Abstract
The mechanism by which protein functional diversity expands is an important evolutionary issue. Studies of recently evolved chimeric genes permit direct investigation of the origin of new protein functions before they become obscured by subsequent evolution. Found in several African Drosophila species, jingwei (jgw), a recently evolved gene with a domain derived from the still extant short-chain alcohol dehydrogenase (ADH) through retroposition, provides an opportunity to examine this previously undescribed process directly. We expressed JGW proteins in a microbial expression system and, after purification, investigated their enzymatic properties. We found that, unexpectedly, positive Darwinian selection for amino acid replacements outside the active site of JGW produced a novel dehydrogenase with altered substrate specificity compared with the ancestral ADH. Instead of detoxifying and assimilating ethanol like its Adh parental gene, we observe that JGW efficiently utilizes long-chain primary alcohols found in hormone and pheromone metabolism. These data suggest that protein functional diversity can expand rapidly under the joint forces of exon shuffling, gene duplication, and natural selection.
The ultimate source of all biological diversity can be found in the functional diversity of macromolecules. Consequently, the origin of new genes and the mechanisms by which they acquire new functions are central to an understanding of molecular evolutionary diversification. These mechanisms include the molecular mechanisms that create the initial structures of new genes and the subsequent evolutionary genetic processes that fix mutations and improve functions. Much is known about the origins of new genes (1) by means of exon shuffling, gene duplication, retroposition, recruitment of transposable elements, horizontal transfer, gene fission/fission, and the generation of coding regions from noncoding regions of the genome, each with many examples. However, the evolution of new functions remains an interesting problem.
Conventional comparative analyses of genes with diverged functions and the creation of new genes in the laboratory have contributed greatly to our understanding of evolutionary forces and functional divergence (2, 3). Building on these approaches, an examination of recently evolved genes provides a window through which both the origin and subsequent divergence are directly observable. This method avoids difficulties with the conventional approach as applied to old genes, where the signatures of early evolutionary processes become obscured by later ones (4). A number of young genes have been identified in various organisms, ranging from protozoa to fruit flies to primates (1-5). These young genes reveal recently acquired functions and make it possible to explore the details of how these functions originated.
Jingwei (jgw) is a young chimeric processed gene that first arose 2.5 million years ago in the common ancestor of two African Drosophila species, Drosophila yakuba and Drosophila teissieri (6-8). Its 3′ exon is a retroposed Alcohol dehydrogenase (Adh) that inserted downstream of the 5′ regulatory region and the first three exons of yande (ynd), the other parental gene of jgw (Fig. 1A) that existed only as an intact gene in the ancestral stage before divergence of D. yakuba and D. teissieri. A molecular characterization of the homologous genomic region in Drosophila melanogaster revealed that ynd is a gene duplicate of another related gene that is expressed specifically in testis, Yellow emperor (Ymp) (6-8). It is clear that insertion of a retroposed Adh sequence into the third intron of ynd produced the chimerical gene structure of jgw, and recruitment of the regulatory regions of Ymp caused the expression pattern of jgw to differ from that of Adh (1). The resulting JGW protein thus consists of two domains, a smaller ynd-derived domain at the amino terminus and a larger Adh-derived domain at the carboxyl terminus. This chimeric structure is a form of exon shuffling in which the Adh coding region was recombined with the Ymp-derived domain of the Ymp gene.
Fig. 1.
Origin and evolution of jgw. (A) Origins of genomic structure of jgw. jgw is a chimera of Yellow emperor (Ymp)- and Alcohol dehydrogenase (Adh) -derived regions. Boxes symbolize exons, and the lines between exons represent introns. The jgw locus still contains degenerate sequences of exons 4-12 of yande gene that was a duplicate copy of Ymp. (B) Phylogenetic distribution of ancestral amino acid substitutions in JGW; the graph represents the origination of jgw in two closely related species, D. teissieri and D. yakuba, and their subsequent evolution. All nine substitutions that took place in ancestral jgw before species divergence are replacement changes (circles), whereas no synonymous substitutions occurred in the same period. The melanogaster outgroup species include D. melanogaster, D. simulans, D. sechellia, and D. mauritiana.
