Abstract
Whole blood transcriptional profiling offers great diagnostic and prognostic potential. While studies have identified signatures for pulmonary tuberculosis (TB) and transcripts that predict the risk of developing active TB in humans, the early transcriptional changes immediately following Mycobacterium tuberculosis (Mtb) infection have not been evaluated. We evaluated the gene expression changes in the cynomolgus macaque model of TB, which recapitulates all clinical aspects of human Mtb infection, using a human microarray and analytics platform. We performed genome-wide blood transcriptional analysis on 38 macaques at 11 post-infection time points during the first 6 months of Mtb infection. Of 6,371 differentially expressed transcripts between pre-and post-infection, the greatest change in transcriptional activity occurred 20–56 days after infection, where fluctuation of innate and adaptive immune response related transcripts was observed. Modest transcriptional differences between active TB and latent infection were observed over the time course with substantial overlap. The pattern of module activity previously published for human active TB was similar in macaques with active disease. Blood transcript activity was highly correlated with lung inflammation (lung FDG avidity) measured by PET CT at early time points post infection. The differential signatures between animals with high and low lung FDG were stronger than between clinical outcomes. Analysis of pre-infection signatures of macaques revealed that interferon signatures could influence eventual clinical outcomes and lung FDG avidity even before infection. Our data support that transcriptional changes in the macaque model are translatable to human Mtb infection and offer important insights into early events of Mtb infection.
Introduction
Despite major global health efforts, 9.6 million new cases of tuberculosis (TB) occurred in 2014 (1) with increasing rates of multi-drug resistant TB annually. Those infected with M. tuberculosis (Mtb) can develop either symptomatic (active) TB disease (5% of people) or asymptomatic infection, called latent infection (LTBI). There is growing body of evidence (2–4) recognizing that Mtb infection outcome is not binary but rather a spectrum of clinical states with varying severity of active TB, as well as LTBI that ranges from low grade or subclinical infection to bacterial clearance. Those with LTBI serve as a reservoir for reactivation TB, which can occur years to decades after initial infection. The spectrum of latent infection likely contributes to the risk of reactivation for an individual(2, 5). We recently showed that the high lung FDG avidity is associated with risk of reactivation in NHP with latent infection (6). There are a variety of factors that could influence the outcome of infection, including Mtb strain type, exposure dose and duration, host genetics and immune response. In human Mtb infection, very little is known about the early events after infection, as current diagnostics are not sensitive enough to determine time of infection, and the early course of infection is relatively clinically silent. However, the early innate and adaptive responses are likely to be crucial in the ultimate outcome of infection (4, 7). In humans and in animal models, various types of immune compromise exacerbate primary infection (reviewed in (8–11)). A better understanding of early host responses to the infection is necessary to improve vaccine development and determine those at greatest risk of disease.
Blood transcriptional profiling provides a global perspective on the dynamics of complex molecular and cellular events in specific human diseases and has improved the diagnosis and biological understanding of diseases (12). Over the past decade, several groups have published PBMC or whole blood transcriptional signatures associated with human active TB or LTBI. Whole genome blood transcriptional profiles from a large scale study of subjects recruited from high and moderate TB endemic countries showed gene expression differences that could discriminate individuals with active TB from those with LTBI and healthy controls (reviewed in (12)). Transcriptional profiles could also distinguish response to anti-TB drug treatment as early as 2 months into treatment (13). Finally, a recent study reported transcriptional signatures that correlated with risk of active TB in adolescents who had latent infection at baseline (14), suggesting a pre-Mtb infection signature might play an important role in determining the outcome of infection. These studies underscored the potential of transcriptional profiling as a diagnostic and predictive tool in TB. Such studies have provided key insights into immune factors that distinguish host immune responses between diseases with similar clinical manifestations.
Here, we used an established non-human primate (NHP) model to interrogate early transcriptional changes after Mtb infection. Cynomolgus macaques infected with low dose Mtb develop the full clinical spectrum of pathology and outcomes seen in humans including latent infection (4, 15). As reported by our group previously (4, 15, 16), active TB disease is defined by clinical signs of active TB (e.g., cough, weight loss, anorexia), presence of Mtb growth in the BAL or gastric aspirate fluid, and/or elevated systemic inflammatory markers (by erythrocyte sedimentation rate). Animals with LTBI are asymptomatic, without Mtb growth in BAL or gastric aspirate and no evidence of systemic inflammation. Latent infection is not declared until 6 months after infection whereas active disease can be declared before 6 months. In this infection model, approximately 40–50% of infected animals progress to active disease with the remainder maintaining latent infection. We have shown outcome of infection (active TB or latent infection) can be distinguish as early as 6 weeks post-infection based on serial PET-CT imaging (16) and, to a lesser extent, by immunological assays (4). Given the close genetic similarities between humans and non-human primates (NHP), this model provides an exceptional platform for in-depth investigations of host-pathogen interactions. Unlike humans, the use of this model allows us to control the inoculation dose, timing, and strain while conducting serial, high frequency sampling to examine post-infection responses.
In this model we evaluated serial whole genome blood transcriptional signatures in NHP from pre-infection to 6 months post Mtb infection using a human microarray platform. We used linear mixed model analysis, k-means clustering and a pre-existing human modular gene expression framework to interrogate the data. The greatest dynamic change occurs between days 20–56 post infection, regardless of the clinically defined outcome. We extended our evaluation to identify signatures that were associated with outcome using both clinical definitions of active TB and LTBI as well as total lung inflammation as measured by the 18F-fluorodeoxyglucose (FDG) avidity in PET-CT, a surrogate marker for disease severity (17). Finally we show that the signature associated with active TB in NHP is comparable to that observed in human TB, suggesting the NHP signature is translational to humans.
Materials and Methods
Animals
Adult (>4 years of age) cynomolgus macaques (Macacca fascicularis) (Valley Biosystems, Sacramento, CA) were housed within a Biosafety Level 3 (BSL-3) primate facility as previously described(4, 18, 19). Monkeys were infected with low dose M. tuberculosis (Erdman strain) via bronchoscopic instillation of ~25 colony-forming units (CFUs) per monkey to the lower lung lobe. Infection was confirmed by tuberculin skin test conversion and/or lymphocyte proliferation assay six weeks post-infection (19). Serial clinical, microbiologic and immunologic examinations were performed, as previously described. Based on defined clinical criteria, radiographic, and microbiologic assessments during the course of infection monkeys were classified as having latent infection or active disease as late as 6 months post infection as described previously (4, 15, 18, 19). A total of 38 animals were included in this study over a period of 2 years from 2011–2013. Due to logistical constraints the study was divided into two sets composed of 19 animals each. 16 animals were declared to have active disease while 22 remained latently infected. After infection outcome was declared, these animals were dedicated to other on-going studies.
Ethics Statement
All experimental manipulations, protocols, and care of the animals were approved by the University of Pittsburgh School of Medicine Institutional Animal Care and Use Committee (IACUC). The protocol assurance number for our IACUC is A3187-01. Our specific protocol approval numbers for this project are 11090030 and 1105870. The IACUC adheres to national guidelines established in the Animal Welfare Act (7 U.S.C. Sections 2131 - 2159) and the Guide for the Care and Use of Laboratory Animals (8th Edition) as mandated by the U.S. Public Health Service Policy.
Blood
Longitudinal blood sampling included: 2 pre-Mtb infection time points, days 3, 7, 10, 20, 30, 42 (6 weeks), 56 (8 weeks), 90 (12 weeks), 120 (16 weeks), 150 (20 weeks) and 180 (24 weeks) post Mtb infection. For animals that developed active disease prior to 180 days post infection (Figure S1), the last time point was at the time at which the active disease was declared.
One milliliter of macaque whole blood was drawn into heparin blood collection tubes (BD Biosciences) and mixed well. Then, 500 µl of this blood was immediately aliquoted into a cryovial containing 1.5 ml of Tempus reagent. All samples were thoroughly vortexed for 15 seconds to facilitate RNA stabilization. Samples were stored at −80°C until RNA extraction.
