Abstract
Nik-related kinase (Nrk) is a Ser/Thr kinase and was initially discovered as a molecule that was predominantly detected in skeletal muscles during development. A recent study using Nrk-null mice suggested the importance of Nrk in proper placental development; however, the molecular mechanism remains unknown. In this study, we demonstrated that differentiated trophoblasts from murine embryonic stem cells (ESCs) endogenously expressed Nrk and that Nrk disruption led to the enhanced proliferation of differentiated trophoblasts. This phenomenon may reflect the overproliferation of trophoblasts that has been reported in enlarged placentas of Nrk-null mice. Furthermore, we demonstrated that AKT phosphorylation at Ser473 was upregulated in Nrk-null trophoblasts and that inhibition of AKT phosphorylation cancelled the enhanced proliferation observed in differentiated Nrk-null trophoblasts. These results indicated that the upregulation of AKT phosphorylation was the possible cause of enhanced proliferation observed in Nrk-null trophoblasts. The upregulation of AKT phosphorylation was also confirmed in enlarged Nrk-null placentas in vivo, suggesting that proper regulation of AKT by Nrk was important for normal placental development. In addition, our detailed analysis on phosphorylation status of AKT isoforms in newly established trophoblast stem cells (TSCs) revealed that different levels of upregulation of AKT phosphorylation were occurred in Nrk-null TSCs depending on AKT isoforms. These results further support the importance of Nrk in proper development of trophoblast lineage cells and indicate the possible application of TSCs for the analysis of differently regulated activation mechanisms of AKT isoforms.
Introduction
The placenta is a unique and complicated organ that plays an essential role in supporting foetal development through its involvement in various functions such as gas exchange, nutrient supply and hormone production during gestation. Because placental defects are associated with many pregnancy-related disorders [1, 2], a better understanding of placental development is important to overcome these clinical problems. Animal models have been used to elucidate the underlying mechanisms of human disorders. From the 1990s to date, numerous gene knockout mice have been generated. Among them, >100 mutant mice have placental defects and have substantially contributed to the advancing of our knowledge regarding the genetic control of placental development [1, 3]. However, further investigations are necessary for the complete understanding of placental development.
Placentomegaly is a major placental abnormality that is associated with several foetal disorders and is present in various gene-manipulated mice, including those with Igf2, H19, Cdkn1 and Phlda2 malfunctions [4–6]. These genes are well conserved in humans and are included in a large cluster of imprinted genes that are located on mouse chromosome 7. Dysregulation of this imprinted region may be associated with Beckwith–Widemann syndrome, which is also known as an overgrowth syndrome that exhibits placentomegaly [5–8]. In addition, previous studies suggested important roles of X-linked genes, such as Gpc3 [9], Esx1 [10], Plac1 [11] and Nrk [12], in regulating normal placental development because mice that lacked any of these X-linked genes had enlarged placentas [9–12]. The human GPC3 gene is responsible for the type 1 Simpson–Golabi–Behmel syndrome, which is another overgrowth syndrome that is characterised by placentomegaly [9]. Moreover, cloned mice that were generated by somatic cell nuclear transfer exhibited placentomegaly with a reduced expression of a number of X-linked genes [13, 14]. Because these previous reports revealed that many placentomegaly-related genes were present on either chromosome 7 or the X chromosome in mice, further studies of the genes on these chromosomes are important to understand the pathogenesis of pregnancy-related disorders.
One of the placentomegaly-related X-linked genes, Nik-related kinase (Nrk), encodes a Ser/Thr kinase that belongs to the germinal centre kinase family. Nrk was originally cloned as a gene that was expressed only in the skeletal muscle during development [15, 16]. However, Denda et al. [12] recently reported that Nrk was also expressed in the spongiotrophoblast layer of the murine placenta and that NRK expression was also detected in the human placenta. This study further demonstrated a high frequency of neonatal death, delivery defects and placental overgrowth in Nrk-null mice, suggesting a novel role of Nrk in reproduction.
Because placentomegaly has been closely related to pregnancy-related disorders, we aimed to elucidate the mechanism through which Nrk deficiency induced placental enlargement. Previous studies regarding overexpression experiments in cultured cells have suggested that Nrk participated in JNK signalling pathway activation through the MEKK1 and MKK4 kinase cascade [16], promotion of apoptosis through JNK activation [17] and polymerization of actin through cofilin phosphorylation [18]. Because these phenomena have been observed in cells that ectopically expressed Nrk, further studies using placental trophoblast cells that endogenously express Nrk are desirable to better understand the molecular functions of Nrk in placental development. The caudal-related homeobox protein CDX2 is the earliest trophoblast-specific transcription factor, and Cdx2 overexpression can induce trophoblast lineages from pluripotent murine embryonic stem cells (ESCs) [19, 20]. Furthermore, Hayashi et al. [21] recently reported that ESCs were able to differentiate into trophoblast lineages on laminin-coated dishes in a BMP4-containing medium through Cdx2 expression. Therefore, this culture system could be applicable for examining the role of Nrk in trophoblasts during placental development. In the present study, we further modified this culture system and demonstrated that Nrk deficiency resulted in enhanced trophoblast proliferation, which could reflect the enlarged placenta observed in Nrk-null mice. Furthermore, we found that enhanced proliferation in Nrk-null trophoblasts was associated with the upregulation of AKT phosphorylation at Ser473. We also established murine trophoblast stem cells (TSCs) culture system that is known as an excellent tool for deciphering gene function in trophoblasts [22] and explored the involvement of Nrk in AKT activation and trophoblast development. Our results indicated that NRK expression was responsible for the regulation of proper AKT activation in TSCs, which was similarly observed in ESC-derived trophoblasts. Moreover, the upregulation of AKT phosphorylation was confirmed in enlarged placentas that were observed in Nrk-null mice in vivo, suggesting an important role of Nrk in maintaining proper AKT activation during placental development.
Methods
Cell culture
EGR-G101 ESC line was derived from a male C57BL/6N mouse blastocyst that expressed a green fluorescent protein [23]. A feeder-dependent EGR-G101 was routinely cultured [23] and later adapted for a feeder-independent culture as previously reported [24]. Feeder-independent ESCs were maintained on type I collagen-coated (Nitta gelatin, Osaka, Japan) dishes in a defined ESF7 medium containing a supernatant of mLIF-producing cells. Inducing differentiation of feeder-independent ESCs to trophoblasts was performed as previously reported [21]. Briefly, feeder-independent ESCs were seeded on laminin-coated (Sigma-Aldrich, St. Louis, MO) dishes in a defined ESF5 medium containing rhBMP4 (BioLegend, San Diego, CA). After 4 days of culture, the cells reached confluency and were then subcultured for an additional 4 days before finally differentiating to trophoblasts.
Mouse TSC lines from wild-type and Nrk-null embryos were established from expanded blastocysts as described previously [22]. Undifferentiated TSCs were cultured in TSC medium [RPMI1640 (Nacalai Tesque, Kyoto, Japan) supplemented with 20% fetal bovine serum (Biowest, Riverside, MO), 1 mM sodium pyruvate (Nacalai Tesque), 0.1 mM β-mercaptoethanol (Thermo Fisher Scientific, Waltham, MA), 2 mM L-glutamine (Nacalai Tesque) and 100 U/ml penicillin and 0.1 mg/ml streptomycin (Nacalai Tesque)] containing 70% mouse embryonic fibroblast-conditioned medium (MEF-CM), 25 ng/ml hFGF4 (BioLegend) and 1 μg ml/ml heparin (Sigma-Aldrich). Differentiation of TSCs was induced by culturing the cells in TSC medium without MEF-CM, hFGF4 and heparin for 6 days.
RNA isolation and RT-PCR analysis
Total RNA was extracted using TRIzol (Thermo Fisher Scientific) from three independent cultures of each cell type, then 0.5 μg of each RNA sample was reverse transcribed using the ReverTraAce (Toyobo, Osaka, Japan). One twentieth of each cDNA sample was subjected to PCR analysis. Conventional PCR followed by gel electrophoresis was performed using the KOD FX (Toyobo) on the Thermal Cycler Dice Touch (Takara bio, Shiga, Japan). Real-time PCR was performed using the THUNDERBIRD SYBR qPCR Mix (Toyobo) on the StepOne Real-Time PCR System (Thermo Fisher Scientific). Data were analyzed by Comparative CT Method using StepOne software and the relative expression level of each target gene mRNA was normalized to the amount of Gapdh control. Each reaction was performed in triplicate and data are presented as the mean plus SD. The primer sets used in this study are shown in Table 1.