Previous sequence analyses of between-species divergence and within-species polymorphism reveal rapid accumulation of non-synonymous substitutions at jgw, suggesting adaptive protein evolution under positive Darwinian selection (6, 9). The early evolution of jgw, before the African species diverged, saw the accumulation of nine amino acid replacements in the complete absence of silent substitutions (Figs. 1B and 2A). Later evolution, after the African species diverged, saw the rapid accumulation of an additional 21 amino acid replacements. These observations suggest that the early evolution of jgw was characterized by positive Darwinian selection, and that selection may still be ongoing in the D. yakuba lineage. In the absence of functional studies, however, one could not even speculate on the structural basis of molecular adaptation. Here, we examine the functional consequences of amino acid replacements accumulated during the evolution of jgw. Taking advantage of the available protein structure of ADH in Drosophila species in the Protein Data Bank, we first mapped the amino acid substitutions in JGW onto the three-dimensional structures of the ancestral Adh-derived domain of JGW. We then hypothesized functional consequences by examining positions of the substitutions on the protein structure. We expressed jgw in an Escherichia coli expression system to obtain a large amount of JGW enzymes to test the predictions derived from comparative analysis of the protein structure and amino acid substitutions. By using the purified enzymes of JGW, finally, we investigated enzymatic properties with major classes of alcohol substrates.
Fig. 2.
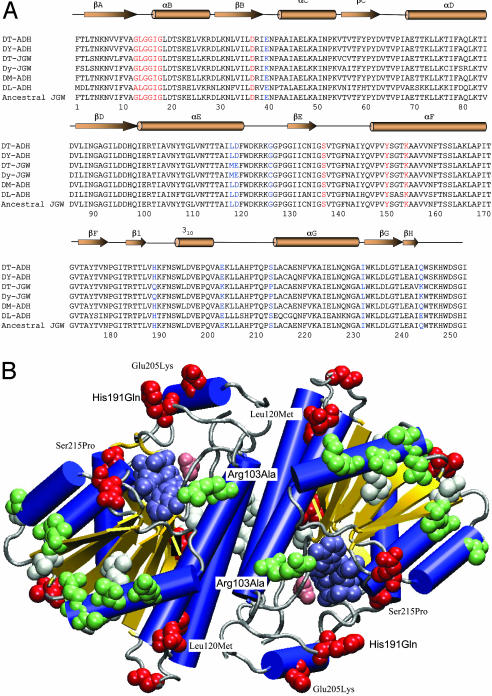
Structural analysis of JGW-specific amino acid substitutions. (A) Multiple sequence alignments of JGWs and related Drosophila ADHs (DT, D. teissieri; DY, D. yakuba; DM, D. melanogaster; and DL, D. lebanonensis). The parsimony inferred ancestral JGW sequence is unambiguous because the outgroup ADH sequences are highly similar to JGW. The secondary structure elements of DM-ADH (PDB ID code 1MG5), shown above the sequence alignments, include β-sheets, α-helices, and 310 helix. Red residues denote key positions in coenzyme binding. Blue residues denote nine positions with amino acid replacements accumulated early in the evolution of jgw. (B) Distribution of JGW-specific amino acid substitutions in the three-dimensional structure of ADH (blue helices, yellow sheets, and gray coils and turns) with bound acetate (pink van der Waals spheres) and NAD (steel van der Waals spheres). Of the nine amino acid replacements accumulated early in the evolution of JGW before the African species diverged (red van der Waals spheres), only two, His191Gln and Glu205Lys, lie in the flap region known to be important for substrate specificity. Two others, Ser215Pro and Leu120Met, contact residues within this region. All later replacements in the D. teissieri lineage (white van der Waals spheres) and in the D. yakuba lineage (green van der Waals spheres) lie outside the active site region.
