Abstract
Lung infection with Pseudomonas aeruginosa is the leading cause of death among cystic fibrosis patients. To initiate infection, P. aeruginosa assembles a protein nanomachine, the type III secretion system (T3SS) to inject bacterial proteins directly into target host cells. An important regulator of the P. aeruginosa T3SS is the chaperone protein PcrG, which forms a complex with the tip protein, PcrV. In addition to its role as a chaperone to the tip protein, PcrG also regulates protein secretion. PcrG homologs are also important in the T3SS of other pathogens such as Yersinia pestis, the causative agent of bubonic plague. The atomic structure of PcrG or any member of the family of tip protein chaperones is currently unknown. Here, we show by CD and NMR spectroscopy that PcrG lacks a tertiary structure. However, it is not completely disordered but contains secondary structures dominated by two long α-helices from residues 16–41 and 55–76. NMR backbone dynamics data show that the helices in PcrG have semi-rigid flexibility and they tumble as a single entity with similar backbone dynamics. NMR titrations show that the entire length of PcrG residues from 9–76 is involved in binding to PcrV. Thus the PcrG family of T3SS chaperone proteins is essentially partially folded.
Keywords: Pseudomonas, PcrG, PcrV, T3SS, chaperone
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen that is most frequently associated with nosocomial infections in patients with cystic fibrosis, pneumonia, AIDS, cancer, or other ailments that compromise the immune system(1). Despite treatment with potent antibiotics, mortality rate is approximately 40% in case of acute infection(2). To initiate infection, the bacterium deploys the type III secretion system (T3SS) to inject effector proteins into host cells(3) to modulate host cell biology for the benefit and survival of the bacterium(4, 5). The T3SS consists of structural proteins that assemble the needle complex, effectors and chaperones(3). Two proteins that play critical roles in the assembly and regulation of type III secretion in P. aeruginosa are the tip protein PcrV(6) and its chaperone PcrG(7–9).
Secretion of effector proteins by the T3SS in P. aeruginosa is tightly regulated and is triggered by either host cell contact or calcium depletion(5). Deletion of genes encoding either PcrG or PcrV results in the loss of regulation of effector secretion leading to constitutive secretion of effectors(8). Multiple results confirm that PcrG and PcrV interact directly(8, 10, 11). PcrG can regulate the secretion of effectors even with a point mutation, V16R, that disrupts binding to PcrV(8). Hence, PcrG regulates the secretion of effectors independent of PcrV-binding(8). Indeed, Lee et al.(7) showed that PcrG also interacts with other T3SS components like PscO and PcrD to control the secretion of effectors. This mode of regulation of effector secretion is different from its homolog, LcrG of Yersinia pestis, which requires binding to its cognate tip protein LcrV(12–14). In addition to regulating the secretion of effectors, PcrG facilitates the secretion of PcrV(8), and directs the translocation of effector proteins into the host cell(9).
Chaperones play important roles in the assembly, function and regulation of the T3SS(15). There are over 30 crystal structures of T3SS chaperones in the Protein Data Bank, however, the atomic structure for any T3SS tip protein chaperone remains unknown. It has been suggested that the T3SS tip proteins SipD (Salmonella typhimurium)(16, 17), IpaD (Shigella flexneri)(18) and BipD (Burkholderia pseudomallei)(19) have an N-terminal α- helical hairpin that serves as a self chaperone for these proteins(18, 20). Due to the absence of this α-helical hairpin in the crystal structure of LcrV(21), and presumably its homolog PcrV (for which the crystal structure is unknown), it has been hypothesized that LcrG and PcrG function as the chaperone proteins for their respective tip proteins(8, 12). Others have proposed that LcrG forms an α-helical coiled-coil based on the N-terminal α-helical hairpin domain of IpaD(17, 18, 20). Because of its homology to LcrG, PcrG is also assumed to have a similar coiled-coil topology. Recently, Basu et al.(22) reported a structure of PcrG derived by computer modeling and showed that PcrG residues 13–72 formed a 4-helix bundle. Here we show the NMR-derived atomic conformation of PcrG. Our results are not in agreement with the structure reported by Basu et al.(22). Instead of a 4-helix bundle, our NMR data suggest that PcrG is partially folded and its secondary structure is dominated by two long alpha helices.
EXPERIMENTAL PROCEDURES
Protein expression and purification
Two constructs of PcrG, full length PcrG and a truncated form spanning residues 9–76 (PcrG9–76), were PCR amplified from P. aeruginosa (strain PAO1) and subcloned into pET-21a as fusion proteins with GB1 (the B1 immunoglobulin-binding domain of Streptococcus protein G) serving as expression and purification tag. The fusion constructs were arranged as follows: PcrG or PcrG9–76 at the N-terminus, followed by tobacco etch virus (TEV) protease cleavage site, GB1 domain, and a His6-tag at the C-terminus. To obtain PcrG, the expression plasmid was transformed in E. coli BL21 (DE3)-DNAY and freshly transformed colonies were used to inoculate 10 mL LB starter cultures overnight at 37° C. For unlabeled protein, the 10 ml LB starter culture was used to inoculate a 1 L LB. For 15N/13C or 15N isotopically labeled proteins, the overnight LB starter culture was centrifuged at 4000 rpm for 15 minutes and the cell pellet was resuspended into 1 L M9 minimal media supplemented with 1 g/L 15NH4Cl and/or 2 g/L 13C-glucose. All media contained 30 µg/mL kanamycin and 100 µg/mL carbenicillin. Cells were grown at 37°C, induced at A600 ~0.6–0.8 with 1.0 mM isopropyl-β-D-thiogalactopyrandoside (IPTG), and cell growth was continued overnight in a 15°C shaker to a final A600 of ~2. Cells were harvested by centrifugation, resuspended in binding buffer (500 mM NaCl, 20 mM Tris-HCl, 5 mM imidazole, pH 8.0), and lysed by sonication in the presence of 10 mg phenylmethyl sulfonyl fluoride. The lysate was centrifuged to remove cellular debris and 600 µL of 5% polyethyleneimine was added to the supernatant to precipitate the nucleic acids. After centrifugation, the supernatant was loaded onto a Ni2+-affinity column, and the column was washed with binding buffer, and the protein was eluted with elution buffer (500 mM NaCl, 20 mM Tris, 250 mM imidazole, pH 8.0). Fractions containing the PcrG-GB1 fusion protein were pooled and dialyzed overnight in buffer (50 mM Tris-HCl, 20 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH 8.0) with 250 µL of 0.06 mM recombinant TEV protease(23). The TEV protease digest was dialyzed in binding buffer and passed through a second round of Ni2+-affinity purification. PcrG eluted in the flow through and wash fractions, whereas the GB1 tag was retained in the column. PcrG9–76 was expressed and purified in a similar manner just described. The PcrG and PcrG9–76 constructs used in this study retained an N-terminal methionine and a C-terminal cloning artifact (GSENLYFQ). Proteins were concentrated using Amicon Ultra 3K (Millipore) and protein concentration was estimated by absorbance at 280 nm.
