely, the Minkowski sum of eight copies of the earlier four-dimensional polytope for (σ1, σ2, σ3) ∈ {0, 1}3. For a concrete numerical example, fix the parameters 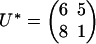 and
and 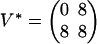 ). We find the following:
). We find the following:
If the observed string at Y1Y2Y3 is
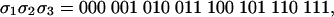 |
then the Viterbi sequence at X1X2X3 is
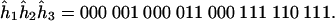 |
The set of all parameters (U, V) leading to the same conclusions as (U*, V*) is the cone defined by the following:
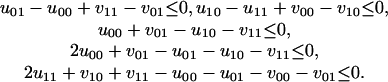 |
Our solution to the parametric inference problem with respect to all observations simultaneously consists of 398 such cones. The tropical HMM is the union of the images of these cones under the piecewise-linear map g: (U, V) ↦ δ. This image is a piecewise-linear set of dimension 7. The cone that contains the chosen parameters (U*, V*) is mapped to a seven-dimensional cone in the tropical HMM (it spans the hyperplane δ010 = δ100), but most of the other 397 cones are mapped to lower-dimensional cones by the map g. The question of how the number 398 grows as the length n increases is addressed in Corollary 10.
3. Positivity and Morphisms in Tropical Geometry
We have shown that a graphical model is the image of a polynomial map f from the space of parameters to the space of joint probability distributions on the observed random variables. Furthermore, we have shown that the tropicalization of f arises naturally in solving problem 4. In this section, we study the geometry of tropicalization in the more general setting where f: Rd → Rm is an arbitrary polynomial map. In statistical applications, each coordinate fσ of the map f is usually a polynomial with positive coefficients. If this condition holds, then the polynomial map f is called positive. We consider f to be surjectively positive if, in addition, f maps the positive orthant surjectively onto the positive points in the image, in symbols,
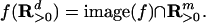 |
2 |
The set of all polynomial functions that vanish on the image of f is a prime ideal If in the polynomial ring R[p1,..., pm]. The closure of the image of f is the variety of the prime ideal If.
In tropical geometry, we replace the variety of If by a piecewise-linear set as follows. The tropical variety 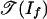 is the set of all weight vectors w ∈ Rm such that the initial ideal inw(If) contains no monomial (4, 13). By following ref. 14, we define the positive tropical variety
is the set of all weight vectors w ∈ Rm such that the initial ideal inw(If) contains no monomial (4, 13). By following ref. 14, we define the positive tropical variety 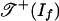 as the set of all weight vectors w ∈ Rm such that the initial ideal inw(If) contains no polynomial with only positive coefficients. The tropical variety
as the set of all weight vectors w ∈ Rm such that the initial ideal inw(If) contains no polynomial with only positive coefficients. The tropical variety 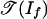 is a polyhedral fan in Rm, and
is a polyhedral fan in Rm, and 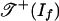 is a polyhedral subcomplex of
is a polyhedral subcomplex of 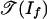 . This observation means that
. This observation means that 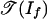 is a finite union of closed convex polyhedral cones that fit together nicely, and
is a finite union of closed convex polyhedral cones that fit together nicely, and 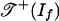 is the union of a subset of these cones. The tropicalization of the polynomial map f is the piecewise-linear map g: Rd → Rm defined by replacing products by sums and sums by minima in the evaluation of f. We consider g to be a tropical morphism. Examples of tropical morphisms appear in Eqs. 1, 3, 4, 9, and 10.
is the union of a subset of these cones. The tropicalization of the polynomial map f is the piecewise-linear map g: Rd → Rm defined by replacing products by sums and sums by minima in the evaluation of f. We consider g to be a tropical morphism. Examples of tropical morphisms appear in Eqs. 1, 3, 4, 9, and 10.
The following theorem describes the geometry of this situation. We define the Newton polytope of a polynomial map f: Rd → Rm as the Minkowski sum in Rd of the Newton polytopes of its coordinates f1,..., fm. For basic information on Newton polytopes and their normal fans, see section 1 of ref. 13.