Materials and Methods
Cloning jgw and Adh cDNAs from Drosophila. cDNA libraries from D. teissieri and D. yakuba were constructed with titers of 1010 and 109 plaque-forming units (pfu) per milliliter, respectively (Uni-ZAP XR vector, Stratagene). We extracted 0.5 mg of total RNA from adult flies. After purification, 0.5 μg of mRNA was applied to synthesize cDNA. The EcoRI adapter was ligated to the cDNA, the adapter was phosphorylated, and the construct was digested by XhoI and fractionated. Only the fragments that were >500 bp were recombined into the Uni-ZAP XR vector. cDNA libraries were titered with host bacteria XL1-Blue and stored at -80°C in 7% DMSO after amplification. cDNAs of jgw and Adh from D. teissieri and D. yakuba were cloned and confirmed by sequence comparison and gene structural analysis. Nonradioactive methods were used for screening cDNA libraries (unpublished manual of the M.L. laboratory). cDNA libraries ware plated in a density of 10,000 plaques per 150-mm Petri dish. After lifting, DNA was denatured, neutralized, and fixed onto nylon membrane (Amersham Pharmacia Biotech). The membrane was prehybridized for 4 h and hybridized at 65°C overnight with a digoxigenin-labeled probe comprising the first three exons of Ymp of D. melanogaster. The Adh cDNA was isolated by using a probe of a PCR-amplified fragment of the D. teissieri Adh gene. Positive clones were selected from examination of the exposed films and confirmed by PCR. A second round of screening was performed to obtain the single positive clone. After in vitro excision of cDNA in SORL E. coli (Stratagene), plasmids were obtained that contained the required gene as confirmed by DNA sequencing.
Expression of jgw and Adh in E. coli. The pGEX protein expression and purification system was used to express jgw and Adh genes (Amersham Pharmacia Biotech). The GST-jgw fusions were sequenced to ensure the correct ORF and intact codon region. These fusions were transformed into the host E. coli strain BL21-CodonPlus-RIL (Stratagene). Gene expression was induced with isopropyl β-d-thiogalactoside at logarithmic phase of the host bacteria at 25°C; cell culture lysate was acquired by sonication for small samples (for larger quantities, a French press was used to break up the cells). The lysate was centrifuged at 20,000 × g, and the supernatant was collected. The fusion protein was purified with glutathione-Sepharose 4B. Denatured protein was removed by centrifugation at 20,000 × g for 15 min at 4°C. The protein concentration was measured by the Bio-Rad protein assay kit.
Assay of Enzymatic Activity. PAGE was performed by using a discontinuous buffer system, and 3 μg of JGW was loaded. After electrophoresis, the native polyacrylamide gels were stained for ADH activity by incubation at 25°C for 2 h in the dark in the following solution: 100 ml of 0.1 M Tris·HCl buffer (pH 8.5), 1 ml of 95% alcohols, 0.5 mg of NAD, 5 mg of p-nitroblue tetrazolium, and 5 mg of phenazine methosulfate. The enzymes were located by tetrazolium reduction (10). Thirty-four alcohols and several derivatives, representative of main classes of alcohols and their derivatives, were chosen as substrates of JGW, as follows: (i) primary alcohols: methanol, ethanol, 1-propanol, 1-butanol, 3-butyn-1-ol, 2-methyl-1-propanol, 3-methyl-1-butanol, 1-pentanol, 1-hexanol, 1-heptanol, 2-ethyl-1-hexanol, 1-octanol, (1S)-cis-verbenol, 1-dodecanol, oleyl alcohol, dibenzo-16-crown-5-alcohol, geraniol, and farnesol; (ii) secondary alcohols: isopropyl alcohol, 2-butanol, 2-pentanol, 2-heptanol, 1,3,3-trimethyl bicyclo[2,2,1] heptan-2-ol, and 2-phenylethyl alcohol; (iii) aromatic alcohols: phenol, benzyl alcohol, 4-methoxybenzyl alcohol, α-methyl-2,3-dimethyl-4-methoxybenzyl alcohol, and 4-hydroxybenzyl alcohol; (iv) diols: ethylene glycol, 1,3-propanediol, 1,2-propandiol, and 2-methyl-2,4-pentanediol; (v) other polyols: 1,2,3-propanetriol (glycerol), adonitol, and xylitol; and (vi) other compounds: 1,1-dimethylethanol, formaldehyde, hexanal, formamide, sodium 3-hydroxybutyrate, and 3-hydroxyheptanenitrile.
Substrate specificity was determined by comparing the rate given by a 1 mM solution of each alcohol with that of 1 mM ethanol. These experiments were performed in duplicate, and each pair of replicates was within 5% of its mean.
Two standard measurements of enzymatic kinetics, the Michaelis constant Km and the maximum rate of reaction Vmax, were determined by double-reciprocal plot (11). Kinetics measurements were performed with various concentrations of alcohols, and a fixed concentration of 500 μM NAD+ was used throughout. Buffer with 0.025 M Mops (pH 7.4) and 0.025 M 2-(N-cyclohexylamino)ethanesulfonic acid (pH 9.4) with 0.1 M NaCl was applied during the kinetics measurement.
