Abstract
Inorganic polyphosphate (poly P), in chains of tens to hundreds of phosphate residues, linked by high-energy bonds, is environmentally ubiquitous and abundant. In prebiotic evolution it could have provided a flexible, polyanionic scaffold to assemble macromolecules. It has been conserved in every cell in nature. In prokaryotes, a major poly P synthetic enzyme is poly P kinase 1 (PPK1), which is found in 100 bacterial genomes, including numerous pathogens. Null mutants of PPK1, with low poly P levels, are defective in survival: namely, they show defective responses to physical/chemical stresses and predation. Pathogens with a PPK1 deletion are defective in biofilm formation, quorum sensing, general stress and stringent responses, motility, and other virulence properties. With the exception of Dictyostelium, PPK1 is absent in eukaryotes and provides a novel target for chemotherapy that would affect both virulence and susceptibility to antibacterial compounds. Remarkably, another PPK in Dictyostelium discoideum (PPK2) is an actin-related protein (Arp) complex that is polymerized into an actin-like filament, concurrent with its reversible synthesis of a poly P chain from ATP.
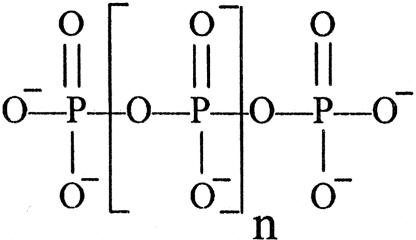
Mineral and organic constituents essential for cellular growth and survival are generally well known; an exception is inorganic polyphosphate (poly P). Likely a key agent in evolution from prebiotic time, poly P is found as a chain of tens to hundreds of phosphate residues linked by high-energy, phosphoanhydride bonds as in ATP.
Figure 1.
This polymer, generated simply by dehydration of orthophosphate at elevated temperatures, is found in volcanic condensates and deep oceanic steam vents. Most remarkably, poly P has been conserved in every cell in nature: bacterial, fungal, plant, and animal (1). In eukaryotes, it is found in subcellular organelles and in yeast vacuoles at levels as high as 20% of the cell dry weight. Yet, for lack of proven function, poly P has been regarded as a “molecular fossil” and not mentioned in textbooks of biology and chemistry (2).
As the large number and variety of physiologic functions of poly P have become known, its unique chemical features as a biomolecule have also become evident. First is the advantage of its being ionized. For example, the orthophosphate ion is maintained in cells within a narrow range near 5 mM in the face of levels outside the cell that may differ by 5 orders of magnitude: 0.01-1,000 mM. The second is that as an anion it can chelate a wide variety of cations, specifically Mg2+, Mn2+, Ca2+, Zn2+, and Fe3+, and influence their biologic roles; also important is the capacity of poly P to sequester toxic metals, such as Cd2+ and Hg+, in their staged removal. Finally, as a polyanion, poly P can be localized in the cell in a flexible, oriented, and relatively stable form and yet be used in response to a wide variety of metabolic needs. In prebiotic evolution, ubiquitous and abundant poly P could have provided a polyanionic, high-energy scaffold of varying polymer lengths to orient or assemble macromolecules. Lacking any inherent tertiary structure, its chelating capacity could provide further options for shape and orientation. Different substrata such as self-assembled lipid vesicles, clay, or rock similarly offer options for polymer shape. As a phosphorylating agent, poly P could donate to alcohols, including sugars, nucleosides, and proteins, and also react with activated alkyl and amino acids to generate fatty acids and polypeptides. As a scaffold, it may have facilitated the assembly and orientation of the major polymers in the biotic world: phospholipids, nucleic acids, and proteins.
Although observed in cells as meta-chromatically stained “volutin granules” 100 years ago and identified as poly P 50 years later, the functions of poly P remained largely unknown or ignored (3). Toward our goal to discover its functions, we identified and purified the enzymes that make and use poly P and then manipulated the genes that encode each of them. These studies have revealed a large number and variety of functions of poly P. Prominent among them are the bacterial responses to each of many stresses and stringencies, the weapons in the constant battle between predators and prey, and ultimately, the means for the emergence and survival of a species (2).