Theorem 2. The tropical morphism g is linear on each cone in the normal fan of the Newton polytope of f. Its image is a fan contained in 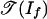 . If f is positive, then image(g) is a subset of
. If f is positive, then image(g) is a subset of 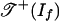 , but it is generally not a polyhedral subcomplex. If f is surjectively positive, then
, but it is generally not a polyhedral subcomplex. If f is surjectively positive, then 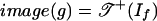 .
.
Proof: Let Pi denote the Newton polytope of the polynomial fi = fi(s1,..., sd). By definition, Pi is the convex hull in Rd of all nonnegative lattice points a = (a1,..., ad) ∈ Nd such that the monomial 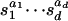 appears with a nonzero coefficient in fi. The piecewise-linear concave function gi is the support function of the polytope Pi. Thus, gi(w) is the minimum value attained on Pi by the linear functional a ↦ w·a. In particular, the function gi: Rd → R is linear on each cone in the normal fan of Pi.
appears with a nonzero coefficient in fi. The piecewise-linear concave function gi is the support function of the polytope Pi. Thus, gi(w) is the minimum value attained on Pi by the linear functional a ↦ w·a. In particular, the function gi: Rd → R is linear on each cone in the normal fan of Pi.
The Newton polytope of the map f is the Minkowski sum P1 +· · ·+ Pm = {a1 +· · ·+ am: ai ∈ Pi}. The normal fan of P1 +· · ·+ Pm is the common refinement of the normal fans of P1,..., Pm. This observation shows that the function f = (f1,..., fm): Rd → Rd is linear on each cone of the normal fan of the Newton polytope of f. Because g is continuous, the image of g is a closed polyhedral fan in Rm.
Consider any vector w ∈ Rd. We must show that g(w) lies in 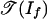 , and if f is positive, then g(w) lies in
, and if f is positive, then g(w) lies in 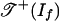 . Let φ be any polynomial in the ideal If. If we substitute p1 = f1,..., pm = fm into φ = φ(p1,..., pm), then the result is zero. Consequently, if we substitute the initial forms p1 = inw(f1),..., pm = inw(fm) into the initial form ing(w) (φ), then the result is zero (see equation 11.2 in ref. 13, p. 100). This fact implies that ing(w) (φ) is not a monomial. Moreover, if f is positive, then φ must have two terms whose coefficients have opposite signs.
. Let φ be any polynomial in the ideal If. If we substitute p1 = f1,..., pm = fm into φ = φ(p1,..., pm), then the result is zero. Consequently, if we substitute the initial forms p1 = inw(f1),..., pm = inw(fm) into the initial form ing(w) (φ), then the result is zero (see equation 11.2 in ref. 13, p. 100). This fact implies that ing(w) (φ) is not a monomial. Moreover, if f is positive, then φ must have two terms whose coefficients have opposite signs.
The following example shows that image (g) need not be a subcomplex of 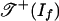 . If f is assumed to be surjectively positive, then it follows from proposition 2.5 in ref. 14 that
. If f is assumed to be surjectively positive, then it follows from proposition 2.5 in ref. 14 that 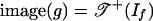 .
.
Example 3: Let d = 3 and m = 4, and consider the linear map
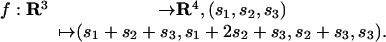 |
Then, If is the principal ideal generated by the linear form p1 - p2 + p3 - p4, and 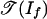 is essentially the normal fan of a tetrahedron. We identify
is essentially the normal fan of a tetrahedron. We identify 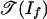 with the complete graph K4. The six edges of K4 are labeled with six monomial-free initial ideals of If, namely, 〈p1 + p3〉, 〈-p2 - p4〉, 〈p1 - p2〉, 〈p1 - p4〉, 〈-p2 + p3〉, 〈p3 - p4〉. The first two of these six initial ideals contain a polynomial with positive coefficients. Hence, the positive tropical variety
with the complete graph K4. The six edges of K4 are labeled with six monomial-free initial ideals of If, namely, 〈p1 + p3〉, 〈-p2 - p4〉, 〈p1 - p2〉, 〈p1 - p4〉, 〈-p2 + p3〉, 〈p3 - p4〉. The first two of these six initial ideals contain a polynomial with positive coefficients. Hence, the positive tropical variety 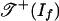 is the four-cycle in K4 formed by the remaining four edges.
is the four-cycle in K4 formed by the remaining four edges.
