SUMMARY
The unprecedented 2013–16 outbreak of Ebola virus (EBOV) resulted in over 11,300 human deaths. Host resistance to RNA viruses requires RIG-I-like receptor (RLR) signaling through the adaptor protein, mitochondrial antiviral signaling (MAVS), but the role of RLR-MAVS in orchestrating anti-EBOV responses in vivo is not known. Here, we apply a systems approach to MAVS−/− mice infected with either wild-type or mouse-adapted EBOV. MAVS controlled EBOV replication through expression of IFNα, regulation of inflammatory responses in the spleen, and prevention of cell death in the liver, with macrophages implicated as a major cell-type influencing host resistance. A dominant role for RLR signaling in macrophages was confirmed following conditional MAVS deletion in LysM+ myeloid cells. These findings reveal tissue-specific MAVS-dependent transcriptional pathways associated with resistance to EBOV, and demonstrate that EBOV adaptation to cause disease in mice involves changes in two distinct events, RLR-MAVS antagonism and suppression of RLR-independent IFN-I responses.
Graphical Abstract
INTRODUCTION
In 2013, the largest recorded outbreak of Ebola virus disease (EVD) caused by Ebola virus (EBOV) began in Guinea and spread to neighboring countries including Sierra Leone, Liberia, Mali, Nigeria and Senegal. Over 28,000 people are estimated or confirmed to have been infected, with more than 11,300 deaths (World Health Organization, 2016) and an estimated cost of $3.6 billion USD in international efforts to stop the outbreak (USAID, 2016). EVD is characterized by rapid and systemic virus replication associated with dysregulated innate and adaptive immune responses. Disease progression is due to cytolytic damage from virus replication, and induction of a massive inflammatory response that leads to vascular leakage and hemorrhage, liver damage, and multi-organ failure associated with a septic shock-like syndrome (Feldmann and Geisbert, 2011). However, little is known regarding the precise role of specific innate signaling pathways in determining the outcome of infection. Following RNA virus infection, viral RNA is recognized by endosomal toll-like receptors (TLRs) or by the RIG-I like receptors (RLR), retinoic acid-inducible gene-I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) (Loo and Gale, 2011). The RLR pathway in particular is implicated as being important to EBOV resistance because EBOV-encoded antagonists of type I interferon (IFN-I) responses are directed towards multiple factors in this pathway (Messaoudi et al., 2015). Activated RIG-I or MDA5 interact with the adaptor protein, mitochondrial antiviral signaling protein (MAVS). Activated MAVS recruits key kinases, IκB kinase epsilon (IKKε) and TANK-binding kinase 1 (TBK1), that activate transcription factors, NFκB and interferon regulatory factor (IRF)-3 and IRF-7. Nuclear translocation of these transcription factors drives expression of pro-inflammatory cytokines and early IFN molecules including IFNβ. Secreted IFN-I signals through the cognate receptor (IFNAR) to activate Janus kinase and signal transducer and activation of transcription (JAK/STAT) signaling and further upregulate expression of IFNs and IFN-stimulated genes (ISGs), the latter of which are responsible for the biological effects of IFN-I (MacMicking, 2012).
The high levels of EBOV replication are thought to be a direct result of a comprehensive and potent ability of the virus to antagonize host antiviral responses, particularly the production and signaling of IFN-I. This is mediated by two viral IFN antagonists, VP35 and VP24. VP35 blocks production of IFN-I by multiple mechanisms at the level of RIG-I activation and downstream of this by inhibiting IKKε and TBK1 activation as well as IRF3 and IRF7 (reviewed in (Messaoudi et al., 2015)). VP24 inhibits signaling through the IFNAR by preventing nuclear localization of STAT1 following IFN stimulation of infected cells that in turn inhibits ISG upregulation (Reid et al., 2007). Recombinant EBOV containing mutations in VP35 grows normally in Vero cells defective for IFN production, but is attenuated in mouse or guinea pig models of infection (Hartman et al., 2008; Prins et al., 2010), demonstrating a critical role for IFN antagonism in virus virulence. This antagonism of RLR sensing implicates MAVS-dependent pathways of IFN production as critical for limiting virus infection. However, the precise contribution of MAVS signaling in coordinating antiviral and inflammatory responses to EBOV infection in vivo is not known.