Mapping Substitutions on the Three-Dimensional Structures of ADH. Molecular modeling procedures were performed on a Macintosh G-3 computer. The programs vmd (www.ks.uiuc.edu/Research/vmd) and rasmol (www.umass.edu/microbio/rasmol) were used to generate a primary model, as well as to verify all model structures shown in this work. Molecular graphics were created with the program vmd. The template used to build the D. teissieri and D. yakuba three-dimensional model was the crystal structure of the orthologous enzyme D. melanogaster ADH (Protein Data Bank ID code 1MG5), and D. lebanonensis ADH (Protein Data Bank ID codes 1B14, 1B15, 1B16, and 1B2L) (12). When needed, data about the position of the coenzyme (NAD+) and the substrate were extracted from the crystal structures of binary and ternary enzyme complexes.
Results and Discussion
Drosophila ADH belongs to the short-chain dehydrogenase/reductase (SDR) family (10). SDRs share a common protein fold (Fig. 2A), consisting of a central β-sheet surrounded by α-helices and a typical nicotinamide coenzyme binding βαβαβ subdomain with a characteristic Gly-Xaa-Gly-Xaa-Xaa-Gly motif (position 13-18). Asp-37 confers specificity toward NAD binding, whereas the active site is characterized by a Ser-Tyr-Lys catalytic triad (12-14). These and other conserved SDR features are preserved in JGW, as shown in Fig. 2A. We predict that JGW will retain NAD-specific dehydrogenase activity.
All amino acid replacements accumulated early in the evolution of the JGW map at or near the surface of the three-dimensional structure of Drosophila ADH (Fig. 2B) (12, 13). Two replacements, His-191 → Gln (His191Gln) and Glu-205 → Lys (Glu205Lys), lie between Thr-186 and Pro-210, a region that forms the flap guarding the entrance to the active site and that is known to be responsible for the different specificities among related enzymes (12, 13). Two additional replacements, Ser215Pro and Leu120Met, contact residues within the Thr-186-Pro-210 region and also may influence specificity. The remaining five early replacements lie outside the active site, farther than 12.0 Å from the bound substrate. All later amino acid replacements, accumulated after the African species diverged, lie outside the active site (Fig. 2B). Twelve are solvent exposed, including the Arg103Ala replacement in D. yakuba that removes a contact with the coenzyme adenine ring NH2. One is completely buried (Ile52Leu of D. teissieri), and two others are partially buried (Val86Ile of D. teissieri and Val88Ile in D. yakuba).
We propose that amino acid replacements accumulated by JGW affect function as a consequence of adaptive evolution. Although only one (Arg103Ala) contacts the coenzyme and none contact the substrate, amino acid replacements in JGW nevertheless might affect function by causing subtle conformational changes, altering hydrogen bond networks, influencing protein electrostatics, and affecting conformational flexibility (15, 16). Such amino acid replacements are anticipated to affect specificity toward the alcohols, which are diverse in structure, rather than specificity toward the coenzymes, which are very similar in structure.
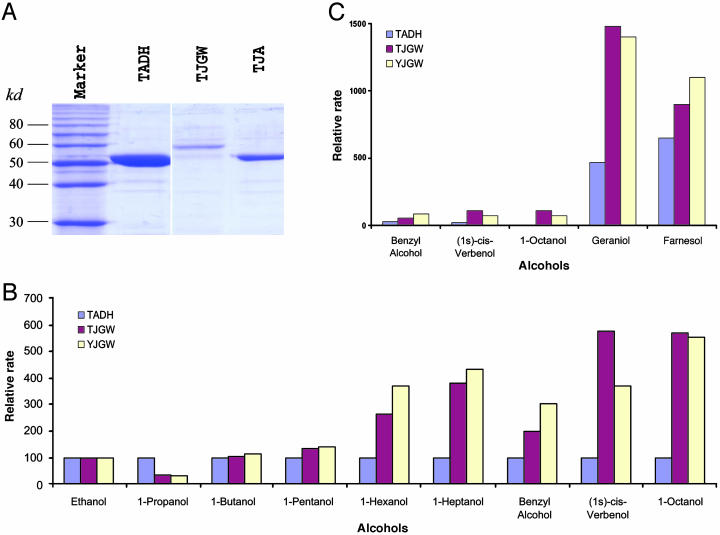
We tested these functional predictions by surveying the activity of in vitro expressed JGW toward both coenzymes and alcohols. Full-length jgw and Adh clones, obtained by screening cDNA libraries from D. teissieri and D. yakuba, were subcloned into the pGEX and expressed in E. coli strain BL21-RIL. Fusion to GST permitted JGW to be purified readily by using affinity chromatography. Preparations are typically 90% pure as judged by Coomassie blue staining of SDS/PAGE gels (Fig. 3A). Native PAGE gels stained for dehydrogenase activity reveal that JGW uses NAD, but not NADP, to oxidize various alcohols (17). Enzyme kinetics studies, performed at 25°C (pH 7.4), by monitoring formation of NADH at 340 nm in a Cary 300 spectrophotometer (Varian), reveal that, compared with ADH, JGW displays altered specificity toward short-chain alcohols (Fig. 3B) and catalyzes ethanol oxidation with lower reaction rates (Vmax), as expected.