P. aeruginosa PcrV (residues 25–294) was subcloned into a pET-21a expression vector as a GB1 fusion protein arranged as follows: N-terminal His6-tag, GB1 domain, a TEV protease cleavage site, and PcrV residues 25–294. The N-terminal 24 residues of PcrV were truncated based on the apparent disorder of the homologous region in the crystal structure of LcrV(21). PcrV was expressed and purified similarly as described above. After digestion with TEV protease, the PcrV construct retained an N-terminal cloning artifact (GHM).
NMR Spectroscopy
NMR data were acquired at 25° C using a Bruker Avance 800 MHz spectrometer equipped with a cryoprobe, processed with NMRPipe(24), and analyzed using NMRView(25). Protein samples were dialyzed overnight in NMR buffer (10 mM NaHPO4, 10 mM NaCl, pH 7.0) and the NMR samples contained 10% (v/v) D2O. 15N-PcrG (0.8 mM) and truncated 15N-PcrG9–76 (0.7 mM) were used to acquire initial 2D 1H-15N HSQC spectra(26). For backbone assignments of 15N-PcrG9–76, 0.7 mM 15N/13C-PcrG9–76 was used to acquire 2D 1H-15N HSQC, 3D HNCA, 3D HNCACB, and 3D CBCA(CO)NH(26–28). Secondary structure was determined using the secondary chemical shifts of Cα and Cβ (29).
Backbone relaxation parameters were collected with a 0.7 mM 15N-PcrG9–76 sample in NMR buffer. The steady-state heteronuclear {1H}-15N NOE was acquired as described(30) with 2048 (1H) * 128 (15N) complex points, 60 scans per point and a 5 s recycle delay. The heteronuclear {1H}-15N NOE was calculated as the ratio of intensities of each peak in the two 2D datasets(30). Error bars were estimated using the standard deviation of the background signal of each spectrum(31). The 15N backbone relaxation rates R1 and R2 were acquired as described(31). The time delays used to determine R1 were 10, 60, 120, 240, 400, 900 and 1100 ms and the time delays used to determine R2 were 20, 40, 50, 60, 70, 90, 100, 120 and 150 ms. Peak intensities were obtained using NMRView(25) and fitted using GNUPLOT(32). Deviations from fitting were reported as error bars. Due to peak overlap, residues 10, 13, 14, 21, 24, 29, 33, 36, 37, 38, 48, 50, 51, 56, 57, 69, 70 and 74 were not used in the analysis.
For NMR chemical shift mapping, 2D 1H-15N HSQC spectra were acquired for 15N-PcrG9–76 and 15N-PcrG9–76-GB1-His6 titrated with increasing amounts of unlabeled PcrV. Six samples were prepared in NMR buffer with 0.13 mM 15N-PcrG9–76 and varying PcrV concentration in molar ratios of 1:0, 1:0.25, 1:0.5, 1:1, 1:2 and 1:4. Four samples were prepared in NMR buffer with 0.39 mM 15N-PcrG9–76-GB1-His6 and varying PcrV concentrations in molar ratios of 1:0, 1:0.5, 1:1 and 1:2.
13C-ILV labeled PcrG9–76 where the methyl groups of isoleucine, leucine and valine were 13C labeled was prepared by overexpression of PcrG9–76 in E. coli minimal media supplemented with 2-ketobutyric acid-4-13C (Isotec, # 571342; to 13C-label the isoleucine Cδ1 methyl group) and 2-keto-3-(methyl-13C)-butyric acid-4-13C (Isotec # 571334; to 13C-label the leucine Cδ and valine Cγ methyl groups) following published protocols(33). The keto acids were added to the minimal media when OD600 reached 0.4 – 0.5. After 45–60 minutes the culture was induced with 1.0 mM IPTG and cell growth was continued overnight in 15°C shaker. Cells were harvested and the proteins were purified as described above. PcrV labeled only with isoleucine 13Cδ1 methyl group was overexpressed and purified in a similar manner. Isoleucine 13Cδ1-labeled PcrV (0.23 mM) was titrated with increasing amounts of unlabeled PcrG9–76 or PcrG in molar ratios of 1:0, 1:0.5, 1:1, 1:2 and 1:3. NMR spectra for the titrations were acquired at 30°C using a Bruker Avance 600 MHz NMR.
Circular dichroism spectroscopy
PcrG and PcrG9–76 (5.8 µM) were dialyzed in buffer (10 mM NaHPO4, 10 mM NaCl, pH 7. 0) for CD spectroscopy and thermal denaturation experiments. CD spectra were acquired using a JASCO J-815 Spectropolarimeter. CD spectra were collected in triplicate from 195 to 260 nm at scan rate of 50 nm/min. Thermal denaturation curves from 20–80° C were obtained by recording the molar ellipticity at 222 nm at a temperature ramp rate of 2° C/min.
Surface plasmon resonance spectroscopy
Apparent dissociation constants (Kd) between PcrG or PcrG9–76 and PcrV were measured by surface plasmon resonance (SPR) using a Biacore 3000 instrument (Biacore, Uppsala, Sweden). A 1:1 (v/v) mixture of 400 mM 1-ethyl-3- (3-dimethylaminopropyl) carbodiimide (EDC) (Sigma) and 100 mM N-hydroxysuccinimide (NHS) (Sigma) was prepared and immediately injected into two flow cells of a CM5 chip to activate the carboxymethylated dextran surface. Lyophilized anti-polyhistidine antibodies (R&D Systems, Inc.) were resuspended to 25 µg/mL in immobilization buffer (10 mM sodium acetate, pH 5.0) and injected into the 2 activated flow cells for amine coupling. The remaining uncoupled surfaces were inactivated by injection of blocking buffer (1 M ethanolamine, pH 8.5) (Sigma). Activation, coupling, and blocking steps were performed for 7 min at a flow rate of 10 µL/min resulting in anti-polyhistidine direct coupling of ~14,000 response units (RUs) in both flow cells.
Biacore assays were performed at 25°C using freshly prepared and degassed running buffer (10 mM HEPES, 150 mM NaCl, 0.005% (v/v) Surfactant P-20, pH 7.4). Unlabeled PcrG or PcrG9–76 (fused to C-terminal GB1-His6 tag) was dialyzed overnight in running buffer and immobilized to the anti-polyhistidine coated surface of flow cell 2 by injection of 6–9 µL of 1 µg/mL protein at 10 µL/min. Varying concentrations (2–32 nM) of PcrV were then injected in both flow cells at 40 µL/min for 3 mins over immobilized PcrG-GB1-His6 and PcrG9–76-GB1-His6. Immobilization of PcrG and binding of PcrV was repeated for each PcrV concentration followed by surface regeneration using 20 µL pulses of regeneration buffer (10 mM glycine, pH 2.0). Non-specific binding of PcrV to the anti-His6 antibody coated surface and the bulk effect from the buffer were determined from flow cell 1 and subtracted from the raw data. The dissociation constant (Kd) values were calculated using BIAevaluation 4.1 software. As a control, the above steps were repeated using the GB1-His6 tag immobilized to the anti-polyhistidine coated surface to verify that the fusion tag did not contribute in binding PcrV.
