Abstract
In humans perturbations of centriole number are associated with tumorigenesis and microcephaly, therefore appropriate regulation of centriole duplication is critical. The C. elegans homolog of Plk4, ZYG-1, is required for centriole duplication, but our understanding of how ZYG-1 levels are regulated remains incomplete. We have identified the two PP1 orthologs, GSP-1 and GSP-2, and their regulators I-2SZY-2 and SDS-22 as key regulators of ZYG-1 protein levels. We find that down-regulation of PP1 activity either directly, or by mutation of szy-2 or sds-22 can rescue the loss of centriole duplication associated with a zyg-1 hypomorphic allele. Suppression is achieved through an increase in ZYG-1 levels, and our data indicate that PP1 normally regulates ZYG-1 through a post-translational mechanism. While moderate inhibition of PP1 activity can restore centriole duplication to a zyg-1 mutant, strong inhibition of PP1 in a wild-type background leads to centriole amplification via the production of more than one daughter centriole. Our results thus define a new pathway that limits the number of daughter centrioles produced each cycle.
Author Summary
The centrosomes are responsible for organizing the mitotic spindle a microtubule-based structure that centers, then segregates, the chromosomes during cell division. When a cell divides it normally possesses two centrosomes, allowing it to build a bipolar spindle and accurately segregate the chromosomes to two daughter cells. Appropriate control of centrosome number is therefore crucial to maintaining genome stability. Centrosome number is largely controlled by their regulated duplication. In particular, the protein Plk4, which is essential for duplication, must be strictly limited as an overabundance leads to excess centrosome duplication. We have identified protein phosphatase 1 as a critical regulator of the C. elegans Plk4 homolog (known as ZYG-1). When protein phosphatase 1 is down-regulated, ZYG-1 levels increase leading to centrosome amplification. Thus our work identifies a novel mechanism that limits centrosome duplication.
Introduction
In mitotic cells the centrosome serves as the primary microtubule-organizing center and consists of two centrioles surrounded by a proteinaceous pericentriolar matrix (PCM). During mitosis the centrosomes organize the poles of the spindle, therefore maintaining appropriate centrosome numbers promotes spindle bipolarity and faithful chromosome segregation. Regulated centrosome duplication is the primary mechanism by which centrosome number is controlled, and involves building a new daughter centriole adjacent to each pre-existing mother centriole. Two features of centriole duplication maintain appropriate centrosome numbers: first, centriole duplication is limited to occurring only once per cell cycle. Second, only a single daughter centriole is assembled in association with each pre-existing mother centriole.
A conserved set of five centriole duplications factors, SPD-2/CEP192, ZYG-1/Plk4, SAS-6, SAS-5/STIL/Ana2, and SAS-4/CPAP are required for daughter centriole assembly and their individual loss results in centriole duplication failure (reviewed in [1]). Conversely, individual over expression of a subset of these duplication factors, Plk4, SAS-6 and STIL/SAS-5, leads to centriole over-duplication (the production of more than a single daughter) leading to a condition known as centriole amplification (the accumulation of an excess number of centrioles) [2–6]. Interestingly, the three factors whose overexpression leads to centriole amplification have been identified as key players in the initial steps of centriole duplication. In human cells Plk4 phosphorylates STIL to trigger centriolar recruitment of SAS-6, which initiates formation of the cartwheel, the central scaffolding structure of the new centriole [7–12]. Similarly in C. elegans the Plk4 homolog ZYG-1 recruits a complex of SAS-5 and SAS-6 through direct physical association with SAS-6 to initiate centriole duplication [13,14].
Because Plk4 overexpression causes the formation of extra daughter centrioles, Plk4 protein levels must be tightly regulated in vivo. One mechanism that regulates Plk4 levels is its SCF-mediated degradation promoted by autophosphorylation, whereby the active kinase induces its own destruction [15–17]. Degradation of Plk4 homologs in Drosophila, (plk4/Sak) and C. elegans (ZYG-1) are similarly regulated by their SCF-mediated targeting for degradation [18,19]. In addition, recent studies have shed light on temporal and spatial regulation of Plk4 levels. Plk4 initially localizes in a broad ring around the mother centriole until, coincident with the initiation of duplication, it becomes restricted to a small focus marking the location of daughter centriole assembly [8,20,21]. Emerging evidence suggests that this transition, which seems to be a key step in ensuring only a single daughter centriole is assembled, relies on spatially regulated Plk4 degradation [8,22]. Because STIL can both activate Plk4 [10,22,23], and also protect it from degradation [22] it is proposed that centriolar recruitment of a small focus of STIL at the G1/S transition triggers broad Plk4 activation and degradation via autophosphorylation, while protecting a local focus. Thus STIL limits the centriolar distribution of Plk4, promoting the assembly of a single daughter centriole. These studies reveal the central role that regulated destruction of ZYG-1/Plk4 plays in controlling centriole number, and highlight the importance of better understanding how the stability of Plk4 is regulated.
Protein phosphatase 1 (PP1) is a major cellular phosphatase that plays well-characterized roles in diverse processes including glycogen metabolism, circadian rhythms and cell division. Humans possess a single PP1α gene, a single PP1β gene (also known as PP1δ) and a single PP1γ gene. The different isoforms are >85% identical and show largely overlapping roles, although some functional specialization has been identified [24]. C. elegans has four PP1 catalytic subunits: a single broadly-expressed PP1β homolog, GSP-1, and three PP1α homologs, GSP-2, which is also widely expressed, and GSP-3 and GSP-4, whose expression is limited to the male germ line [25,26]. The core catalytic subunit of PP1 can directly bind a subset of substrates, but functional specificity is largely conferred by its interaction with regulatory proteins. Over 200 PP1 interactors exist, which regulate PP1 through modulating substrate specificity, enzyme localization or by inhibition or activation of phosphatase activity [27]. Two evolutionarily conserved PP1 regulators that play a role in cell division are inhibitor 2 (I-2) and SDS22. I-2 was originally identified as an inhibitor of PP1 and shows potent inhibitory activity in vitro, although interestingly the yeast homolog of I-2, GLC8, can also stimulate PP1 activity [28–30]. Down regulation of I-2 in Drosophila or human cells leads to chromosome mis-segregation, which is proposed to result from mis-regulation of Aurora B [31,32]. Similarly SDS22 antagonizes Aurora B autophosphorylation, downregulating Aurora B kinase activity [33]. Although it has been noted that PP1α regulates centrosome cohesion, [34,35], no role for PP1 in regulating centrosome duplication has previously been found.
Here we report a novel PP1-dependent pathway that plays a critical role in ensuring that each mother centriole produces one and only one daughter centriole during each cell cycle. We show that PP1 together with two of the most conserved PP1 regulators, I-2 and SDS-22 downregulates ZYG-1 protein abundance. Our data indicate that PP1 regulates ZYG-1 levels post-translationally and we demonstrate that loss of PP1-mediated regulation leads to ZYG-1 overexpression and centriole amplification through the production of multiple daughter centrioles.
Results
Loss of I-2szy-2, a conserved PP1 regulator, rescues the centriole assembly defect of zyg-1(it25) embryos
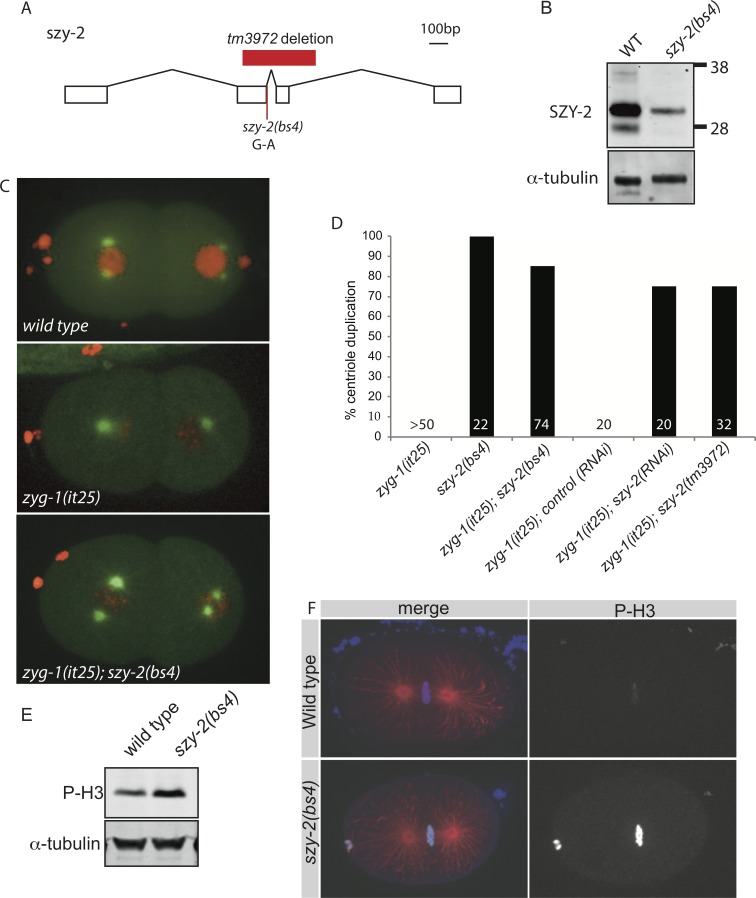
ZYG-1 is essential for centriole assembly: when hermaphrodites carrying the temperature sensitive zyg-1(it25) mutation are grown at the non-permissive temperature of 24°C centriole duplication fails, resulting in 100% embryonic lethality. To identify additional regulators of centriole assembly, we screened for suppressors of the zyg-1(it25) phenotype and isolated mutations in 20 szy (suppressor of zyg-1) genes that rescue embryonic survival [36]. We mapped one of these mutations, szy-2(bs4), to the Y32H12A.4 locus on chromosome III, hereafter referred to as szy-2. The szy-2(bs4) mutation is a single base pair change (G to A) in a splice donor site, which is predicted to alter splicing of the szy-2 transcript resulting in a frame-shift (Fig 1A). Analysis of the SZY-2 sequence shows that it is homologous to inhibitor-2 (I-2), a conserved regulator of protein phosphatase 1 [30]. Using antibodies raised against I-2SZY-2 we confirmed that the szy-2(bs4) mutation significantly reduces I-2SZY-2 protein levels, although residual I-2SZY-2 is still detected, indicating that at least some message is properly spliced in the mutant (Fig 1B). At 24°C the embryonic viability of zyg-1(it25); szy-2(bs4) double mutant embryos is 83%, significantly higher than zyg-1(it25) (0% viability). To determine if suppression of the embryonic lethal phenotype was associated with restoration of centriole duplication, we imaged zyg-1(it25) and zyg-1(it25); szy-2(bs4) embryos expressing GFP::tubulin and mCherry::histone (Fig 1C). Normally at fertilization the sperm delivers a single pair of centrioles to the acentrosomal egg, and during the first cell cycle the centrioles separate, duplicate and form the poles of the mitotic spindle (Fig 1C (top panel), S1A Fig and S1 Movie). Subsequent duplication events ensure the formation of bipolar spindles in later divisions. When the first round of centriole duplication fails, as in zyg-1(it25) mutants, the two sperm-derived centrioles still separate and organize the poles of the first mitotic spindle leading to a normal first division. However as each daughter cell inherits a single centriole, monopolar spindles assemble in the 2-cell embryo (Fig 1C (middle panel), S1B Fig and S2 Movie). As this phenotype is indicative of centriole duplication failure, we scored the presence of monopolar spindles in 2-cell stage embryos and found that >80% of centrioles duplicated during the first round of duplication in zyg-1(it25);szy-2(bs4) double mutant embryos (Fig 1C (bottom panel) and 1D & S3 Movie). In contrast, this first duplication event always failed in zyg-1(it25) control embryos (Fig 1C (middle panel) & D). We confirmed that this effect is specific by using RNAi to deplete SZY-2 in zyg-1(it25) worms; RNAi of szy-2 but not the non-essential gene smd-1 (control RNAi), restored centriole duplication to the zyg-1(it25) mutant (Fig 1D). Moreover the deletion allele szy-2(tm3972), in which the C-terminal 190 residues are removed, was also able to suppress the zyg-1(it25) phenotype (Fig 1D). Together these data demonstrate that reducing I-2SZY-2 function suppresses zyg-1(it25) and suggest that modulating PP1 activity may impact centriole duplication.