Table 1. List of primer sequences.
| Name | Sequence |
|---|---|
| Nrk-F | 5′-tagtggattttggagtgagtgc-3′ |
| Nrk-R | 5′-cttctgtaatcataggaacacc-3′ |
| Plf-F | 5′-aggaacaagccaggctcaca-3′ |
| Plf-R | 5′-ttccggactgcgttgatctt-3′ |
| Tpbpa-F | 5′-acagccagttgttgatgacc-3′ |
| Tpbpa-R | 5′-caggatcccacttgtcagg-3′ |
| Hand1-F | 5′-cgtgagtgcatccccaatg-3′ |
| Hand1-R | 5′-gccagcacgtccatcaagtag-3′ |
| Psx1-F | 5′-cgatggatgggtgtggatga-3′ |
| Psx1-R | 5′-tgacagggctggcactcaag-3′ |
| Mash2-F | 5′-cgggatctgcactcgaggat-3′ |
| Mash2-R | 5′-ggtgggaagtggacgtttgc-3′ |
| Esx1-F | 5′-gagctggaggcctttttcca-3′ |
| Esx1-R | 5′-acacccacagggggactcat-3′ |
| Ets2-F | 5′-ctcggctcaacaccgtcaat-3′ |
| Ets2-R | 5′-agctgtccccaccgttctct-3′ |
| Pl2-F | 5′-ctgctcttccacatgtacct-3′ |
| Pl2-R | 5′-taagagcactgttcttagtgg-3′ |
| Essrb-F | 5′-atggatgaggaacactctcg-3′ |
| Essrb-R | 5′-cagctcatagtcctgcagc-3′ |
| Gapdh-F | 5′-tgtgtccgtcgtggatctga-3′ |
| Gapdh-R | 5′-ttgctgttgaagtcgcaggag-3′ |
| 5′-F | 5′-actggtgaatttggagcagtcaaaacttcc-3′ |
| 5′-R | 5′-cttccccacaacgggttcttctgttagtcc-3′ |
| 3′-F | 5′-ggttgatatctctatagtcgcagtaggcgg-3′ |
| 3′-R | 5′-ttgtggaccggacaagtaatcctgcagtcg-3′ |
| F | 5′-acaccaagtaattcaccgggacatcaaagg-3′ |
| WT-R | 5′-gtcttagatagtctttcagaaagcatcg-3′ |
| KO-R | 5′-cttccccacaacgggttcttctgttagtcc-3′ |
Generation of Nrk-null ESC
The targeting vector PRPGS00061_B_H04 was purchased from the International Knock-Out Mouse Program. Gene targeting in feeder-dependent EGR-G101 ESCs was performed as previously described [25, 26]. Briefly, single cell suspensions (0.8 mL of 107 cells/mL) were mixed with 20 μg of AsiSI-digested linear-targeting vector and were subjected to electroporation at a pulse of 250 V and 500 μF using a Gene Pulser II (Bio-Rad, Hercules, CA). The mutated cells were selected with G418 (150 μg/mL), and the homologous recombination events were detected using PCR amplification with the following primers: 5′-F and 5′-R for the 5′-arm and 3′-F and 3′-R for the 3′-arm. The primer sequences are shown in Table 1.
Cell proliferation assay
Cell proliferation was evaluated by counting the number of cells. Cells were washed with PBS and were then dissociated with TrypLETM Express Enzyme (Thermo Fisher Scientific). The dissociated cell suspensions were mixed with an equal volume of 0.5% trypan blue stain solution (Nacalai Tesque), and the cell numbers were counted using the LunaTM automated cell counter (Logos Biosystems, Annandale, VA). The proliferation rate was calculated relative to the cell numbers of the day 1 controls. The value of day 1 was indicated as 1.
Western blotting
To prepare whole-cell extracts, cells were first washed with ice-cold PBS and were then lysed in RIPA buffer [1% Nonidet P-40, 150 mM NaCl, 50 mM Tris-HCl (pH7.5), 1 mM EDTA and 0.5% sodium deoxycholate] that contained the cOmpleteTM, Mini protease inhibitor cocktail (Roche, Basel, Switzerland) and the PhosSTOPTM phosphatase inhibitor cocktail (Roche) on ice. After being passed through a 26-gauge needle 10 times, cell extracts were centrifuged and the supernatants were collected. To prepare the extracts of E18.5 mouse placentas, which had been stored at −80°C, the tissues were disrupted using a bead crusher μT-01 (TAITEC, Saitama, Japan) for 15 s at maximum speeds in ice-cold RIPA buffer that was supplemented with protease and phosphatase inhibitors. After centrifugation, the supernatants were collected as placental extracts. The protein concentration of each sample was determined using the Quick StartTM Bradford 1x Dye Reagent and the Quick StartTM Bovine Serum Albumin Standard Set (Bio-Rad), according to the manufacturer’s instructions.
Whole-cell extracts (30 μg) or placental extracts (100 μg) were separated using SDS-PAGE with 12% gel and subsequently transferred onto a polyvinylidene fluoride membrane. The membranes were blocked with TBST buffer [25 mM Tris-HCl (pH 7.5), 137 mM NaCl, 2.5 mM KCl and 0.1% Tween-20] that contained 5% skim milk. After washing with TBST, the blocked membranes were incubated overnight at 4°C with a primary antibody in TBST that contained 5% BSA. After washing with TBST, the membranes were incubated for 1 h at room temperature with a secondary antibody in TBST that contained 5% skim milk. After washing with TBST, the protein–antibody complex was detected using the Chemi-Lumi One L (Nacalai Tesque) and the ImageQuantTM LAS 4000 mini imaging system (GE Healthcare Japan, Tokyo, Japan), according to the manufacturer’s instructions. The signal density was analyzed by ImageQuantTM TL software. As needed, membranes were stripped by incubation at 50°C in SDS-based stripping buffer [62.5 mM Tris-HCl (pH 6.8), 2% SDS and 100 mM β-mercapthoethanol] for 30 min to detect other proteins.
Immunoprecipitation
For immunoprecipitation, 1 mg/ml whole-cell extracts were prepared as described above. The 250 μg proteins were incubated for 3 h at 4°C with the indicated antibody under constant rotation. Then Protein A-coated magnetic beads (Bio-Rad) were added and mixed samples were incubated overnight at 4°C under constant rotation. Magnetic beads were washed 3 times with ice-cold RIPA buffer supplemented with phosphatase inhibitors and the binding proteins were eluted using SDS-PAGE loading buffer for western blotting.
Antibodies
Anti-NRK rabbit polyclonal primary antibodies were generated against a synthetic peptide (CGAAGYNGGDVGGNHGAAFN), and then purified with a column immobilized with antigen peptides (Sigma-Aldrich). We confirmed that the newly established polyclonal antibodies are able to detect several proteins in wild-type cells, but not in Nrk-null cells. Because it has been reported that ectopically expressed NRK in culture cells is cleaved by caspases [17], the largest protein probably corresponds to full-length NRK and others are judged to be the cleaved fragments of NRK proteins. The antibodies also recognize several non-specific proteins that are present in both wild-type and Nrk-null cells. These anti-NRK antibodies were used at a 1:4,000 dilution for western blotting. The commercially available primary and secondary antibodies used in this study are summarized in Table 2.
Table 2. List of antibodies.
| Primary antibodies | |||
| Name | Company | Catalog No. | Dilution |
| Phospho-Akt (Ser473) | CST | #4060 | 1:2,000 (WB)1:50 (IH) |
| Phospho-Akt (Thr308) | CST | #13038 | 1:1,000 (WB) |
| Akt (pan) | CST | #4691 | 1:1,000 (WB)1:50 (IH) |
| Akt1 | CST | #2938 | 1:1,000 (WB)1:50 (IP) |
| Akt2 | CST | #3063 | 1:1,000 (WB)1:100 (IP) |
| Akt3 | CST | #14982 | 1:1,000 (WB)1:100 (IP) |
| Phospho-Erk1/2 (Thr202/Tyr204) | CST | #4370 | 1:2,000 (WB) |
| Erk1/2 | CST | #4695 | 1:1,000 (WB) |
| Phospho-SAPK/JNK (Thr183/Tyr185) | CST | #9251 | 1:1,000 (WB) |
| SAPK/JNK | CST | #9252 | 1:1,000 (WB) |
| β-Actin | Sigma-Aldrich | A5441 | 1:5,000 (WB) |
| Secondary antibodies | |||
| Name | Company | Catalog No. | Dilution |
| HRP-conjugated goat anti-rabbit IgG | Jackson IR | 111-035-003 | 1:5,000 |
| HRP-conjugated donkey anti-mouse IgG | Jackson IR | 715-036-151 | 1:20,000 |
CST: Cell Signaling Technology, Jackson IR: Jackson ImmunoResearch, WB: western blotting, IH: immunohistochemistry, IP: immunoprecipitation
Inhibition of AKT phosphorylation
The AKT inhibitor MK-2206 (Cayman Chemical, Ann Arbor, MI) was dissolved in DMSO solution, stored at −80°C as a stock solution (5 mM) and then diluted with a culture medium prior to use. Undifferentiated ESCs were seeded on a laminin-coated 24-well plate at 106 cells/well in an rhBMP4-supplemented ESF5 medium, and the adherent cell number was counted 1 day after seeding. The cells were treated at 0–500 nM of MK-2206 on day 2 and maintained in this culture condition until day 4 (n = 8). Four days after seeding, the cell number was counted (n = 4) and the proliferation rate was calculated relative to the cell numbers of the untreated day 1 controls. The value of day1 was indicated as 1. The remaining cells (n = 4) were collected together, and western blotting was performed to assess the AKT Ser473 phosphorylation status.