Among the notable poly P enzymes are the poly P kinases (PPKs) that make poly P reversibly from the terminal phosphate of ATP and the poly P-AMP-phosphotransferases that use poly P to phosphorylate AMP to ADP, an immediate source of ATP (3). Other enzymes that influence the accumulation of poly P are the hydrolytic enzymes: exopolyPases that release Pi from the ends of poly P and endopolyPases that cleave poly P to progressively shorter chains.
Most attention in biology has focused on growth and development; relatively little has been paid to “life in the slow lane” (4). In the stationary phase of microbial life cycles, cells must be able to cope with all manner of stresses and stringencies to survive. These hazards include heat, desiccation (commonly preceded by hyperosmolarity), radiation, oxidants, starvation, and predation. By sensing any one or more of these stresses, even at an early stage, bacteria can sound an alarm by accumulating poly P, which can then activate or augment several systems.
The stringent response system, triggered by low phosphate or amino acid levels, leads to the RelA synthesis of the guanosine nucleotides, pppGpp and ppGpp, which in turn activate a large number of genes (5). These nucleotides inhibit the hydrolysis of poly P by exopolyPase and thereby increase its accumulation. Another major player in the regulation of gene expression in the stationary phase is the RNA polymerase sigma factor RpoS. This factor, itself influenced by ppGpp (6, 7), directs transcriptional responses in >50 genes in cascades that contribute to the shifting down of metabolism geared to growth, thereby adjusting the cell to dormancy (8). In Escherichia coli, poly P increases the levels of RpoS and may also act directly at the loci of gene action (9, 10). RpoS has been implicated in the virulence of bacterial pathogens for animals (11) and protozoa (12). Legionella pneumophilus emerges after replication within Acanthamoeba both more virulent (13) and more resistant to chemical attack (14, 15). It would thus seem that poly P has a role in the merged pathways of both virulence and resistance.
A striking example of poly P action in starvation can be elicited in the nutrient downshift of an E. coli culture from a rich to a minimal medium. A lag of ≈2 h ensues before resumption of growth because of a lack of nutrients, principally amino acids; the lag is abolished by addition of amino acids to the medium (16). Downshifted cells accumulate a level of poly P sufficient to complex the quiescent Lon protease and activate it by >20-fold in its degradation of protein subunits released from idle ribosomes (17, 18). The amino acids generated by the protease become available for the biosynthesis of proteins in the cycle now geared to growth in a minimal medium. By contrast, the downshifted ppk1 mutant, which fails to accumulate poly P, takes as many as 10 h to resume growth unless relieved by addition of amino acids to the medium. In the absence of an external nutrient source, these responses could direct metabolism away from growth toward dormancy.
The PPK sequences, first identified in E. coli (PPK1), are found in 100 bacterial genomes, including 17 pathogens responsible for a variety of severe human diseases. The role of poly P in any of these organisms is best inferred from the phenotypic deficiencies in the PPK1-null mutants in which the poly P level is severely depressed. Among the pathogens, defects have been observed in growth, motility (19), quorum sensing, biofilm formation, virulence (20), and predation (unpublished results). In terms of human infection, PPK deletion resulted in extreme serum sensitivity in Neisseria species (21), perhaps because the Neisseria capsule has a lot of poly P.
The widespread conservation of the protein kinase site of PPK1 (22), a site not observed in eukaryotes apart from Dictyostelium (see below), prompted the extensive screening of libraries of compounds for inhibitors of the site. A number of promising antibiotics have been found that produce the same phenotypic defects found in the null ppk1 mutants and are now available for drug development (Sam Lee, personal communication).
Mobility (swimming, swarming, and twitching) serve the planktonic organism in seeking nutrients, avoiding toxins, and finding a suitable surface for aggregation. Signals of increasing cell density (quorum sensing) set the stage for the aggregation that precedes the development of biofilms, the niche that includes nearly all of the bacteria on Earth. The biofilm exopolymer reduces UV penetration (23) and the hydrophilic surface reduces the risk of predation (24). Being hygroscopic, the biofilm reduces drying, and the high cell density enhances quorum sensing and may in part explain high resistance to antimicrobials (25).