Fig 1. Loss-of-function mutations in the PP1 regulator I-2SZY-2 rescues the zyg-1(it25) phenotype.
A) Scale diagram of the structure of the szy-2 gene, indicating the location of the tm3972 deletion and the szy-2(bs4) splice site mutation. B) Western blot of embryo extracts from wild-type and szy-2(bs4) mutants showed a reduced level of the SZY-2 protein. C) Stills from movies of embryos expressing mCherry::H2B and GFP::tubulin grown at 24°C. Top, wild type; Middle, zyg-1(it25) mutant at the 2-cell stage following centriole duplication failure; bottom, zyg-1(it25); szy-2(bs4) double mutants at 2 cell-stage showing a rescue of the centriole duplication. D) Quantification of centriole duplication failure at 24°C in zyg-1(it25) mutants and in zyg-1(it25) mutants in which szy-2 activity has been down-regulated by mutation or RNAi. Number of centriole duplication events analyzed is indicated. E) Western blot of embryo extracts, showing levels of phospho-histone H3 in wild-type and szy-2(bs4) embryos. F) Phospho-histone H3 staining of wild-type and szy-2(bs4) embryos at first metaphase. Left, merge: DNA blue, Microtubules red, Phospho-histone H3 green. Right, phospho-histone H3.
Loss of szy-2 decreases PP1 activity
Because szy-2 encodes the C. elegans homolog of I-2, this suggested that the szy-2(bs4) allele may alter PP1 activity, resulting in the observed suppression of centriole duplication failure. I-2 is conserved from yeast through humans and has been described as both an inhibitor and an activator of PP1 [28–30]. Analyses in vitro have shown that C. elegans I-2SZY-2 can inhibit rabbit PP1 activity, but the in vivo role of szy-2, in particular in relation to endogenous worm PP1 subunits, has not previously been investigated [30]. We sought to determine whether the important function of I-2SZY-2 with respect to zyg-1(it25) suppression was as an inhibitor or an activator of PP1. To determine whether PP1 activity is up- or down-regulated in the szy-2(bs4) mutants we wanted to monitor the phosphorylation levels of a known PP1 substrate. One such substrate is histone H3, which is phosphorylated in the early stages of mitosis; this modification is removed by PP1 [37,38]. To investigate whether I-2SZY-2 is an activator or inhibitor of PP1 we analyzed phospho-histone levels in szy-2(bs4) mutant embryos. We used a phospho-histone H3 antibody to detect phosphorylated histone H3 levels in embryos and found them to be greatly elevated in szy-2(bs4) when compared with the wild type (Fig 1E & 1F), a result reminiscent of PP1 depletion (see below; [25,39]). Phospho-histone levels were elevated in both mixed stage embryos (Fig 1E) as well as during the metaphase stage of mitosis (Fig 1F). This result indicates that the szy-2(bs4) mutation reduces PP1 activity and suggests that I-2SZY-2 normally acts as an activator of PP1. Consistent with I-2SZY-2 being a positive regulator of PP1, szy-2(bs4) mutant embryos exhibit a chromosome mis-segregation defect [36] that is similar to that of embryos depleted of PP1 (S4 Movie). There is precedent for I-2 acting as an activator of PP1: in yeast I-2GLC8 binds and inactivates PP1GLC7, but upon phosphorylation of I-2GLC8, PP1 becomes activated [29]. The phosphorylated residue is conserved in all I-2 homologs including C. elegans, although it is not clear whether I-2SZY-2 is similarly regulated in the worm [30].
Loss of a second PP1 regulator, SDS-22, rescues the centriole duplication failure of zyg-1(it25) embryos
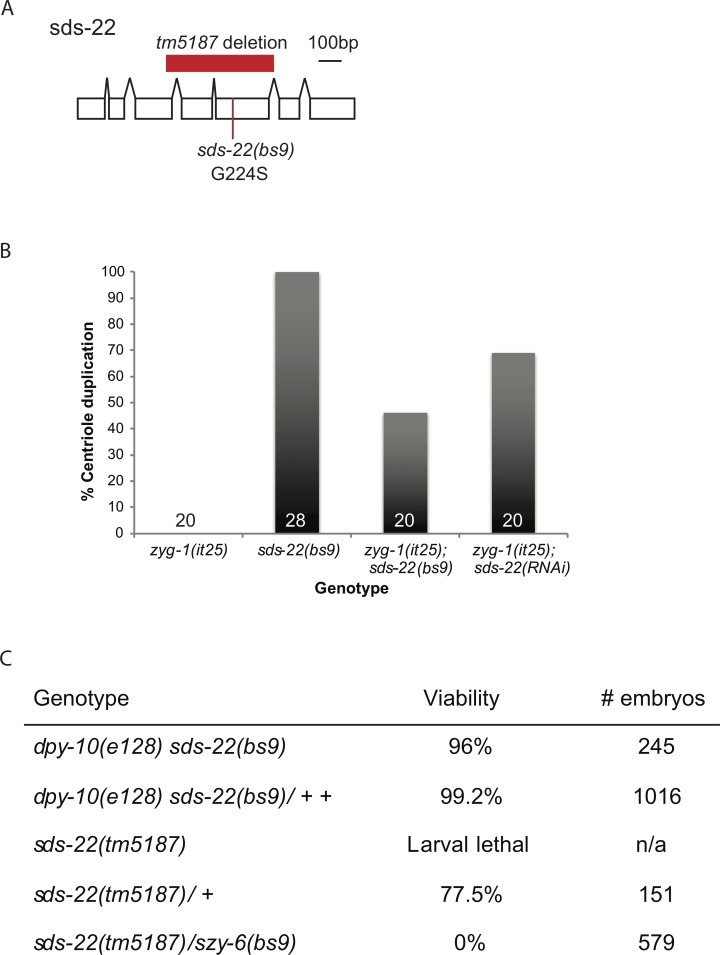
Because we had identified a mutation in I-2szy-2 as a suppressor of zyg-1(it25), we speculated that loss of additional PP1 regulatory proteins may have a similar effect. To test this hypothesis we identified seven C. elegans proteins which show a high degree of conservation with human PP1 regulators. We then used RNAi to deplete each factor in zyg-1(it25) worms, and monitored embryonic viability (S2 Fig). RNAi of only the sds-22 gene significantly increased embryonic viability of the zyg-1(it25) embryos. SDS-22 is a leucine-rich repeat (LRR) domain containing protein that localizes PP1 during mitosis, regulating mitotic progression [33,40–42]. Although sds-22(RNAi) had only a modest effect on zyg-1(it25) embryonic viability, direct observation of centriole duplication in zyg-1(it25); sds-22(RNAi) embryos, revealed 70% of centriole duplication events occurred normally (Fig 2B). The disparity between the strength of suppression of embryonic lethality and the strength of suppression of centriole duplication failure is likely explained by a combination of two factors. First, we find that on average, 30% of the duplication events in zyg-1(it25); sds-22(RNAi) embryos fail; such a high degree of cell division failure during early development would result in embryonic lethality. Second, since sds-22 is an essential gene, RNAi knockdown may cause embryonic lethality even though centriole duplication is normal. Since reducing SDS-22 function is able to suppress the zyg-1(it25) phenotype we hypothesized that one of the suppressors of zyg-1(it25) recovered in our genetic screen [36] may have a mutation in sds-22. We therefore sequenced the sds-22 coding region in those szy mutants that mapped close to the sds-22 genetic locus. The szy-6(bs9) strain contained a G-to-A missense mutation in the coding region of sds-22 which results in a single amino-acid substitution (G224S) within the eighth LRR (Fig 2A). To confirm that bs9 was an allele of sds-22 we performed a complementation test with sds-22(tm5187) a deletion allele that exhibits a fully-penetrant larval-lethal phenotype (Fig 2A & 2C). While the szy-6(bs9) homozygotes exhibited minimal embryonic lethality, the trans-heterozygotes (bs9/ tm5187) exhibited 100 percent embryonic lethality confirming that bs9 and tm5187 are allelic (Fig 2C). We have therefore renamed the szy-6(bs9) allele sds-22(bs9). Although by itself, the sds-22(bs9) mutation does not appear to affect centriole duplication, zyg-1(it25); sds-22(bs9) double mutant embryos exhibit an approximately 50% success rate of centriole duplication, comparable to levels seen after SDS-22 RNAi depletion (Fig 2B). Together these data show that reducing the function of either of two PP1 regulatory proteins, I-2SZY-2 or SDS-22, is able to partially suppress the failure of centriole duplication caused by the zyg-1(it25) mutation.
Fig 2. Reducing SDS-22 activity rescues the zyg-1(it25) phenotype.
A) Scale diagram of the structure of the sds-22 gene, indicating the location of the tm5187 deletion and the sds-22(bs9) substitution. B) Quantification of centriole duplication when zyg-1(it25) is rescued by the sds-22(bs9) mutation or sds-22(RNAi). Number of centriole duplication events analyzed is indicated. C) Complementation test showing bs9 and tm5187 are allelic. Hermaphrodites of the indicated genotype were shifted to 25°C at the L4 stage and embryonic lethality was determined over the next 24 hours. The szy-6(bs9) allele was marked with the closely linked dpy-10(e128) mutation.