The tropicalization of the linear map f is the tropical morphism:
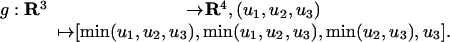 |
3 |
The image of g is the set of all vectors (a, a, b, c) with a ≤ b ≤ c. Each vector (a, a, b, c) with a < b < c has the initial ideal 〈p1 - p2〉, so it lies on a particular edge of K4. But the same edge also accounts for all vectors (a, a, b, c) with a < c < b, none of which is in the image of g. Thus, image(g) is a closed segment that covers only half of the edge of K4 indexed by 〈p1 - p2〉.
In the rest of this section, we examine Theorem 2 for a small but important graphical model, namely, the naive Bayes model with two features (ref. 1, section 7). There are two observed random variables Y1 and Y2 that depend on one hidden binary random variable X. The two observed variables take k and l possible values, respectively. The parameterization f of this model is the map f: R2(k+l) ↦ Rkl given by pij = si0t0j + si1t1j. Thus, the model consists of all k × l matrices P = (pij) of the form P = S·T, where S is a k × 2 matrix and T is a 2 × l matrix (i.e., the model consists of precisely the k × l matrices of rank ≤2).
Proposition 4. The parameterization f of the naive Bayes model with two features is surjectively positive. The ideal If is generated by the 3 × 3 subdeterminants of the k × l matrix P = (pij).
Proof: The map f being positive means that, if P is any positive matrix of rank 2, then S and T can be chosen to be positive. This is a known result in linear algebra (e.g, see ref. 15). The same statement is false for rank ≥3 (i.e., the parameterization of the naive Bayes model with three or more features is not surjectively positive). A well known result in commutative algebra states that the (r + 1) × (r + 1) minors of a k × l matrix generate a prime ideal. The variety of this ideal is the set of k × l matrices of rank ≤r. This is our ideal If for r = 2.
The objects of Theorem 2 have been studied (3, 16). The tropical variety 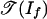 is the set of k × l matrices of tropical rank ≤2, and the tropical variety
is the set of k × l matrices of tropical rank ≤2, and the tropical variety 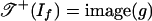 is the set of k × l matrices of Barvinok rank ≤2. Develin (16) determined the combinatorics and topology of these spaces when min(k, l) = 3. He showed that
is the set of k × l matrices of Barvinok rank ≤2. Develin (16) determined the combinatorics and topology of these spaces when min(k, l) = 3. He showed that 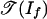 is shellable but that
is shellable but that 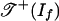 can have torsion in its integral homology groups.
can have torsion in its integral homology groups.
The Newton polytope of the map f is an interesting combinatorial object, namely, it is the (kl - k - l + 2)-dimensional zonotope associated with the complete bipartite graph Kk,l. The Newton polytope of each coordinate fij is a line segment, and the zonotope is their Minkowski sum. The normal fan is the hyperplane arrangement {ui0 - ui1 = v1j - v0j}. Its maximal cones correspond to the acyclic orientations of the complete bipartite graph Kk,l. West (17) showed that the number of facets of such a cone can be any integer between k + l - 1 and kl. The total number of cones equals 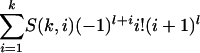 , where S(k, i) is the Stirling number of the second kind. Here, the tropical morphism g is given by the following:
, where S(k, i) is the Stirling number of the second kind. Here, the tropical morphism g is given by the following:
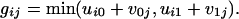 |
4 |
The map g: R2(k+l) ↦ Rkl is piecewise linear with respect to the hyperplane arrangement.