RESULTS
Protein expression
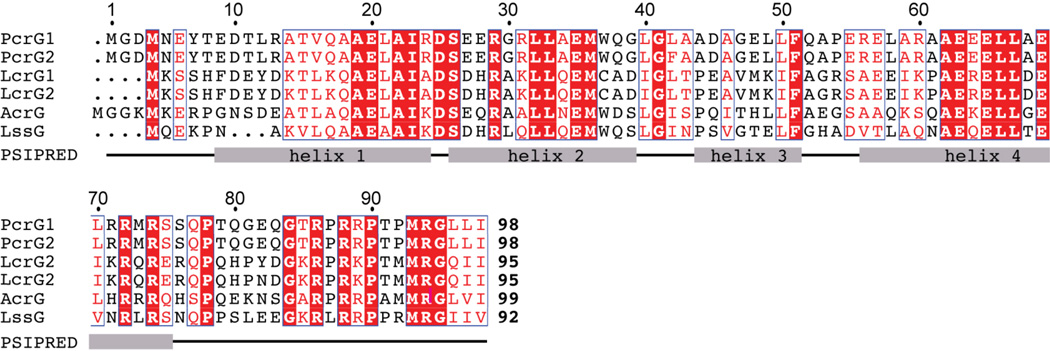
Full length PcrG and a truncated form, PcrG9–76, were expressed and purified as fusion proteins with the GB1 domain as a solubility tag and linked by a cleavable TEV protease site. A PcrG truncation, PcrG9–76 was constructed based on the sequence conservation of the PcrG family of tip protein chaperones (Figure 1), proposed binding regions in the Yersinia homolog LcrG with its cognate tip protein(34), and secondary structure prediction using PSIPRED(35), DSC(36), MLRC(37) and PHD(38) that suggested that the extreme N and C termini of PcrG were random coils (Figure 1B). PcrG and PcrG9–76 were overexpressed in E. coli and purified under native conditions by Ni2+-affinity chromatography and produced yields in millimolar quantities that facilitated NMR studies.
Figure 1. Sequence alignment of the PcrG protein family.
Protein sequences PcrG1 PcrG2 (from different strains of Pseudomonas aeruginosa), LcrG1 (from Yersinia pestis), LcrG2 (from Yersinia enterocolitica), AcrG1 (from Aeromonas hydrophila) and LssG (from Photorhabdus luminescens). Secondary structure prediction from PSIPRED(35) denoting the 4 helices are indicated.
Cicular Dichroism and Thermal Denaturation
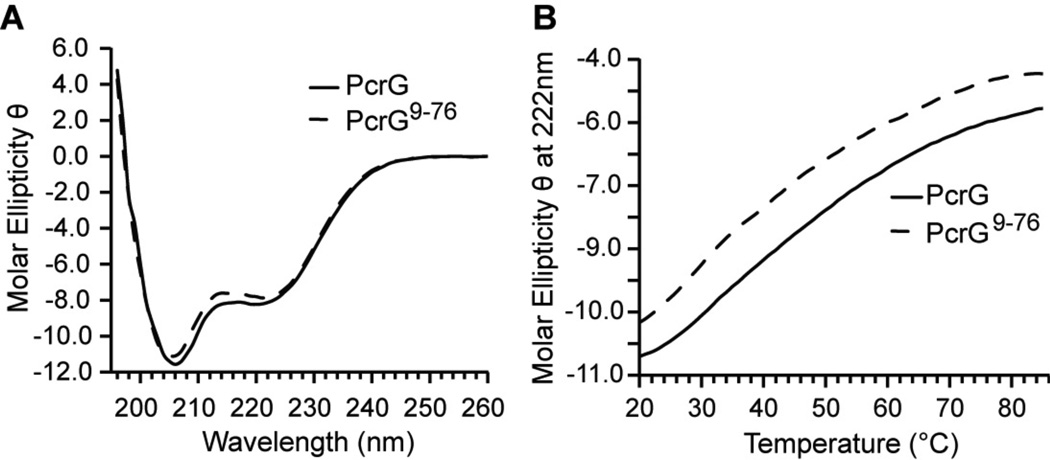
Circular dichroism (CD) spectra of PcrG and PcrG9–76 showed alpha helical content based on the minima at 222 nm and a significant amount of protein disorder based on the predominant minima at 208 nm. Nevertheless, comparison of the two CD spectra (Figure 2A) confirmed that PcrG and PcrG9–76 have nearly identical secondary structures because their CD spectra were nearly superimposable. Thus the truncation in PcrG9–76 did not produce any appreciable changes to the secondary structure of PcrG. The thermal denaturation curves of PcrG and PcrG9–76 (Figure 2B) showed that they unfolded in a non-cooperative manner without showing any clear inflection points. Nanao et. al(10) also observed that PcrG does not display a clear Tm, which is consistent with our observation. Additionally, the θ222/208 ratio of 0.7 for PcrG and PcrG9–76 suggested partially folded α-helical proteins with non-interacting helices(39). The results of CD and thermal denaturation suggested that PcrG has secondary α-helical structure but lacked a tertiary structure.
Figure 2. Circular Dichroism and thermal denaturation spectra of PcrG constructs.
(A) CD spectra for PcrG and PcrG9–76. (B) CD thermal denaturation plots of PcrG and PcrG9–76. [Molar ellipticity θ = (deg·cm2·dmol−1) × 10−5].