Cellular composition of the blood was monitored by complete blood counts including differential counts (analyzed by the clinical lab at UPMC) and by flow cytometry at the same time points. PBMCs were isolated via Percoll gradient centrifugation as previously described(20). PBMC were stained for cell surface markers: T cell markers include CD3 APC-Cy7 (Clone SP34-2, BD Horizon), CD4FiTc (Clone: L200, BD Pharmigen), CD8 Pac Blue (Clone DK25,Dako); B cell CD20 PE Cy7 (Clone 2H7, ebioscience) and for NK cells CD16 PE (Clone 3G8, BD Pharmingen) along with respective isotypes for flow cytometry. Data acquisition was performed using an LSR II (BD) and analyzed using FlowJo Software v.9.7 (Treestar Inc, Ashland, OR).
PET-CT Imaging and analysis
At the time that infection outcome was declared, animals underwent PET-CT scan. Animals were sedated, intubated and imaged by 2-deoxy-2-18F-D-deoxyglucose (FDG) PET imaging (microPET Focus 220 preclinical PET scanner, Seimens Molecular Solutions) and CT scanner (Neurologica Corp) within our biosafety level 3 facility as previously described (16, 18). The total lung FDG avidity was analyzed using Osirix viewer, an open-source PACS workstation and DICOM viewer. The whole lung was segmented on CT by using the Growing region algorithm on the Osirix viewer to create a ROI of normal lung (Hounsfield units ≤ 200). The closing tool was used to include individual nodules and other pulmonary disease. The ROI was transferred to the co-registered PET scan and manually edited to ensure all pulmonary disease was included. All extrapulmonary structures and disease, including mediastinal lymph nodes, were excluded. Voxels outside the ROI were set to zero and voxels with an SUV greater than or equal to normal lung (SUV ≥ 2.3) were isolated. Finally, the "Export ROIs" plug-in was then used to export the data from these isolated ROIs to a spreadsheet, where the total SUV per voxel were summed to represent the total lung FDG avidity. PET-CT imaging data (Lung FDG avidity) was available only for 37 animals.
RNA isolation and Microarray hybridization
Total RNA was isolated from whole blood lysate using the MagMax-96 Blood RNA Isolation Kit (Applied Biosystems) according to the manufacturer's instructions. All post-extraction RNA yields were measured using a Nanodrop 8000 (Nanodrop Technologies). RNA integrity (RIN) values were assessed on an Agilent 2100 Bioanalyzer (Agilent). Both RIN and yield data were managed using a LIMS system for quality control and sample tracking. Samples with RNA integrity values >5.5 were retained for further processing. Globin mRNA was depleted from a portion of each total RNA sample using the GLOBINclear™-Human kit (Ambion, Austin, TX, USA). Following Globin-reduction RNA integrity was assessed a second time (RIN cutoff = 5.5, average RNA integrity values = 7.5, standard deviation = 0.83). Globin-reduced RNA was amplified and labeled using the Illumina TotalPrep RNA Amplification Kit (Ambion). The RNA input for this reaction was 250ng and 750ng of amplified labeled RNA was hybridized overnight to Illumina HT12 V4 beadchips (Illumina). Following hybridization each chip was washed, blocked, stained and scanned on an Illumina BeadStation 500 following the manufacturer's protocols. Microarray data can be downloaded via the GEO Omnibus GSE84152 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ansjeswmhnsvzud&acc=GSE84152).
Data Analysis
Normalization and Batch Correction
Illumina's Genome Studio version HT12 software was used to generate signal intensity values from the scans. After background subtraction, the average normalization recommended by the Genome software was used to rescale the difference in overall intensity to the median average intensity for all samples across multiple arrays and chips. All data below 10 was set to 10 and then the dataset was log2 transformed. In total, 489 microarray samples passed stringent quality control standards following hybridization and microarray scanning. Principle Variance Component Analysis (PVCA) was preformed using JMP Genomics 6.0 (SAS Institute, Inc.) analysis software to identify major sources of variability within a combined dataset. PVCA revealed dataset membership (training or test) and hybridization chamber (chamber in which batches of arrays were hybridized) as major sources of technical variation. In order to correct for this technical variation and increase sensitivity to sources of biologic variability, batch correction (Figure S1B) using the ComBat R package was employed(21). After batch correction for technical variables the overall sensitivity to biologic variables was increased, which accounted for 35,082 probes expressed in at least one sample. Next a filter selecting the 60% most variable transcripts across all time points for all animals was used to further increase the signal to noise ratio (n= 21,049 probes).
Unsupervised analysis
PVCA was used as an unsupervised analysis tool to explore the contribution of biologic variables and outcome groups to the variance attributed to the first three principal components. In addition to PVCA analyses, unsupervised hierarchical clustering and classic PCA analysis were conducted at each time point to assess sample clustering and association with biologic variables (data not shown).
Linear Mixed-Model Analysis (LMMA) of Longitudinal Data
For each macaque, two pre-infection baseline samples were used and combined to create a shared baseline to which all post-challenge time points were be compared. Statistical analyses of log2-transformed microarray data were performed using SAS software (v9.3) and JMP Genomics (v6) (SAS Institute, Inc.). A linear mixed-model analysis (LMMA), accounting for repeated measures (repeated within each macaque) and unequal time-point spacing, was employed to the top 60% variable transcripts to test for significant differences in gene expression between each post-challenge time point and the shared baseline. Benjamini-Hochberg multiple testing correction was applied (FDR=0.05) for all hypothesis testing resulting in the selection of 6,371 differentially expressed probes. These probes were hierarchically clustered using the Ward clustering method. The cubic clustering criterion (CCC) was used to identify k=20 for k-means clustering. Ingenuity Pathway Analysis (Ingenuity Systems) and MetaCore Pathway Analysis (Thompson Reuters) network analysis tools were used to annotate the k=20 clusters. Furthermore, the Novartis Gene Atlas was queried for modules containing genes with expression limited to particular cell types. Additionally, we employed cross matching of probes within the K=20 clusters with the predefined human disease modules (22) to further inform the annotation of clusters. Annotated modules contained multiple genes with similar function, cellular localization, or membership in known biologic pathways.
Percent Cluster Activity and Cluster Activity Scores
To assess the longitudinal activity of each of the k-means (k=20) clusters relative to pre-challenge baseline, a percent activity score was determined for each cluster at all time points post-challenge. Briefly, this activity score represents the percent of all genes within each cluster that are differentially expressed at each time point using LMMA. The sign associated with the mean estimate of the LMMA for the significant genes within each cluster, at each time point, was used to assign increased or decreased activity for each cluster.
A cluster activity score was calculated for each animal at each post-infection time point and was used to correlate CBC, flow cytometry and outcome. This score was calculated for each animal by summing the difference in probe-level post-infection expression values and the average pre-infection expression value for all probes within a cluster. This sum of differences was then divided by the number of probes within each cluster plus the number of samples within each time group.
Molecular Distance to Health (MDTH) Analysis
MDTH was performed as previously using a Microsoft Excel 2010 visual basic add-on (23) Briefly, in this approach a score is computed to represent the molecular distance of a given sample relative to a baseline, i.e., pre-infection time points. This is performed by determining whether the expression of a given sample lies inside or outside two standard deviations from the mean of the baseline. The MDTH (Figure 1C) was calculated only for those 6,371 probes identified in the LMMA analysis.