Example 5: Let k = l = 3, so the two observed random variables are ternary. The prime ideal is the following:
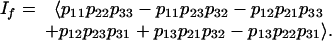 |
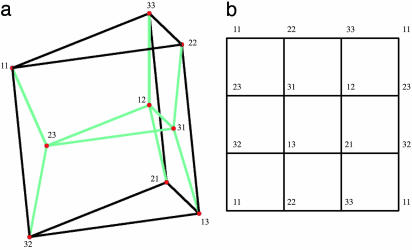
The tropical variety 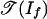 is the fan over a two-dimensional polyhedral complex consisting of six triangles and nine quadrangles. This complex is the 2-skeleton of the product of two triangles, labeled as in Fig. 2a. This complex is shellable. The positive tropical variety
is the fan over a two-dimensional polyhedral complex consisting of six triangles and nine quadrangles. This complex is the 2-skeleton of the product of two triangles, labeled as in Fig. 2a. This complex is shellable. The positive tropical variety 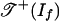 is the subcomplex consisting of the nine quadrangles shown in Fig. 2b. Note that
is the subcomplex consisting of the nine quadrangles shown in Fig. 2b. Note that 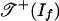 is a torus.
is a torus.
Fig. 2.
The tropical variety and positive-tropical variety of the 3 × 3 determinant.
The Newton polytope of f is a five-dimensional zonotope with 230 vertices, one for each acyclic orientation of the complete bipartite graph K3,3. The map g is linear on each of the 230 cones in the corresponding hyperplane arrangement, but it is rank-deficient on 68 of the cones. The remaining 162 = 18 × 9 cones are mapped onto the nine quadrangles of the torus 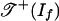 . Thus, the general fiber of g involves 18 cones. Of these 18 cones, 8 cones have five facets, 8 cones have six facets, and 2 cones have eight facets.
. Thus, the general fiber of g involves 18 cones. Of these 18 cones, 8 cones have five facets, 8 cones have six facets, and 2 cones have eight facets.
4. Newton Polytopes of Graphical Models and their Complexity
Consider a graphical model with E edges and n observed random variables Y1,...,Yn, each taking l values. Such a model is given by a positive polynomial map f: Rd → Rln. Each coordinate fσ of f is a polynomial of degree e in the model parameters s1,..., sd. In this section, we discuss the statistical meaning and the computational complexity of the mathematical objects introduced in section 3.
We write 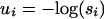 for the negative logarithms of the model parameters. Consider any of the ln possible observations σ. The quantity fσ(s1,..., sd) is the probability of making this particular observation [i.e., it is Prob(Y = σ)]. The quantity gσ(u1,..., ud) is the negative logarithm of the conditional probability Prob(X = h|Y = σ), where h maximizes Prob(X = ĥ|Y = σ) for the parameters (s1,..., sd). Clearly, the function gσ: Rd → R is piecewise linear and concave on the logarithmic parameter space.
for the negative logarithms of the model parameters. Consider any of the ln possible observations σ. The quantity fσ(s1,..., sd) is the probability of making this particular observation [i.e., it is Prob(Y = σ)]. The quantity gσ(u1,..., ud) is the negative logarithm of the conditional probability Prob(X = h|Y = σ), where h maximizes Prob(X = ĥ|Y = σ) for the parameters (s1,..., sd). Clearly, the function gσ: Rd → R is piecewise linear and concave on the logarithmic parameter space.
The domains of linearity of the function gσ are the cones in the normal fan of the Newton polytope of fσ. Each maximal cone C is indexed by the hidden data ĥ that maximizes Prob(X = h|Y = σ) for any of the parameters (u1,..., ud) ∈ C. The hidden data ĥ that arise in this manner, for some choice of logarithmic parameters u, are called the possible explanations of the observation σ. For example, for the HMM described in section 2, the explanations are the Viterbi sequences.
We vary the observations as follows. Each logarithmic parameter vector u defines an inference function σ ↦ ĥ from the set of observations to the set of explanations. For the HMM, each inference function {1,..., l}n → {1,..., k}n takes an observed sequence σ to the corresponding Viterbi sequence ĥ. There are 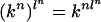 such functions, but most of these are not inference functions. For example, consider the binary HMM of length three. There are 88 = 16,777,216 Boolean functions {0, 1}3 → {0, 1}3, but as we show at the end of section 2, only 398 of these are inference functions for the HMM.
such functions, but most of these are not inference functions. For example, consider the binary HMM of length three. There are 88 = 16,777,216 Boolean functions {0, 1}3 → {0, 1}3, but as we show at the end of section 2, only 398 of these are inference functions for the HMM.