Animals
Wild-type C57BL/6N and ICR mice were purchased from Japan SLC (Shizuoka, Japan) and were maintained under specific-pathogen free conditions. The mice were housed in plastic cages with wire mesh lids and water bottles (CLEA Japan, Tokyo, Japan), and were put on the open racks. Mice were fed Labo MR Stock diet (Nosan, Kanagawa, Japan) and maintained on wood chip bedding (Iwakura, Hokkaido, Japan). All animals were observed daily for signs of illness or abnormal behavior. All animal experiments were conducted in accordance with the Hokkaido University Guide for the Regulation on Animal Experimentation. All experiments were reviewed and approved by the Animal Care and Use Committee of the Hokkaido University (Permit Number: 10–0114 and 14–0143). All surgery was performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering. Prior to our experimental endpoint, no mice became severely ill or died. In our experimental endpoint, all mice were euthanized by a high-dose (>120 mg/kg) intraperitoneal administration of sodium pentobarbital.
Generation of Nrk-null mice
We generated chimeric mice by aggregation of Nrk-null ESCs and 8-cell embryos, followed by transplantation into pseudopregnant ICR females as previously described [27]. Briefly, 8-week-old wild-type ICR females were superovulated and then mated with wild-type ICR males. Two-cell stage embryos were collected from the females at 1.5 days after copulation and then incubated in KSOM-AA medium for 1 day to obtain 8-cell embryos. The zona pellucida of the 8-cell embryos was removed with acidic Tyrode’s solution (Sigma-Aldrich) [28]. These 8-cell embryos without zona pellucida were individually incubated in 5 μL of KSOM-AA microdrops that contained 5–10 ESCs for 1 day to obtain blastocysts. We transplanted 10 blastocysts into each horn of the uterus of pseudopregnant ICR females. The chimeric male mice were mated with wild-type C57BL/6N females, and the germ line transmission was confirmed by observing green fluorescence in the neonates [29]. Mice were genotyped using PCR amplification with the following primers: F and WT-R for detection of the wild-type allele. F and KO-R for detection of the targeted allele. The primer sequences are shown in Table 1.
Histological and immunohistochemical analysis
E18.5 placentas were fixed in 4% paraformaldehyde in PBS and were processed for paraffin embedding. For histological analysis, the 4-μm paraffin sections were stained with periodic acid-Schiff (PAS) and then counterstained with hematoxylin. For immunohistochemical analysis, the 4-μm paraffin sections were incubated overnight at 4°C with indicated primary antibodies. After incubation with secondary antibodies conjugated with horseradish peroxidase, sections were stained with diaminobenzidine tetrahydrochloride and then counterstained with hematoxylin.
Statistics
Data were presented as mean plus SD. Groups were compared using the two-tailed Student’s t-test. We considered P values of <0.01 to be significant.
Results
Establishment of an in vitro culture system for investigating the role of Nrk in trophoblast proliferation and differentiation
To investigate the precise roles of Nrk in trophoblast proliferation and differentiation, we first attempted to establish an in vitro culture system, wherein we could observe Nrk expression during trophoblast differentiation. As shown in Fig 1A, Nrk mRNA expression was not detected in undifferentiated ESCs. After initiating trophoblast differentiation, Nrk mRNA expression was faintly detected on the fourth day of culture and was clearly induced after 8 days of culture. These results indicated that this culture system could be applicable for examining the role of Nrk during ESC differentiation to trophoblasts.
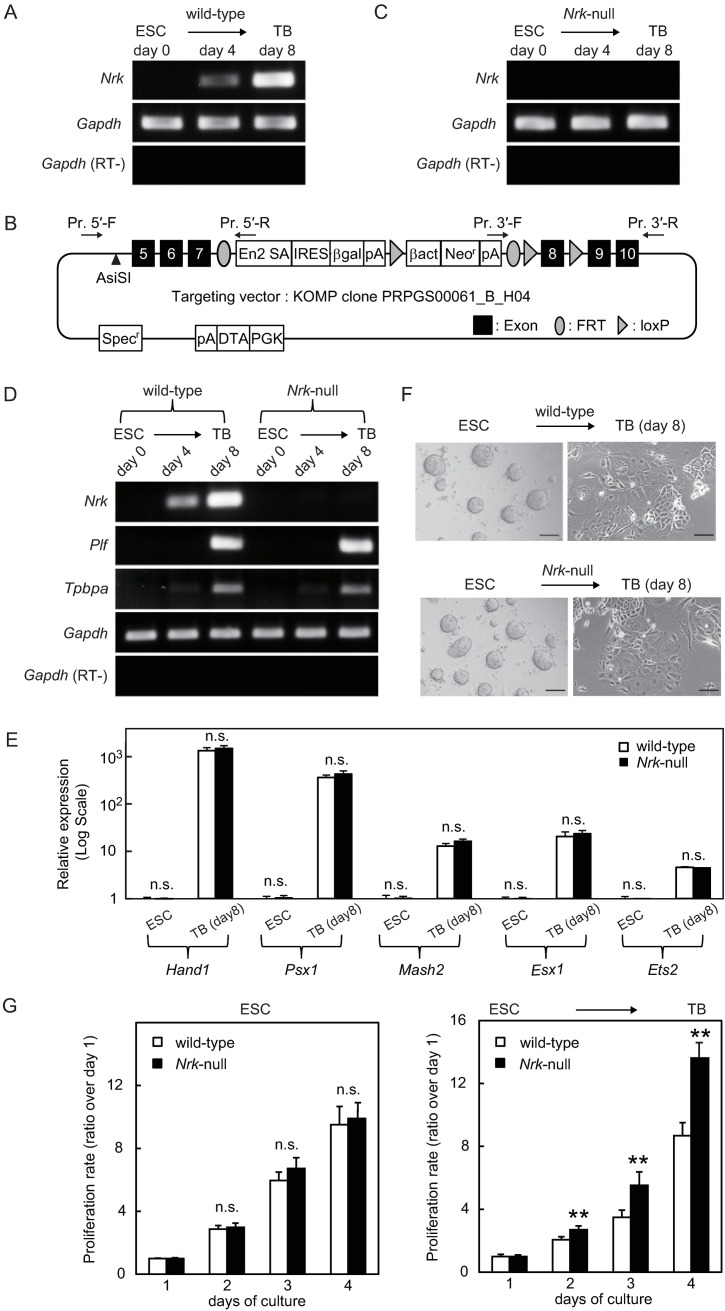
Fig 1. Disruption of Nrk resulted in enhanced proliferation of trophoblasts without obvious influence on their differentiation.
(A) Nrk expression in wild-type cells was examined using RT-PCR analysis. Total RNAs were isolated from undifferentiated ESCs (day 0) and differentiated trophoblasts (day 4 and day 8) and were subjected to RT-PCR amplification. As an internal control, Gapdh was also amplified. RT-PCR amplification of Gapdh was also performed in the absence of reverse transcriptase (RT-). (B) Construct of the gene-targeting vector for Nrk disruption. The endogenous Nrk expression was disturbed by a gene-trapping cassette inserted between exon 7 and 8 of Nrk gene. The arrowhead indicates AsiSI recognition site for linearization of the vector. The arrows indicate PCR primers for detection of the homologous recombination events. (C) Deficiency of Nrk expression in Nrk-null cells was confirmed using RT-PCR analysis. Total RNAs were isolated from Nrk-null cells and were subsequently subjected to RT-PCR amplification as described in (A). (D) RT-PCR analysis of trophoblast marker gene expressions in wild-type and Nrk-null cells. Total RNAs were isolated from undifferentiated ESCs (day 0) and differentiated trophoblasts (day 4 and day 8) and were subsequently subjected to RT-PCR amplification as described in (A). (E) Real-time PCR analysis of transcription factor expressions in wild-type and Nrk-null cells. Undifferentiated ESCs and differentiated trophoblasts (day 8) were analyzed (n = 3). The examined gene expressions were normalized to the amount of Gapdh control. Each gene expression in undifferentiated wild-type ESCs is indicated as 1. Data are presented as mean plus SD. n.s. = no significant difference. (F) Phase-contrast photomicrographs of undifferentiated ESCs (left) and differentiated trophoblasts at day 8 (right) with or without Nrk. Scale bars indicate 200 μm. (G) Proliferation of undifferentiated ESCs and differentiated ESCs into trophoblasts. Undifferentiated ESCs were seeded on a type I collagen-coated 24-well plate at 106 cells/well in a LIF-supplemented ESF7 medium (left) in which undifferentiated state of ESCs was maintained. Undifferentiated ESCs were seeded on a laminin-coated 24-well plate at 106 cells/well in an rhBMP4-supplemented ESF5 medium (right) in which ESCs were induced to differentiate into trophoblasts. The cells were counted every 24 h (n = 6). The value of day1 is indicated as 1. Data are presented as mean plus SD. **P<0.01 compared to wild-type. n.s. = no significant difference. Shown are cropped gels. The gels with indicated cropping lines are shown in S1 Fig. ESC: embryonic stem cell, TB: trophoblast.