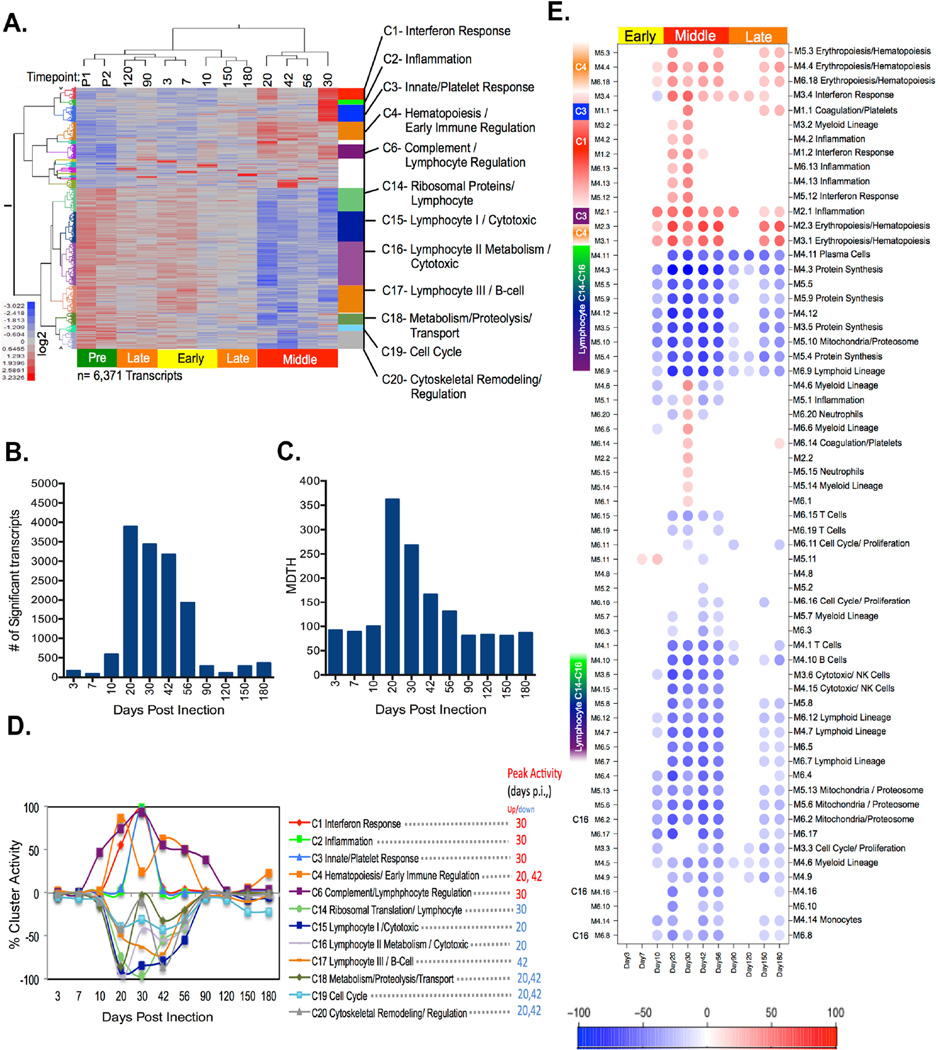
Figure 1. Supervised LMMA reveals three distinct transcriptional phases following Mtb infection.
(A) Two-way hierarchical clustering of LMMA selected transcripts (columns) (FDR=0.05) followed by k-means clustering (kn=20) were used to compare each post-infection time point (rows) with the pre-infection baseline. Those clusters (12/20) with significant ontology and network associations determined by Ingenuity Pathway Analysis (IPA) are annotated. (B) The total number of differentially expressed genes determined by LMMA at each post-infection time point are shown. (C) The amplitude of post-infection transcript perturbations as measured by the Molecular Distance to Health are shown. (D) Cluster activity at each time point post-infection is represented as the percentage of all genes within each cluster that are differentially expressed using LMMA. (E) Transcripts from NHP were also evaluated using pre existing human expression modular framework. Module activity scores representing the percentage of transcripts that are either over-expressed (red) or under-expressed (blue) in 62 predefined human co-expression modules. Module activity was hierarchically clustered to group modules with similar activity patterns over time. Those clusters with >15% representations are presented here. Red color represents over expression over pre-infection, whereas blue color represent under expression. Color intensity represents the percent of each module that is differentially expressed from pre-infection baseline. K-means clusters from 1D were mapped to those framework modules which share >15% similar transcript constituents.
Module Maps
A previously defined and annotated framework of 260 gene expression modules with coordinate expression across 1-9 human disease datasets was used to assess post infection changes(22). Gene expression levels of the top 60% variable transcripts were compared between a shared pre-infection baseline across all animals and healthy controls on a module-by-module basis. The percentage of transcripts showing significant differences (Unpaired T-test, p<0.05) in expression was used as an indicator of module activity.
Module activity scores
For correlation analysis with CBC, flow cytometry and outcome a module activity score was calculated for each animal at each post-infection time point. This score was calculated for each animal by summing the difference in probe-level post-infection expression values and the average pre-infection expression value for all probes within a module. This sum of differences was then divided by the number of probes within each module plus the number of samples within each time group.
Cross-Correlation analyses
Cluster activity scores (k=20) and Module activity scores were cross-correlated with absolute cell counts, flow-cytometry counts, or lung FDG avidity (as continuous variable) using JMP Genomics (SAS Institute Inc.) For all cross-correlations, non-parametric spearman correlation was employed. Significant correlations were identified using Benjamin- Hochberg multiple testing correction (FDR=0.05).
Statistical analysis and Data Visualization
Molecular distance to health and modular frame work analysis calculations were performed using Microsoft Excel 2010 and associated visual basic add-on. For statistical analysis JMP genomics 6.0, GraphPad Prism v6.0h and GeneSpringv12.6.1 was used for fold change analysis. Cross-correlation heat maps were created by exporting statistical results from JMP Genomics and then visualized using Plotly (Plotly Inc.).
Results
Greatest transcriptional activity occurs between 20 and 56 days post Mtb infection
To investigate blood transcriptome dynamics over the course of Mtb infection, whole blood transcripts were compared between pre-Mtb infection and 11 serial time points following Mtb infection among 38 NHPs (Figure S1A). We performed Linear Mixed Model Analysis (LMMA), an adjusted supervised analysis to assess transcripts with differential expression between the pre-infection and each post-infection (p.i.) time point. Overall, 6,371 transcripts were differentially expressed in at least one post-infection time point, regardless of infection outcome. The hierarchical clustering of time points based on differentially expressed transcripts (Figure 1A), number of differentially expressed transcripts at each time point (Figure 1B) and molecular distance to health (MDTH) (Figure 1C) in comparison to the pre-infection baseline revealed evidence of 3 phases of transcriptional activity post Mtb infection. These phases include early (3–10 days p.i.), middle (20–56 days p.i.) and late (90–180 days p.i.) (Figure 1A-C). Modest transcript perturbations were observed between days 3–10 and days 90–180 p.i. (Figure 1A and 1B). The greatest differential expression from pre-infection time points was observed at 20–56 days p.i. and the highest number of differentially expressed transcripts was observed at 20 days p.i. (Figure 1B). Furthermore, MDTH, a metric conveying the magnitude of deviation from baseline for each list of significant genes by time point, confirmed that the greatest gene expression perturbations occur 20 days p.i. (Figure 1C).
To identify functional components of the transcriptional host response, we investigated clusters of differentially expressed transcripts with similar expression patterns across time points using K-means clustering (Kn=20) (Figure 1A,D). Pathway analysis (MetaCore) was used to determine functional annotations that identified ontologies for 12 of the 20 clusters (Table S I). A cluster activity score for each cluster was determined to assess the dynamic changes over time. (Figure 1A,D). Clusters exhibited both mono and bi-phasic activity with peak cluster activity observed between 20–56 days p.i. Each cluster reached maximum change from baseline at either day 20 or 30 p.i. Compared to baseline, increased cluster activity was observed with Interferon response (C1), Inflammation (C2), Innate/Platelet response (C3), Haematopoiesis (C4) and Complement/lymphocyte regulation (C6) clusters, while Ribosomal translation/Lymphocyte (C14), Lymphocyte related clusters (C15, C16 & C17) and those clusters related to Metabolism/Transport (C18), Cell cycle (C19) and Cytoskeletal remodeling (C20) (Figure 1D) had decreased activity. By 90–120 days p.i, the whole blood signature returned to near baseline expression levels.
In a complementary approach, aimed at gaining increased biologic resolution and greater downstream interpretability, we performed an independent analysis using a pre-existing human modular gene expression framework (24, 25) to assess changes in transcript abundance following Mtb challenge (Figure 1E). This analysis facilitates the direct comparison of this macaque Mtb signature and previously described human signatures (3) using the same array platform and a similar module-based analysis approach. In this analysis the differential expression of 14,424 gene probes constituting the module framework were assessed comparing pre- and post-infection time points for each module of the co-expressed transcript (% up or down). One-way hierarchical clustering was used to cluster modules with similar modular activity patterns over time (Figure 1E). Consistent with the k-means cluster analysis, the greatest number and intensity of module activity (overexpressed or under-expressed) peaked between 20–56 days p.i. Similarly, the modules that represent innate immune responses were significantly over-expressed from baseline and those that represent adaptive immune responses were under-expressed in p.i time points. In contrast to the cluster analysis, modular framework analysis revealed modest modular activity (both over and under expression) in the late phase (150–180 days p.i.), especially in modules representing erythropoiesis/haematopoiesis, protein synthesis, and adaptive immune responses.