NMR Spectroscopy
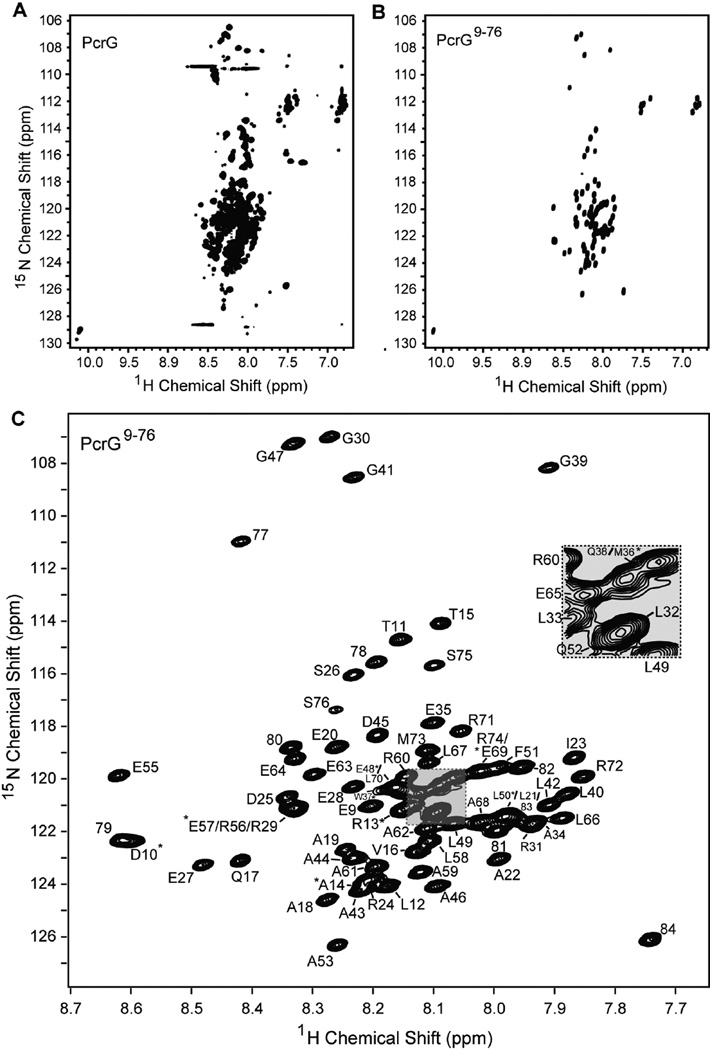
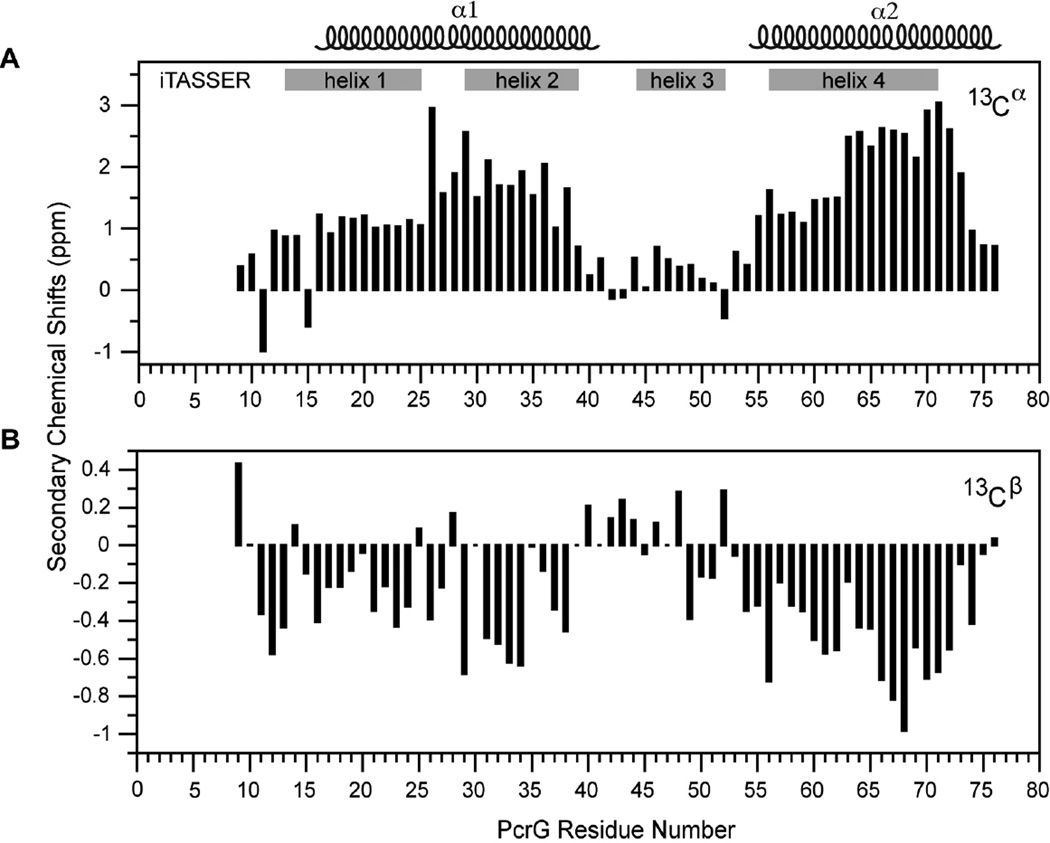
The 2D 1H-15N HSQC spectrum (Figure 3A) for full length PcrG showed a non-ideal NMR spectrum with broad and overlapped peaks with many more peaks expected from a 98-residue protein. This suggested multiple conformations or oligomerization of PcrG, which would have made further characterization by 3D NMR challenging. On the other hand, PcrG9–76 afforded a spectrum suitable for further NMR characterization based on the presence of sharp and well-resolved peaks with the correct number of peaks expected for a 68-residue protein (Figure 3B). Consistent with the CD data (Figure 2), the narrow proton chemical shift range of less than 1 ppm for the backbone amides of PcrG (Figure 3A) or PcrG9–76 (Figure 3B) suggested that the protein might be highly α-helical or unstructured. Additional 3D NMR datasets were acquired on PcrG9–76 to assign the backbone resonances of PcrG9–76 to identify the alpha-helical regions by Cα and Cβ chemical shifts(29). The backbone amides of the 67 non-proline residues of PcrG9–76 were assigned as well as all 8 residues of the C-terminal cloning artifact GSENLYFQ (Figure 3C). The secondary Cα and Cβ chemical shifts suggested PcrG9–76 contained two long α-helices spanning residues 16–41 and 54–76 and linked by a short region from residues 42–53 that lacked any apparent secondary structure (Figures 4). Because the CD spectra (Figure 2A) showed nearly identical secondary structures for the truncated and full length PcrG, the NMR results (Figure 4) suggested that the secondary structure of PcrG consist of two long alpha helices from residues 16–41 and 54–76 (Figure 4).
Figure 3.
Two-dimensional 1H-15N HSQC spectra of (A) PcrG and (B) truncated PcrG9–76. (C) Assigned 2D 1H-15N HSQC spectrum of PcrG9–76. Overlapped peaks are indicated with asterisk. The W37 side chain peak is not shown. Residues 77–84 (G77SENLYFQ84) are cloning artifacts.
Figure 4. The (A) Cα and (B) Cβ secondary chemical shifts suggest PcrG9–76 contains two α-helical regions.