To further understand this role, we generated Nrk-mutated EGR-G101 cells by homologous recombination with the gene-targeting vector shown in Fig 1B. We believed that a gene-trapping cassette that was inserted between exon 7 and 8 of Nrk gene would disrupt the endogenous Nrk expression. EGR-G101 cells with an Nrk-destructive mutation on the X chromosome were found to be incapable of producing NRK proteins because EGR-G101 cells have an XY genotype. To select Nrk-mutated cells, we assessed the homologous recombination using PCR analysis and then examined the ability of the mutated cells to express Nrk mRNA using RT-PCR analysis. In contrast to the clear induction of Nrk mRNA in wild-type EGR-G101 cells (Fig 1A), mutant EGR-G101 cells did not express Nrk mRNA even after 8 days of culture under differentiation-inducing conditions (Fig 1C), indicating Nrk deficiency in the mutant cells. Thus, we created an in vitro culture system wherein we could observe trophoblast proliferation and differentiation both in the presence and absence of Nrk.
Disruption of Nrk did not show obvious influence on the differentiation from ESC to trophoblast
To examine whether ESC differentiation to trophoblasts was influenced by Nrk expression, we next compared trophoblast marker gene (Plf [30] and Tpbpa [31]) expressions, which were absent in ESCs, between wild-type and Nrk-null cells during trophoblast differentiation using RT-PCR analysis. As shown in Fig 1D, we found that the expression of both marker genes was similarly induced during trophoblast differentiation, regardless of the presence or absence of Nrk. In addition, we performed real-time PCR analysis to assess transcription factor (Hand1 [32], Psx1 [33], Mash2 [34], Esx1 [35] and Ets2 [36]) expressions. These genes had been detected even in undifferentiated ESCs and had been enhanced during trophoblast differentiation. Our results demonstrated that the relative mRNA levels were increased approximately 4- to 1,500-fold in the differentiated trophoblasts on day 8 as compared with those in ESCs and that a similar degree of increase in each transcription factor was induced in both wild-type and Nrk-null cells during differentiation (Fig 1E). Moreover, we observed that the morphological changes during differentiation were not affected by Nrk deficiency (Fig 1F). These results suggested that Nrk did not play a critical role in the differentiation process from ESCs to trophoblasts.
Disruption of Nrk resulted in enhanced proliferation of trophoblasts, which was associated with the upregulation of AKT Ser473 phosphorylation
Next, we examined whether Nrk deficiency affected cell proliferation. After having seeded ESCs, the cell numbers were counted every day until day 4 of culture, and the proliferation rate was calculated relative to the cell numbers on day 1. To avoid cell passaging during a proliferation assay, we thus decided to detect cell proliferation until day 4. Indeed, we confirmed Nrk expression on day 4 in wild-type cells (Fig 1A) and thus believed that a culture period of 4 days is enough to detect the difference of the cell proliferation between wild-type and Nrk-null cells. Our analysis of undifferentiated ESCs revealed equivalent levels of proliferation regardless of Nrk genotype (Fig 1G, left), which was consistent with the fact that Nrk was not detected in ESCs as shown in Fig 1A. In contrast, when we cultivated ESCs under differentiation-inducing conditions, a significant increase in proliferation was observed in Nrk-null cells on day 2 as compared with that in wild-type cells and was maintained until day 4 of culture (Fig 1G, right).
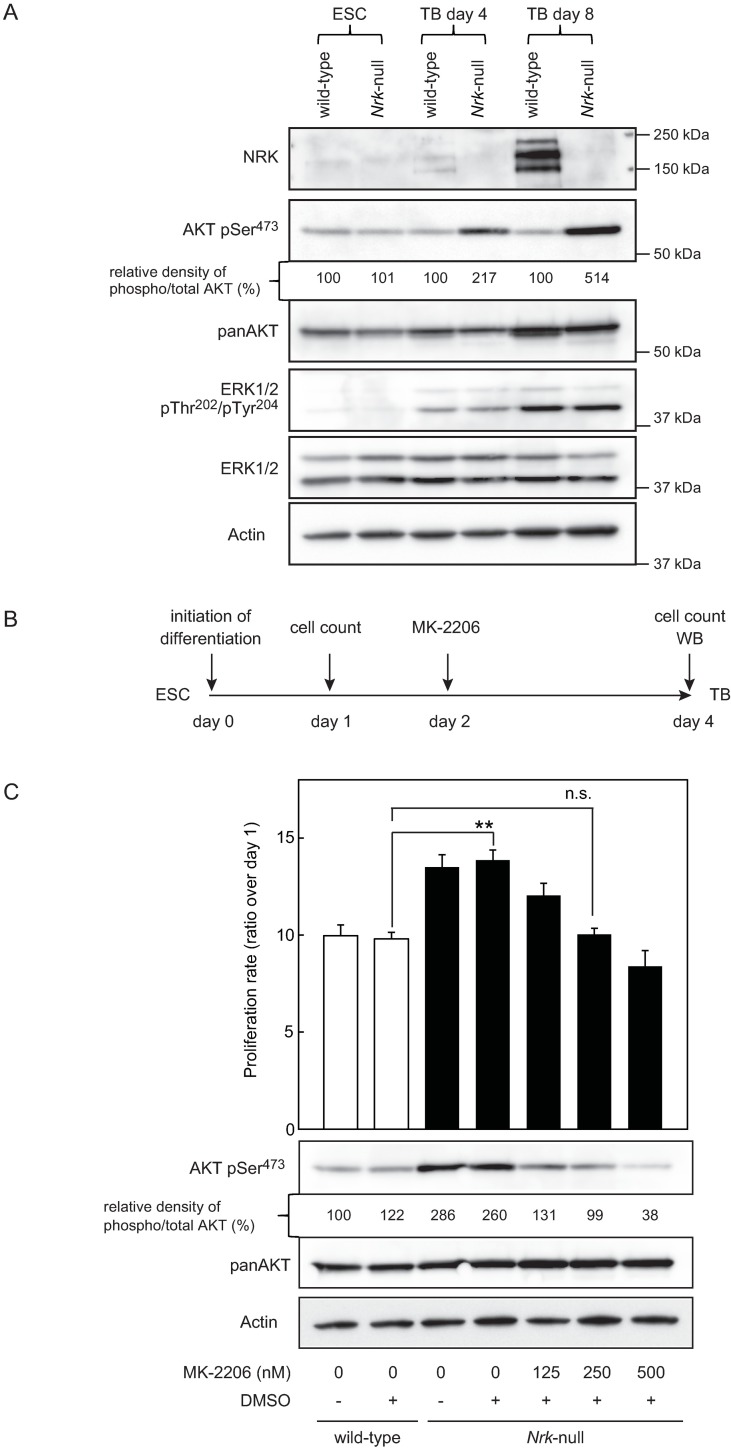
To explore the molecular mechanism of enhanced proliferation that was observed in Nrk-null trophoblasts, we first examined the phosphorylation status of JNK using western blotting. Although previous studies have suggested that Nrk participated in JNK activation [16–17], we detected no significant difference in JNK phosphorylation between wild-type and Nrk-null cells as shown in S2 Fig. This was consistent with a previous report by Denda et al. describing that the levels of JNK phosphorylation in placentas were equivalent between wild-type and Nrk-null embryos [12]. Next, we examined the phosphorylation statuses of AKT and ERK1/2 that have both been associated with placental development [37, 38] using western blotting. As shown in Fig 2A, we confirmed the induction of NRK protein expression in differentiated trophoblasts that were derived from wild-type ESCs but not from Nrk-null ESCs; this was consistent with the mRNA expression patterns shown in Fig 1D. Under these conditions, we found that AKT phosphorylation at Ser473 was upregulated in Nrk-null trophoblasts as compared with that in the wild-type controls. This was not due to the enhanced expression of total AKT proteins because we assessed the total AKT protein levels and failed to detect any significant differences between wild-type and Nrk-null trophoblasts. An anti-panAKT monoclonal antibody used in this study recognises all three AKT isoforms (AKT1, AKT2 and AKT3). Calculation of relative densities of phosphorylated vs total AKT revealed 2- to 5-fold increment of AKT Ser473 phosphorylation in Nrk-null trophoblasts as compared with that in wild-type control. We further assessed ERK1/2 phosphorylation and found no significant differences between wild-type and Nrk-null cells during the experiment, although a similar induction of ERK1/2 phosphorylation was observed in both types of cells during trophoblast differentiation. Thus, ERK1/2 phosphorylation may also be associated with trophoblast differentiation but may not be involved in the enhanced trophoblast proliferation that was observed in Nrk-null cells. Altogether, our results strongly suggested that enhanced proliferation observed in Nrk-null trophoblasts was caused by the upregulation of AKT Ser473 phosphorylation.
Fig 2. Enhanced proliferation observed in differentiated Nrk-null trophoblasts was associated with the upregulation of AKT Ser473 phosphorylation.
(A) Western blot analysis of whole-cell extracts from ESCs and trophoblasts. Expressions of NRK, phosphorylated AKT at Ser473, total AKT, phosphorylated ERK1/2 at Thr202/Tyr204 and total ERK1/2 were compared between wild-type and Nrk-null cells. As an internal control, β-Actin was also detected. The relative density of phosphorylated vs total AKT was analyzed and was normalized to the value of wild-type. (B) The schematic representation of the experimental procedure to examine the effect of AKT inhibitor MK-2206. (C) Dose dependent effect of MK-2206 on the proliferation and AKT Ser473 phosphorylation in the differentiated Nrk-null trophoblasts. The proliferation rate of the trophoblasts on day 4 was calculated relative to the untreated day 1 control (n = 4) (top). Data are presented as mean plus SD. **P<0.01compared to wild-type. n.s. = no significant difference. Expression of phosphorylated AKT at Ser473 and total AKT in trophoblasts on day 4 was analyzed by western blotting (bottom). As an internal control, β-Actin was also detected. The relative density of phosphorylated vs total AKT was analyzed and was normalized to the value of untreated wild-type control. Shown are cropped blots. The blots with indicated cropping lines are shown in S3 Fig. WB: western blotting, ESC: embryonic stem cell, TB: trophoblast.