Loss of PP1βGSP-1 activity rescues zyg-1(it25)
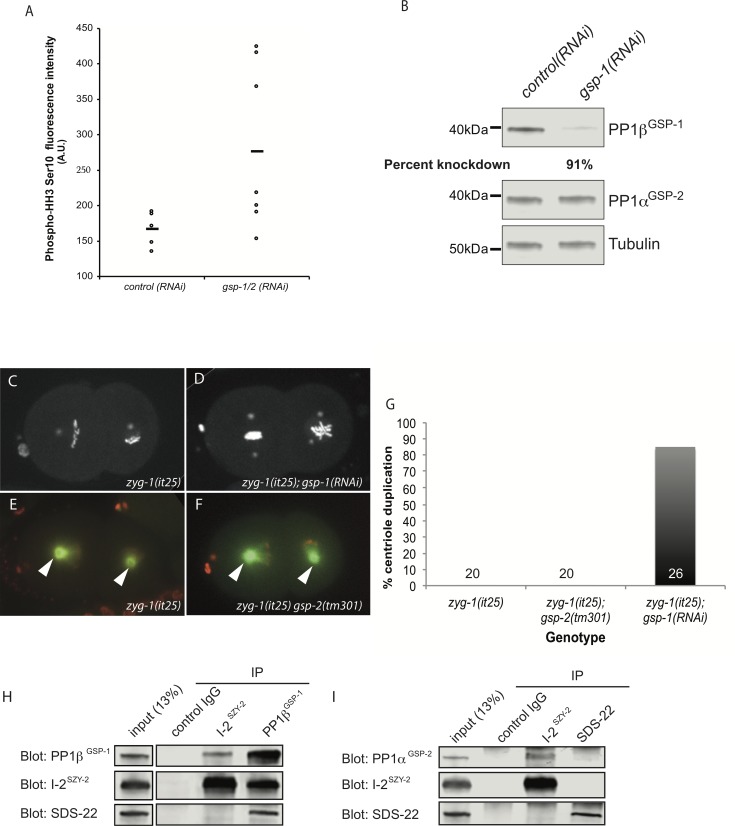
Our data suggest that I-2SZY-2 is an activator of PP1, and work in vertebrate cells has shown that the SDS-22 ortholog positively regulates PP1 activity [33]. Because the szy-2(bs4) and sds-22(bs9) mutations rescue zyg-1(it25) phenotypes it followed that directly lowering PP1 activity should also suppress the zyg-1(it25) phenotype. Embryos express two PP1 catalytic subunits encoded by the genes gsp-1 and gsp-2. Similar to what is seen in the szy-2(bs4) mutant (Fig 1E & 1F), co-depletion of GSP-1 and GSP-2 by RNAi resulted in an increase in phospho-histone H3 staining of mitotic chromosomes (Fig 3A). We next tested whether individually reducing the activity of either PP1βGSP-1 or PP1αGSP-2, could suppress the centriole duplication failure seen in zyg1(it25) embryos. Although the gsp-1(tm4378) deletion allele is sterile, precluding its use in our analysis, we were able to specifically deplete PP1βGSP-1 by RNAi (Fig 3B). When PP1βGSP-1 was depleted in zyg-1(it25) embryos we saw a robust restoration of centriole duplication (90% of centrioles; Fig 3B, 3C, 3D & 3G). In contrast, the putative null allele gsp-2(tm301) [25] did not suppress the zyg-1(it25) phenotype and we never observed centriole duplication in the zyg-1(it25); gsp-2(tm301) double mutant embryos (Fig 3E, 3F & 3G). The C. elegans PP1αGSP-2 and PP1βGSP-1 isoforms show a high degree of identity and are both expressed in the early embryo [25]. Since depletion of only the PP1βGSP-1 isoform can suppress the zyg-1(it25) allele this suggests either 1) a divergence in function of the two enzymes, with only PP1βGSP-1 being involved in the regulation of centriole duplication, or 2) that, despite partial redundancy between the two enzymes, zyg-1(it25) mutants provides a sensitized background where loss of PP1βGSP-1 alone is sufficient to subvert normal controls. Indeed, two observations support the later possibility. First, we have only observed defects in chromosome segregation after co-depletion of both PP1βGSP-1 and PP1αGSP-2 suggestive of some functional redundancy (S4 Movie). Second, we show below that co-depletion of PP1βGSP-1and PP1αGSP-2 disrupts the normal pattern of centriole duplication whereas single depletions of either phosphatase have no effect (see below & S5 Movie).
Fig 3. PP1 functions with I-2SZY-2 and SDS-22 to regulate centriole duplication.
A) Comparison of phospho-HH3 levels in control and PP1βGSP-1 & PP1αGSP-2 co-depleted embryos as determined by quantitative immunofluorescence microscopy. The mean is indicated by the bar. B) Western blot demonstrating specificity of gsp-1(RNAi). C&D) Stills from time-lapse recordings of embryos of the indicated genotypes, expressing GFP::histone and GFP::SPD-2 grown at the normally restrictive temperature of 24°C. E&F) Stills from time-lapse recordings of embryos of the indicated genotypes, expressing mCherry::histone and GFP::tubulin grown at the normally restrictive temperature of 24°C; centrosomes are indicated by arrow heads. G) Quantitation of centriole duplication in indicated strains. Number of centriole duplication events scored is indicated. H) Western blots of immunoprecipitated material from whole worm extracts using control IgG, I-2SZY-2, or PP1βGSP-1 antibodies, and probed with I-2SZY-2, PP1βGSP-1, or SDS-22 antibodies as indicated. I) Western blots of immunoprecipitated material from whole worm extracts using control IgG, I-2SZY-2, or SDS-22 antibodies, and probed with I-2SZY-2 or PP1αGSP-2, or SDS-22 antibodies.
Our data indicate that individually reducing the function of either I-2SZY-2, SDS-22 or PP1βGSP-1 can partially suppress the zyg-1(it25) centrosome duplication failure. Using GFP fusions we were, however, unable to detect centrosome localization of any of the proteins (S3 Fig). Nevertheless we reasoned that all three proteins work in a common process and sought to determine whether they physically interact. When we immunoprecipitated I-2SZY-2 from C. elegans embryonic extracts, we found by immunoblotting that we could detect co-precipitated PP1βGSP-1 and reciprocally when we immunoprecipitated PP1βGSP-1 we were able to pull down I-2SZY-2 (Fig 3H), confirming that C. elegans I-2SZY-2 is a PP1 binding protein in vivo. Similarly, antibodies against PP1βGSP-1 co-precipitated SDS-22 (Fig 3H). However, we were unable to detect any interaction between I-2SZY-2 and SDS-22, suggesting that a complex simultaneously containing all three proteins does not form. Additional IP-western experiments demonstrated that I-2SZY-2 also interacts with PP1αGSP-2, but did not reveal an interaction between SDS-22 and PP1αGSP-2 (Fig 3I). To confirm and extend these results we also individually immunoprecipitated I-2SZY-2, SDS-22, and PP1βGSP-1 and analyzed the immunoprecipitated material by mass spectrometry (S4 Fig). Using this approach we found that I-2SZY-2 and SDS-22 could independently interact with both PP1βGSP-1 and PP1αGSP-2 but not with each other (S4 Fig). We conclude that components of the PP1-dependent regulatory pathway physically interact but that I-2SZY-2 and SDS-22 do not appear to reside in the same complex.
Finally, we have found that although both I-2SZY-2 and SDS-22 interact with PP1 and positively regulate PP1’s function in controlling centriole duplication, the two regulators do not always function together to control PP1 activity. Specifically we have found that while loss of I-2SZY-2 activity results in an increase in the level of phospho-histone H3 in mitotic chromosomes (Fig 1F), no such increase is observed upon loss of SDS-22 activity (S5B Fig). Thus I-2SZY-2 appears to function independently of SDS-22 to mediate the role of PP1 in regulating histone H3 phosphorylation. Curiously, loss of SDS-22 resulted in an elevated phospho-histone H3 signal as detected by immunoblotting (S5A Fig). Because SDS-22 does not control the chromatin content of phospho-histone H3, we speculate that loss of SDS-22 leads to a mitotic delay; this would explain the elevated signal of phospho-histone H3 in mixed-stage worm extracts.
Aurora B, Aurora A and Plk1 are not required for szy-2(bs4) suppression of zyg-1(it25)
Given that PP1 is a mitotic phosphatase that antagonizes many known mitotic kinases we wanted to investigate whether these relationships contribute to the regulation of centriole duplication. In Drosophila and human cells, I-2 is implicated in regulation of Aurora B and its depletion results in chromosome mis-segregation [31,32]. Similarly in humans, SDS22 antagonizes Aurora B autophosphorylation, downregulating Aurora B kinase activity [33]. PP1, I-2 and SDS22 therefore seem to cooperatively regulate chromosome segregation by modulating Aurora B activity. In C. elegans PP1 also regulates Aurora B, contributing to its correct localization during meiosis and to chromosome segregation in mitosis [39,43]. Although Aurora B has not been implicated in centriole duplication we wanted to determine whether szy-2(bs4) suppresses centriole duplication failure through upregulation of Aurora B activity. We therefore RNAi-depleted Aurora BAIR-2 in szy-2(bs4); zyg-1(it25) worms and monitored centriole duplication (S6A, S6B & S6C Fig). Although we observed failures in chromosome segregation and cytokinesis consistent with successful Aurora B depletion (S6C Fig;[44]), centriole duplication was unperturbed (S6A Fig), indicating that szy-2(bs4) does not regulate centriole duplication by modulating Aurora B activity.
Since PP1 and I-2 are associated with regulation of Aurora A activity [45] and Aurora A plays a conserved role in centrosome separation and maturation [46,47] we tested whether Aurora AAIR-1 activity is required for suppression of zyg-1 (it25) by szy-2(bs4). After exposing double mutants to air-1(RNAi) we observed reduced SPD-2 localization and incomplete centrosome separation, consistent with loss of Aurora A activity (S7H Fig), however we did not see a perturbation of centriole duplication (S6A, S6Fa & S6Fb Fig), suggesting that Aurora A is not required for suppression of zyg-1(it25) by szy-2(bs4). In vertebrate cells PP1 also has a recognized role in antagonizing Plk1 [48], a kinase with an established role in centriole duplication [49]. However, depletion of plk-1 in zyg-1(it25), szy-2(bs4) embryos did not affect centriole duplication (S6A Fig) even though PLK-1 activity was clearly compromised (S6D Fig). In summary our data suggest that reducing PP1 activity does not restore centriole duplication in the zyg-1(it25) mutant by relieving antagonism, and thus increasing the relative activity, of Aurora B, Plk1 or Aurora A, which are known PP1 antagonists.