Sporulation is one of the most effective devices for long survival in the face of extreme stresses and for rapid responses to signals for germination. Inasmuch as spores have not been known to store ADP or ATP, another source of energy has now been revealed to be poly P. Spores of Myxobacteria (e.g., Myxobacteria xanthus), Bacillus sp. (e.g., Bacillus cereus), and the slime mold, Dictyostelium discoideum, contain large stores of poly P, as well as the enzymes (e.g., poly P-AMP-phosphotransferases and PPK) to convert poly P to ATP (unpublished data). Also, with the action of exopolyPase, poly P can be a reservoir for Pi for the start of metabolic cycles.
Aside from the activation of Lon protease in response to a nutrient downshift, the many physiologic functions of poly P have yet to be explained at a molecular level. For example, as noted, mutants of Pseudomonas aeruginosa in PPK1 with depressed levels of poly P are defective in motility, quorum sensing, biofilm formation, virulence, and other features. That poly P may exert its regulatory effects upstream, perhaps at the level of gene expression, is suggested by several observations that follow.
A striking compaction of the nucleoid of the P. aeruginosa PPK1 mutant is observed in the electron microscope as compared with the extended state of the WT. Not surprising, in view of the major phenotypic changes, global changes are observed in the mRNA levels in the DNA arrays of this PPK1 mutant: ≈240 genes are up-regulated and ≈460 are down-regulated. By contrast, in the P. aeruginosa mutant PPK2, another PPK especially proficient in the poly P-driven synthesis of GTP from GDP, only 20 genes are up- or down-regulated. A compelling finding with regard to gene expression is the very strong affinity of poly P for the histone-like proteins, such that it can displace them from their attachments to the DNA of the nucleoid. Experiments are needed to explore the role that poly P may have in the structural organization of the nucleoid that exposes specific genes for expression or repression. It is noteworthy that calcium and poly P, enveloped by poly β-hydroxybutyrate, contribute to the structure and function of membrane channels (26, 27).
Searches of genomic databases showed that among eukaryotes the social slime mold D. discoideum is the only example that possesses a PPK1 highly homologous to the bacterial enzyme. Possibly it was acquired by horizontal gene transfer from prokaryotes. The enzymes involved in synthesizing poly P in other eukaryotes remain unknown. A null mutant of D. discoideum (DdPPK1) is deficient in growth and sporulation but still retains a significant level of poly P attributable to a PPK2 in a vacuole, rich in calcium and poly P and responsible for calcium flux in the cell. Such an “acidocalcisome” vacuole is also conserved from bacteria to humans: in several pathogenic protozoal species (Trypanosoma, Leishmania, and Toxoplasma), in the green alga Chlamydomonas, and in human platelets (28). The enzyme, freed from the vacuole of D. discoideum by strong salt, was purified ≈1,000-fold and proved to be an actin-related-protein (Arp) complex. It has 60% homology in sequence to the common muscle actin and similar properties and globular-filamentous transitions (29). The globular enzyme [Arp(G)] is polymerized into a long, actin-like filament [Arp(F)] concurrent with its synthesis of a poly P chain from ATP; the reaction is reversible.
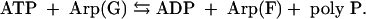 |
Thus, unlike the wasteful ATPase activity attributed to actins, the energy is conserved by DdPPK2 as poly P; the contractility associated with the vacuole may well depend on the growth of the Arp filament in conjunction with other motor elements in the cell. Synthesis of poly P from ATP in the vacuoles may differ from that of the solubilized DdPPK2, inasmuch as it is inhibited by carbonyl cyanide m-chlorophenylhydrazone (29), the proton-motive force inhibitor; in the membranous location, poly P may be assembled directly from Pi, as in the synthesis of inorganic pyrophosphate and of ATP from ADP (30).
In summary, poly P and the enzymes that make and use it play numerous and vital roles in metabolism, those involved in the survival of a species being the most notable so far. Because poly P has been present in all cells from early in evolution, and plausibly from the beginning, it is clear that it came to be used in diverse ways. Among these uses are its capacity to stimulate proliferation of mammary cancer cells (31) and participate with actin filaments in the operations of the cell (29). Surely a host of functions still remain to be discovered for this ubiquitous, abundant, and versatile polyanion.
Acknowledgments
We are grateful to The Ellison Medical Foundation for supporting a sabbatical visit by M.R.W.B.