Gray bars indicate the four helices of the iTASSER computer predicted structure of PcrG reported by Basu et al.(22)
PcrG Backbone Dynamics
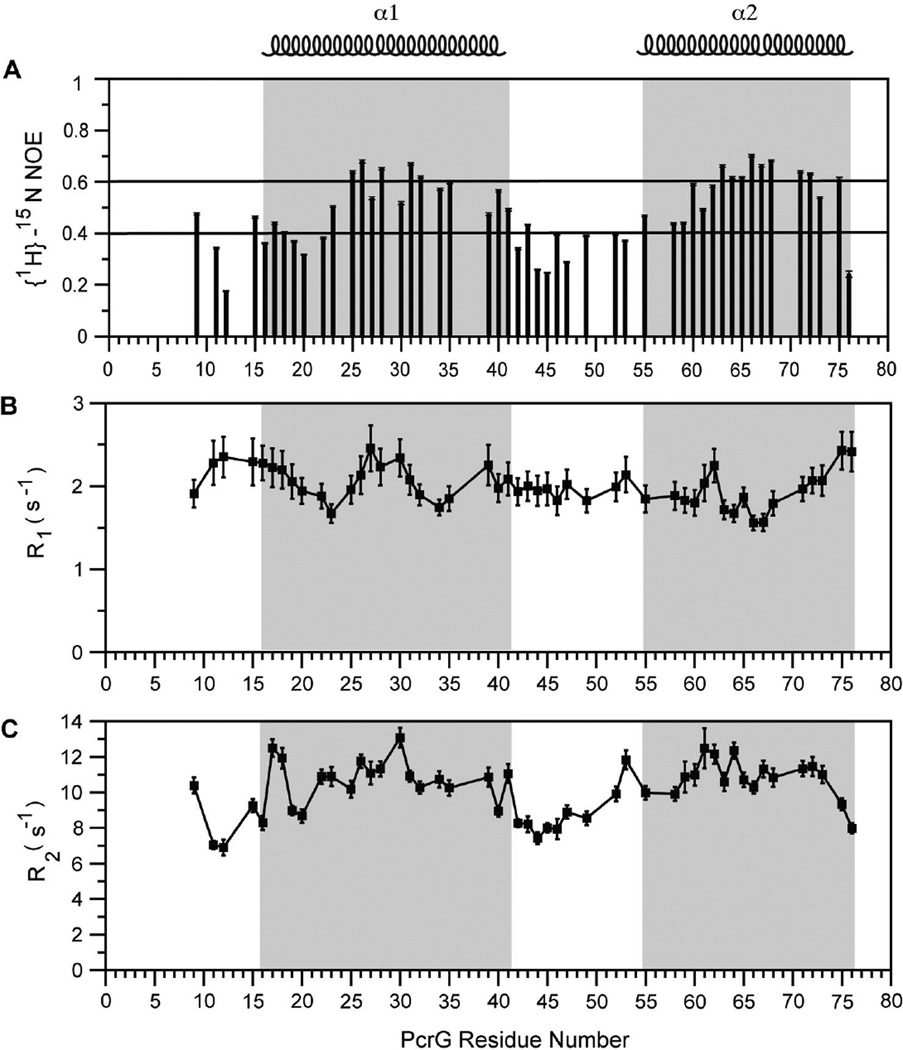
To assess the backbone dynamics of PcrG9–76, we acquired the steady state heteronuclear {1H}-15N NOE and 15N backbone R1 and R2 relaxation rates for PcrG9–76. The heteronuclear {1H}-15N NOE provides insight into the backbone dynamics of proteins at picosecond to nanosecond time scales. Heteronuclear {1H}-15N NOE values above 0.6 indicate well-structured regions and can be qualitatively described as being rigid. Heteronuclear {1H}-15N NOE values between 0.4–0.6 can be qualitatively described as semi-rigid flexibility, whereas {1H}-15N NOE values below 0.2 indicate completely unstructured regions as those found in random coils. Most of the residues in the two helices showed {1H}-15N NOEs between 0.4 to 0.6 (Figure 5A) in the range that qualitatively can be described as having semi-rigid flexibility. Residues found in the middle of the helices showed {1H}-15N NOE values above 0.6, in the rigid range, similar to those found in helices of well-structured proteins. The middle of the helices could be described as fully formed with the ends showing more flexibility. Residues flanking the helices were more flexible and showed {1H}-15N NOEs of 0.4 or below with a few residues (Glu9 and Ala-43) showing {1H}-15N NOEs falling into the semi-rigid range. Only one residue (Leu12) had a {1H}-15N NOE of slightly below 0.2 indicating random coil flexibility. The 15N backbone relaxation rates (R1 and R2) indicated that the two helices in PcrG behaved with nearly similar amide backbone dynamics (Figure 5B, 5C). For both the datasets, there was no appreciable difference in the relaxation rates from the beginning to the end of the protein. This indicated that PcrG tumbles as a single entity in solution, as opposed to for example, helix α1 tumbling at a different rate compared to helix α2.
Figure 5. (A) Heteronuclear {1H}-15N NOEs, (B) R1 and (C) R2 relaxation data for PcrG.
The two helical regions are shaded and gaps in the datasets are due to overlapped and missing peaks.
NMR Titrations
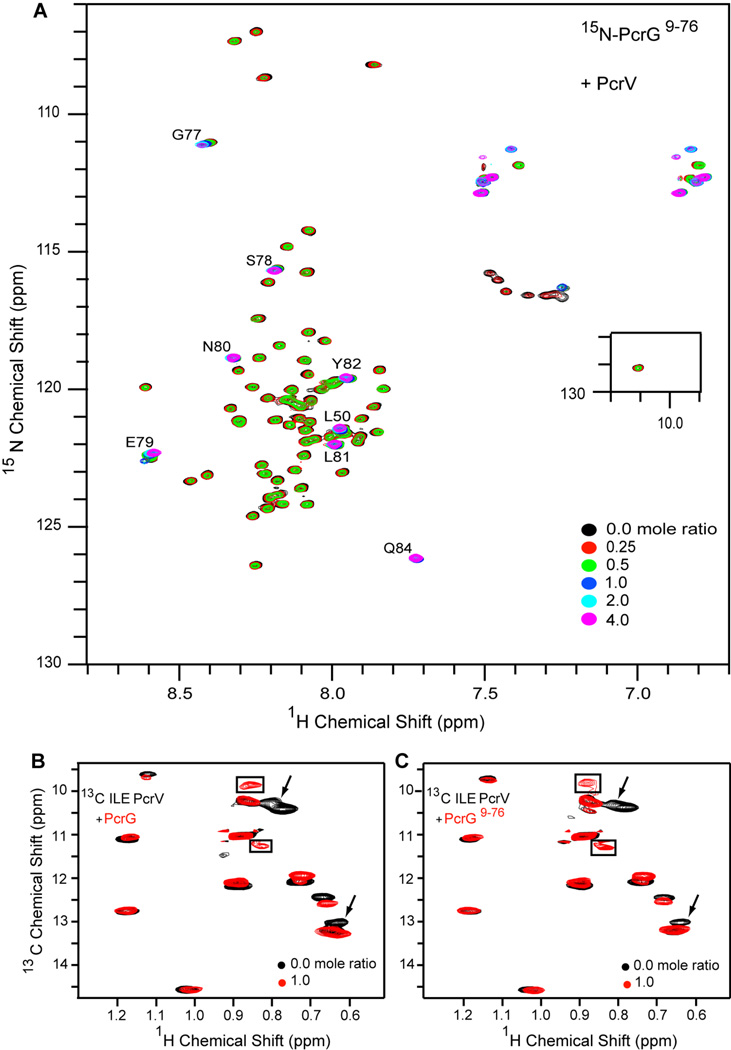
NMR chemical shift mapping was used to characterize the PcrG-PcrV interaction. Purified 15N-labeled PcrG9–76 was titrated with PcrV at increasing PcrV:PcrG molar ratios (of 0.0, 0.25, 0.5, 1.0, 2.0 and 4.0) in NMR buffer and monitored by acquiring 2D 1H-15N HSQC spectra (Figure 7A). With the exception of 7 of the 75 backbone amide peaks including the C-terminal cloning artifact, nearly all peaks showed significant reduction in peak intensities with increasing molar ratios of PcrV. The 7 peaks that did not show any change in peak intensities throughout the titration corresponded to the cloning artifact residues (Ser78, Glu79, Asn80, Leu81, Tyr82, Gln84) and one PcrG residue Leu50. However, one of the cloning artifact residues Gly-77 appeared to be in fast exchange time scale on titration with PcrV.