Published blood transcriptional studies suggested that transcript abundance often reflects the changes in cellular composition of the peripheral blood occur after infection (3, 10, 12, 26–30). In blood, a significant increase in monocyte numbers was observed at 30 days p.i and neutrophil numbers at 20 and 30 days p.i compared to baseline (Figure S2A-B) that positively correlated with increases in transcriptional activity of clusters (and modules) related to interferon, inflammation, and innate immune response (monocytes, neutrophils, myeloid lineage) (Figure S2 A-B and Table S II). A reduction in lymphocyte numbers positively correlated with reduced transcriptional activity of modules and clusters relating to lymphoid lineage, cytotoxic/NK T cells, T cell and B (Figure S2C and Table S II). Thus, the dynamic changes in the cellular composition of the blood contribute to the total transcriptional abundance. In summary, the early blood signature of Mtb infection in macaques, irrespective of final disease outcome, indicates three distinct phases of transcriptional activity that are composed of dynamic immune related pathways driven by changes in both gene expression and underlying cellular composition of the blood.
Modest signatures differentiate active TB and LTBI at the time of diagnosis
To determine whether there was a differential signature that distinguished active disease and LTBI outcomes, we analyzed whole blood transcriptional signatures of animals at the time of clinical diagnosis. Animals that developed active TB (n=16) were diagnosed between 90–180 days p.i., whereas LTBI (n=22) was declared at 180 days p.i. (4, 15, 19). Significant fold change differences (>1.5) between active TB and LTBI were observed in 109 transcripts, with 84 transcripts up-regulated in active TB (Mann-Whitney un-paired T-test with Benjamini-Hochberg for multiple testing correction) (Figure 2A and B, Table S III). Canonical pathway analysis (IPA Metacore) identified interferon signaling and dendritic cell maturation to be significantly represented in this signature (Figure S3A-B). When two-way hierarchical clustering of these 109 transcripts was performed (Figure 2B), 10 of the 16 animals with active disease clustered together while only 8 of the 22 animals with latent infection clustered together; the remaining 6 animals with active TB and 14 with LTBI were interspersed. This interspersed clustering likely reflects the spectrum of Mtb infection observed in both humans and NHP (2–4).
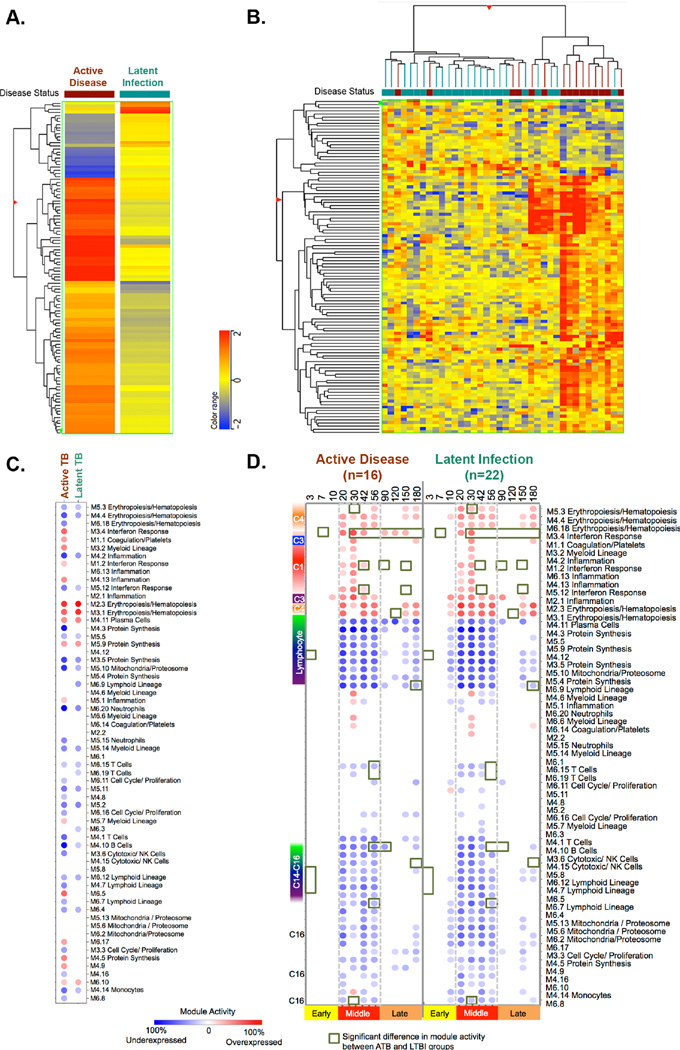
Figure 2. Differential gene expression at the time of clinical diagnosis.
At the time of clinical diagnosis 109 transcripts were differentially expressed between animals with active disease and latent infection (Fold Change >1.5). (A) The mean differential signature was hierarchically clustered based the genes with similar differential expression between active disease and latent infection as clinical groups. (B) Two-way hierarchical clustering was used to cluster animals and transcripts with similar patterns of expression. NHPs with latent infection are marked in green, while those that developed active disease are in maroon (C) Transcripts from NHP at the time of clinical diagnosis were evaluated using a pre-existing human gene expression module framework for animals that either developed active disease (maroon) or remained latently infected (green). (D) Transcripts from NHP were evaluated using a pre-existing human gene expression module frame work for animals that either developed active disease (on the left) or remained latently infected over the course of Mtb infection. Module activity scores representing the percent of genes that are either over-expressed (red) or under-expressed (blue) relative to the pre-infection baseline are calculated. Red color represents over expression compared to pre-infection, whereas blue color represent under expression. The intensity of the color represents the degree of differential expression from baseline. Module activity >15% are visualized. Green squares indicate significant differential expression (Module activity) between animals with active disease and latent infection.
A complementary module based analysis of the transcriptional changes between pre-infection and at the post-infection time of clinical diagnosis (Figure 2C) was performed for animals as clinical groups. The pattern of module activity was overlapped between active disease and latent infection. Nevertheless, when evaluated as clinical group differences in module activity was observed in macaques with different clinical outcomes. Over-expression of interferon response, inflammation, myeloid lineage and coagulation/platelets modules and under expression of T cells, lymphoid lineage, cytotoxic T cells/ NK cells and cell cycle modules were seen in macaques with active disease, compared to those with LTBI.
Active disease signatures in Macaques are similar to those observed in humans
To accurately assess the relationship of macaques signatures at the time of clinical diagnosis to those previously reported in humans with active disease and latent infection, we compared the modular signatures presented here with those derived from Berry et al, 2010. This prior assessment of human TB signatures utilized much of the same sample collection, processing, and analytic infrastructure as the current study thus facilitating this cross-species signature comparison.
Utilizing module based analysis (Figure S4A-B, D-E) we observed high concordance between active TB signatures in both humans (3) and macaque (r=0.72, p <0.001; Figure S4A-B). We further compared and validated gene-level signature of human pulmonary TB (Berry et al. 393 gene set) to the 2,048 transcripts differentially expressed in macaques with active disease compared to pre-infection baseline (Figure S4D-E). This comparative analysis identified 102 of the 393 differentially expressed genes observed in human active TB that are also differentially expressed in the macaque when comparing pre-infection to the signature at the time of clinical diagnosis (closest approximation of sample comparison in human Healthy controls vs. Active TB diagnosis) (Figure S4D). Together these comparative analyses suggest that the macaque signature of active TB infection presented here is homologous to previously validated human signatures of active pulmonary TB. The signature for LTBI in humans and macaques also exhibited some similarities but the lower intensity of transcriptional perturbations from pre-infected and healthy baselines coupled with heterogeneity within the groups yielded lower concordance than that observed when comparing active signatures (Figure S4C).
Modest signatures discriminate active TB and LTBI outcome early in the course of Mtb infection
To determine whether there were transcriptional differences observed at early post-infection time-points, i.e., prior to clinical diagnosis that could distinguish outcome, we evaluated module activity of animals that would develop active TB or LTBI over each of the time points post-infection (Figure 2D). Interestingly, we observed a significant decrease in the activity of modules associated with lymphoid lineage and increased activity of Interferon response at 3 and 7 days p.i respectively (Table S IV) in animals that would develop latent infection compared to active disease. However, by 30 days post infection, the Interferon response becomes significantly higher in animals that would develop active disease and remained higher in later time points. Similarly, modules such as lymphoid lineage, T cell and B cell were significantly higher by 56 days p.i in animals that develop latent infection (Table S IV). This suggests the timing of the innate or adaptive response influences the outcome of Mtb infection. Overall, while these differences in module activity were statistically significant, the magnitude of activity often fell below the 15% minimum cutoff for visualization in Figure 2D, suggesting that the heterogeneity among LTBI and active TB among animals dampens the differential signature. Only modest transcript signatures discriminate infection outcome early in the course of Mtb infection, despite the fact that early differences in disease outcome can be measured by other parameters (e.g., PET-CT)(16).