Proposition 6. The inference functions σ ↦ ĥ of a graphical model f are in bijection with the vertices of the Newton polytope of the map f. The explanations ĥ for a fixed observation σ in a graphical model are in bijection with the vertices of the Newton polytope of the polynomial fσ.
In applications of graphical models, the number d of parameters and the number l of values of the observed random variables are small and fixed, but the number n of observed random variables is large. Recall that the model is the image of the map f: Rd → Rln. Hence, the dimension of the model remains fixed, but the dimension of its ambient space grows exponentially in n. Therefore, it is algorithmically infeasible to compute the full tropical variety 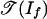 . However, we can efficiently compute the Newton polytopes of the fσ, or even the Newton polytope of f. This insight allows us to glean information about the tropical variety from the domains of linearity of its “coordinate functions” gσ.
. However, we can efficiently compute the Newton polytopes of the fσ, or even the Newton polytope of f. This insight allows us to glean information about the tropical variety from the domains of linearity of its “coordinate functions” gσ.
Our next goal is to derive an upper bound on the number of vertices of the Newton polytopes.
Theorem 7. Consider graphical models f whose number of parameters d is fixed and whose number n of observed random variables and number of edges E varies. (Typically, E is a linear function of n.) Then, the number of vertices of the Newton polytope NP(fσ) of fσ is bounded above by the following:
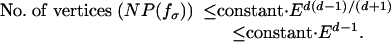 |
For many important families of graphical models, the number E of edges is bounded by a linear function in terms of the number n of observed nodes, and in these cases, we can replace E by n. Hence, for any given observation σ, the number of explanations grows polynomially in n. For example, in the HMM described in section 2, we have E = 2n - 1, and a similar relationship holds in the tree model of section 5.
Corollary 8. For any fixed observation in the homogeneous HMM, the number of explanations is at most Ck,l·nk(k+l). If all random variables are binary, then the upper bound C·n10/3 holds.
The proofs of Theorem 7 and Corollary 8 are derived from the following classical result on lattice polytopes by Andrews (18). The necessary observation is that the Newton polytope of fσ is contained in the cube [0, E]d, and the volume of this cube equals Ed.
Proposition 9. For every fixed integer d, there exists a constant Cd such that the number of vertices of any lattice polytope P in Rd is bounded above by Cd·volume(P)(d-1)/(d+1) (18).
The Newton polytope of the map f was defined as the Minkowski sum of the ln smaller Newton polytopes in Theorem 7. We infer the following to be naive bound on its number of vertices.
Corollary 10. The number of inference functions of a graphical model is at most lnCdEd-1; hence, this number scales at most singly exponentially in the complexity (n, E) of the graphical model.
Consider the homogeneous HMM on binary random variables. Each inference function is a Boolean function {0, 1}n → {0, 1}n, but not conversely. The number of all Boolean functions is 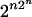 , which grows doubly exponentially in n. However, the number of inference functions is at most 2polynomial(n).
, which grows doubly exponentially in n. However, the number of inference functions is at most 2polynomial(n).
In practical applications of graphical models, it may be infeasible to compute all (singly exponentially many) inference functions. Nonetheless, we believe that important insight can be gained by computing and classifying the Newton polytopes of graphical models f on few random variables. Such a study would be the polyhedral analog to the algebraic classification of ref. 1.
However, for a fixed observation σ, the size of the Newton polytope of fσ grows polynomially with the size of the graphical model, and therefore, there is hope that the polytopes can be computed efficiently. Despite the fact that the Newton polytope of fσ has polynomially many vertices in the size of the graphical model, the number of terms in fσ grows exponentially. This is a potential problem because the computation of the Newton polytope requires the inspection of these terms. The following result states that the convex hull computations scale with the running time of the sum-product algorithm, which for many models of interest scales polynomially with the size of the graphical model.
Proposition 11 (Polytope Propagation). The Newton polytopes of the polynomials fσ can be computed recursively by using the decomposition of fσ according to the sum-product algorithm.