References
- 1.Kulaev, I. S. & Vagabov, V. M. (1983) Adv. Microb. Physiol. 24, 83-171. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg, A., Rao, N. N. & Ault-Riché, D. (1999) Annu. Rev. Biochem. 68, 89-125. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg, A. (1995) J. Bacteriol. 177, 491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegele, D. A. & Kolter, R. (1992) J. Bacteriol. 174, 345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cashel, M., Gentry, D. R., Hernandez, V. J. & Vinella, D. (1996) in Escherichia coli and Salmonella typhimurium, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), pp. 1458-1496.
- 6.Gentry, D. R., Hernandez, V. J., Nguyen, D. B., Jensen, D. B. & Cashel, M. (1993) J. Bacteriol. 175, 7982-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch, M. & Elliott, T. (2002) J. Bacteriol. 184, 5077-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hengge-Aronis, R. (2002) Microbiol. Mol. Biol. Rev. 66, 373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao, N. & Kornberg, A. (1996) J. Bacteriol. 178, 1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiba, T., Tsutsumi, K., Yano, H., Ihara, Y., Kameda, A., Tanaka, K., Takahashi, H., Munekata, M., Rao, N. N. & Kornberg, A. (1997) Proc. Natl. Acad. Sci. USA 94, 11210-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nickerson, C. A & Curtiss, R., III (1997) Infect. Immun. 65, 1814-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hales, L. M. & Shuman, H. A. (1999) J. Bacteriol. 181, 4879-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cirillo, J. D., Falkow, S. & Tompkins, L. S. (1994) Infect. Immun. 62, 3254-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barker, J., Brown, M. R. W., Collier, P. J., Farrell, I. D. & Gilbert, P. (1992) Appl. Environ. Microbiol. 58, 2420-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker, J., Scaife, H. & Brown, M. R. W. (1995) Antimicrob. Agents Chemother. 39, 2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda, A., Tanaka, S., Ikeda, T., Kato, J., Takiguchi, N. & Ohtake, H. (1999) Proc. Natl. Acad. Sci. USA 96, 14264-14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda, A., Nomura, K., Ohtomo, R., Kato, J., Ikeda, T., Takiguchi, N., Ohtake, H. & Kornberg, A. (2001) Science 293, 705-708. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman, S. & Maurizi, M. R. (2001) Science 293, 614-615. [DOI] [PubMed] [Google Scholar]
- 19.Rashid, M. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rashid, M. H., Rumbaugh, K., Passador, L., Davies, D. G., Hamood, A. N., Iglewski, B. H. & Kornberg, A. (2000) Proc. Natl. Acad. Sci. USA 97, 9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tinsley, C. R. & Gotschlich, E. C. (1995) Infect. Immun. 63, 1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tzeng, C.-M. & Kornberg, A. (1998) Mol. Microbiol. 29, 381-382. [DOI] [PubMed] [Google Scholar]
- 23.Elasri, M. O. & Miller, R. V. (1999) Appl. Environ. Microbiol. 65, 2025-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Oss, C. J. (1978) Annu. Rev. Microbiol. 32, 19-39. [DOI] [PubMed] [Google Scholar]
- 25.Brown, M. R. W. & Smith, A. W. (2003) in Medical Implications of Biofilms, eds. Wilson, M. & Devine, D. (Cambridge Univ. Press, Cambridge, U.K.), pp. 36-58.
- 26.Reusch, R. N., Huang, R. & Kosk-Kosicka, D. (1997) FEBS Lett. 412, 592-596. [DOI] [PubMed] [Google Scholar]
- 27.Zakharian, E. & Reusch, R. N. (2004) Biochem. Biophys. Res. Commun. 322, 1059-1065. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz, F. A., Lea, C. R., Oldfield, E. & Docampo, R. (2004) J. Biol. Chem. 279, 44250-44257. [DOI] [PubMed] [Google Scholar]
- 29.Gómez-García, M. R. & Kornberg, A. (2004) Proc. Natl. Acad. Sci. USA 101, 15876-15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyer, P. (1997) Annu. Rev. Biochem. 66, 717-749. [DOI] [PubMed] [Google Scholar]
- 31.Wang, L., Fraley, C. D., Faridi, J., Kornberg, A. & Roth, R. A. (2003) Proc. Natl. Acad. Sci. USA 100, 11249-11254. [DOI] [PMC free article] [PubMed] [Google Scholar]