Although non-human primates (NHP) are regarded as the gold standard animal model for EBOV pathogenesis, mouse models have clear advantages in BSL4 containment including relative ease of handling and genetic manipulation (Bradfute et al., 2012). However, these models also have limitations in that immunocompetent adult mice are resistant to infection with wild-type EBOV (WT-EBOV). WT-EBOV causes lethal disease in mice deficient for IFNAR (Bray, 2001) or STAT1 (Raymond et al., 2011), again implicating the host IFN response as a critical determinant of resistance to infection. Mouse-adapted EBOV (MA-EBOV) causes disease in some mouse strains such as Balb/c wherein it is associated with increased virus replication and induction of inflammatory cytokines (Bray, 2001; Gibb et al., 2001). Infection with MA-EBOV can recapitulate the hemorrhagic manifestations of disease observed in humans or NHP, dependent on the mouse genotype (Rasmussen et al., 2014). Therefore, MA-EBOV infection of mice provides a useful model to understand the contribution of various immune responses and genetic diversity in control of virus replication that cannot readily be studied in NHPs or other models. MA-EBOV differs from WT-EBOV at 13 nucleotide positions, which includes mutations in both VP35 and VP24 (Ebihara et al., 2006). However, mutations conferring virulence in mice map to the nucleoprotein (NP) and VP24, but not VP35. In addition, adaptation of VP24 does not confer an increased ability to antagonize murine IFN signaling (Reid et al., 2007). Thus, the basis of mouse adaptation is not fully understood.
In the present study, we determined the contribution of MAVS signaling to control of both WT-EBOV and MA-EBOV in a mouse model using transcriptional profiling of two target organs, the spleen and liver. Virulence of MA-EBOV was linked to two separate events: suppression of MAVS-dependent signaling as well as increased antagonism of MAVS-independent IFN-I. Furthermore, examination of cell-type specific gene signatures suggested that RLR signaling associated with host resistance to EBOV affected macrophages, T cells, and plasmacytoid dendritic cells (pDCs). We also generated mice with a conditional deletion of MAVS. Infection of mice deleted for MAVS in LysM-positive macrophages/monocytes confirmed a dominant role of RLR sensing specifically in myeloid cells in host resistance to lethal EBOV infection.
RESULTS
RLR signaling restricts EBOV replication, controls expression of IFNα, and reveals differences in pathogenesis of WT-EBOV and MA-EBOV
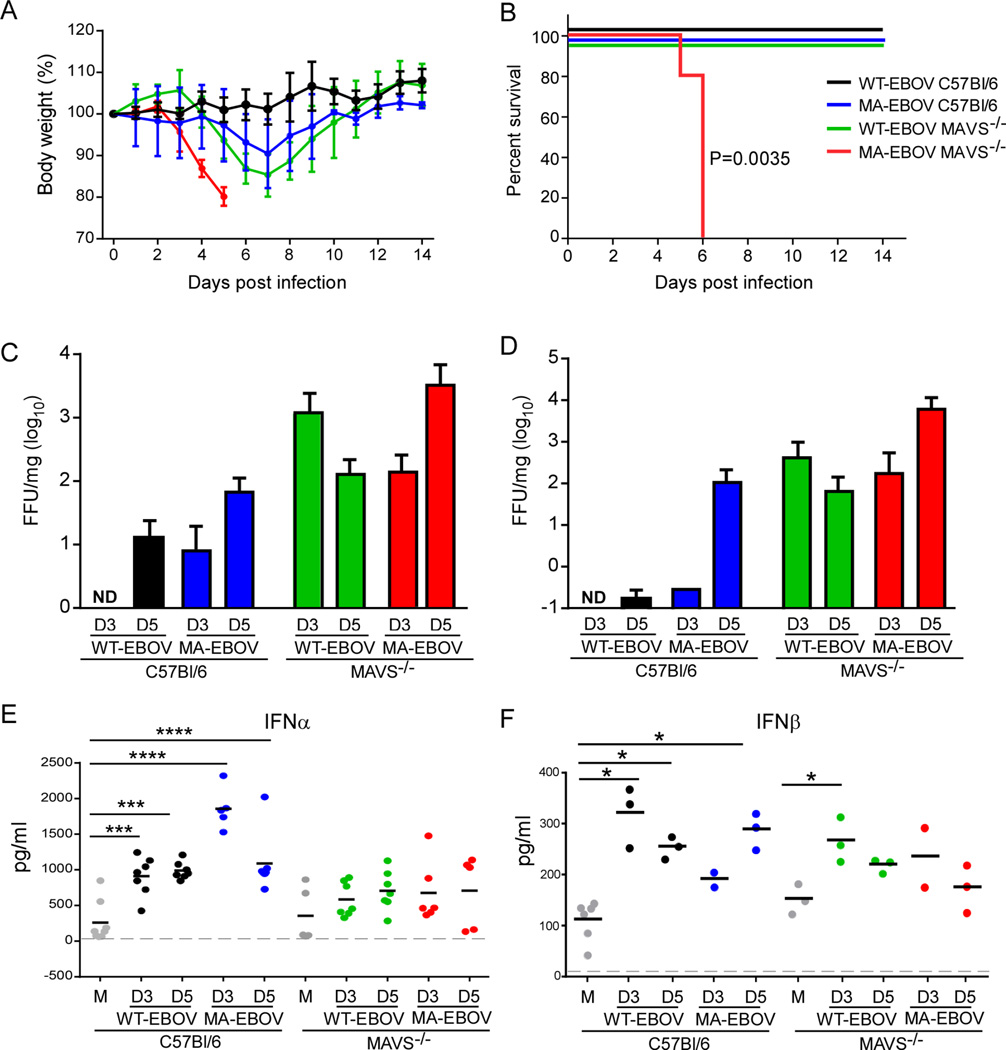