I-2SZY-2 controls SPD-2 levels at the centrosome
During the course of our analyses we noted that the level of the coiled-coil protein SPD-2 was elevated at the centrosome in szy-2(bs4) mutant embryos. SPD-2 is a component of both the PCM and centrioles and is required for centriole duplication and PCM assembly [50,51]. We quantified the effect on SPD-2 levels in szy-2(bs4) embryos expressing SPD-2::GFP and found a 1.5-fold increase in centrosomal SPD-2 levels throughout the cell cycle (S7A Fig). When we compared levels of endogenous SPD-2 at the centrosome, we found a similar increase in centrosome-associated SPD-2 in szy-2(bs4) embryos (S7B–S7D Fig). Since SPD-2 plays a positive role in centriole duplication, we reasoned that its increased levels at the centrosome in the szy-2(bs4) mutant might be responsible for PP1-mediated suppression. To test this possibility, we utilized a codon-optimized spd-2::gfp transgene [52] to overexpress SPD-2 protein in the zyg-1(it25) mutant. Despite a large increase in centrosome-localized SPD-2 (S7E & S7F Fig) we did not see any suppression of zyg-1(it25) embryonic lethality (S7G Fig), indicating that centriole duplication failure persisted. Thus by itself, a general elevation of the level of SPD-2 at the centrosome is not sufficient for suppression of zyg-1(it25). Conversely, we also find that the elevated level of centrosome-associated SPD-2 is not required for szy-2(bs4)-mediated suppression: Aurora AAIR-1 is required for SPD-2 recruitment to the centrosome [50], and depletion of aurora AAIR-1 in the zyg-1(it25); szy-2(bs4) double mutant drastically reduced the amount of SPD-2 at the centrosome, (S7H Fig) nevertheless, centriole duplication was not perturbed (S6A, S6E & S6F Fig). Thus, I-2SZY-2 regulates both centriole duplication and PCM assembly, but the elevated SPD-2 levels at the centrosome observed in szy-2(bs4) mutants do not constitute the primary mechanism by which szy-2(bs4) suppresses the centriole duplication defect of zyg-1(it25) mutants.
Reducing PP1 activity elevates ZYG-1 protein levels
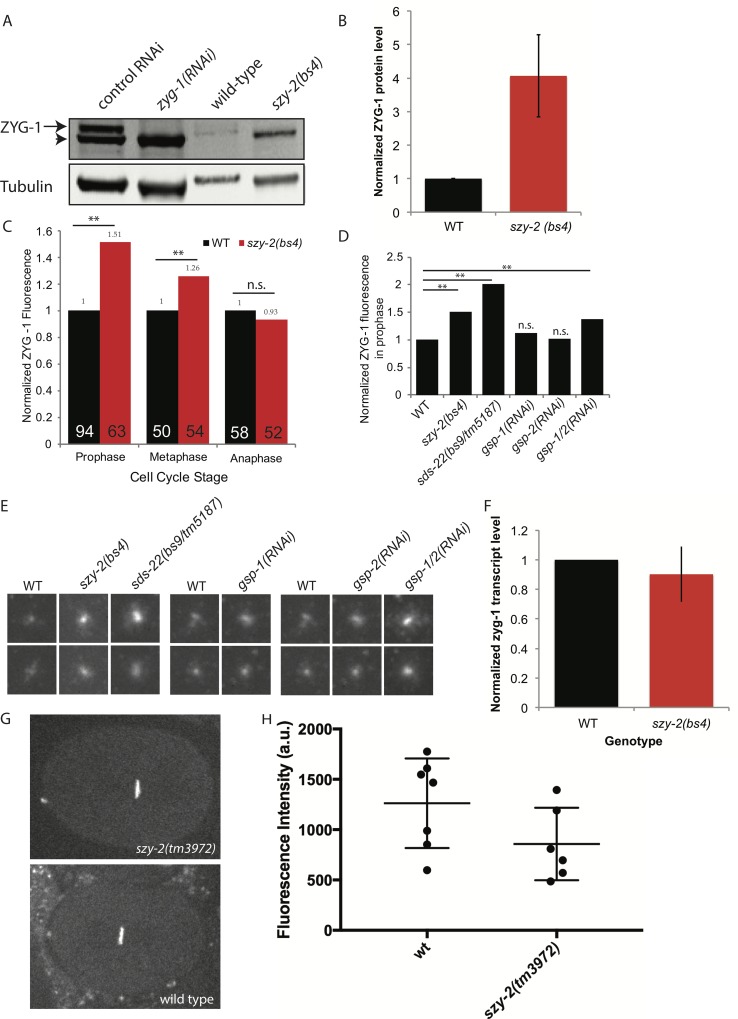
Previous work has shown that the failure of centriole duplication in zyg-1(it25) mutants can be rescued by increasing centriole-associated levels of the mutant ZYG-1 protein [19,53]. We therefore sought to determine if decreasing PP1 activity affects ZYG-1 protein levels or localization. To determine whether the szy-2(bs4) mutation affects ZYG-1 abundance, we measured total ZYG-1 levels in early mixed-stage embryos using quantitative immunoblotting. Strikingly, in comparison to the wild type, szy-2(bs4) embryos consistently possessed approximately four-fold more ZYG-1 at the restrictive temperature of 25°C (Fig 4A & 4B). We then used quantitative immunofluorescence to see if the elevated level of total ZYG-1 protein resulted in increased levels of ZYG-1 at centrioles. As expected, the overall increase in ZYG-1 levels in embryos was associated with elevated ZYG-1 at the centrioles (Fig 4C, 4D & 4E). This increase however was cell cycle stage specific with increases observed in prophase and metaphase but not anaphase, of the first cell cycle (Fig 4C). We further sought to determine whether decreasing SDS-22 or PP1 similarly increased ZYG-1 levels. Quantification of centrosome-associated ZYG-1 in prophase revealed elevated levels of ZYG-1 in the sds-22(bs9/tm5187) trans-heterozygous mutant but a minimal increase after RNAi-depletion of PP1βGSP-1 (Fig 4D & 4E). Similarly depletion of PP1αGSP-2 had little effect on ZYG-1 levels at centrioles (Fig 4D & 4E). Since we only observed chromosome segregation defects when we depleted both PP1βGSP-1 and PP1αGSP-2 (S4 Movie), we wondered whether they also acted redundantly to regulate ZYG-1. We therefore co-depleted PP1βGSP-1 and PP1αGSP-2 and found an increase in centrosome-associated ZYG-1, consistent with the existence of redundancy between the two PP1 isoforms (Fig 4D & 4E). Overall our results suggest that reducing PP1 activity leads to an elevation of both total ZYG-1 levels, and of centriole-associated ZYG-1, providing a likely mechanism for the suppression of the zyg-1(it25) centriole duplication defect. Notably, this effect, of reduced PP1 activity leading to increased ZYG-1 levels, seems to be specific as we did not see a similar increase in SAS-6 levels in szy-2(bs4) embryos (S8A, S8B & S8C Fig). Finally, because ZYG-1 has also been implicated in positively regulating centrosome size [53], elevated ZYG-1 levels may also account for the observed increase in centrosomal SPD-2.
Fig 4. ZYG-1 protein levels are elevated in the szy-2(bs4) mutant.
A) Western blot of embryo extracts from wild-type and szy-2(bs4) embryos probed with a ZYG-1 antibody. The ZYG-1 band (arrow) is identified by its absence in zyg-1(RNAi) extracts. Non-specific proteins (arrowhead) are also bound by the ZYG-1 antibody and probably represent contaminants from the E. coli strain used for RNAi feeding experiments. B) Normalized ZYG-1 protein levels from two independent experiments. Error bars indicate standard deviation. C) Relative levels of centriole-localized ZYG-1 in wild-type and szy-2(bs4) one-cell embryos made by quantitative immunostaining. Levels at each stage are normalized to the wild-type control. Number of centrosomes analyzed indicated inside each bar. **p<0.01; n.s. no significant difference (Student’s t-test). D) Centriole-localized ZYG-1 protein levels for embryos of indicated genotypes at first mitotic prophase. Levels normalized to the wild-type control. Number of centrosomes analyzed indicated inside each bar. **p<0.01; n.s. no significant difference from WT (Student’s t-test). E) Representative images of centrosomal ZYG-1 levels in prophase. Centrosomes from 1-cell stage embryos of the indicated genotypes are grouped with a wild type sample from the same experiment. F) Quantification of zyg-1 transcript levels in embryos by qRT-PCR. Shown is the average of two independent experiments. Error bars indicate standard deviation. G) Representative images of wild-type and szy-2(tm3972) embryos expressing GFP::histone driven by the zyg-1 promoter and 3’-UTR. H) Quantitation of GFP intensities with mean and standard deviations indicated. No difference between strains was found (Student’s t-test p>0.1).
PP1 regulates ZYG-1 levels post-translationally
We next sought to determine how ZYG-1 levels are regulated by PP1. First, we measured zyg-1 transcript levels by quantitative RT-PCR and found that wild-type and szy-2(bs4) embryos possessed similar levels of zyg-1 mRNA (Fig 4F). The observed increase in ZYG-1 protein levels was not, therefore, due to an effect on transcription or mRNA stability. To test whether decreasing PP1 activity resulted in an effect on ZYG-1 translation efficiency, we crossed the szy-2(tm3972) allele into a strain carrying a GFP::histone reporter expressed under the control of the zyg-1 regulatory sequences (promoter and UTRs) [54]. Since this reporter contains zyg-1 regulatory sequences, but lacks the zyg-1 coding sequence, it allows us to determine whether elevated ZYG-1 protein levels stem from alterations at the level of gene expression (transcription/translation) or from post-translational controls. Quantitative fluorescence intensity measurements of GFP::histone revealed that the szy-2(tm3972) mutation did not increase expression of the reporter relative to that observed in the wild-type strain (Fig 4G & 4H; Student’s t-test p>0.1), indicating that reduced PP1 activity does not increase zyg-1 translation via the 3′-UTR. In order to determine whether PP1 might directly regulate ZYG-1 levels we tested for an interaction between the two proteins. Immunoprecipitation of I-2SZY-2, SDS-22, and PP1βGSP-1 from worm extracts, followed by western blotting or mass spectrometry failed to detect an interaction between any of the three proteins and ZYG-1. Because ZYG-1 is a low abundance protein [14] and interactions between PP1 and its substrates may be transient we cannot however rule out a direct interaction between ZYG-1 and PP1. Cumulatively, therefore, our data point to PP1 regulating ZYG-1 protein levels either directly or indirectly via a post-translational mechanism.