5. The General Markov Model on a Binary Tree
We conclude by illustrating the concepts that we have developed in the context of tree Markov models. These models are directed graphical models in which the graph is a directed tree τ with observed random variables Y1,...,Yn at the leaves. The naive Bayes model in section 3 is the special case in which n = 2. Each edge e has a different transition matrix 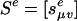 . We consider the general model given by Allman and Rhodes (8), which means that the Se are arbitrary distinct l × l matrices. In most applications, the transition matrices are from a special model family (for example, in phylogenetics, these may be Jukes-Cantor model or the Hasegawa-Kishino-Yano model). As before, we relax the hypothesis that transition probabilities are nonnegative and sum to 1. Hence, the
. We consider the general model given by Allman and Rhodes (8), which means that the Se are arbitrary distinct l × l matrices. In most applications, the transition matrices are from a special model family (for example, in phylogenetics, these may be Jukes-Cantor model or the Hasegawa-Kishino-Yano model). As before, we relax the hypothesis that transition probabilities are nonnegative and sum to 1. Hence, the 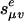 are distinct unknowns. For simplicity, we further assume that the tree τ is binary.
are distinct unknowns. For simplicity, we further assume that the tree τ is binary.
Proposition 12. The general Markov model for the binary tree τ is the image of a map 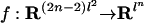 , where each coordinate of f is a multilinear polynomial in the unknowns {(
, where each coordinate of f is a multilinear polynomial in the unknowns {(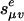 ), e edge of τ}.
), e edge of τ}.
If we denote an edge between nodes i and j by (ij), and τ′ is the tree τ without the leaves, then the coordinate of the multilinear map f indexed by an observed sequence (σ1,...,σn) can be written as follows:
 |
5 |
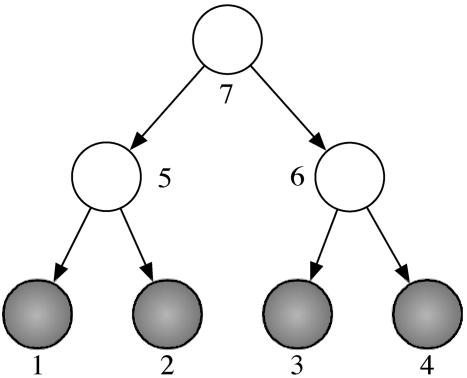
Here, h ranges over all colorations h = (hi)i∈τ of the nodes such that hj = σj for all leaves j. Our running example in this section is the binary tree in Fig. 3 with binary random variables (l = 2).
Fig. 3.
A directed binary tree with n = 4 leaves.
In this example, the coordinates of the multilinear map f: R24 → R16 are given by the following formula:
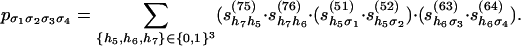 |
6 |
The prime ideal If of polynomial invariants is generated by the 3 × 3 subdeterminants of the following matrix:
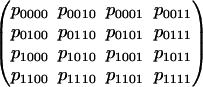 |
7 |
Thus, this particular model is the k = l = 4 instance of the determinantal variety in Proposition 4.
We generalize the determinantal presentation in this example by proposing the following explicit solution to problem 5 for arbitrary binary trees τ. Every edge of τ induces a split of the set of leaves {1, 2,..., n}, corresponding to the two connected components of the tree obtained by removing that edge. The unrooted tree underlying τ is uniquely determined by the set of these splits.
Conjecture 13. The ideal If of phylogenetic invariants of the general Markov model for any binary tree τ on binary random variables is generated by the 3 × 3-determinants of all two-dimensional matrices obtained by flattening the 2 × · · · × 2-table (pσ1...σn) according to the splits induced by the edges of τ.
We need to explain the meaning of the word “flattening.” If (A, B) is any split of the set {1,..., n}, then this term refers to the 2#(A) × 2#(B) matrix whose rows and columns are indexed by the functions A → {0, 1} and B → {0, 1}, respectively, and whose entries are the 2n probabilities pσ1...σn.
The sum-product algorithm is used in practice to evaluate the polynomial of Eq. 5. Its running time is linear in n, despite the fact that the number ln-1 of terms in Eq. 5 grows exponentially. This reduction in complexity is achieved by recursively grouping subsums. For example, Eq. 6 becomes the following:
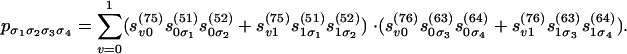 |
8 |
Remember the following rule: Polynomials are evaluated recursively as sums of products of smaller polynomials. This is the solution to problem 1. For details on the tree case, see ref. 19.