To evaluate the role of MAVS in control of EBOV, C57Bl/6 or MAVS−/− mice were inoculated intraperitoneally with 102 focus forming units (ffu) of either WT-EBOV or MA-EBOV and monitored for weight loss (Figure 1A) and survival (Figure 1B). C57Bl/6 mice were resistant to infection with EBOV as WT-EBOV did not cause any weight loss while MA-EBOV-infected mice lost some body weight (~8%) but survived. MAVS-deficient mice infected with WT-EBOV lost considerable (~15%) weight but also recovered. In contrast, MAVS−/− mice infected with MA-EBOV reached the pre-determined criteria for euthanasia including 20% weight loss by 5–6 days post infection (dpi) (Figure 1B). Virus titer was examined in two target organs (Gibb et al., 2001), the spleen (Figure 1C) and liver (Figure 1D). As expected, MA-EBOV replicated to higher titers than WT-EBOV in C57Bl/6 mice. Deletion of MAVS enabled greater replication of both viruses but interestingly WT-EBOV replicated to equal levels as MA-EBOV in the liver of C57Bl/6 mice at 3 dpi, suggesting that early differences between WT- and MA-EBOV replication are dependent on MAVS signaling. However, even in the absence of MAVS, control of WT-EBOV replication was apparent by 5 dpi consistent with mouse survival, suggesting that MAVS-independent pathways are important for late control of WT-EBOV in mice. In contrast, MAVS was required for late control of MA-EBOV replication as titers continued to increase by 5 dpi. These results demonstrate the importance of RLR signaling in host resistance to EBOV infection and establish models to examine correlates of MAVS-dependent and –independent resistance.
Figure 1. MAVS is required for control of EBOV replication and resistance associated with production of IFNα.
(A) Percent change in body weight following infection with WT-EBOV or MA-EBOV (mean ± SD). (B) Survival curves of C57Bl/6 or MAVS−/− mice infected with 100 ffu WT-EBOV or MA-EBOV. N=5 per group from 2 experiments. P value of Kaplan-Meier survival curve analysis is indicated. (C–D) Virus titer in spleen (C) and liver (D) from mice infected with WT-EBOV or MA-EBOV and euthanized at day 3 (D3) or day 5 (D5) post infection. Data is represented as mean ± SD from 3–5 mice per group from 2 experiments. ND, none detected. (E–F) IFNα or IFNβ concentrations in the sera of mice measured by ELISA (n=3–7 from 2 experiments performed). M, mock infected. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 determined by one-way ANOVA with Dunnett’s multiple comparison test. Dashed line indicates limit of detection.