Fig. 3.
Purification of JGW and its enzymatic properties. (A) Purification of JGW and ADH revealed by Coomassie blue staining of SDS/PAGE gels. TADH, ADH of D. teissieri; TJA, ADH-derived domain in JGW of D. teissieri; TJGW, JGW of D. teissieri. (B) The changes of Vmax, relative to ethanol, which is the abundant natural substrate of ancestral ADH, of JGW toward short- and medium-chain alcohols measured according to the method of Waller (35) in three proteins: ADH of D. teissieri (TADH, blue), JGW of D. teissieri (TJGW, red), and JGW of D. yakuba (YJGW, yellow) (these abbreviations and colors are the same in C). Substrates shown are a series of primary alcohols, including benzyl alcohol. (C) The changes of Vmax, relative to ethanol, of JGW toward long-chain alcohols (see Materials and Methods). As we explained in Materials and Methods, with variation in activity among replicate assays <5% of the average rates between repeats of experiments, the variation in the rates between enzymes is statistically significant. We observed that the differences between JGW and ADH in the activities shown in this figure for long-chain alcohol substrates (B, 1-hexanol, 1-heptanol, benzyl alcohol, (1S)-cis-verbenol, and 1-octanol; C, geraniol and farnesol) are at least 7 times greater than standard deviation, indicating that these differences between JGW and ADH are statistically significant (P < 0.001).
Substrate specificity of JGW was further characterized in a survey of 34 alcohols that included representatives from all major classes found in nature (see above). Like ADH, JGW shows activities toward a broad range of alcohols. However, compared with ADH, JGW also shows a systematic preference for long-chain primary alcohols and increased specificity (for example, see Fig. 3B), including farnesol and geraniol (Fig. 3C). These results confirm that jgw has evolved altered specificity after diverging from its parental genes, Adh and ynd.
The discovery that farnesol and geraniol are substrates is intriguing. In insects, farnesol is the starting material for biosynthesis of juvenile hormone (18), whereas geraniol is a primary recruitment pheromone (19) used by the honeybee, Apis mellifera, to mark the nest as well as food sources. These observations, together with the testes expression pattern of jgw (6, 8), suggest that JWG may be involved in hormone and pheromone biosynthesis/degradation processes in Drosophila.
JGW has a chimeric structure with two distinct domains, posing a question of whether the Ymp-derived domain would have impacted the positions of substitutions or structure in Adh-derived domains. It is known that protein packing of subunits and domains is dominated by hydrophobic interactions (20). It is thus unlikely that the Ymp-derived domain, if indeed it folds at all, would pack against the hydrophilic surface of the Adh-derived domain. Without packing against the Adh-derived domain, it seems unlikely that the Ymp-derived domain could greatly influence substrate specificity. Indeed, the ability to fuse proteins without affecting the function of either forms the very basis of the phage and cell display techniques used to select proteins of novel function (21).
Kinetic analyses and structural modeling reveal that JGW is a new member of the SDR family with expanded biochemical properties. ADH is normally a dimer (12, 13), as shown in Fig. 2B. Our observation of no amino acid substitution in the interface within dimers suggests that JGW protein, like ADH, is a dimer as well. In addition, one interpretation for the diverged substrate specificity of JGW from ADH is that JGW in E. coli expression system might not necessarily fold properly, leading to changed substrate specificity in the assay of enzymatic properties. However, this scenario is unlikely because JGW readily forms a functional dimer like ADH. In addition, ADH in this expression system functions normally, precluding any possibilities of abnormal folding.