Reduction of PP1 activity causes centriole amplification
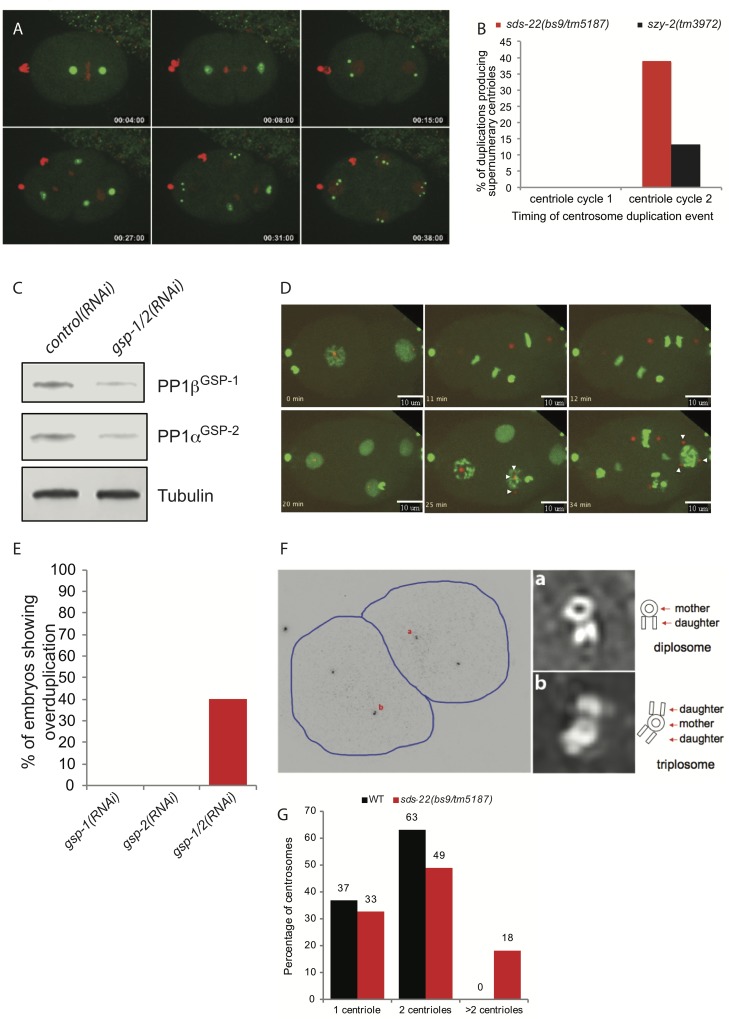
Elevated levels of the human and Drosophila homologs of ZYG-1 are associated with centriole amplification [2,5,6]. Although centriole amplification has not previously been observed in C. elegans embryos, given the increases in ZYG-1 levels we observed in the szy-2(bs4) and sds-22(bs9/tm5187) mutants we were intrigued whether this would be sufficient to subvert the normal regulation of centriole duplication, resulting in supernumerary centrosomes. We therefore performed time-lapse confocal microscopy of embryos expressing GFP::SPD-2 and mCherry::histone. Initial inspection of szy-2 and sds-22 mutants did not reveal any obvious defects during the first two cell cycles; centriole duplication proceeded normally and bipolar spindles assembled in all cells. Unexpectedly however, in embryos strongly impaired for either I-2SZY-2 or SDS-22 function, extra centrosomes were frequently observed at the four-cell stage (Fig 5A and 5B & S6 Movie). Closer inspection of the movies indicated that the extra centrosomes arose approximately synchronously from the spindle poles as the PCM dispersed near the completion of the second mitotic divisions (S6 Movie & S7 Movie). In a single case (out of 79 analyzed) we were not able to trace the origin of one of these centrosomes to a spindle pole. Thus, it is possible that very infrequently in this mutant a centriole arises spontaneously in the cytoplasm via a de novo pathway. The production of extra centrosomes was most prevalent in sds-22(bs9/tm5187) trans-heterozygotes where nearly 40% of the spindle poles gave rise to more than two centrosomes (Fig 5B). Although less frequent, an identical defect was observed in szy-2(tm3972) embryos where supernumerary centrosomes arose with the same timing and spatial pattern as those observed in the sds-22 mutant (Fig 5B). These results suggest that extra centrioles form during the second round of centriole duplication (which occurs in the 2-cell embryo) such that the extra centrioles only become apparent by confocal microscopy when mother and daughter centrioles separate as cells enter the third cell cycle (S1C Fig).
Fig 5. Decreasing PP1 activity leads to centriole overduplication.
A) Frames from a time-lapse recording of an sds-22(bs9/tm5187) trans-heterozygous embryo expressing GFP::SPD-2 and mCherry::histone. Supernumerary centrosomes appear at the 4-cell stage. B) Quantification of centrosome over-duplication in sds-22(bs9/tm5187) and szy-2(tm3972) embryos during the first and second rounds of centriole duplication. For sds-22(bs9/tm5187), n = 31/77 (first round/second round) and for szy-2(tm3972), n = 42/68 (first round/second round) C) Immunoblot showing extent of knockdown of PP1βGSP-1 and PP1αGSP-2 in gsp-1; gsp-2 double RNAi embryos. D) Selected frames from a time-lapse recording of a gsp-1(RNAi); gsp-2(RNAi) embryo expressing GFP::histone and mCherry::SPD-2. Arrowheads indicate extra centrosomes. E) Quantitation of the frequency of supernumerary centrioles in embryos depleted of the indicated PP1 genes. F) Structured illumination microscopy (SIM) of a late 2-cell stage sds-22(bs9/tm5187) trans-heterozygous embryo reveals the presence of excess centrioles. The low magnification image on the left shows the positions of the centrioles stained for the centriole marker SAS-4. Images on the right are enlargements of the indicated centrosomes. G) Quantification of SIM data to indicate the number of centrioles observed per centrosome comparing wild type and sds-22(bs9/tm5187) transheterozygous mutants.
We next followed the fate of 79 centrosomes in sds-22(bs9/tm5187) trans-heterozygotes and found that all eventually accumulated PCM and participated in spindle assembly, often giving rise to multipolar spindles (S7 Movie). A minority (4/79 centrosomes), however, exhibited a one-cell-cycle delay before accumulating normal levels of PCM and participating in spindle assembly (S9 Fig and S7 Movie). This is somewhat reminiscent of the reversible “inactivation” of centrioles observed in Drosophila embryos following over-expression of Plk4 [55]. The production of excess centrosomes continued in the ensuing cell cycles. We quantified the rate of over-duplication following the third centriole cycle of sds-22(bs9/tm5187) embryos and found a similar frequency of over-duplication (35%, n = 96 centrosomes). Thus our results suggest that over-duplication begins during the second centriole duplication event and continues through embryonic development.
To confirm that the excess centrosomes observed in szy-2 and sds-22 mutant embryos arose due to a reduction of PP1 activity, we also followed centrosomes during the first several cell cycles of embryos depleted for one or both PP1 catalytic subunits. Surprisingly, neither depletion of PP1βGSP-1 (n = 10 embryos) nor of PP1αGSP-2 (n = 11 embryos) resulted in the appearance of extra centrosomes during the first three cell cycles (Fig 5E). We therefore co-depleted both catalytic subunits (Fig 5C) and found that excess centrosomes appeared at the four-cell stage in 4/10 embryos (Fig 5D and 5E & S5 Movie). Further, these extra centrosomes could function as microtubule-organizing centers and directed the formation of tripolar spindles (S8 Movie). Interestingly, we also observed occasional anaphase chromatin bridges in these embryos (S4 Movie), whereas in singly depleted embryos chromosomes always segregated normally. We conclude that PP1βGSP-1 and PP1αGSP-2 exhibit some level of functional redundancy in their roles in chromosome segregation and centriole duplication.
To further investigate the origin of the excess centrosomes we analyzed sds-22(bs9/tm5187) embryos by structured illumination microscopy (SIM), which has proven an effective means to observe centriole arrangement and number within the centrosome [56,57]. SIM imaging of SAS-4-stained embryos allowed us to resolve the basic structure of worm centrioles and we could clearly detect the normal arrangement of mother and daughter centrioles (Fig 5Fa). Strikingly, in sds-22(bs9/tm5187) embryos we were also able to find mother centrioles bearing more than one daughter (Fig 5Fb). We first detected extra daughter centrioles at the late 2-cell stage, consistent with the appearance of excess centrosomes at the four-cell stage. Furthermore, the appearance of extra daughter centrioles in association with a single mother is reminiscent of what is seen after Plk4 overexpression [2] and indicates that the excess centrosomes we observe in the 4-cell embryo originate from the formation of extra daughters during centriole duplication. Quantification of our SIM data reveals that while wild-type embryos never exceed two centrioles per centrosome, 18% of sds-22(bs9/tm5187) centrosomes contain more than 2 centrioles (Fig 5G). We did not observe any other unusual centriole configurations such as mother-daughter-granddaughter arrangements indicative of reduplication of centrioles during a single cell cycle. In summary, our data indicate that loss of PP1 activity results in overexpression of ZYG-1 and consequently centriole amplification. Amplification appears to be largely, if not entirely, driven by the production of multiple daughter centrioles. However, we cannot rule out that centriole reduplication and de novo formation also contribute to the excess centrioles observed after PP1 inhibition. In conclusion, we have described a new PP1-dependent mechanism that limits centriole duplication so that each mother produces one and only one daughter per cell cycle.
Discussion
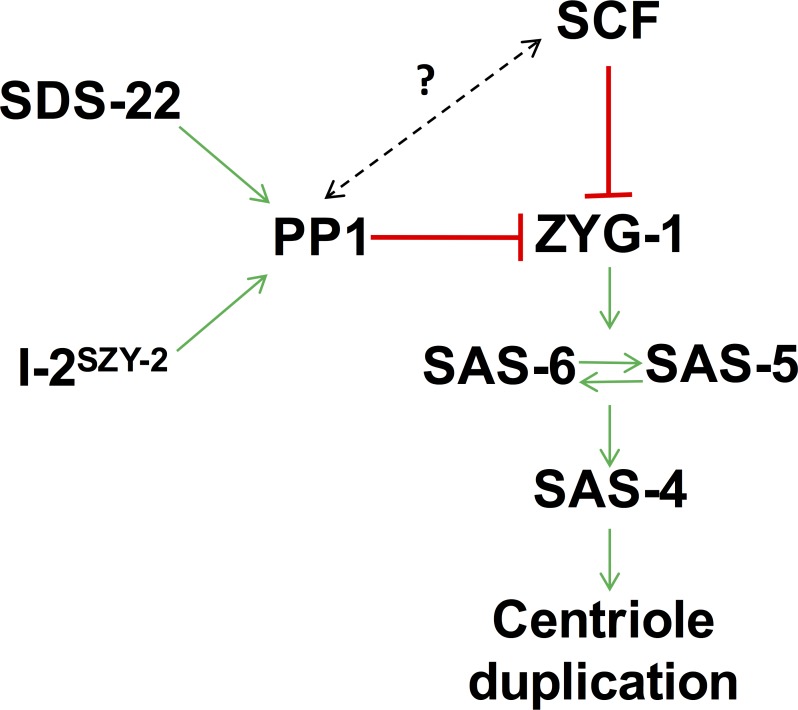
We have shown that PP1 activity is an important regulator of ZYG-1 levels in the C. elegans embryo and therefore that it is a critical regulator of centriole duplication. Although PP1α has previously been implicated in the regulation of centrosome separation at the beginning of mitosis [34], this is the first indication that PP1 regulates centriole duplication. Our data indicate that PP1 inhibits centriole duplication by restraining the accumulation of ZYG-1 in the embryo either directly or indirectly (Fig 6). Previous work has shown that Plk4 levels can be regulated by SCF-mediated degradation promoted by autophosphorylation [15–17]. Similarly in C. elegans the SCF complex is involved in regulating ZYG-1 levels and depletion of SCF components leads to an increase in centrosome-associated ZYG-1 [19]. Our finding that down-regulation of PP1 activity leads to centrosome amplification implicates PP1 as an additional regulator of ZYG-1 levels.