Problem 2 is known in phylogeny as the joint ancestral reconstruction problem, which asks for the MAP ancestral assignments ĥi given the observations (σ1,..., σn) at the leaves. An efficient method for solving this problem is given in ref. 20. This method is nothing but the sum-product algorithm with ordinary arithmetic (+, ×) replaced by tropical arithmetic (min, +). The σ coordinate of the tropicalization 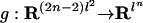 of the map in Eq. 5 is
of the map in Eq. 5 is
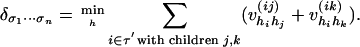 |
9 |
This expression can be evaluated efficiently by the same scheme as used previously. The rule is now the following: Piecewise-linear concave functions are evaluated recursively as minima of sums of smaller such functions. A simple example illustrating this rule is the following tropicalization of Eq. 8:
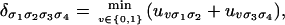 |
10 |
where 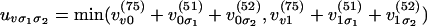 and similarly for uvσ3σ4.
and similarly for uvσ3σ4.
In section 4, we showed that the number of vertices of the Newton polytopes of the coordinate polynomials fσ is critical for efficient parametric inference. That number grows polynomially in n if the number of parameters is fixed (because of Theorem 7), but it may grow exponentially if the number of parameters is not bounded. For the general Markov model on a tree τ, the growth will be exponential unless we restrict the number of parameters. This can be done, for example, by considering the homogeneous tree model as follows, where the transition matrices along all edges are identical: 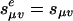 is independent of the edge e. By using Theorem 7, we obtain the following result analogous to Corollary 8.
is independent of the edge e. By using Theorem 7, we obtain the following result analogous to Corollary 8.
Proposition 14. The number of vertices of the Newton polytope of any coordinate fσ in the homogeneous tree model is bounded above by 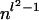 times a constant depending only on l.
times a constant depending only on l.
For tree models that are used in applications, such as phylogenetics, the number of parameters is likely to be reduced even further. In such cases, the parametric joint ancestral reconstruction problem can be solved efficiently by using the polytope propagation algorithm techniques given in Proposition 11.
6. Summary: A Statistics-Geometry Dictionary
The algebraic representation for graphical models with hidden variables leads naturally to an interpretation of a parameterized model as a point on an algebraic variety. Marginal probabilities are coordinates of points on the variety. Varieties can be tropicalized, and the statistical meaning is that the MAP probabilities (calculated with logarithms of the parameters) can be interpreted as coordinates of points on the positive part of the tropical variety. Hence, the tropical model is fundamental for understanding MAP probabilities. Although we have not addressed it in this article, the logarithms of the marginal probabilities are coordinates of points on the amoeba (21) of the model. Amoebas are likely to be important for understanding the geometry of maximum-likelihood estimation.
The sum-product algorithm for graphical models is an efficient method for evaluating the coordinate polynomials of a graphical model. This algorithm works in exactly the same way for classical arithmetic (+, ×) and for tropical arithmetic (min, +). The same method is used to evaluate coordinates of points on the variety and of points on the tropical variety.
An explanation for an observation σ is a vertex of the Newton polytope of fσ. Thus, the parametric inference problem is solved by finding the normal fans of the Newton polytopes of the coordinate polynomials. For many important applications, the number of vertices of the polytopes is polynomial in the size of the graphical model. The polytope propagation algorithm, which is a geometric analog of the sum-product algorithm, finds the Newton polytopes and is efficient when the sum-product algorithm is fast and the number of vertices on the Newton polytopes is small.
In our companion article (10), we show that polytope propagation is practical and useful in the important application of biological sequence analysis. In particular, existing parametric alignment methods (22-24) can be viewed as special cases of parametric inference for a pair HMM. The computation of the Newton polytopes is also useful for Bayesian computations, where we have priors on the parameters and it is of interest to integrate over the maximal cones in the normal fan of the Newton polytope (ref. 10, section 5).