Our investigations of JGW evolution cast a different light on the process of functional divergence in newly evolved proteins. Conventional theory emphasizes changes in function produced by amino acid replacements in active sites (22). Our data demonstrate that natural selection for amino acid replacements outside active sites can produce unexpected functional changes in a new gene. This finding is likely a general phenomenon (16). Previous results of related studies, including in vitro protein engineering (23), enzyme breeding by directed evolution (24, 25), pseudoreversions of catalytically compromised enzymes (26), catalytic antibodies (27), and spectral tuning in opsins (28), all are consistent with the notion that amino acid replacements outside active sites can affect specificity and catalytic efficiency.
JGW provides an example of how functional diversity in a new protein can be expanded under the joint forces of exon shuffling, gene duplication, and natural selection. The biochemical functions of JGW were evolved by changing substrates under positive selection, whereas the ADH reaction was maintained by purifying selection. A previous investigation of evolution of function through analysis of protein structure by Todd et al. (29) reported an interesting observation in the evolution of protein superfamilies: Substrate specificity is usually diverse in different members of a superfamily, but the reaction chemistry is maintained throughout the evolution of the superfamily. The origin and evolutionary process of JGW functions may represent a general evolutionary mechanism that governs evolution of such protein superfamilies.
Progress has been made for new gene evolution at various levels of biological diversity, including how new expression patterns arise after gene fusion (30), how similar biochemical functions are maintained in a changed physiological environment (31-33), and, more broadly, how gene-expression patterns evolve during development (34). JGW demonstrates that protein functional diversity also can evolve, both rapidly and under positive selection. The emergence of new functions provides a basis for further disposition by the changes in regulatory systems in new developmental stages and tissues. For example, jgw in D. teissieri is expressed only in testis, an ancestral character that was inherited from ynd, the parental gene of jgw (1, 4, 6-8). However, jgw in D. yakuba evolved a new expression pattern in which the novel biochemical functions were executed in tissues and stages beyond the testis tissue in male adults.
Acknowledgments
We thank all members in the Long laboratory at the University of Chicago and the Dean laboratory at the University of Minnesota for experimental assistance and helpful discussions; L. Li for technical advice on overexpression experiments; A. Llopart for assistance in cloning jgw from D. teissieri; and two anonymous reviewers for their valuable suggestions. This study was supported by grants to M.L. from the National Science Foundation and the National Institutes of Health, a National Science Foundation CAREER award, and a Packard Fellowship in Science and Engineering.
Author contributions: M.L. designed research; J.Z. performed research; A.M.D. contributed new reagents/analytic tools; J.Z., A.M.D., F.B., and M.L. analyzed data; and J.Z., A.M.D., and M.L. wrote the paper.
Abbreviations: ADH, alcohol dehydrogenase; SDR, short-chain dehydrogenase/reductase.
References
- 1.Long, M., Betran, E., Thornton, K. & Wang, W. (2003) Nat. Rev. Genet. 4, 865-875. [DOI] [PubMed] [Google Scholar]
- 2.Patthy, L. (1999) Protein Evolution (Blackwell, Oxford).
- 3.Long, M., ed. (2003) Origin and Evolution of New Gene Functions, Contemporary Issues in Genetics and Evolution (Kluwer, Dordrecht, The Netherlands) Vol. 10.