Figure 7. NMR titrations of PcrG and PcrV.
(A) Six overlaid 2D 1H-15N HSQC spectra of 15N-labeled PcrG9–76 titrated with increasing amounts of PcrV. (B, C) Two overlaid 2D 1H-13C HSQC spectra of isoleucine 13Cδ1 methyl labeled PcrV in complex with (B) PcrG and (C) PcrG9–76 in a 1:1 ratio. The slow-exchange peaks (boxed), and peaks in fast-exchange (arrows) are indicated.
Determination of PcrG-PcrV interaction by surface plasmon resonance (see below) required that the PcrG construct retain the His-tagged GB1 domain. To assess the effect of having the GB1 tag, 15N-labeled PcrG9–76-GB1-His6 fusion protein was titrated with increasing amounts of PcrV and the titration was monitored by acquiring 2D 1H-15N HSQC spectra (Figure S1). The HSQC peaks of PcrG9–76 (Figure 3B) were superimposable on the HSQC peaks of PcrG9–76-GB1-His6 fusion protein, thus, readily identifying the peaks belonging to the GB1-His6 tag. As the concentration of PcrV increased, the GB1-His6 peaks remained unperturbed while the same subset of PcrG9–76 peaks reduced in intensity as seen in the titration of PcrG9–76 without the GB1-His6 tag. Thus, the GB1-His6 tag did not alter the ability of PcrG to bind to PcrV, also highlighting the specificity of PcrG-PcrV binding in these assays.
To determine if truncating PcrG affected its binding to PcrV, NMR titrations were carried out using PcrV labeled with isoleucine 13Cδ1 methyl group and complexed with increasing amounts of unlabeled PcrG9–76 or full length PcrG. The isoleucine residues are represented throughout the different regions of PcrV and served as probes of protein-protein interactions of the different regions of PcrV with PcrG. For comparison of the results of PcrG and PcrG9–76, spectra from two molar ratios (0.0 and 1.0) are shown for clarity (Figure 7B, 7C). In both the datasets, there were two new peaks (shown as boxes) and two peaks that disappeared from the spectra (shown with arrows) upon complex formation (Figure 7B and Figure 7C). Although the NMR assignments for the isoleucine 13Cδ1 methyl group of PcrV are currently unavailable, the results of the titrations indicate that the same set of PcrV isoleucine residues were affected when titrated with either full length PcrG or truncated PcrG9–76. Thus, both PcrG and PcrG9–76 bound to the same surface of PcrV, and the truncation did not alter the ability of PcrG9–76 to bind to PcrV.
Surface Plasmon Resonance Spectroscopy
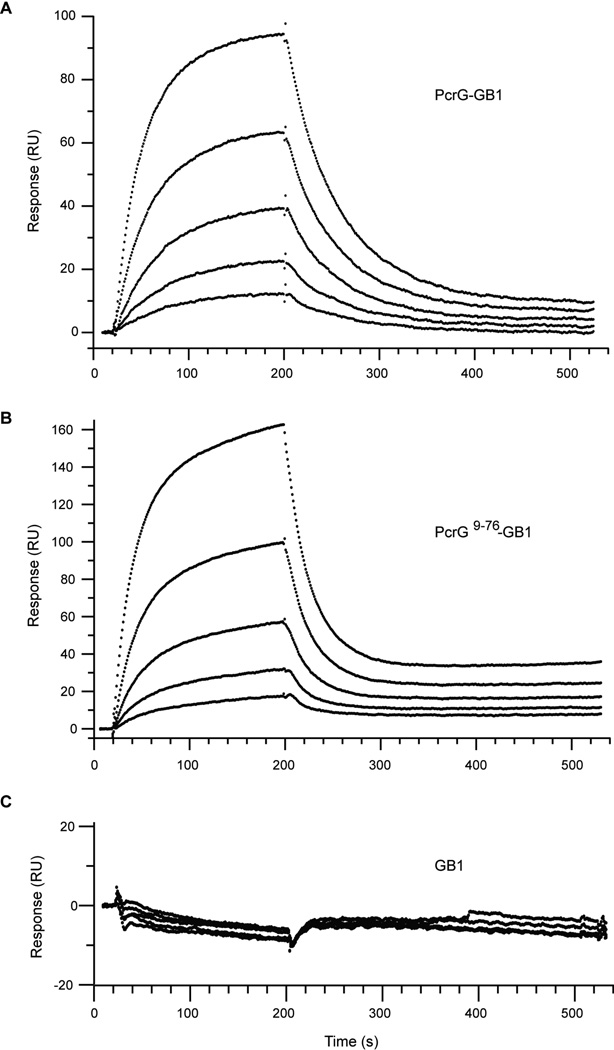
Surface plasmon resonance (SPR) was used to determine the binding affinity and binding kinetics between PcrV and PcrG. PcrG-GB1-His6 and PcrG9–76-GB1-His6 fusion proteins were used as ligands immobilized on CM5 flow cells coated with anti-polyhistinine antibodies for SPR studies. We used the GB1-His6 tag to increase the distance between the surface of the CM5 chip and PcrG to allow better sampling of possible binding sites upon interaction with PcrV. The sensorgrams in Figure 6A and 6B show that the optical responses increased upon injection of increasing concentrations of PcrV on immobilized PcrG-GB1. The SPR data for the interaction between PcrV and PcrG-GB1-His6 was best fit to a 1:1 Langmuir model (χ2 = 0.314) (Figure 6A). The dissociation constant (Kd) for this interaction was calculated to be 26 nM. This high affinity binding is similar to that previously described by Nanao et. al(10) in which SPR was used to evaluate binding between full length proteins, PcrG and PcrV. Thus, the N-terminal 24-residue truncation of PcrV has little effect on the interaction. The binding occurred with rapid kinetics, with ka = 5.6 × 105 M−1s−1 and kd = 1.5 × 10−2 M−1s−1.
Figure 6. SPR sensorgrams for PcrV binding to (A) PcrG-GB1-His6, (B) PcrG9–76-GB1-His6 and (C) GB1-His6 fusion tag.