To determine the consequence of RLR signaling to EBOV-induced production of IFN-I, IFNα (subtypes IFN-α1, -α2, -α4, -α5 and -α6) and IFNβ were measured in the serum. IFNα induced by both WT- and MA-EBOV was highly dependent on signaling through MAVS (Figure 1E), despite the higher virus burden in MAVS−/− mice that would result in more viral RNA available for sensing by MAVS-independent means. MAVS was not required for early (3 dpi) serum IFNβ levels in WT-EBOV infected mice. However, the IFNβ concentration tended to be lower in MAVS−/− mice infected with MA-EBOV despite uncontrolled virus replication (Figure 1F). Thus, RLR signaling represents a dominant pathway in vivo for production of IFNα throughout infection and for efficient production of IFNβ. However, additional pattern recognition receptors (PRRs) such as TLRs likely contribute to production of IFNβ in response to EBOV.
MAVS-dependent signaling in the spleen is associated with control of liver pathology
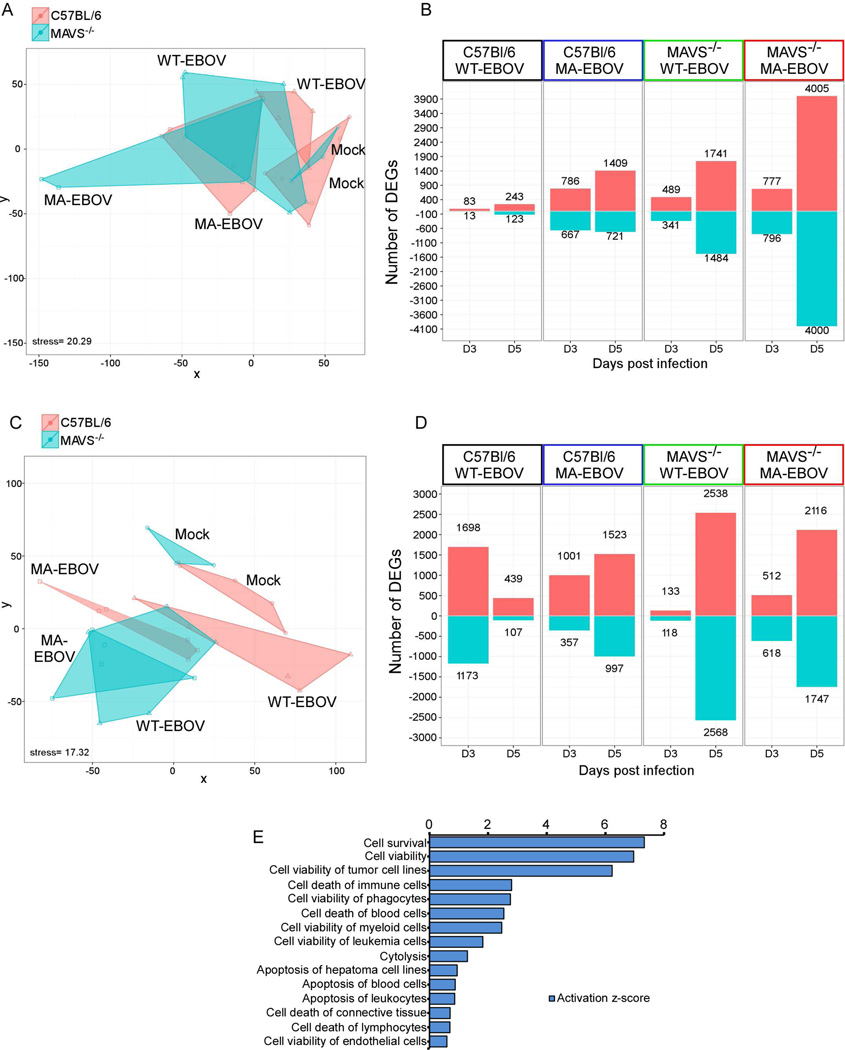
Mouse whole-genome microarrays were used to profile global gene expression changes at 3 and 5 dpi in spleen and liver from C57Bl/6 and MAVS−/− mice infected with WT-EBOV or MA-EBOV. Differentially expressed genes (DEG) (defined as ≥1.5-fold change in expression with paired Student's t-test P≤0.01) were identified by comparing infected spleen or liver to that of strain- and age-matched mock-infected controls. Multidimensional scaling (MDS) used to visualize the transcriptomic profile demonstrated significant overlap in the liver between all conditions with resolution of each transcriptomic profile restricted to one dimension (virus infection) (Figure 2A). Nevertheless, gene expression profiles associated with the pathogenic outcome of weight loss fell in-between profiles of lethal outcome (MA-EBOV infected MAVS−/− mice) at one extreme and mock-infected at the other (Figure 2A). In contrast, transcriptomic profiles in the spleen were both significantly different between conditions and diverged on two dimensions according to virus strain (WT- or MA-EBOV) and the presence of MAVS (Figure 2C), a finding supported by overall higher numbers of total DEG in infected spleen in each condition compared to the liver (Figure 2B and D). However, spleen DEG profiles in WT- and MA-EBOV-infected mice converged in the absence of MAVS (Figure 2C), suggesting that host-specific interactions of these viruses occurring early in the spleen are highly dependent on MAVS signaling.