Fig 6. Model of how I-2SZY-2, SDS-22 and PP1 might cooperate with the SCF to regulate centriole duplication.
Our work shows that PP1, I-2SZY-2 and SDS-22 down regulate ZYG-1 levels to constrain centriole duplication. We speculate that this is a new mechanism of regulation that operates independently of the previously documented SCF-mediated regulation. We cannot rule out, however, an interplay between the two pathways (dotted line).
How does PP1 regulate ZYG-1 levels? Although many PP1 targets have been identified none are known regulators of centrosome duplication. A major substrate of PP1, I-2 and SDS22 is Aurora B kinase and activity of the C. elegans homolog, AIR-2, is inhibited by PP1 [25,39,43]. Down regulation of AIR-2 activity in the zyg-1(it25); szy-2(bs4) double mutant however did not have any measurable effect on the level of centriole duplication suggesting that PP1 is not regulating centriole duplication by antagonizing Aurora B activity. Identical results were obtained from two additional mitotic kinases, Plk1 or Aurora A. Our data indicate that PP1 regulates ZYG-1 levels through a post-translational mechanism. In theory PP1 could regulate the recognition of ZYG-1 by the SCF, however we think that this is unlikely as our results do not match with the canonical mechanism for SCF regulation: substrate recognition by the SCF is regulated by phosphorylation, but the increased phosphorylation associated with PP1 down-regulation would be expected to increase proteosomal degradation, decreasing substrate accumulation. We, however, find an increase in ZYG-1 levels when PP1 activity is reduced. Furthermore, evidence in Drosophila and C. elegans indicates that degradation of Plk4/ZYG-1 is antagonized by PP2A [58,59]. Therefore, our favored hypothesis is that PP1 regulates ZYG-1 protein levels through an SCF-independent mechanism, perhaps by removing a stabilizing phosphorylation, however the identity of the opposing kinase remains unknown. We cannot, however, rule out the possibility that PP1 functions through the SCF pathway in a non-canonical fashion to mediate ZYG-1 degradation. Of note, a KVXF consensus site for PP1 binding [60] does exist in ZYG-1, however it is not conserved even in closely related nematode species. Nevertheless, although this is a common motif by which PP1 interacts with its substrates the presence of this motif is not an absolute requirement for PP1 binding.
Our data suggest that PP1, I-2SZY-2 and SDS-22 cooperate to regulate overall cellular levels of ZYG-1. Down regulation of PP1 activity leads to accumulation of excess ZYG-1 and in embryos expressing a wild-type version of ZYG-1, the elevated activity is sufficient to drive centriole over-duplication. This is similar to the situation in humans and Drosophila where overexpression of homologs of ZYG-1 causes centriole over-duplication [2,5,6]. In agreement with this previous work [2] the appearance of supernumerary centrosomes in C. elegans results from the formation of extra daughter centrioles in association with a single mother. Intriguingly, we find that when PP1 activity is decreased, the initial duplication event (that occurs during the first cell cycle following the female meiotic division) is unaffected, yielding bipolar spindles at the two-cell stage. However, beginning with the second centriole duplication event (that occurs at the two cell stage), centrioles commence over-duplicating resulting in multipolar spindles, abnormal divisions, and ultimately lethality.
Why is the first duplication event unaffected by the loss of PP1-mediated regulation while the ensuing duplication events go awry? One unique feature of the first duplication event is that it involves paternally-inherited (sperm) centrioles while later events involve centrioles assembled in the embryo. The sperm-derived centrioles are not, however, permanently immune to elevated ZYG-1 levels, as we have observed cases where, during the second centriole cycle, all four mother centrioles (the two original sperm-derived centrioles plus the two centrioles produced during the first duplication event) produce multiple daughters. Thus, it seems that the elevated level of ZYG-1 present in the embryo only triggers over-duplication of the sperm-derived centrioles following a one-cell-cycle delay. There are at least three possibilities to explain the pattern of over-duplication in PP1-compromised embryos. First, loss of PP1 activity might only lead to overexpression of ZYG-1 beginning with the second centriole duplication event. This however seems unlikely, as we have shown that loss of PP1 activity results in elevated ZYG-1 levels even in the first cell cycle (Fig 4C & 4D), and that it can suppress the failure of the first duplication event in zyg-1(it25) embryos (Figs 1C, 1D, 2B and 3G). Another possibility is that the critical period for exposure to elevated ZYG-1 is one cell cycle prior to the actual over-duplication event. Thus the sperm centrioles, which presumably first encounter elevated ZYG-1 in the zygote, would only over-duplicate during the second centriole cycle. This model is however inconsistent with previous work showing that when zyg-1 is overactive in the male germ line, centrioles will over-duplicate during spermatogenesis, but when these centrioles are introduced into a wild-type egg they duplicate normally [61]. Thus the prior exposure of sperm centrioles to elevated ZYG-1 activity does not program them to over-duplicate in the zygote. Finally, a third and more likely possibility is that the centrioles need to be exposed to elevated ZYG-1 for two consecutive cell cycles before they will over-duplicate. Thus sperm-derived centrioles are first exposed to elevated ZYG-1 in the zygote, but do not overduplicate until the second cell cycle, after they have been exposed to elevated ZYG-1 activity for two consecutive cell cycles. There is precedent for this type of regulation; during centriole maturation in human cells, Plk1 activity is needed over the course of two successive cell cycles in order for centrioles to fully mature and become capable of organizing PCM and serving as basal bodies [62]. Thus, this mode of regulation might be a common feature of how polo like kinases operate, at least when it comes to the control of centriole function.
Why do increased ZYG-1 levels lead to centriole overduplication in the C. elegans embryo? Normally, levels of centriole-associated Plk4 peak in mitosis before being reduced to a single focus marking the site of procentriole assembly at the G1/S transition [8,20,21]. Centrosome-associated ZYG-1 levels also peak in mitosis [63] and we speculate that elevated ZYG-1 levels prevent it from becoming restricted to a single focus during the ensuing cell cycle, leading to the assembly of multiple daughter centrioles. Ensuring that only a single ZYG-1/Plk4 focus is maintained seems to be a key regulatory step in centriole duplication, however our understanding of how this is achieved remains limited. Clearly excess ZYG-1/Plk4 is sufficient to subvert the normal controls. Since Plk4/ZYG-1 plays an important role in the recruitment of SAS-5/ana2/STIL and SAS-6 at the initiation of procentriole assembly [13,14] we envision elevated ZYG-1 levels may be required at this time. However whether continual exposure to elevated ZYG-1 through two cell cycles is required for overduplication remains an open question.
In summary, we have identified I-2SZY-2, SDS-22, and PP1 as novel regulators of centriole duplication. The involvement of PP1 in regulating centriole duplication has not previously been described, but interestingly centrosome amplification was reported after depletion of I-2 from human cells, suggesting that the function of PP1 in regulating centriole duplication may indeed be conserved [31]. We show that the key function of PP1 is in limiting the availability of ZYG-1. When PP1 activity is decreased, excessive accumulation of ZYG-1 leads to the formation of extra daughter centrioles in association with a single mother, resulting in centriole amplification. Appropriate regulation of PP1 activity is therefore crucial to maintaining correct centrosome numbers. Although the requirement for PP1 in chromosome segregation due to its function at the kinetochores is well documented, our work suggests a novel requirement for PP1 in the maintenance of genome stability by regulating centriole duplication.
Materials and Methods
Worm strains and RNAi
All worm strains were maintained at 20°C on MYOB plates seeded with OP50. The strains used in these experiments are listed in supplemental S1 Table. To monitor the effect of the sds-22(bs9) mutation on centriole duplication sds-22(bs9) homozygotes were selected from the OC626 strain using the visible dpy marker which is closely linked to the sds-22 gene. RNAi of SZY-2 was carried out by soaking worms in dsRNA [64], all other RNAi experiments were carried out by feeding worms bacteria expressing dsRNA as previously described [65]. Briefly, bacteria containing the RNAi construct were grown overnight and seeded onto MYOB plates supplemented with 25ug/ml ampicillin and 1mM IPTG. For double RNAi a 50:50 mix of overnight cultures of the two bacterial strains was plated. Worms were placed on RNAi at the L4 stage for 28h before analysis. The sequence contained in the GSP-1 RNAi construct was evaluated using the Clone mapper tool (http://bioinformatics.lif.univ-mrs.fr/RNAiMap/index.html)[66], which confirmed likely specificity for only the gsp-1 gene. For control RNAi we used an smd-1-containing vector.
Immunoprecipitation and western blotting
Embryonic extracts for western blots were prepared and analyzed as detailed previously [59]. Whole worm extracts for the immunoprecipitation (IP) experiments were made according to [67]. Total protein concentration was determined using the Biorad Protein Assay Dye Reagent (Bio-Rad). 1.6 mg of total protein from N2 worms was used for performing each IP. Briefly, for each IP, 30 μl of Dynabeads Protein A (Life Technologies) were incubated with 10 μg of each respective antibody at 4°C for 2 hours. Beads were washed three times in worm lysis buffer (50 mM HEPES (pH 7.4), 1 mM EGTA, 1 mM MgCl2, 100 mM KCl, 10% Glycerol, 0.05% NP-40), re-suspended in 2X Laemmli Buffer (Bio-Rad), boiled at 100°C for 2 minutes and analyzed by western blotting. The following antibodies/reagents were used in this study: polyclonal SZY-2 antibodies were raised and purified against the entire szy-2 ORF (Covance); GSP-1 antibodies were raised and purified against the peptide CQYQGMNSGRPAVGGGRPGTTAGKK (YenZym Antibodies LLC); ZYG-1 [53], phospho-histone H3 (Abcam); GSP-2 [68]; DM1A (Sigma). Mass spectrometry analysis was carried out by the NIDDK mass spec core facility.
qRT-PCR
RNA was extracted from wild-type and szy-2(bs4) embryos, DNase treated and cDNA made using a superscriptIII first strand synthesis kit (Invitrogen). Forward (ACAGTACGCGGAAGAAATGG) and reverse (CACAGCAACCATCTTTTGGA) primers were used to amplify zyg-1. Primers against ama-1 were used as a control [69]. qRT reactions used iQ SYBR green supermix (Bio-Rad) as directed by the manufacturer.
Imaging
Fixation and staining of embryos was carried out as described previously [36]. The following antibodies were used at a 1/1000 dilution: DM1A (Sigma), phospho-histone H3 (Abcam), ZYG-1 [19], SPD-2 [50] and anti-SAS-4 [53]. For live and fixed imaging we used a spinning disk confocal microscope which has been described previously [61]. To determine whether PP1 affects translation of the ZYG-1 transcript we shifted worms carrying the reporter construct to 25°C as L4s and imaged embryos the next day. Intensities of chromatin GFP at first metaphase were measured using Metamorph. Levels of ZYG-1 or SPD-2 at the centrosome were determined by quantification of average pixel intensity at the centrosome. Maximal projections of the centrosome were used for quantification of fluorescence in ImageJ 1.40g and background fluorescence was subtracted. Centrosome fluorescence was normalized to controls such that control intensity is 1. For structured illumination microscopy, embryos were immuno-labeled as usual and mounted in Vectashield (Vector Laboratories, Inc.). Samples were imaged with a DeltaVision OMX4 SIM Imaging System (Applied Precision).