The truncations made in PcrG9–76 did not have a significant effect in its capacity to bind PcrV. The sensorgrams for PcrV binding to immobilized PcrG9–76-GB1-His6 was fit to a 1:1 Langmuir model (χ2 = 2.53) and the calculated dissociation constant (Kd) of 24 nM was essentially similar to that of full length PcrG (Figure 6B). Also, the binding kinetics were somewhat more rapid than seen for full length PcrG, although less than an order of magnitude, with ka = 1.0 × 106 M−1s−1 and kd = 2.4 × 10−2 M−1s−1. These minimal changes in binding suggested that truncated PcrG9–76 retained the regions needed for tight-binding interaction with PcrV. The SPR data was consistent with the results of our NMR titrations using isoleucine 13Cδ1-labeled PcrV (Figure 7), which showed that both the truncated and full length PcrG bound similarly to PcrV.
To account for possible interference of the GB1-His6 tag, control sensorgrams were acquired under identical conditions in SPR assay but with GB1-His6 as the ligand (Figure 6C). Results show that the optical responses with increasing concentrations of PcrV were insignificant. Therefore, the GB1-His6 tag remained inert in the interaction. This result was also consistent with the results of NMR titration using 15N PcrG9–76-GB1-His6, which showed that the GB1-His6 residues remain unchanged and the same set of PcrG9–76 residues were affected on binding to PcrV. This observation confirmed the specificity of the PcrG-PcrV and PcrG9–76-PcrV interaction.
DISCUSSION
The atomic structure of PcrG or any member of the PcrG family of tip protein chaperones (Figure 1) is currently unknown. Our CD (Figure 2) and NMR (Figure 3) results suggested that crystallization of full length PcrG will not be feasible because of the significant amount of protein disorder as indicated by CD spectroscopy (Figure 2) and the possible multiple conformations or oligomerization of PcrG as indicated by its 2D HSQC spectrum (Figure 3). Nevertheless, a truncated form, PcrG9–76, yielded an ideal NMR spectrum (Figure 3C) that retained nearly identical secondary structures of full length PcrG (Figure 2A). Our NMR and CD data showed that PcrG lacks a tertiary structure and it only consists of two alpha-helical secondary structures that do not interact with each other. Our results presented here represent the current knowledge on the atomic-level structure of PcrG derived by NMR. Our results suggest that other members of the PcrG family of chaperone proteins will lack tertiary structures as well.
Basu et al.(22) reported a 3D structure for PcrG derived by I-TASSER computer modeling. In their structure, PcrG formed a compact bundle of four helices spanning residues 13–25, 29–39, 44–52, and 56–71 (depicted in Figure 4). Our secondary Cα chemical shifts (Figure 4A) could support the presence of their helix 3 (and its attendant flanking linker regions) because of the consistently positive, albeit weak, secondary Cα chemical shifts of residues 46–51 (Figure 4A). We deemed, however, that the magnitude of the secondary Cα chemical shifts of residues 46–51 to be too small compared to helix α1 and β2 and we therefore did not assign residues 46–51 to form a helix. The major disagreement between our experimental data and their proposed structure is that their structure would require breaking the middle of helix α1 at residues Ser26, Glu27 and Glu28 to form a linker between their first two helices. In our case, residues Ser26, Glu27 and Glu28 showed strong alpha-helical secondary Cα chemical shifts (Figure 4A) suggesting that they are part of a helix. Additionally, a protein with a tertiary structure consisting of packed helices would show a sigmoidal curve in a CD denaturation experiment to show the cooperative melting of helices that interact with each other; unlike the CD melting curves we obtained for full length and truncated PcrG shown in Figure 2B. Our NMR and CD results therefore do not support the reported 3D structure of PcrG by Basu et al(22) derived by computer modeling. Basu et al(22) then used their 3D structure of PcrG to model the atomic structure of the PcrG-PcrV complex. In light of contrary NMR and CD experimental data presented above, caution is necessary in using the predicted structure of PcrG(22) to elucidate the mechanism of PcrG-PcrV interaction with respect to type III secretion.
Direct binding between PcrG and PcrV has been demonstrated previously(8, 10, 11), however, the crystal or NMR structure of the PcrG-PcrV complex remains unknown. Studies based on its closest homolog LcrG suggest that the N-terminus of LcrG is involved in LcrV binding(12, 34). Results of yeast two-hybrid suggested that the LcrG N-terminal residues 7–40 (corresponding to residues 10–43 in PcrG) are required for interaction with LcrV(34). This region is highly conserved between LcrG and PcrG with a 47% sequence identity. Nanao et. al(10) and Lee et. al(8) also suggested that the N-terminus of PcrG binds to PcrV. Basu et al(22) showed that PcrG residues 13–72 interacts with PcrV. Our NMR titrations using PcrG9–76 indicate that PcrV induces a global change in the structure of PcrG that may likely result from a combination of both PcrV-binding interactions and binding-induced conformational changes (Figure 7A). This is not surprising considering the sizes of the two proteins PcrV (33 kDa) and PcrG9–76 (8 kDa). It is possible that the entire PcrG wraps around PcrV, as it is not uncommon for flexible proteins to interact with a larger surface area as compared to globular proteins(40) to stabilize the protein-protein interaction.
Earlier results have pointed out multiple putative roles for PcrG towards control of secretion and translocation of effector proteins in P. aeruginosa T3SS, in addition to its role as a chaperone for PcrV(7–11, 22). Due to its inherent functional versatility therefore, it is possible that the structure of PcrG is conformationally interchangeable and dynamic. Interaction with a wide variety of protein targets is also a trademark of disordered and flexible proteins(40, 41). The inherent flexibility enhances the functional diversity of PcrG and allows it to adopt transiently stable conformations to perform its specific functions, for example, by binding to PcrV(8, 10, 11), or by binding to PscO and PcrD(7). This is not unprecedented for natively unstructured proteins, which in most cases undergo binding induced folding transition(42). In fact, these kinds of protein folding transitions are not uncommon in T3SS either. It is applicable to both the structural components of T3SS and in case of chaperone-effector interactions(43–46).
Supplementary Material
Acknowledgments
We acknowledge support from NIH for grants 74856 (R.N.D.) and P30GM110761 (KU Bio NMR Facility) that made this research possible. We are grateful to Asokan Anbanandam for assistance in the NMR experiments.