The degree of lung inflammation correlates with the blood transcriptional signature
Excessive inflammation is associated with poor clinical outcomes in TB (reviewed in(31)). We stratified NHP in this study using a quantitative measure of lung inflammation determined by PET-CT (using 18F-flurodeoxyglucose (FDG) probe). FDG is taken up and retained by metabolically active host cells, including inflammatory cells in the lungs. We and others have previously shown that total FDG avidity in the lungs correlates with Mtb disease severity and bacterial burden, and is reduced upon successful drug treatment (16, 18, 32). We theorized that total lung FDG avidity (measured as standardized uptake volume, SUV) could more accurately reflect the severity of infection, revealing the spectrum of infection. Rather than relying on the binary clinical definitions (active disease or LTBI) this measurement could provide us with a continuous variable to which we could correlate gene expression and module activity. To determine the association and difference in gene expression in animals with high and low lung FDG avidity, we initially classified macaques into two groups: high or lung FDG avidity (described below). However, to determine the degree of gene expression change that correlates with extent of the lung disease, we also used lung FDG avidity as a continuous variable in all animals and also within those with high or low FDG avidity.
The range of total lung FDG avidity for macaques in this study at the time of clinical diagnosis was 0–106 SUV (Figure 3A); the median was 1820 SUV and macaques were classified as low FDG (≤ 1820 SUV) or high FDG (>1820 SUV) (Figure 3A). Most of the animals with active disease (12 of 16) had high lung FDG avidity (>1820) and most animals with latent infection (15 of 22) had low lung FDG avidity (<1820). However, there is overlap of lung inflammation between the clinical classifications of these macaques (Figure 3A), reflecting the spectrum of Mtb infection.
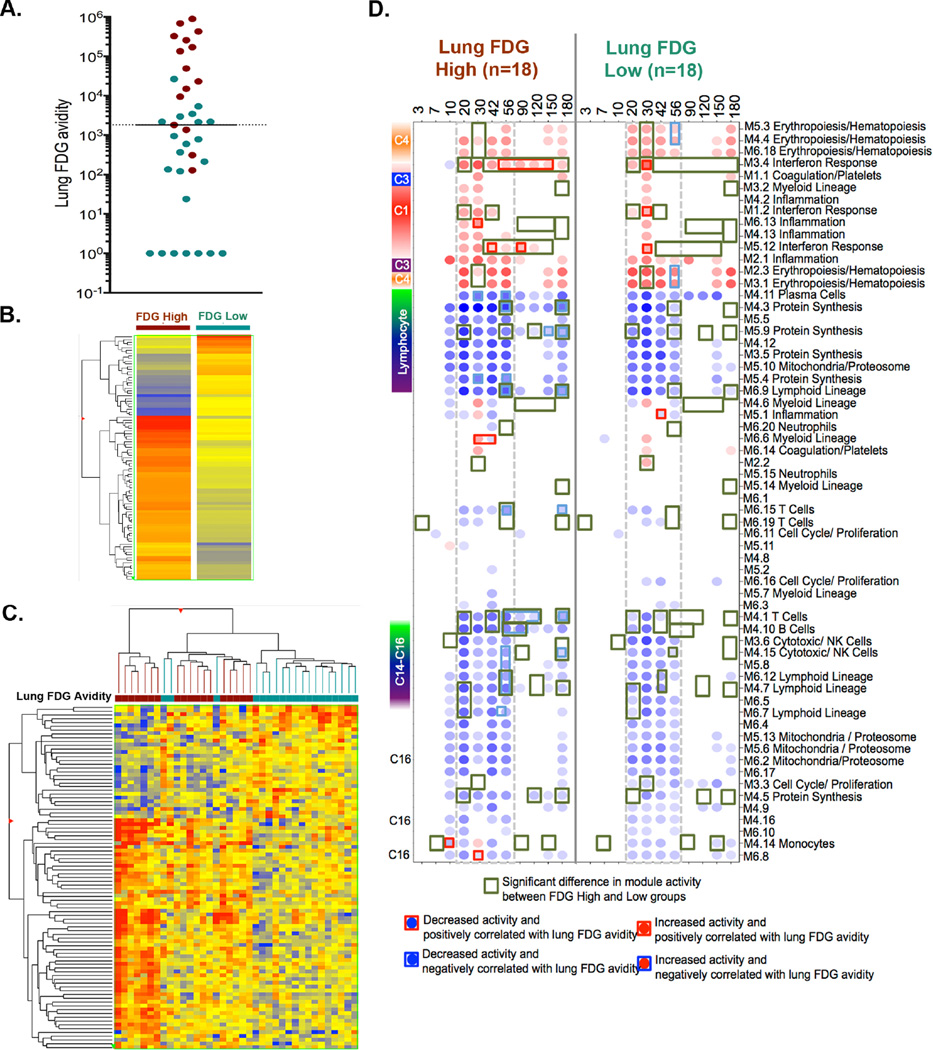
Figure 3. Differential gene expression based on lung inflammation measured by PET-CT.
(A) Lung FDG avidity of 37 animals after diagnosis represented in a log scale. Each symbol represents an animal and the color indicates the clinical status of the animal (maroon=active TB, green=latent infection). Median FDG avidity was 1820 (indicated by a line). We classified animals above the median FDG value as “High” and those at and below as “low” FDG avidity groups for further evaluation. At the time of clinical diagnosis 91 transcripts (Fold Change >1.5) were differentially expressed between animals with high and low lung FDG avidity (B and C). (B) Overall differential signature was hierarchically clustered based on genes with similar differential expression between high and low FDG avidity of the animals. The intensity of the heat map signifies the range of differential expression from blue (under expression) to red (over expression). (C) Differential transcripts were hierarchically clustered based on individual animal and the pattern of expression. NHPs with low FDG avidity are marked in green, while those with high FDG avidity are in maroon. (D) Transcripts from NHP were evaluated using pre-existing human gene expression modular framework for animals with high (on the right) or low (left) lung FDG avidity over the course of Mtb infection. Module activity scores representing the percent of genes that are either over-expressed (red) or under-expressed (blue) relative to the pre-infection baseline are calculated. The intensity of the color represents the degree of differential expression from baseline. Module activities that are significantly different between animals with high and low FDG avidity are marked with green boxes. Modules that are overexpressed and positively correlated with lung FDG avidity are represented as red filled circles with red squares, while those are overexpressed and negatively correlated are represented as red filled circles and a blue squares. Similarly, modules that are under expressed and positively correlated are represented by blue filled circles with red squares, and those that are under expressed and negatively correlated are represented by blue filled circles with blue squares.
First, we determined whether there was a differential signature between animals that have high and low lung FDG avidity at the time of clinical diagnosis. Ninety-one transcripts were differentially expressed (fold change >1.5) between animals with high versus low lung FDG avidity (Figure 3 B-C, Table S III). In contrast to differential signatures based on clinical status (active vs LTBI), only 3 animals with low lung FDG avidity clustered with animals with high FDG avidity (Figure 3C) in two-way hierarchical clustering. The Ingenuity canonical pathway that was most dominant based on lung inflammation in these 91 differential transcripts was TREM1 signaling (Figure S3C), which was previously reported humans (27).
Secondly, we investigated whether any differential signatures were observed earlier in the course of Mtb infection that reflect lung FDG avidity measured at the time of diagnosis. Animals classified as high or low FDG avidity in lungs at clinical diagnosis (Figure 3D, S5A-B and Table S IV) had many differences in transcriptional activity over the course of Mtb infection using module based functional analysis. At 10 days p.i there was higher B cell (M4.10) activity in animals with low FDG avidity (Figure S5B). After 20 days p.i., animals with high lung FDG avidity had significantly higher Interferon response, Inflammation, Myeloid lineage and neutrophils modules while animals with low lung FDG avidity had significantly higher T cell, B cell, cytotoxicity/NK cells, lymphoid lineage and protein synthesis modules at various p.i time points (Figure 3D, S5 A-B and Table S IV). Of note, interferon response modules were the only dominant differential signatures observed 42 and 150 days post infection in animals with high lung FDG avidity. Overall, we observed a higher number of modules that are differentially activated between high and low lung FDG avidity (71 vs 27 modules) compared to the classification based on clinical status, suggesting that evaluating Mtb infection based on the total lung inflammation may more closely reflect the changes in whole blood transcript abundance.