Inhibition of AKT Ser473 phosphorylation cancelled the enhanced proliferation observed in differentiated Nrk-null cells
To further confirm whether excessive and non-physiological AKT Ser473 phosphorylation was responsible for enhanced proliferation of Nrk-null trophoblasts, we assessed the effect of an AKT-specific allosteric inhibitor, MK-2206, on Nrk-null trophoblast proliferation. MK-2206 binds to AKT, resulting in a decrease in AKT phosphorylation in a non-ATP competitive manner [39]. The schematic representation of the experimental procedure is shown in Fig 2B. The effects of MK-2206 were evaluated on day 4 by cell counting and western blotting. When we added various doses of MK-2206 dissolved in DMSO to the culture of differentiating Nrk-null trophoblasts on day 2, a dose-dependent proliferation inhibition was clearly observed on day 4 (Fig 2C, top). It was of note that this decrease in cell proliferation was accompanied by a reduction in AKT Ser473 phosphorylation. In particular, it appeared that 250 nM of MK-2206 was enough to reduce the level of AKT Ser473 phosphorylation in the Nrk-null cells to the level in the wild-type trophoblasts in which only DMSO was added (Fig 2C, bottom). Interestingly, similar levels of cell proliferation were observed in these two cell populations with equivalent levels of AKT Ser473 phosphorylation. These results further indicated that the upregulation of AKT activation was the possible cause of enhanced proliferation observed in Nrk-null trophoblasts.
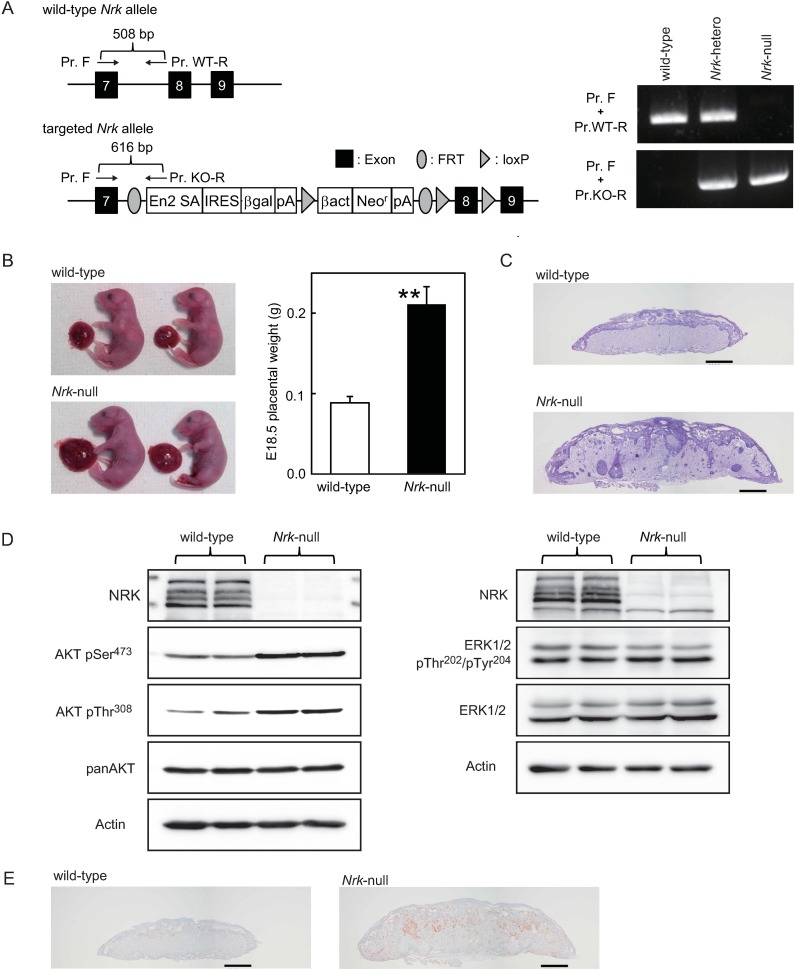
AKT was highly activated in enlarged placentas derived from Nrk-null mice
To study whether Nrk deficiency led to aberrant activation of AKT in placentas, we developed Nrk-null mice from Nrk-null EGR-G101 ESCs. Genotypes of mice were confirmed using PCR analysis (Fig 3A). As shown in Fig 3B, placentas from Nrk-null embryos were significantly larger than those from wild-type controls, which was consistent with the previous observation [12]. In addition, histological analysis by PAS staining indicated the increase of glycogen positive trophoblast cells labelled by PAS in the junctional zone of Nrk-null placenta and the abnormal invasion of these PAS-positive cells into the labyrinth layer. Thus, NRK deficiency resulted not only in hypertrophy but also in the disorder of internal structure of placentas (Fig 3C). A western blot analysis revealed that NRK proteins were absent in Nrk-null placentas that were isolated from two individual embryos at E18.5 (Fig 3D). Furthermore, phosphorylation of AKT at both Ser473 and Thr308 were clearly upregulated in Nrk-null placentas, while total levels of AKT remained constant. In contrast, the levels of phosphorylated and total ERK1/2 proteins were similar in both wild-type and Nrk-null placentas. To confirm whether the cells in placenta express phosphorylated AKT at Ser473, E18.5 placental sections were stained with anti-AKT Ser473 antibodies (Fig 3E). Though wild-type placenta showed few expressions of phosphorylated AKT at Ser473, strong expressions of phosphorylated AKT at Ser473 were detected in a part of junctional zone of Nrk-null placenta. Thus, these results indicated that enhanced AKT phosphorylation was associated with the placentomegaly that was observed in Nrk-null mice and suggested that Nrk-regulating AKT activation was important for normal placental development.
Fig 3. Upregulation of AKT phosphorylation in enlarged placentas derived from Nrk-null E18.5 mouse embryos.
(A) The wild-type Nrk allele and the targeted Nrk allele are shown (left). The arrows indicate PCR primers for detection of each allele. PCR genotyping of mice (right). (B) Wild-type and Nrk-null mouse placentas and foetuses at E18.5 are shown (left). The placental weight was measured at E18.5 in wild-type (n = 24) and Nrk-null (n = 25) embryos (right). Data are presented as mean plus SD. **P<0.01 compared to wild-type. (C) PAS staining of E18.5 placental sections. Scale bars indicate 1 mm. (D) The lysates were prepared from wild-type (n = 2) and Nrk-null (n = 2) placentas at E18.5 and were subjected to western blot analysis. Expressions of NRK, phosphorylated AKT at Ser473, phosphorylated AKT at Thr308 and total AKT were compared between wild-type and Nrk-null placentas (left). Expressions of NRK, phosphorylated ERK1/2 and total ERK1/2 were compared between wild-type and Nrk-null placentas (right). As an internal control, β-Actin was also detected. (E) Immunostaining of E18.5 placental sections with anti-AKT pSer473 antibodies. Scale bars indicate 1 mm. Shown are cropped gels/blots. The gels/blots with indicated cropping lines are shown in S4 Fig.
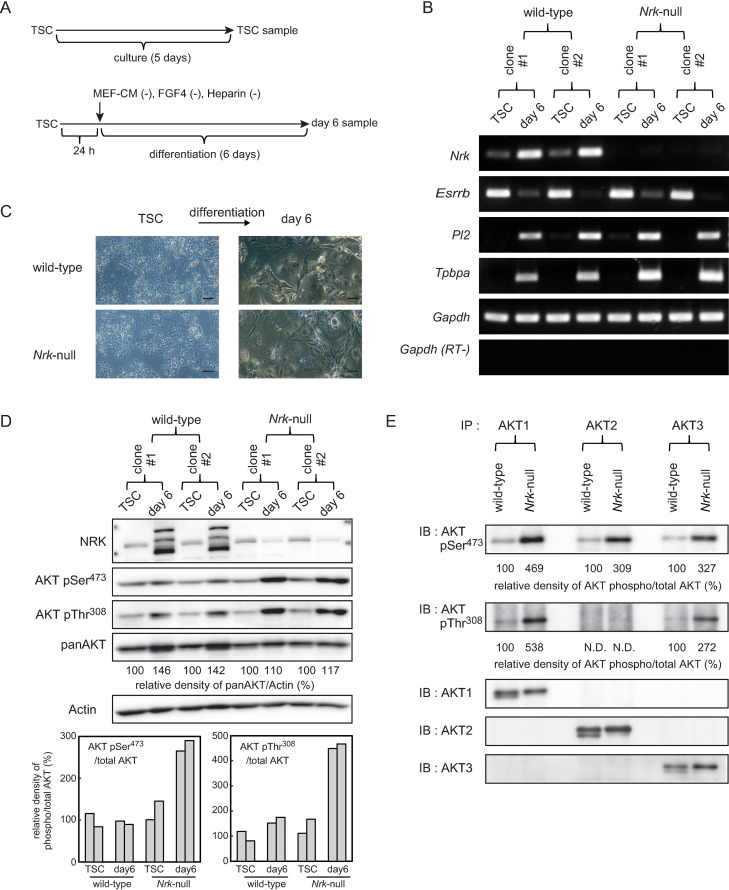
Establishment efficiency of trophoblast stem cell (TSC) lines from wild-type and Nrk-null blastocyst was equivalent
We then tried to establish TSC lines from the blastocysts obtained from Nrk heterozygous mutant X¯X female mice that were mated with wild-type XY male mice. TSCs are derived from trophectoderm that is a precursor of placenta that makes the outermost layer of blastocysts. Theoretically, Nrk deficient X¯Y TSC lines should be established at the rate of 25%, because the expected genotypes would occur in a 1:1:1:1 ratio of X¯X: XX: X¯Y: XY. Indeed, we could establish 16 TSC lines from 36 blastocysts, in which 5 (31%), 4 (25%), 4 (25%) and 3 (19%) lines were identified as genotype X¯X, XX, X¯Y or XY, respectively. These results indicated that Nrk deficiency did not affect the establishment efficiency of TSC lines from mouse blastocysts.