REFERENCES
- 1.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 2.Engel J, Balachandran P. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 2009;12:61–66. doi: 10.1016/j.mib.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 2009;7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13899–13904. doi: 10.1073/pnas.95.23.13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dacheux D, Epaulard O, de Groot A, Guery B, Leberre R, Attree I, Polack B, Toussaint B. Activation of the Pseudomonas aeruginosa type III secretion system requires an intact pyruvate dehydrogenase aceAB operon. Infect. Immun. 2002;70:3973–3977. doi: 10.1128/IAI.70.7.3973-3977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goure J, Pastor A, Faudry E, Chabert J, Dessen A, Attree I. The V antigen of Pseudomonas aeruginosa is required for assembly of the functional PopB/PopD translocation pore in host cell membranes. Infect. Immun. 2004;72:4741–4750. doi: 10.1128/IAI.72.8.4741-4750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee PC, Zmina SE, Stopford CM, Toska J, Rietsch A. Control of type III secretion activity and substrate specificity by the cytoplasmic regulator PcrG. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2027–E2036. doi: 10.1073/pnas.1402658111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee PC, Stopford CM, Svenson AG, Rietsch A. Control of effector export by the Pseudomonas aeruginosa type III secretion proteins PcrG and PcrV. Mol. Microbiol. 2010;75:924–941. doi: 10.1111/j.1365-2958.2009.07027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundin C, Thelaus J, Broms JE, Forsberg A. Polarisation of type III translocation by Pseudomonas aeruginosa requires PcrG, PcrV and PopN. Microb. Pathog. 2004;37:313–322. doi: 10.1016/j.micpath.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Nanao M, Ricard-Blum S, Di Guilmi AM, Lemaire D, Lascoux D, Chabert J, Attree I, Dessen A. Type III secretion proteins PcrV and PcrG from Pseudomonas aeruginosa form a 1:1 complex through high affinity interactions. BMC Microbiol. 2003;3:21. doi: 10.1186/1471-2180-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allmond LR, Karaca TJ, Nguyen VN, Nguyen T, Wiener-Kronish JP, Sawa T. Protein binding between PcrG-PcrV and PcrH-PopB/PopD encoded by the pcrGVH-popBD operon of the Pseudomonas aeruginosa type III secretion system. Infect. Immun. 2003;71:2230–2233. doi: 10.1128/IAI.71.4.2230-2233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matson JS, Nilles ML. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J. Bacteriol. 2001;183:5082–5091. doi: 10.1128/JB.183.17.5082-5091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilles ML, Fields KA, Straley SC. The V antigen of Yersinia pestis regulates Yop vectorial targeting as well as Yop secretion through effects on YopB and LcrG. J. Bacteriol. 1998;180:3410–3420. doi: 10.1128/jb.180.13.3410-3420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilles ML, Williams AW, Skrzypek E, Straley SC. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J. Bacteriol. 1997;179:1307–1316. doi: 10.1128/jb.179.4.1307-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis GR. The type III secretion injectisome. Nat. Rev. Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell SW, De Guzman RN. The Crystal Structure of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Deoxycholate and Chenodeoxycholate. Protein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawton DG, Longstaff C, Wallace BA, Hill J, Leary SE, Titball RW, Brown KA. Interactions of the type III secretion pathway proteins LcrV and LcrG from Yersinia pestis are mediated by coiled-coil domains. J. Biol. Chem. 2002;277:38714–38722. doi: 10.1074/jbc.M203632200. [DOI] [PubMed] [Google Scholar]
- 18.Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J. Biol. Chem. 2007;282:4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erskine PT, Knight MJ, Ruaux A, Mikolajek H, Wong Fat Sang N, Withers J, Gill R, Wood SP, Wood M, Fox GC, Cooper JB. High Resolution Structure of BipD: An Invasion Protein Associated with the Type III Secretion System of Burkholderia pseudomallei. J. Mol. Biol. 2006;363:125–136. doi: 10.1016/j.jmb.2006.07.069. [DOI] [PubMed] [Google Scholar]
- 20.Blocker AJ, Deane JE, Veenendaal AK, Roversi P, Hodgkinson JL, Johnson S, Lea SM. What's the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U.S.A. 2008;105:6507–6513. doi: 10.1073/pnas.0708344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derewenda U, Mateja A, Devedjiev Y, Routzahn KM, Evdokimov AG, Derewenda ZS, Waugh DS. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure. 2004;12:301–306. doi: 10.1016/j.str.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Basu A, Das U, Dey S, Datta S. PcrG protects the two long helical oligomerization domains of PcrV, by an interaction mediated by the intramolecular coiled-coil region of PcrG. BMC Struct Biol. 2014;14:5. doi: 10.1186/1472-6807-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geisbrecht BV, Bouyain S, Pop M. An optimized system for expression and purification of secreted bacterial proteins. Protein Expr. Purif. 2006;46:23–32. doi: 10.1016/j.pep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 26.Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 27.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 1992;114:6291–6293. [Google Scholar]
- 28.Wittekind M, Mueller L. HNCACB, a high sensitivity 3D NMR experiment to correlate amide proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J. Magn. Reson. 1993;101 B:201–205. [Google Scholar]
- 29.Wishart DS, Nip AM. Protein chemical shift analysis: a practical guide. Biochem. Cell Biol. 1998;76:153–163. doi: 10.1139/bcb-76-2-3-153. [DOI] [PubMed] [Google Scholar]
- 30.Stone MJ, Fairbrother WJ, Palmer AG, III, Reizer J, Saier MH, Jr, Wright PE. Backbone dynamics of the Bacillus subtilis glucose permease IIA domain determined from 15N NMR relaxation measurements. Biochemistry. 1992;31:4394–4406. doi: 10.1021/bi00133a003. [DOI] [PubMed] [Google Scholar]
- 31.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 32.Williams T, Kelley C. Gnuplot 4.4: an interactive plotting program. 2010 http://gnuplot.sourceforge.net/ [Google Scholar]
- 33.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 34.Matson JS, Nilles ML. Interaction of the Yersinia pestis type III regulatory proteins LcrG and LcrV occurs at a hydrophobic interface. BMC Microbiol. 2002;2:16. doi: 10.1186/1471-2180-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 36.King RD, Sternberg MJ. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 1996;5:2298–2310. doi: 10.1002/pro.5560051116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guermeur Y, Geourjon C, Gallinari P, Deleage G. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics. 1999;15:413–421. doi: 10.1093/bioinformatics/15.5.413. [DOI] [PubMed] [Google Scholar]
- 38.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 39.Lau SY, Taneja AK, Hodges RS. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J Biol Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- 40.Babu MM, van der Lee R, de Groot NS, Gsponer J. Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Poyraz O, Schmidt H, Seidel K, Delissen F, Ader C, Tenenboim H, Goosmann C, Laube B, Thunemann AF, Zychlinsky A, Baldus M, Lange A, Griesinger C, Kolbe M. Protein refolding is required for assembly of the type three secretion needle. Nat. Struct. Mol. Biol. 2010;17:788–792. doi: 10.1038/nsmb.1822. [DOI] [PubMed] [Google Scholar]
- 44.Rodgers L, Gamez A, Riek R, Ghosh P. The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J. Biol. Chem. 2008;283:20857–20863. doi: 10.1074/jbc.M802339200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L, Balabanidou V, Remeta DP, Minetti CA, Portaliou AG, Economou A, Kalodimos CG. Structural instability tuning as a regulatory mechanism in protein-protein interactions. Mol. Cell. 2011;44:734–744. doi: 10.1016/j.molcel.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong D, Lefebre M, Kaur K, McDowell MA, Gdowski C, Jo S, Wang Y, Benedict SH, Lea SM, Galan JE, De Guzman RN. The Salmonella type III secretion system inner rod protein PrgJ is partially folded. J. Biol. Chem. 2012;287:25303–25311. doi: 10.1074/jbc.M112.381574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.