Supporting Information
Schematic showing how centrioles behave and how their numbers are established at each cell stage through the first three cell cycles in A) wild-type embryos, B) zyg-1(it25) mutant embryos and in C) embryos with reduced PP1 function.
(PDF)
Quantification of embryonic viability among the progeny of worms grown at the semi-permissive temperature of 24°C and depleted for the indicated PP1 regulators. smd-1(RNAi) targets a nonessential gene and serves as a negative control. Only reduction of sds-22 led to substantial rescue of zyg-1(it25) lethality.
(PDF)
GFP fusions of the indicated protein were expressed in the embryo. I-2SZY-2 is enriched in the nuclei. Pronuclei meeting during the first cell cycle is shown. PP1βGSP-1 is enriched on the chromatin throughout mitosis. Anaphase of the first cell cycle is shown. SDS-22 is weakly localized to the spindle. Metaphase of the first cell cycle is shown.
(PDF)
Each of the indicated proteins was immunoprecipitated from worm extracts and co-purifying proteins identified by mass spec. Shown are the top five hits based on peptide number. In cases where PP1GSP-1, SDS-22 or I-2SZY-2 were not among the top hits, they are also shown along with their rank and number of identifying peptides.
(PDF)
A) Western blot showing elevated total phospho-histone levels present in extract from mixed stage sds-22(b9/tm5187) embryos. This elevation is likely due to an increase in the length of mitosis in sds-22 mutant embryos as B) quantitative immunofluorescence microscopy shows that mitotic chromatin in sds-22(bs9/tm5187) embryos is not enriched for phospho-histone H3 relative to the wild type, and in fact, appears reduced. (a.u. = arbitrary units).
(PDF)
Each of the specified kinases were RNAi depleted in zyg-1(i25); szy-2(bs4) worms and centriole duplication monitored by microscopy. A) Quantification of centriole duplication. Numbers above bars indicate the percentage of successful centriole duplication events and the number within the bars indicate the number of events scored. B-D) Representative stills from time-lapse recordings of zyg-1(it25); szy-2(bs4) embryos expressing GFP::tubulin and mCherry::histone and treated with control RNAi or RNAi against one of the three indicated mitotic kinases. B) Embryos treated with control RNAi duplicate centrioles, proceed to the 2 cell stage, and build bipolar spindles. C) Embryos treated with air-2 RNAi duplicate centrioles, but fail in chromosome segregation, and thus display 4 centrosomes associated with a single enlarged nucleus. D) Embryos treated with plk-1 RNAi show a variety of cell division defects yet present with 4 centrosomes indicating centriole duplication has occurred. E&F) Representative stills from time-lapse recordings of zyg-1(it25); szy-2(bs4) embryos expressing GFP::SPD-2 and treated with control RNAi (E) or RNAi against air-1 (F). Centrioles from F are enlarged in a&b. Embryos are at the early 2-cell stage and duplicated centrioles are visible (arrow heads). Note, in this example only one centriole of the control embryo has duplicated.
(PDF)
A) Average GFP::SPD-2 levels at the centrosome during the first cell cycle were calculated and normalized to control. B) Average levels of endogenous SPD-2 at the centrosome during the first metaphase in szy-2(bs4) embryos were normalized to controls. C&D) Representative wild-type and szy-2(bs4) embryos stained for DNA (blue), tubulin (red) and SPD-2 (green). SPD-2 staining at centrosomes is enlarged in a and b. E&F) Comparison of SPD-2::GFP levels at the centrosome in an embryo expressing a codon-optimized version of SPD-2::GFP (OE = overexpression) (E) and a strain expressing GFP::SPD-2 from the native sequence (F). G) Quantification of embryonic viability among the indicated strains to determine whether overexpression of SPD-2 is sufficient to rescue the zyg-1(it25) phenotype. In each case n = 15 worms, >1000 embryos. H) GFP::SPD-2 levels at the centrosome in zyg-1(it25); szy-2(bs4) embryos treated with negative control smd-1(RNAi) or air-1(RNAi).
(PDF)
A) Western blot demonstrating equivalent total levels of SAS-6 in wild-type and szy-2(bs4) embryos. B) Quantitation of western blot data (n = 2). C) Average levels of centrosome-localized GFP::SAS-6 in wild-type and szy-2(bs4) embryos were determined throughout the first cell cycle. Values shown are normalized to the average wild-type value at each stage. Error bars represent standard error.
(PDF)
Select frames from a time-lapse recording of an sds-22(bs9/tm5187) embryo expressing GFP::SPD-2 and mCherry::histone. The arrowhead shows a spindle pole (frame 18:00) giving rise to multiple centrosomes during the next cell cycle (frame 28:00). White arrows indicate centrosomes that become inactive during the round of division following their birth (frames 10:00 and 18:00) but become active again during the next cell cycle (frames 22:00–34:00). Note that both centrosomes were not always visible in these frames.
(PDF)
(DOCX)
Note bipolar spindle formation at the two-cell stage.
(AVI)
Note monopolar spindle formation at the two-cell stage.
(AVI)
Note restoration of bipolar spindle formation at the two-cell stage.
(AVI)
(AVI)
(AVI)
(AVI)
The extra centrosomes recruit PCM and participate in spindle assembly.
(AVI)
(AVI)
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center (CGC), and the National BioResource Project (Japan). We thank M. Colaiacovo for C. elegans GSP-2 antibody, and E. Anderson for assistance with mass spectrometry.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Diabetes and Digestive and Kidney Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fu J, Hagan IM, Glover DM. The centrosome and its duplication cycle. Cold Spring Harb Perspect Biol. 2015;7: a015800 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Habedanck R, Stierhof Y-D, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7: 1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- 3.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gönczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13: 203–213. 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arquint C, Sonnen KF, Stierhof Y-D, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci. 2012;125: 1342–1352. 10.1242/jcs.099887 [DOI] [PubMed] [Google Scholar]
- 5.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17: 834–843. 10.1016/j.cub.2007.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316: 1046–1050. 10.1126/science.1142950 [DOI] [PubMed] [Google Scholar]
- 7.Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, Fu J, et al. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr Biol. 2014;24: 2526–2532. 10.1016/j.cub.2014.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, et al. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat Commun. 2014;5: 5267 10.1038/ncomms6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kratz A-S, Bärenz F, Richter KT, Hoffmann I. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol Open. 2015;4: 370–377. 10.1242/bio.201411023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J Cell Biol. 2015;209: 863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitagawa D, Flückiger I, Polanowska J, Keller D, Reboul J, Gönczy P. PP2A Phosphatase Acts upon SAS-5 to Ensure Centriole Formation in C. elegans Embryos. Dev Cell. 2011;20: 550–562. 10.1016/j.devcel.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 12.van Breugel M, Hirono M, Andreeva A, Yanagisawa H-A, Yamaguchi S, Nakazawa Y, et al. Structures of SAS-6 Suggest Its Organization in Centrioles. Science. 2011. [DOI] [PubMed] [Google Scholar]
- 13.Delattre M, Canard C, Gönczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16: 1844–1849. 10.1016/j.cub.2006.07.059 [DOI] [PubMed] [Google Scholar]
- 14.Lettman MM, Wong YL, Viscardi V, Niessen S, Chen S-H, Shiau AK, et al. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev Cell. 2013;25: 284–298. 10.1016/j.devcel.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sillibourne JE, Tack F, Vloemans N, Boeckx A, Thambirajah S, Bonnet P, et al. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell. 2010;21: 547–561. 10.1091/mbc.E09-06-0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188: 191–198. 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J Cell Sci. 2010;123: 2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- 18.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184: 225–239. 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peel N, Dougherty M, Goeres J, Liu Y, O'Connell KF. The C. elegans F-box proteins LIN-23 and SEL-10 antagonize centrosome duplication by regulating ZYG-1 levels. J Cell Sci. 2012;125: 3535–3544. 10.1242/jcs.097105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim T-S, Park J-E, Shukla A, Choi S, Murugan RN, Lee JH, et al. Hierarchical recruitment of Plk4 and regulation of centriole biogenesis by two centrosomal scaffolds, Cep192 and Cep152. Proceedings of the National Academy of Sciences. 2013;110: E4849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnen KF, Gabryjonczyk A-M, Anselm E, Stierhof Y-D, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J Cell Sci. 2013;126: 3223–3233. 10.1242/jcs.129502 [DOI] [PubMed] [Google Scholar]
- 22.Arquint C, Gabryjonczyk A-M, Imseng S, Böhm R, Sauer E, Hiller S, et al. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klebba JE, Buster DW, McLamarrah TA, Rusan NM, Rogers GC. Autoinhibition and relief mechanism for Polo-like kinase 4. Proceedings of the National Academy of Sciences. 2015;112: E657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchner J, Gross S, Bennett D, Alphey L. Essential, overlapping and redundant roles of the Drosophila protein phosphatase 1 alpha and 1 beta genes. Genetics. 2007;176: 273–281. 10.1534/genetics.106.069914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sassa T, Ueda-Ohba H, Kitamura K-I, Harada S-I, Hosono R. Role of Caenorhabditis elegans protein phosphatase type 1, CeGLC-7 beta, in metaphase to anaphase transition during embryonic development. Exp Cell Res. 2003;287: 350–360. [DOI] [PubMed] [Google Scholar]
- 26.Wu J-C, Go AC, Samson M, Cintra T, Mirsoian S, Wu TF, et al. Sperm development and motility are regulated by PP1 phosphatases in Caenorhabditis elegans. Genetics. 2012;190: 143–157. 10.1534/genetics.111.135376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33: 537–545. 10.1016/j.molcel.2009.02.015 [DOI] [PubMed] [Google Scholar]
- 28.Huang FL, Glinsmann WH. Separation and characterization of two phosphorylase phosphatase inhibitors from rabbit skeletal muscle. Eur J Biochem. 1976;70: 419–426. [DOI] [PubMed] [Google Scholar]
- 29.Tung HY, Wang W, Chan CS. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol Cell Biol. 1995;15: 6064–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Satinover DL, Brautigan DL. Phosphorylation and functions of inhibitor-2 family of proteins. Biochemistry. 2007;46: 2380–2389. 10.1021/bi602369m [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Stukenberg PT, Brautigan DL. Phosphatase inhibitor-2 balances protein phosphatase 1 and aurora B kinase for chromosome segregation and cytokinesis in human retinal epithelial cells. Mol Biol Cell. 2008;19: 4852–4862. 10.1091/mbc.E08-05-0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Cronmiller C, Brautigan DL. Maternal phosphatase inhibitor-2 is required for proper chromosome segregation and mitotic synchrony during Drosophila embryogenesis. Genetics. 2008;179: 1823–1833. 10.1534/genetics.108.091959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posch M, Khoudoli G, Swift S, King E, DeLuca J, Swedlow J. Sds22 regulates aurora B activity and microtubule–kinetochore interactions at mitosis. J Cell Biol. 2010;191: 61 10.1083/jcb.200912046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114: 3749–3757. [DOI] [PubMed] [Google Scholar]