To evaluate the association between the degree of gene expression and the extent of lung disease, by using the lung FDG avidity as a continuous variable, we determined correlation between module activity and lung FDG avidity within the high and low FDG groups and also overall correlation with all animals. Multiple modules exhibited significant correlation with lung FDG avidity, as highlighted in Figure 3D (Table S V). For example, in the low lung FDG avidity animals, interferon modules (M3.4 and M5.12) are overexpressed at 30 days p.i and positively correlated with lung FDG avidity at the time of clinical diagnosis. In the animals with high lung FDG avidity, positive correlation with Interferon modules is seen later in infection (i.e., 42–150 days p.i.). At days 56–120 p.i., T (M4.1) and B (M4.10) cell modules had decreased activity and negatively correlated with lung inflammation in high FDG avidity macaques, while no changes with these modules were observed in PET low animals. Overall the timing of the correlation and their respective modules vary between high and low lung FDG avidity groups (Figure S5). We also investigated overall association module activity and the lung FDG avidity of all animals as one group. Overall, significant negative correlation was observed in modules associated with B cells, T cell, cytotoxic/ NK cells, lymphoid lineage and erythropoiesis from day 10–180 p.i. Positive correlation was observed in modules associated interferon, inflammation, monocytes, myeloid lineage post day 20 p.i (Figure S5C and Table S V). Some of the correlations observed when analyzed as high or low groups were no longer observed in the combined evaluation. This could be due to the spectrum of module activity amongst each macaque (Figure 3D and S5 A-B).
Higher interferon and inflammation signatures before infection are associated with poor outcome
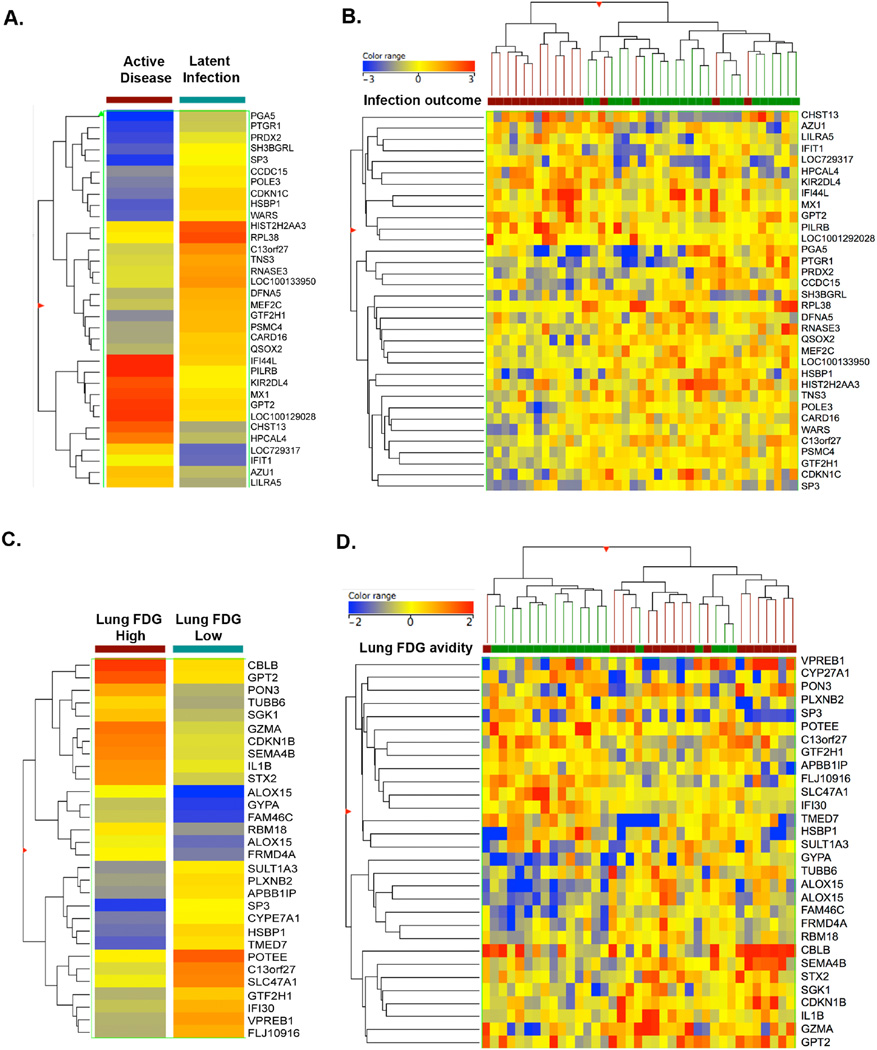
Our study also provides an opportunity to examine whether a pre-infection immune status (signature) in blood is associated with outcome. We first evaluated differences in transcripts at the pre-infection time point between animals that would later develop active disease or latent infection. Thirty-four pre-infection transcripts were differentially regulated (defined as >1.5 fold change, Mann-Whitney un-paired T-test with Benjamini-Hochberg correction) between those animals that would develop active disease or LTBI (Table S VI). Of these, 12 transcripts were up-regulated and 22 down-regulated in animals that would eventually present with active disease, compared to those animals that would develop LTBI (Figure 4A-B). The majority of these up-regulated transcripts are associated with interferon, cell cycle and inflammation functions. Stratifying outcome based on lung FDG avidity identified 30 pre-infection transcripts (14 down-regulated, 16 up-regulated) that were different between high and low FDG avidity animals (Figure 4C-D and Table S VI). Only 5% overlap was found with the specific genes that are differentially regulated between the groups of either clinical status or lung FDG avidity, but the pathways associated with the genes are similar. In both the clinical outcome and FDG avidity analysis, the majority of the differential transcripts belong to pathways associated with interferon and inflammation, suggesting that the host’s inherent up-regulation of these pathways may predispose to poor infection outcome.
Figure 4. Differential gene expression among active and latent outcomes observed before Mtb infection.
At baseline, 34 transcripts were differentially expressed between animals that developed active disease or latent infection (A &B) (Fold Change >1.5). (A) The mean differential signature before Mtb infection was hierarchically clustered based on the genes with similar differential expression between animals that later developed active disease and latent infection as clinical groups. (B) Two-way hierarchical clustering was used to cluster animals and transcripts with similar patters of expression to be clustered together. Green depicts latently infected NHP while maroon depicts animals with active TB. Similarly, 30 transcripts were differentially expressed at baseline between animals that had high or low lung FDG avidity (C and D). (C) Overall differential signature was hierarchically clustered based the genes with similar differential expression between high and low FDG avidity of the animals. The intensity of the heat map shows the range of under expression (blue) and over expression (red). (D) Differential transcripts were hierarchically clustered based on individual animal and the pattern of expression. NHPs with low FDG avidity are marked in green, while those with high FDG avidity are in maroon.
Discussion
Using a unique animal model (NHP) that mimics human Mtb infection in pathology and variability of outcome, we are able to detect time specific, transcriptional changes in the blood before and during the course of low dose Mtb infection. We show that the greatest fluctuation at the transcript level occurs early during infection (days 20–56), long before Mtb infected animals develop overt clinical symptoms of TB. The greatest change is observed among innate and adaptive pathways and this correlates to the time of initial detection of a systemic inflammatory response (measured by erythrocyte sedimentation rate) (Figure S1C), initiation of adaptive immunity, and initial lung granuloma formation seen by PET-CT (16). Early innate transcriptional changes are temporally associated with increased monocytes and neutrophils circulating in the blood. Similarly, adaptive response transcripts are down-regulated when lymphocytes are reduced in the blood at the early time points, likely due to lymphocytes trafficking to the lung as infection is established. The early signatures within 6 months p.i. seen here are consistent with the human signatures in adults seen during diagnosis of clinical disease, which occurs at much later time points (33). In fact, for the IFN and inflammation pathways, signatures prior to infection were associated with a poor outcome, suggesting that these pathways could contribute to increased susceptibility to TB disease. These data also suggest that the signatures in the human literature that distinguish active TB may not necessarily reflect the direct host response to Mtb infection but rather the predisposition of outcome.