Figure 2. Global analysis of the transcriptional profile of WT-EBOV and MA-EBOV infections, see also Figure S1.
(A) Multidimensional scaling (MDS) representation of the similarities in transcriptional profiles elicited by viral infection over time in liver. Each biological replicate is represented by shape denoting virus infection and color denoting mouse strain. The quality of the representation is provided by the Kruskal stress value, with a low percentage of Kruskal stress (15.02%) suggesting a faithful two-dimensional representation of global transcriptional differences between viral strains. (B) Most highly DEGs following virus infection relative to time-matched mocks in the liver. DEG cutoff was set to a fold change >1.5 and a q-value < 0.05 calculated using a moderated t test with subsequent Benjamini-Hochberg correction. The number of DEGs is indicated in each direction. (C) MDS representation and (D) DEGs in the spleen of the same animals depicted in (A) and (B). (E) Biofunction enrichment analysis of 4000 DEG in liver of MA-EBOV-infected MAVS−/− mice. Based on Fisher's exact test performed with IPA software, nonredundant biological functions with the top 10 enrichment scores (given by −log10 P values) or absolute activation Z-score values are shown. Bar graphs represent the activation z- scores for each biofunction (bar length) and the number of target genes associated with each biofunction.
Robust changes in transcriptional responses were observed in spleens of C57Bl/6 mice infected with WT-EBOV (survival, no weight loss) at 3 dpi that were markedly reduced by 5 dpi (Figure 2D). This early response in the spleen was of lower magnitude in C57Bl/6 mice infected with MA-EBOV, or in mice deficient in MAVS. Indeed, 90% of the transcriptional response to WT-EBOV infection in the spleen associated with resistance at 3 dpi was dependent on MAVS (Figure 2D). In contrast to the spleen, the changes in DEG in the liver of these mice were relatively small (Figure 2B). DEGs in the liver were lowest in C57Bl/6 mice infected with WT-EBOV (resistant), and highest in MA-EBOV infected MAVS−/− mice (greatest susceptibility) (Figure 2B). To gain further insight into these processes, we analyzed 4000 DEG in MA-EBOV infected liver of MAVS−/− mice at 5 dpi by Ingenuity Pathway Analysis (IPA) (Figure 2E). Functional categories enriched among DEGs (activated or repressed) were dominated by genes involved in cell survival, particularly pathways associated viability or apoptosis of myeloid cells, endothelial cells and hepatic cells. Together, these findings suggest that gene expression in the spleen (and possibly other lymphoid organs) is associated with early MAVS-dependent responses that dictate resistance to EBOV. In contrast, DEGs in the liver are a direct indication of severity of disease and pathologic outcome following EBOV infection in mice.