- 35.Helps NR, Luo X, Barker HM, Cohen PT. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem J. 2000;349: 509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kemp CA, Song MH, Addepalli MK, Hunter G, O'Connell K. Suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in Caenorhabditis elegans. Genetics. 2007;176: 95–113. 10.1534/genetics.107.071803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102: 279–291. [DOI] [PubMed] [Google Scholar]
- 38.Qian J, Lesage B, Beullens M, Van Eynde A, Bollen M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr Biol. 2011;21: 766–773. 10.1016/j.cub.2011.03.047 [DOI] [PubMed] [Google Scholar]
- 39.Kaitna S, Pasierbek P, Jantsch M, Loidl J, Glotzer M. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis. Curr Biol. 2002;12: 798–812. [DOI] [PubMed] [Google Scholar]
- 40.Ohkura H, Yanagida M. S. pombe gene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell. 1991;64: 149–157. [DOI] [PubMed] [Google Scholar]
- 41.Stone EM, Yamano H, Kinoshita N, Yanagida M. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr Biol. 1993;3: 13–26. [DOI] [PubMed] [Google Scholar]
- 42.MacKelvie SH, Andrews PD, Stark MJ. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol Cell Biol. 1995;15: 3777–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers E, Bishop JD, Waddle JA, Schumacher JM, Lin R. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol. 2002;157: 219–229. 10.1083/jcb.200110045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumacher JM, Golden A, Donovan PJ. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satinover DL, Leach CA, Stukenberg PT, Brautigan DL. Activation of Aurora-A kinase by protein phosphatase inhibitor-2, a bifunctional signaling protein. Proc Natl Acad Sci USA. 2004;101: 8625–8630. 10.1073/pnas.0402966101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol. 2001;155: 1109–1116. 10.1083/jcb.200108051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81: 95–105. [DOI] [PubMed] [Google Scholar]
- 48.Yamashiro S, Yamakita Y, Totsukawa G, Goto H, Kaibuchi K, Ito M, et al. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev Cell. 2008;14: 787–797. 10.1016/j.devcel.2008.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsou M-FB, Wang W-J, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17: 344–354. 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6: 511–523. [DOI] [PubMed] [Google Scholar]
- 51.Pelletier L, Ozlü N, Hannak E, Cowan C, Habermann B, Ruer M, et al. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14: 863–873. 10.1016/j.cub.2004.04.012 [DOI] [PubMed] [Google Scholar]
- 52.Decker M, Jaensch S, Pozniakovsky A, Zinke A, O'Connell KF, Zachariae W, et al. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol. 2011;21: 1259–1267. 10.1016/j.cub.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 53.Song MH, Aravind L, Müller-Reichert T, O'Connell KF. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev Cell. 2008;15: 901–912. 10.1016/j.devcel.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller JG, Liu Y, Williams CW, Smith HE, O'Connell KF. The E2F-DP1 Transcription Factor Complex Regulates Centriole Duplication in Caenorhabditis elegans. G3 (Bethesda). 2016;6: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133: 1032–1042. 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2: 120104 10.1098/rsob.120104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1: 965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brownlee CW, Klebba JE, Buster DW, Rogers GC. The Protein Phosphatase 2A regulatory subunit Twins stabilizes Plk4 to induce centriole amplification. J Cell Biol. 2011;195: 231–243. 10.1083/jcb.201107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song MH, Liu Y, Anderson DE, Jahng WJ, O'Connell KF. Protein Phosphatase 2A-SUR-6/B55 Regulates Centriole Duplication in C. elegans by Controlling the Levels of Centriole Assembly Factors. Dev Cell. 2011;20: 563–571. 10.1016/j.devcel.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. EMBO Press; 1997;16: 1876–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters N, Perez DE, Song MH, Liu Y, Müller-Reichert T, Caron C, et al. Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. J Cell Sci. 2010;123: 795–805. 10.1242/jcs.050682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kong D, Farmer V, Shukla A, James J, Gruskin R, Kiriyama S, et al. Centriole maturation requires regulated Plk1 activity during two consecutive cell cycles. J Cell Biol. Rockefeller University Press; 2014;206: 855–865. 10.1083/jcb.201407087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, et al. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105: 547–558. [DOI] [PubMed] [Google Scholar]
- 64.Fernandez AG, Gunsalus KC, Huang J, Chuang L-S, Ying N, Liang H-L, et al. New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res. 2005;15: 250–259. 10.1101/gr.3194805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421: 231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- 66.Thakur N, Pujol N, Tichit L, Ewbank JJ. Clone mapper: an online suite of tools for RNAi experiments in Caenorhabditis elegans. G3 (Bethesda). Genetics Society of America; 2014;4: 2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zanin E, Dumont J, Gassmann R, Cheeseman I, Maddox P, Bahmanyar S, et al. Affinity purification of protein complexes in C. elegans. Methods Cell Biol. 2011;106: 289–322. 10.1016/B978-0-12-544172-8.00011-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tzur YB, Egydio de Carvalho C, Nadarajan S, Van Bostelen I, Gu Y, Chu DS, et al. LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion. PLoS Biol. 2012;10: e1001378 10.1371/journal.pbio.1001378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang H, Spang A, Sullivan MA, Hryhorenko J, Hagen FK. The terminal phase of cytokinesis in the Caenorhabditis elegans early embryo requires protein glycosylation. Int Rev Cytol. 2005;16: 4202–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic showing how centrioles behave and how their numbers are established at each cell stage through the first three cell cycles in A) wild-type embryos, B) zyg-1(it25) mutant embryos and in C) embryos with reduced PP1 function.
(PDF)
Quantification of embryonic viability among the progeny of worms grown at the semi-permissive temperature of 24°C and depleted for the indicated PP1 regulators. smd-1(RNAi) targets a nonessential gene and serves as a negative control. Only reduction of sds-22 led to substantial rescue of zyg-1(it25) lethality.
(PDF)
GFP fusions of the indicated protein were expressed in the embryo. I-2SZY-2 is enriched in the nuclei. Pronuclei meeting during the first cell cycle is shown. PP1βGSP-1 is enriched on the chromatin throughout mitosis. Anaphase of the first cell cycle is shown. SDS-22 is weakly localized to the spindle. Metaphase of the first cell cycle is shown.
(PDF)
Each of the indicated proteins was immunoprecipitated from worm extracts and co-purifying proteins identified by mass spec. Shown are the top five hits based on peptide number. In cases where PP1GSP-1, SDS-22 or I-2SZY-2 were not among the top hits, they are also shown along with their rank and number of identifying peptides.
(PDF)
A) Western blot showing elevated total phospho-histone levels present in extract from mixed stage sds-22(b9/tm5187) embryos. This elevation is likely due to an increase in the length of mitosis in sds-22 mutant embryos as B) quantitative immunofluorescence microscopy shows that mitotic chromatin in sds-22(bs9/tm5187) embryos is not enriched for phospho-histone H3 relative to the wild type, and in fact, appears reduced. (a.u. = arbitrary units).
(PDF)
Each of the specified kinases were RNAi depleted in zyg-1(i25); szy-2(bs4) worms and centriole duplication monitored by microscopy. A) Quantification of centriole duplication. Numbers above bars indicate the percentage of successful centriole duplication events and the number within the bars indicate the number of events scored. B-D) Representative stills from time-lapse recordings of zyg-1(it25); szy-2(bs4) embryos expressing GFP::tubulin and mCherry::histone and treated with control RNAi or RNAi against one of the three indicated mitotic kinases. B) Embryos treated with control RNAi duplicate centrioles, proceed to the 2 cell stage, and build bipolar spindles. C) Embryos treated with air-2 RNAi duplicate centrioles, but fail in chromosome segregation, and thus display 4 centrosomes associated with a single enlarged nucleus. D) Embryos treated with plk-1 RNAi show a variety of cell division defects yet present with 4 centrosomes indicating centriole duplication has occurred. E&F) Representative stills from time-lapse recordings of zyg-1(it25); szy-2(bs4) embryos expressing GFP::SPD-2 and treated with control RNAi (E) or RNAi against air-1 (F). Centrioles from F are enlarged in a&b. Embryos are at the early 2-cell stage and duplicated centrioles are visible (arrow heads). Note, in this example only one centriole of the control embryo has duplicated.
(PDF)
A) Average GFP::SPD-2 levels at the centrosome during the first cell cycle were calculated and normalized to control. B) Average levels of endogenous SPD-2 at the centrosome during the first metaphase in szy-2(bs4) embryos were normalized to controls. C&D) Representative wild-type and szy-2(bs4) embryos stained for DNA (blue), tubulin (red) and SPD-2 (green). SPD-2 staining at centrosomes is enlarged in a and b. E&F) Comparison of SPD-2::GFP levels at the centrosome in an embryo expressing a codon-optimized version of SPD-2::GFP (OE = overexpression) (E) and a strain expressing GFP::SPD-2 from the native sequence (F). G) Quantification of embryonic viability among the indicated strains to determine whether overexpression of SPD-2 is sufficient to rescue the zyg-1(it25) phenotype. In each case n = 15 worms, >1000 embryos. H) GFP::SPD-2 levels at the centrosome in zyg-1(it25); szy-2(bs4) embryos treated with negative control smd-1(RNAi) or air-1(RNAi).
(PDF)
A) Western blot demonstrating equivalent total levels of SAS-6 in wild-type and szy-2(bs4) embryos. B) Quantitation of western blot data (n = 2). C) Average levels of centrosome-localized GFP::SAS-6 in wild-type and szy-2(bs4) embryos were determined throughout the first cell cycle. Values shown are normalized to the average wild-type value at each stage. Error bars represent standard error.
(PDF)
Select frames from a time-lapse recording of an sds-22(bs9/tm5187) embryo expressing GFP::SPD-2 and mCherry::histone. The arrowhead shows a spindle pole (frame 18:00) giving rise to multiple centrosomes during the next cell cycle (frame 28:00). White arrows indicate centrosomes that become inactive during the round of division following their birth (frames 10:00 and 18:00) but become active again during the next cell cycle (frames 22:00–34:00). Note that both centrosomes were not always visible in these frames.
(PDF)
(DOCX)
Note bipolar spindle formation at the two-cell stage.
(AVI)
Note monopolar spindle formation at the two-cell stage.
(AVI)
Note restoration of bipolar spindle formation at the two-cell stage.
(AVI)
(AVI)
(AVI)
(AVI)
The extra centrosomes recruit PCM and participate in spindle assembly.
(AVI)
(AVI)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.