Disruption of Nrk did not show obvious influence on marker gene expressions and cell morphology in undifferentiated and differentiated TSCs
TSCs can maintain their undifferentiated status under defined culture conditions, and can differentiate into multiple trophoblast lineages after withdrawal of FGF4, heparin and MEF-CM for 6 days. To explore the influence of Nrk deficiency on TSCs, samples of undifferentiated TSCs and differentiated TSCs (day 6) were prepared as described in Fig 4A. We first examined mRNA expressions of marker genes in undifferentiated TSCs and differentiated TSCs (day 6) using RT-PCR analysis (Fig 4B). We analyzed two independent clones of both wild-type and Nrk-null cells and confirmed that there was no big difference in the results between two cell lines with the same genotype. In wild-type cells, Nrk mRNA expression was faintly detected in undifferentiated TSC, and was clearly induced after 6 days of culture for differentiation, whereas Nrk mRNA expression was not detected in Nrk-null cells. TSC marker gene Essrb expression was highly detected in undifferentiated TSC and was decreased in differentiated TSCs (day 6) regardless of the presence or absence of Nrk. Both trophoblast giant cell marker gene Pl2 and spongiotrophoblast cell marker gene Tpbpa were not detected in undifferentiated TSCs, and were induced after 6 days of culture for differentiation regardless of the presence or absence of Nrk. Moreover, cell morphology of undifferentiated TSCs and differentiated TSCs (day 6) were not affected by Nrk deficiency (Fig 4C). These results suggested that Nrk did not play a critical role in both the maintenance of undifferentiated TSCs and the differentiation process of TSCs.
Fig 4. Phosphorylation of all AKT isoforms were upregulated in differentiated Nrk-null trophoblast stem cells.
(A) The schematic representation of cell culture conditions. (B) RT-PCR analysis of undifferentiated and differentiated TSC marker gene expressions in wild-type and Nrk-null cells. Total RNAs were isolated from undifferentiated (TSC) and differentiated (day 6) cells and were subjected to RT-PCR amplification. As an internal control, Gapdh was also amplified. RT-PCR amplification of Gapdh was also performed in the absence of reverse transcriptase (RT-). (C) Phase-contrast photomicrographs of undifferentiated TSCs (left) and differentiated TSCs at day 6 (right) with or without Nrk. Scale bars indicate 200 μm. (D) Western blot analysis of whole-cell extracts from undifferentiated (TSC) and differentiated (day 6) cells. Expressions of NRK, phosphorylated AKT at Ser473, phosphorylated AKT at Thr308 and total AKT were compared between wild-type and Nrk-null cells. As an internal control, β-Actin was also detected (top). The relative density of total AKT vs β-Actin was analyzed and was indicated in the blot (top). The value of day 6 was normalized to the value of TSC of each clone. The relative density of phosphorylated vs total AKT was analyzed and was indicated in the graphs (bottom). The relative density was calculated by dividing the percent value for each sample by the mean percent value for wild-type TS cells, which represents 100%. (E) Immunoprecipitation was performed using anti-AKT1, anti-AKT2 and anti-AKT3 antibodies in whole-cell extracts from differentiated (day 6) TSCs. Following immunoprecipitation, immunoblot was performed using anti-AKTpSer473, anti-AKTpThr308, anti-AKT1, anti-AKT2 and anti-AKT3 antibodies. The relative density of phosphorylated vs total AKT was analyzed and was normalized to the value of wild-type. Shown are cropped gels/blots. The gels/blots with indicated cropping lines are shown in S5 and S6 Figs. TSC: trophoblast stem cell, IP: immunoprecipitation, IB: immunoblot, N.D.: not detectable.
AKT phosphorylation was upregulated in differentiated TSCs (day 6) derived from Nrk-null TSCs
To confirm whether Nrk deficiency resulted in activation of AKT in differentiated TSCs (day 6) as well as in differentiated trophoblasts from ESCs (as shown in Fig 2A), western blot analysis was performed (Fig 4D top). Our results showed that NRK protein expression was clearly detected in differentiated wild-type TSCs (day 6), but not in any other cells tested, which was consistent with the mRNA expression patterns shown in Fig 4B. Furthermore, it was revealed that in addition to AKT Ser473 phosphorylation, phosphorylation of AKT Thr308 was also upregulated in differentiated Nrk-null TSCs (day 6). To evaluate the phosphorylation level of each site, we calculated the relative densities of phosphorylated vs total AKT and confirmed 2.8- and 4.6-fold increase of AKT phosphorylation at Ser473 and Thr308, respectively, in Nrk-null TSCs (Fig 4D bottom). We also noticed that the total amount of AKT was increased to 1.4-fold during differentiation in wild-type TSCs. However, the rate of AKT phosphorylation was fairly stable throughout the 6 days of differentiation in these cells. These results indicated that Nrk deficiency resulted in upregulation of AKT phosphorylation in newly established TSCs as well as in trophoblasts derived from ESCs.
Phosphorylation of all AKT isoforms were upregulated in differentiated Nrk-null TSCs
AKT is known to be comprised of three highly homologous isoforms: AKT1, AKT2 and AKT3. Each of these isoforms presents a highly homologous structure and functional redundancy with each other [40]. Though many studies have been focused on the overall role of AKT without regard to isoform specificities, recent studies using gene-manipulated mice which have single or multiple ablation of AKT isoforms indicate the evidence of isoform-specific functions in addition to overlapping functions [41–46]. To examine which AKT isoforms were responsible for the upregulated AKT phosphorylation observed in differentiated Nrk-null TSCs (day 6), immunoprecipitation followed by western blot analysis was performed. As shown in Fig 4E, upregulation of serine phosphorylation was clearly observed in all three AKT isoforms derived from Nrk-null cells compared with those from wild-type cells. The rate of upregulation was 4.7-, 3.1- and 3.3-fold in AKT1, AKT2 and AKT3, respectively. In contrast, threonine phosphorylation was upregulated only in AKT1 (5.4-fold) and AKT3 (2.7-fold) of Nrk-null cells. AKT2 was not phosphorylated at the threonine residue in both wild-type and Nrk-null cells. Antibody-specific immunoprecipitation was properly performed, because equivalent amounts of each AKT isoform were precipitated and detected by the corresponding antibodies.
Discussion
In this study, we found that increased phosphorylation of AKT was associated with enhanced proliferation observed in Nrk-null trophoblasts that were differentiated from ESCs. Furthermore, we indicated that AKT was actually hyperphosphorylated in enlarged placentas of Nrk-null mice. Since AKT has been shown to be important for cell growth and survival [47], it is reasonable to assume that hyperphosphorylation of AKT leads to enhanced proliferation of trophoblasts, resulting in placentomegaly. Indeed, a previous study has demonstrated the overproliferation of the spongiotrophoblast layer cells in enlarged placentas of Nrk-null mice. This result suggested the presence of unidentified factors that were responsible for abnormal trophoblast proliferation [12]. Moreover, it has been reported that AKT1 was widely expressed in murine placentas and that Akt1-deficient placentas exhibited severe hypotrophy, signifying the importance of AKT1 for normal placental development [37]. A recent study has also demonstrated that the deletion of TFAP2C resulted both in severe placental abnormalities with reduced trophoblast population and in the downregulation of AKT phosphorylation; this further indicated the pivotal role of AKT in proper trophoblast proliferation and placental development [47].