Acknowledgments
We thank Komei Fukuda, Michael Joswig, and Kristian Ranestad for their help in obtaining the computational results reported in section 2. L.P. was supported in part by National Institutes of Health Grant R01-HG02362-02. B.S. was supported by a Hewlett Packard Visiting Research Professorship 2003/2004 at the Mathematical Sciences Research Institute (MSRI) at the University of California, Berkley, and in part by National Science Foundation Grant DMS-0200729.
Abbreviations: MAP, maximum a posteriori; HMM, hidden Markov model.
References
- 1.Garcia, L. D., Stillman, M. & Sturmfels, B. (2003) arXiv: math.AG/0301255.
- 2.Pistone, G., Riccomagno, E. & Wynn, H. P. (2001) Algebraic Statistics: Computational Commutative Algebra in Statistics (Chapman & Hall, Boca Raton, FL).
- 3.Develin, M., Santos, F. & Sturmfels, B. (2003) arXiv: math.CO/0312114.
- 4.Richter-Gebert, J., Sturmfels, B. & Theobald, T. (2004) in Idempotent Mathematics and Mathematical Physics, eds. Litvinov, G. L. & Maslov, V. P. (Am. Math. Soc., Providence, RI).
- 5.Jordan, M. I. & Weiss, Y. (2002) in Handbook of Brain Theory and Neural Networks ed. Arbib, M. (MIT Press, Cambridge, MA), 2nd Ed.
- 6.Kschischang, F., Frey, B. & Loeliger, H. A. (2001) IEEE Trans. Inform. Theory 47, 498-519. [Google Scholar]
- 7.Aji, S. & McEliece, R. J. (2000) IEEE Trans. Inform. Theory 46, 325-343. [Google Scholar]
- 8.Allman, E. & Rhodes, J. (2003) Math. Biosci. 186, 113-144. [DOI] [PubMed] [Google Scholar]
- 9.Cavender, J. & Felsenstein, J. (1987) J. Classif. 4, 57-71. [Google Scholar]
- 10.Pachter, L. & Sturmfels, B. (2004) Proc. Natl. Acad. Sci. USA 101, 16138-16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, D., O'Shea, D. & Little, J. (1996) Ideals, Varieties and Algorithms (Springer, New York).
- 12.Rabiner, L. R. (1989) Proc. IEEE 77, 257-286. [Google Scholar]
- 13.Sturmfels, B. (1996) Gröbner Bases and Convex Polytopes (Am. Math. Soc., Providence, RI), Vol. 8.
- 14.Speyer, D. & Williams, L. (2003) arXiv: math.CO/0312297.
- 15.Cohen, J. & Rothblum, U. (1993) Linear Algebra Appl. 190, 149-168. [Google Scholar]
- 16.Develin, M. (2004) arXiv: math.CO/0401224.
- 17.West, D. (1995) Discrete Math. 138, 393-396. [Google Scholar]
- 18.Andrews, G. (1963) Trans. Am. Math. Soc. 106, 270-273. [Google Scholar]
- 19.Durbin, R., Eddy, S., Krogh, A. & Mitchison, G. (1998) Biological Sequence Analysis (Probabilistic Models of Proteins and Nucleic Acids) (Cambridge Univ. Press, Cambridge, U.K.).
- 20.Pupko, T., Pe'er, I., Shamir, R. & Graur, D. (2000) Mol. Biol. Evol. 17, 890-896. [DOI] [PubMed] [Google Scholar]
- 21.Viro, O. (2002) Notices Am. Math. Soc. 49, 916-917. [Google Scholar]
- 22.Fernández-Baca, D., Seppäläinen, T. & Slutzki, G. (2000) in Combinatorial Pattern Matching, Lecture Notes in Computer Science, eds. Giancarlo, R. & Sankoff, D. (Springer, Berlin), Vol. 1,848, pp. 68-82. [Google Scholar]
- 23.Gusfield, D., Balasubramanian, K. & Naor, D. (1994) Algorithmica 12, 312-326. [Google Scholar]
- 24.Waterman, M., Eggert, M. & Lander, E. (1992) Proc. Natl. Acad. Sci. USA 89, 6090-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]