Multiple studies reported up-regulated IFN-inducible genes (type I & II) as the most prominent signature associated active TB. Signatures including myeloid and inflammatory transcripts, Fc-gamma receptor signaling, JAK-STAT pathway, complement, pattern recognition receptors, antigen presentation, B cell markers and CD64 were also shown to be associated with active TB (3, 29, 33–37). An independent integration and meta-analysis of eight independently obtained human TB microarray datasets revealed that the genes associated with pattern recognition receptor signaling, Fc receptors, fibrosis myeloid cell inflammation, and TREM1 signaling were strongly associated with active TB, in addition to T cells, B cells and interferon signaling (27). Our dataset confirms that IFN signaling was the most significant pathway associated with active disease. However, when the macaques were evaluated by the extent of lung inflammation (lung FDG avidity), TREM 1 represented a top canonical pathway analysis with highest log p-value, in concordance with the meta-analysis of human datasets (27).
At the time of clinical diagnosis in macaques, 109 transcripts were differentially expressed between animals with active disease and LTBI that overlapped with 24 transcripts reported in the meta analysis of human TB studies (27). While only 20% of the differentially expressed individual molecules in NHP overlapped with human signatures, the pathways associated with these molecules are similar to those observed human studies (27, 34). The major component of the active TB disease signature in NHP is interferon signaling. Similarly, module based analysis, using the same platform as a previously published human study (3) gave us similar trends in modular activity between animals with active TB and latent infection as humans (Figure S4). Only 4 of the 38 modules between NHP and human with active disease were discordant from human signature and this could be due to differences in infection duration. Active TB in humans can be more severe (requiring medical attention in a resource limited area) and likely more prolonged than NHPs that are regularly monitored. Further, in the NHP model, animals are declared to have latent infection at 6 months post infection whereas humans were likely infected for years and possibly decades. Thus the differences in transcripts between macaques and humans likely reflect more extreme differences in human clinical presentation. Recently, a 16-gene signature was reported to determine risk of TB disease in a prospective cohort of human adolescents (14). We had 43% overlap of this signature in our dataset, and 4 of these were differentially regulated between active disease and latent infection.
Two-way hierarchical clustering of individual NHPs based on the differential transcripts revealed interspersed co-clustering among both active and latently infected animals (Figure 2B, 2G). Similar findings were reported in transcriptional signatures in humans with Mtb infection (3, 34) and likely could reflect the spectrum of Mtb infection in both humans and macaques by pathology (4). Similarly, a combined evaluation as a clinical outcome group revealed only modest differences between the signatures, with a number of modules with <15% activity, suggesting the larger heterogeneity within the clinical outcome groups might hinder the detection of differential expression. Given the overlap observed in clinically defined outcomes, we took advantage of a PET-CT determined measurement of inflammation, total lung FDG avidity, which is positively correlated with bacterial burden (16) and indicates disease severity in this model. Unsupervised cluster analysis of transcripts based on fold change provided some separation between groups classified by clinical data (active disease versus LTBI), while many animals were interspersed. In contrast, a similar analysis using lung FDG avidity to classify the outcome resulted in fewer misclassifications. Further, the intensity of the differential signature was much greater when analyzed by lung FDG avidity than by the clinical outcome. This suggests that lung inflammation quantified by PET-CT FDG avidity may be a more definitive method of estimating disease severity along a spectrum rather than the subjective, variable and binary clinical assays used to designate active and latent outcomes.
Limitations of this study include the use of a human array for hybridization of our NHP samples. While this platform has been used in other macaque studies (38–40), there are sequence homology differences between NHP and humans that reduce total probe hybridization and therefore reduce the strength of transcriptional signature to only modest levels compared to human. Even with a relatively large cohort of 38 macaques the variability within the stratified responses limited our ability to perform classical predictive analysis using independent training and test sets. Therefore, we performed combined analyses to increase the statistical power for interpreting results instead of having small cohorts as test and validation sets. We present here only an overview of the transcriptional profiles during the course of Mtb infection but not an in-depth analysis of specific transcripts associated with infection outcome, as we were not currently able to validate these findings using independent test and validation sets or by RT-PCR.
While we did not observe a definitive signature for clinical outcome early after infection using our clinical classifications of infection outcome, several key findings were made at the early time points using lung FDG avidity as an outcome measure of disease that may warrant further evaluation. In addition, the role of pre-existing interferon activity and inflammation in the evolution of Mtb infection warrants further evaluation. To our knowledge, this is the only longitudinal study of Mtb infection with multiple early time points. We also used a pre-existing human expression module framework for better interpretation of the results in comparison to the existing human TB data. We used lung FDG avidity (PET-CT) for inferring inflammation in the lung and disease severity which is more sensitive than using chest X-rays. Finally, we show that our NHP model not only recapitulates human TB with clinical and radiological presentation, but also in the transcriptional signature of active disease.
Supplementary Material
Acknowledgments
Acknowledgements and funding: We are grateful to veterinary technicians Melanie O’Malley and Paul Johnston for sample collection and veterinary care, Jaime Tomko, Dan Fillmore, and L. James Frye for performing PET-CT scans, Pauline Maiello and M. Teresa Coleman for their PET-CT analysis, and to the former and current members of the Flynn and Lin Laboratories for their intellectual contributions. We would also like to acknowledge the efforts of the Baylor Institute for Immunology Research Genomics Core including Parvathy Vinod and Phuong Nguyen at the for their assistance in sample preparation and Rahul Maurya for microarray processing.
Funding: This study was supported by the following grants from the NIH (JLF: HL106804, HL110811; PLL: AI111871) and the Bill and Melinda Gates Foundation (PLL, JLF).
References
- 1.WHO. Global Tuberculosis Report 2015. World Health Organisation; 2016. [Google Scholar]
- 2.Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O'Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin PL, Rodgers M, Smith L, Bigbee M, Myers A, Bigbee C, Chiosea I, Capuano SV, Fuhrman C, Klein E, Flynn JL. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect Immun. 2009;77:4631–4642. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin PL, Flynn JL. Understanding latent tuberculosis: a moving target. J Immunol. 2010;185:15–22. doi: 10.4049/jimmunol.0903856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin PL, Maiello P, Gideon HP, Coleman MT, Cadena AM, Rodgers MA, Gregg R, O'Malley M, Tomko J, Fillmore D, Frye LJ, Rutledge T, DiFazio RM, Janssen C, Klein E, Andersen PL, Fortune SM, Flynn JL. PET CT Identifies Reactivation Risk in Cynomolgus Macaques with Latent M. tuberculosis. PLoS Pathog. 2016;12:e1005739. doi: 10.1371/journal.ppat.1005739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadena AM, Flynn JL, Fortune SM. The Importance of First Impressions: Early Events in Mycobacterium tuberculosis Infection Influence Outcome. MBio. 2016;7 doi: 10.1128/mBio.00342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisson-Dupuis S, Bustamante J, El-Baghdadi J, Camcioglu Y, Parvaneh N, El Azbaoui S, Agader A, Hassani A, El Hafidi N, Mrani NA, Jouhadi Z, Ailal F, Najib J, Reisli I, Zamani A, Yosunkaya S, Gulle-Girit S, Yildiran A, Cipe FE, Torun SH, Metin A, Atikan BY, Hatipoglu N, Aydogmus C, Kilic SS, Dogu F, Karaca N, Aksu G, Kutukculer N, Keser-Emiroglu M, Somer A, Tanir G, Aytekin C, Adimi P, Mahdaviani SA, Mamishi S, Bousfiha A, Sanal O, Mansouri D, Casanova JL, Abel L. Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol Rev. 2015;264:103–120. doi: 10.1111/imr.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flynn JL, Gideon HP, Mattila JT, Lin PL. Immunology studies in non-human primate models of tuberculosis. Immunol Rev. 2015;264:60–73. doi: 10.1111/imr.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Garra A. Systems approach to understand the immune response in tuberculosis: an iterative process between mouse models and human disease. Cold Spring Harb Symp Quant Biol. 2013;78:173–177. doi: 10.1101/sqb.2013.78.020172. [DOI] [PubMed] [Google Scholar]