NRK induces the activation of the JNK pathway [16], promotes apoptosis [17] and phosphorylates an actin-depolymerizing protein, cofilin [18] in cultured cells that forcibly express NRK. However, these functions were not shown to be directly related to the regulation of AKT phosphorylation that we observed in this study. Moreover, it appeared that NRK did not directly phosphorylate AKT because a disruption in NRK expression resulted in hyperphosphorylation but not hypophosphorylation of AKT (Figs 2A, 3D and 4D). Because AKT phosphorylation was controlled by the balance between protein kinases and phosphatases, NRK may be involved in regulating phosphorylation by either repressing kinases or activating phosphatases in the AKT signal transduction pathway. A study demonstrated that after stimulating the PI3K pathway, AKT was rapidly and directly phosphorylated on the activation loop site residue Thr308 by PDK1 [48] and on the hydrophobic region residue Ser473 by PDK2 (mTORC2 complex) [49]. These two phosphorylation events are required for complete induction of AKT activity. Thus, repressing PDK1 or PDK2 may be a mechanism for regulating AKT activation by NRK. There have also been studies regarding molecules that negatively regulated AKT activity. Lipid phosphatases, PTEN [50] and SHIP [51], have indirectly inhibited PI3K/AKT signal transduction through the enzymatic conversion of PIP3. In contrast, other phosphatases, such as PHLPP [52] and PP2A [53], can exert direct effects by dephosphorylating AKT Thr308 and Ser473, respectively. These phosphatases could also be possible candidates that NRK affects in order to control AKT phosphorylation. In addition, a recent study demonstrated that increased production of ceramide was associated with a decline of AKT phosphorylation in trophoblasts [54]. Thus, NRK may also affect the production of ceramide or other molecules that negatively regulate AKT phosphorylation. Moreover, it is also possible that NRK could affect unknown pathways that regulate AKT phosphorylation. Because the only substrate for NRK identified to date is cofilin, and because cofilin has not been shown to be involved in regulating AKT phosphorylation, it is important to identify novel NRK substrates that affect AKT phosphorylation. Thus, further studies including the assessment of possible candidates that NRK affects in order to control AKT phosphorylation and the identification of the novel substrates for NRK will be necessary to elucidate the signal pathways underlying the regulation of AKT phosphorylation by NRK.
In addition to enhanced AKT Ser473 phosphorylation in Nrk-null trophoblasts, our present results revealed the induction of ERK1/2 phosphorylation in both wild-type and Nrk-null trophoblasts established in this study (Fig 2A). Moreover, phosphorylated ERK1/2 was also present in placentas on E18.5 (Fig 3D), suggesting the involvement of the ERK signal transduction pathway in normal placental development. Indeed, defective placental development accompanied with embryonic lethality has been reported in Erk2-deficient mice [38]. In contrast, Erk1-deficient mice have been demonstrated to be viable, fertile and of normal size [55]. Interestingly, a recent study demonstrated that the transgenic expression of Erk1 rescues the defects in embryonic and placental development that are observed in Erk2-deficient mice, indicating the functional redundancy of Erk1 and Erk2 in mouse development [56]. By genetically modifying Akt and Erk1/2 expression in the in vitro culture system of ESCs that were used in this study, it may be possible to further dissect the precise functions of these factors in trophoblast proliferation and differentiation.
In addition to the analysis in trophoblasts differentiated from ESCs, in this study, we examined the phosphorylation of AKT in newly established TSC lines from both wild-type and Nrk-null mouse blastocysts. The TSC culture system is known as an excellent tool for deciphering gene function in trophoblasts because TSCs can differentiate into trophoblast subtypes in vitro and contribute to trophoblast lineages in vivo [22]. Though ESCs and TSCs are morphologically and molecularly quite different, several studies demonstrated that ESCs can be converted to trophoblast lineages by modifying culture conditions [21] and gene expressions [19, 20]. Because no apparent difference has been shown between differentiated trophoblasts derived from TSCs and those from ESCs [19–21], it is reasonable to apply these two culture systems for better understanding the role of Nrk in trophoblast developments. Indeed, our present study confirmed the similar upregulation of AKT phosphorylation at Ser473 in the two culture systems. We further confirmed that all three AKT isoforms were expressed in differentiated TSCs (day 6) with almost equivalent levels between wild-type and Nrk-null genotypes. However, their phosphorylation levels were quite different. Although the mechanism of AKT2-speficic defect in threonine phosphorylation is not clear, we can at least say that Nrk deficiency strengthened complete activation of AKT1 and AKT3 and partial activation of AKT2 in differentiated TSCs (day 6). Though the precise roles of the three AKT isoforms in trophoblasts have not been elucidated, Haslinger et al. reported that all AKT isoforms were detected in human trophoblast cell lines [57]. They also demonstrated that gene silencing of AKT1 or AKT3 in human trophoblast cell lines resulted in the downregulation of epidermal growth factor-induced phosphorylation of total AKT at both serine and threonine residue. In contrast, gene silencing of AKT2 reduced only total AKT phosphorylation at serine residue without affecting total AKT phosphorylation at threonine residue. These results suggest that AKT2 phosphorylation at threonine residue did not occur in human trophoblasts under ordinary condition, which is consistent with our present data obtained from mouse trophoblasts. Thus, TSCs should be a useful tool to further understand the differently regulated activation mechanisms of AKT isoforms in trophoblasts.
In conclusion, this was the first study to demonstrate the involvement of Nrk-regulating AKT phosphorylation in trophoblast proliferation and its possible control of proper placenta formation. These findings provide important implications for our understanding of placental development. Moreover, because controlled trophoblast proliferation and differentiation are essential for placental and foetal development, our experimental systems established in this study could be useful for gaining further insights into disorders that may occur during pregnancy.
Supporting Information
(TIF)
Expression of total and phosphorylated JNK showed no significant differences between wild-type and Nrk-null cells.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
We are very grateful to Dr. Futoshi Suizu, Hokkaido University, for his fruitful discussions and helpful comments. We would like to thank Enago (www.enago.jp) for the English language review.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Japan Society for Promotion of Science (https://www.jsps.go.jp) KAKENHI Grant Number 24592454 (Y.M.); Japan Society for Promotion of Science (https://www.jsps.go.jp) KAKENHI Grant Number 15K15591 (Y.M.); Kanzawa medical research foundation (http://www.kissei.co.jp) (Y.M.); Takeda Science Foundation (http://www.takeda-sci.or.jp) (Y.M.); Ichiro Kanehara Foundation (http://www.kanehara-zaidan.or.jp) (Y.M.); and Inamori Foundation (http://www.inamori-f.or.jp) (Y.M.). The founders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice—a review. Placenta. 2005; 26 Suppl A: S3–9. [DOI] [PubMed] [Google Scholar]
- 2.Fisher SJ. Why is placentation abnormal in preeclampsia? Am J Obstet Gynecol. 2015; 213 (4 Suppl): S115–122. 10.1016/j.ajog.2015.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001; 2: 538–548. 10.1038/35080570 [DOI] [PubMed] [Google Scholar]
- 4.Frank D, Fortino W, Clark L, Musalo R, Wang W, Saxena A, et al. Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci USA. 2002; 99: 7490–7495. 10.1073/pnas.122039999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tunster SJ, Van de Pette M, John RM. Fetal overgrowth in the Cdkn1c mouse model of Beckwith-Wiedemann syndrome. Dis Model Mech. 2011; 4: 814–821. 10.1242/dmm.007328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggenschwiler J, Ludwig T, Fisher P, Leighton PA, Tilghman SM, Efstratiadis A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev. 1997; 11: 3128–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerrato F, Sparago A, Di Matteo I, Zou X, Dean W, Sasaki H, et al. The two-domain hypothesis in Beckwith-Wiedemann syndrome: autonomous imprinting of the telomeric domain of the distal chromosome 7 cluster. Hum Mol Genet. 2005; 14: 503–511. 10.1093/hmg/ddi047 [DOI] [PubMed] [Google Scholar]
- 8.Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, et al. Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2005; 13: 1025–1032. 10.1038/sj.ejhg.5201463 [DOI] [PubMed] [Google Scholar]
- 9.Chiao E, Fisher P, Crisponi L, Deiana M, Dragatsis I, Schlessinger D, et al. Overgrowth of a mouse model of the Simpson-Golabi-Behmel syndrome is independent of IGF signaling. Dev Biol. 2002; 243: 185–206. 10.1006/dbio.2001.0554 [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Behringer RR. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nat Genet. 1998; 20: 309–311. 10.1038/3129 [DOI] [PubMed] [Google Scholar]