- 4.Long, M., Deutsch, M., Wang, W., Betran, E., Brunet, F. G. & Zhang, J. (2003) Genetica 118, 171-182. [PubMed] [Google Scholar]
- 5.Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. (2004) Nature 430, 569-573. [DOI] [PubMed] [Google Scholar]
- 6.Long, M. & Langley, C. H. (1993) Science 260, 91-95. [DOI] [PubMed] [Google Scholar]
- 7.Long, M., Wang, W. & Zhang, J. (1999) Gene 238, 135-141. [DOI] [PubMed] [Google Scholar]
- 8.Wang, W., Zhang, J., Alvarez, C., Llopart, A. & Long, M. (2000) Mol. Biol. Evol. 17, 1294-1301. [DOI] [PubMed] [Google Scholar]
- 9.Yang, Z. & Bielawski, J. P. (2000) Trends Ecol. Evol. 15, 496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, F. M. & Denniston, C. (1964) Nature 204, 906-907. [DOI] [PubMed] [Google Scholar]
- 11.Winberg, J. O., Thatcher, D. R. & McKinley-McKee, J. S. (1982) Biochim. Biophys. Acta 704, 17-25. [DOI] [PubMed] [Google Scholar]
- 12.Benach, J., Atrian, S., Gonzàlez-Duarte, R. & Ladenstein, R. (1998) J. Mol. Biol. 282, 383-399. [DOI] [PubMed] [Google Scholar]
- 13.Benach, J., Atrian, S., Gonzàlez-Duarte, R. & Ladenstein, R. (1999) J. Mol. Biol. 289, 335-355. [DOI] [PubMed] [Google Scholar]
- 14.Benach, J., Atrian, S., Fibla, J., Gonzàlez-Duarte, R. & Ladenstein, R. (2000) Eur. J. Biochem. 267, 3613-3622. [DOI] [PubMed] [Google Scholar]
- 15.Watt, W. B. & Dean, A. M. (2000) Annu. Rev. Genet. 34, 593-622. [DOI] [PubMed] [Google Scholar]
- 16.Golding, G. B. & Dean, A. M. (1998) Mol. Biol. Evol. 15, 355-369. [DOI] [PubMed] [Google Scholar]
- 17.Jornvall, H., Persson, B., Krook, M., Atrian, S., Gonzàlez-Duarte, R., Jeffery, J. & Ghosh, D. (1995) Biochemistry 34, 6003-6013. [DOI] [PubMed] [Google Scholar]
- 18.van Tamelen, E. E. & McCormick, J. P. (1970) J. Am. Chem. Soc. 92, 737-738. [DOI] [PubMed] [Google Scholar]
- 19.Bhagavan, S. & Smith, B. H. (1997) Physiol. Behav. 61, 107-117. [DOI] [PubMed] [Google Scholar]
- 20.Branden, C. & Tooze, J. (1999) Introduction to Protein Structure (Garland, New York).
- 21.Benhar, I. (2001) Biotechnol. Adv. 19, 1-33. [DOI] [PubMed] [Google Scholar]
- 22.Bishop, J. G., Dean, A. M. & Mitchell-Olds, T. (2000) Proc. Natl. Acad. Sci. USA 97, 5322-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, R., Greer, A. & Dean, A. M. (1995) Proc. Natl. Acad. Sci. USA 92, 11666-11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano, T., Oue, S. & Kagamiyama, H. (1998) Proc. Natl. Acad. Sci. USA 95, 5511-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oue, S., Okamoto, A., Yano, T. & Kagamiyama, H. (1999) J. Biol. Chem. 274, 2344-2349. [DOI] [PubMed] [Google Scholar]
- 26.Blacklow, S. C., Liu, K. D. & Knowles, J. R. (1991) Biochemistry 30, 8470-8476. [DOI] [PubMed] [Google Scholar]
- 27.Wedemayer, G. J., Patten, P. A., Wang, L. H., Schultz, P. G. & Stevens, R. C. (1997) Science 276, 1665-1669. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama, S. (1997) Annu. Rev. Genet. 31, 315-336. [DOI] [PubMed] [Google Scholar]
- 29.Todd, A. E., Orengo, C. A. & Thornton, J. M. (2001) J. Mol. Biol. 307, 1113-1143. [DOI] [PubMed] [Google Scholar]
- 30.Nurminsky, D. I., Nurminskaya, M. V., De Aguiar, D. & Hartl, D. L. (1998) Nature 396, 572-575. [DOI] [PubMed] [Google Scholar]
- 31.Messier, W. & Stewart, C. B. (1997) Nature 385, 151-154. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J., Zhang, Y. P. & Rosenberg, H. F. (2002) Nat. Genet. 30, 411-415. [DOI] [PubMed] [Google Scholar]
- 33.Trabesinger-Ruef, N., Jermann, T., Zankel, T., Durrant, B., Frank, G. & Benner, S. A. (1996) FEBS Lett. 382, 319-322. [DOI] [PubMed] [Google Scholar]
- 34.Wray, G. A., Hahn, M. W., Abouheif, E., Balhoff, J. P., Pizer, M., Rockman, M. V. & Romano, L. A. (2003) Mol. Biol. Evol. 20, 1377-1419. [DOI] [PubMed] [Google Scholar]
- 35.Waller, G. R. (1965) Nature 207, 1389-1390. [DOI] [PubMed] [Google Scholar]