- 11.O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 12.Blankley S, Berry MP, Graham CM, Bloom CI, Lipman M, O'Garra A. The application of transcriptional blood signatures to enhance our understanding of the host response to infection: the example of tuberculosis. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130427. doi: 10.1098/rstb.2013.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bloom CI, Graham CM, Berry MP, Wilkinson KA, Oni T, Rozakeas F, Xu Z, Rossello-Urgell J, Chaussabel D, Banchereau J, Pascual V, Lipman M, Wilkinson RJ, O'Garra A. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One. 2012;7:e46191. doi: 10.1371/journal.pone.0046191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff TH, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J, Shankar S, Parida SK, Kaufmann SH, Walzl G, Aderem A, Hanekom WA Acs, groups GCcs. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016 doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capuano SV, 3rd, Croix DA, Pawar S, Zinovik A, Myers A, Lin PL, Bissel S, Fuhrman C, Klein E, Flynn JL. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–5844. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman MT, Maiello P, Tomko J, Frye LJ, Fillmore D, Janssen C, Klein E, Lin PL. Early Changes by (18)Fluorodeoxyglucose positron emission tomography coregistered with computed tomography predict outcome after Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2014;82:2400–2404. doi: 10.1128/IAI.01599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coleman MT, Chen RY, Lee M, Lin PL, Dodd LE, Maiello P, Via LE, Kim Y, Marriner G, Dartois V, Scanga C, Janssen C, Wang J, Klein E, Cho SN, Barry CE, 3rd, Flynn JL. PET/CT imaging reveals a therapeutic response to oxazolidinones in macaques and humans with tuberculosis. Sci Transl Med. 2014;6:265ra167. doi: 10.1126/scitranslmed.3009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin PL, Coleman T, Carney JP, Lopresti BJ, Tomko J, Fillmore D, Dartois V, Scanga C, Frye LJ, Janssen C, Klein E, Barry CE, 3rd, Flynn JL. Radiologic responses in cynomolgous macaques for assessing tuberculosis chemotherapy regimens. Antimicrob Agents Chemother. 2013 doi: 10.1128/AAC.00277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin PL, Pawar S, Myers A, Pegu A, Fuhrman C, Reinhart TA, Capuano SV, Klein E, Flynn JL. Early events in Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2006;74:3790–3803. doi: 10.1128/IAI.00064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pawar SN, Mattila JT, Sturgeon TJ, Lin PL, Narayan O, Montelaro RC, Flynn JL. Comparison of the effects of pathogenic simian human immunodeficiency virus strains SHIV-89.6P and SHIV-KU2 in cynomolgus macaques. AIDS Res Hum Retroviruses. 2008;24:643–654. doi: 10.1089/aid.2007.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 22.Banchereau R, Jordan-Villegas A, Ardura M, Mejias A, Baldwin N, Xu H, Saye E, Rossello-Urgell J, Nguyen P, Blankenship D, Creech CB, Pascual V, Banchereau J, Chaussabel D, Ramilo O. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 2012;7:e34390. doi: 10.1371/journal.pone.0034390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankla R, Buddhisa S, Berry M, Blankenship DM, Bancroft GJ, Banchereau J, Lertmemongkolchai G, Chaussabel D. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol. 2009;10:R127. doi: 10.1186/gb-2009-10-11-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, Bennett L, Allantaz F, Mejias A, Ardura M, Kaizer E, Monnet L, Allman W, Randall H, Johnson D, Lanier A, Punaro M, Wittkowski KM, White P, Fay J, Klintmalm G, Ramilo O, Palucka AK, Banchereau J, Pascual V. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, Edens M, Cepika AM, Acs P, Turner J, Anguiano E, Vinod P, Khan S, Obermoser G, Blankenship D, Wakeland E, Nassi L, Gotte A, Punaro M, Liu YJ, Banchereau J, Rossello-Urgell J, Wright T, Pascual V. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell. 2016;165:1548–1550. doi: 10.1016/j.cell.2016.05.057. [DOI] [PubMed] [Google Scholar]
- 26.Berry MP, Blankley S, Graham CM, Bloom CI, O'Garra A. Systems approaches to studying the immune response in tuberculosis. Curr Opin Immunol. 2013;25:579–587. doi: 10.1016/j.coi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Joosten SA, Fletcher HA, Ottenhoff TH. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PLoS One. 2013;8:e73230. doi: 10.1371/journal.pone.0073230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joosten SA, Goeman JJ, Sutherland JS, Opmeer L, de Boer KG, Jacobsen M, Kaufmann SH, Finos L, Magis-Escurra C, Ota MO, Ottenhoff TH, Haks MC. Identification of biomarkers for tuberculosis disease using a novel dual-color RT-MLPA assay. Genes Immun. 2012;13:71–82. doi: 10.1038/gene.2011.64. [DOI] [PubMed] [Google Scholar]
- 29.Maertzdorf J, Ota M, Repsilber D, Mollenkopf HJ, Weiner J, Hill PC, Kaufmann SH. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS One. 2011;6:e26938. doi: 10.1371/journal.pone.0026938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, Walzl G, Kaufmann SH. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12:15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 31.Kaufmann SH, Dorhoi A. Inflammation in tuberculosis: interactions, imbalances and interventions. Curr Opin Immunol. 2013;25:441–449. doi: 10.1016/j.coi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Chen RY, Dodd LE, Lee M, Paripati P, Hammoud DA, Mountz JM, Jeon D, Zia N, Zahiri H, Coleman MT, Carroll MW, Lee JD, Jeong YJ, Herscovitch P, Lahouar S, Tartakovsky M, Rosenthal A, Somaiyya S, Lee S, Goldfeder LC, Cai Y, Via LE, Park SK, Cho SN, Barry CE., 3rd PET/CT imaging correlates with treatment outcome in patients with multidrug-resistant tuberculosis. Sci Transl Med. 2014;6:265ra166. doi: 10.1126/scitranslmed.3009501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y, Xu Z, Wilkinson KA, Wilkinson RJ, Kendrick Y, Devouassoux G, Ferry T, Miyara M, Bouvry D, Valeyre D, Gorochov G, Blankenship D, Saadatian M, Vanhems P, Beynon H, Vancheeswaran R, Wickremasinghe M, Chaussabel D, Banchereau J, Pascual V, Ho LP, Lipman M, O'Garra A. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cliff JM, Kaufmann SH, McShane H, van Helden P, O'Garra A. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol Rev. 2015;264:88–102. doi: 10.1111/imr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, Mollenkopf HJ, Ziegler A, Kaufmann SH. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med (Berl) 2007;85:613–621. doi: 10.1007/s00109-007-0157-6. [DOI] [PubMed] [Google Scholar]
- 36.Kaforou M, Wright VJ, Oni T, French N, Anderson ST, Bangani N, Banwell CM, Brent AJ, Crampin AC, Dockrell HM, Eley B, Heyderman RS, Hibberd ML, Kern F, Langford PR, Ling L, Mendelson M, Ottenhoff TH, Zgambo F, Wilkinson RJ, Coin LJ, Levin M. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med. 2013;10:e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mistry R, Cliff JM, Clayton CL, Beyers N, Mohamed YS, Wilson PA, Dockrell HM, Wallace DM, van Helden PD, Duncan K, Lukey PT. Gene-expression patterns in whole blood identify subjects at risk for recurrent tuberculosis. J Infect Dis. 2007;195:357–365. doi: 10.1086/510397. [DOI] [PubMed] [Google Scholar]
- 38.Lu YR, Wang LN, Jin X, Chen YN, Cong C, Yuan Y, Li YC, Tang WD, Li HX, Wu XT, Li YP, Wang L, Cheng JQ. A preliminary study on the feasibility of gene expression profile of rhesus monkey detected with human microarray. Transplant Proc. 2008;40:598–602. doi: 10.1016/j.transproceed.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 39.Nieto-Diaz M, Pita-Thomas W, Nieto-Sampedro M. Cross-species analysis of gene expression in non-model mammals: reproducibility of hybridization on high density oligonucleotide microarrays. BMC Genomics. 2007;8:89. doi: 10.1186/1471-2164-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skinner JA, Zurawski SM, Sugimoto C, Vinet-Oliphant H, Vinod P, Xue Y, Russell-Lodrigue K, Albrecht RA, Garcia-Sastre A, Salazar AM, Roy CJ, Kuroda MJ, Oh S, Zurawski G. Immunologic characterization of a rhesus macaque H1N1 challenge model for candidate influenza virus vaccine assessment. Clin Vaccine Immunol. 2014;21:1668–1680. doi: 10.1128/CVI.00547-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.