- 11.Jackman SM, Kong X, Fant ME. Plac1 (placenta-specific 1) is essential for normal placental and embryonic development. Mol Reprod Dev. 2012; 79: 564–572. 10.1002/mrd.22062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denda K, Nakao-Wakabayashi K, Okamoto N, Kitamura N, Ryu JY, Tagawa Y, et al. Nrk, an X-linked protein kinase in the germinal center kinase family, is required for placental development and fetoplacental induction of labor. J Biol Chem. 2011; 286: 28802–28810. 10.1074/jbc.M111.258160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010; 330: 496–499. 10.1126/science.1194174 [DOI] [PubMed] [Google Scholar]
- 14.Wakayama T, Yanagimachi R. Cloning of male mice from adult tail-tip cells. Nat Genet. 1999; 22: 127–128. 10.1038/9632 [DOI] [PubMed] [Google Scholar]
- 15.Kanai-Azuma M, Kanai Y, Okamoto M, Hayashi Y, Yonekawa H, Yazaki K. Nrk: a murine X-linked NIK (Nck-interacting kinase)-related kinase gene expressed in skeletal muscle. Mech Dev. 1999; 89: 155–159. [DOI] [PubMed] [Google Scholar]
- 16.Nakano K, Yamauchi J, Nakagawa K, Itoh H, Kitamura N. NESK, a member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and is expressed during the late stages of embryogenesis. J Biol Chem. 2000; 275: 20533–20539. 10.1074/jbc.M001009200 [DOI] [PubMed] [Google Scholar]
- 17.Kakinuma H, Inomata H, Kitamura N. Enhanced JNK activation by NESK without kinase activity upon caspase-mediated cleavage during apoptosis. Cell Signal. 2005; 17: 1439–1448. 10.1016/j.cellsig.2005.03.004 [DOI] [PubMed] [Google Scholar]
- 18.Nakano K, Kanai-Azuma M, Kanai Y, Moriyama K, Yazaki K, Hayashi Y, et al. Cofilin phosphorylation and actin polymerization by NRK/NESK, a member of the germinal center kinase family. Exp Cell Res. 2003; 287: 219–227. [DOI] [PubMed] [Google Scholar]
- 19.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005; 123: 917–929. 10.1016/j.cell.2005.08.040 [DOI] [PubMed] [Google Scholar]
- 20.Tolkunova E, Cavaleri F, Eckardt S, Reinbold R, Christenson LK, Scholer HR, et al. The caudal-related protein cdx2 promotes trophoblast differentiation of mouse embryonic stem cells. Stem cells. 2006; 24: 139–144. 10.1634/stemcells.2005-0240 [DOI] [PubMed] [Google Scholar]
- 21.Hayashi Y, Furue MK, Tanaka S, Hirose M, Wakisaka N, Danno H, et al. BMP4 induction of trophoblast from mouse embryonic stem cells in defined culture conditions on laminin. In Vitro Cell Dev Biol Anim. 2010; 46: 416–430. 10.1007/s11626-009-9266-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998; 282: 2072–2075. [DOI] [PubMed] [Google Scholar]
- 23.Fujihara Y, Kaseda K, Inoue N, Ikawa M, Okabe M. Production of mouse pups from germline transmission-failed knockout chimeras. Transgenic Res. 2013; 22: 195–200. 10.1007/s11248-012-9635-x [DOI] [PubMed] [Google Scholar]
- 24.Furue M, Okamoto T, Hayashi Y, Okochi H, Fujimoto M, Myoishi Y, et al. Leukemia inhibitory factor as an anti-apoptotic mitogen for pluripotent mouse embryonic stem cells in a serum-free medium without feeder cells. In Vitro Cell Dev Biol Anim. 2005; 41: 19–28. 10.1290/0502010.1 [DOI] [PubMed] [Google Scholar]
- 25.Okada Y, Ueshin Y, Hasuwa H, Takumi K, Okabe M, Ikawa M. Targeted gene modification in mouse ES cells using integrase-defective lentiviral vectors. Genesis. 2009; 47: 217–223. 10.1002/dvg.20469 [DOI] [PubMed] [Google Scholar]
- 26.Morioka Y, Fujihara Y, Okabe M. Generation of precise point mutation mice by footprintless genome modification. Genesis. 2014; 52: 68–77. 10.1002/dvg.22727 [DOI] [PubMed] [Google Scholar]
- 27.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo: a laboratory manual Cold Spring Harbor Laboratory Press; NY, USA: 2003. [Google Scholar]
- 28.Nicolson GL, Yanagimachi R, Yanagimachi H. Ultrastructural localization of lectin-binding sites on the zonae pellucidae and plasma membranes of mammalian eggs. J Cell Biol. 1975; 66: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. 'Green mice' as a source of ubiquitous green cells. FEBS Lett. 1997; 407: 313–319. [DOI] [PubMed] [Google Scholar]
- 30.Simmons DG, Rawn S, Davies A, Hughes M, Cross JC. Spatial and temporal expression of the 23 murine Prolactin/Placental Lactogen-related genes is not associated with their position in the locus. BMC genomics. 2008; 9: 352 10.1186/1471-2164-9-352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lescisin KR, Varmuza S, Rossant J. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes Dev. 1988; 2: 1639–1646. [DOI] [PubMed] [Google Scholar]
- 32.Cross JC, Flannery ML, Blanar MA, Steingrimsson E, Jenkins NA, Copeland NG, et al. Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development. 1995; 121: 2513–2523. [DOI] [PubMed] [Google Scholar]
- 33.Chun JY, Han YJ, Ahn KY. Psx homeobox gene is X-linked and specifically expressed in trophoblast cells of mouse placenta. Dev Dyn. 1999; 216: 257–266. [DOI] [PubMed] [Google Scholar]
- 34.Guillemot F, Caspary T, Tilghman SM, Copeland NG, Gilbert DJ, Jenkins NA, et al. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet. 1995; 9: 235–242. 10.1038/ng0395-235 [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Lemaire P, Behringer RR. Esx1, a novel X chromosome-linked homeobox gene expressed in mouse extraembryonic tissues and male germ cells. Dev Biol. 1997; 188: 85–95. 10.1006/dbio.1997.8640 [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998; 12: 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang ZZ, Tschopp O, Hemmings-Mieszczak M, Feng J, Brodbeck D, Perentes E, et al. Protein kinase B alpha/Akt1 regulates placental development and fetal growth. J Biol Chem. 2003; 278: 32124–32131. 10.1074/jbc.M302847200 [DOI] [PubMed] [Google Scholar]
- 38.Hatano N, Mori Y, Oh-hora M, Kosugi A, Fujikawa T, Nakai N, et al. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells. 2003; 8: 847–856. [DOI] [PubMed] [Google Scholar]
- 39.Kettle JG, Brown S, Crafter C, Davies BR, Dudley P, Fairley G, et al. Diverse heterocyclic scaffolds as allosteric inhibitors of AKT. J Medic Chem. 2012; 55: 1261–1273. [DOI] [PubMed] [Google Scholar]
- 40.Liao Y, Hung MC. Physiological regulation of Akt activity and stability. A J Transl Res. 2010; 2: 19–42. [PMC free article] [PubMed] [Google Scholar]
- 41.Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001; 15: 2203–2208. 10.1101/gad.913901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, et al. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J Clinic Invest. 2003; 112: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, et al. Role for Akt3/protein kinase Bgamma in attainment of normal brain size. Mol Cell Biol. 2005; 25: 1869–1878. 10.1128/MCB.25.5.1869-1878.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dummler B, Tschopp O, Hynx D, Yang ZZ, Dirnhofer S, Hemmings BA. Life with a single isoform of Akt: mice lacking Akt2 and Akt3 are viable but display impaired glucose homeostasis and growth deficiencies. Mol Cell Biol. 2006; 26: 8042–8051. 10.1128/MCB.00722-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, et al. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003; 17: 1352–1365. 10.1101/gad.1089403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang ZZ, Tschopp O, Di-Poi N, Bruder E, Baudry A, Dummler B, et al. Dosage-dependent effects of Akt1/protein kinase Balpha (PKBalpha) and Akt3/PKBgamma on thymus, skin, and cardiovascular and nervous system development in mice. Mol Cell Biol. 2005; 25: 10407–10418. 10.1128/MCB.25.23.10407-10418.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007; 129: 1261–1274. 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997; 7: 261–269. [DOI] [PubMed] [Google Scholar]
- 49.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005; 307: 1098–1101. 10.1126/science.1106148 [DOI] [PubMed] [Google Scholar]
- 50.Maehama T, Dixon JE. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999; 9: 125–128. [DOI] [PubMed] [Google Scholar]
- 51.Rohrschneider LR, Fuller JF, Wolf I, Liu Y, Lucas DM. Structure, function, and biology of SHIP proteins. Genes Dev. 2000; 14: 505–520. [PubMed] [Google Scholar]
- 52.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005; 18: 13–24. 10.1016/j.molcel.2005.03.008 [DOI] [PubMed] [Google Scholar]
- 53.Andjelkovic M, Jakubowicz T, Cron P, Ming XF, Han JW, Hemmings BA. Activation and phosphorylation of a pleckstrin homology domain containing protein kinase (RAC-PK/PKB) promoted by serum and protein phosphatase inhibitors. Proc Natl Acad Sci USA. 1996; 93: 5699–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh AT, Dharmarajan A, Aye IL, Keelan JA. Ceramide biosynthesis and metabolism in trophoblast syncytialization. Mol Cell Endocrinol. 2012; 362: 48–59. 10.1016/j.mce.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 55.Pages G, Guerin S, Grall D, Bonino F, Smith A, Anjuere F, et al. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999; 286: 1374–1377. [DOI] [PubMed] [Google Scholar]
- 56.Fremin C, Saba-El-Leil MK, Levesque K, Ang SL, Meloche S. Functional Redundancy of ERK1 and ERK2 MAP Kinases during Development. Cell Rep. 2015; 12: 913–921. 10.1016/j.celrep.2015.07.011 [DOI] [PubMed] [Google Scholar]
- 57.Haslinger P, Haider S, Sonderegger S, Otten JV, Pollheimer J, Whitley G, et al. AKT isoforms 1 and 3 regulate basal and epidermal growth factor-stimulated SGHPL-5 trophoblast cell migration in humans. Biol Reprod. 2013; 88: 54 10.1095/biolreprod.112.104778 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Expression of total and phosphorylated JNK showed no significant differences between wild-type and Nrk-null cells.
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.