To determine if the functional categories of DEG reflect the pathology associated with infection, we evaluated histology (Figure S1) and EBOV antigen (Figure 3 and Figure S2) in the spleen and liver. Splenic changes in C57Bl/6 mice infected with either MA- or WT-EBOV were limited to minimal to mild necrosis of lymphocytes within the white pulp that did not progress over time (data not shown). In contrast, the liver of C57Bl/6 mice infected with MA-EBOV developed multifocal lesions characterized by necrosis of individual hepatocytes bounded by infiltrates of neutrophils and macrophages that increased in number over time producing moderate pathologic change in all mice (Figure S1). MAVS−/− mice infected with either WT- or MA-EBOV developed significant multifocal hepatic lesions at 3dpi similar to those observed in MA-EBOV-infected C57Bl/6 mice. By 5 dpi MAVS−/− mice infected with MA-EBOV had large, coalescing foci of hepatocellular necrosis infiltrated by small to moderate numbers of neutrophils and macrophages (Figure S1). WT-EBOV-infected MAVS−/− mice had less hepatocellular necrosis with a more robust inflammatory infiltration of neutrophils and macrophages (data not shown).
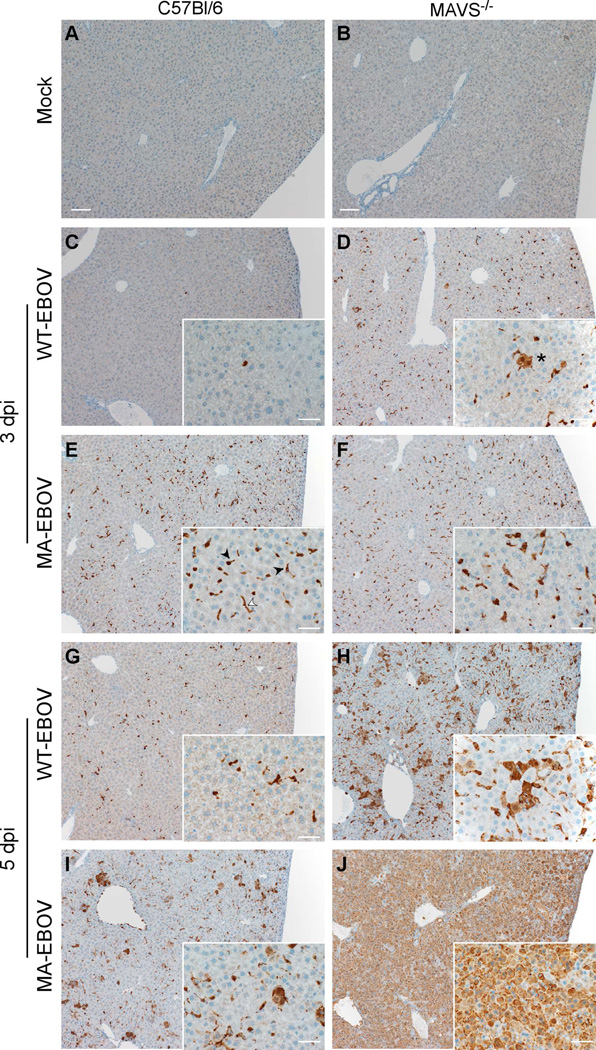
Figure 3. EBOV VP40 antigen in liver of C57Bl/6 and MAVS−/− mice, see also Figure S2.
The presence of EBOV VP40 was examined by immunohistochemistry in C57Bl/6 or MAVS−/− mice that were (A–B) mock infected (scale bars=100µm), (C–D) infected with WT-EBOV at 3 dpi, (E–F) infected with MA-EBOV at 3 dpi, (G–H) infected with WT-EBOV at 5 dpi, or (I–J) infected with MA-EBOV at 3dpi. Insets (scale bars=20µm) show greater detail of infected cells. In all conditions, endothelial (white arrowhead) and Kupffer cells (black arrowheads) contain viral antigen. In the absence of MAVS, hepatocytes (asterisk) also contain EBOV VP40. Data are representative of 5 mice examined from 2 experiments performed.
In the absence of MAVS, WT-EBOV antigen in the spleen was indistinguishable from MA-EBOV at 3dpi (Figure S2). Antigen of both WT-EBOV and MA-EBOV was reduced at 5 dpi in spleens of C57Bl/6 mice, but increased in MA-EBOV MAVS−/− mice, consistent with mouse survival. In the liver, MA-EBOV infection resulted in large amounts of viral antigen present predominantly in endothelial cells and Kupffer cells; rare hepatocytes were also positive particularly at 5 dpi (Figure 3). Conversely, WT-EBOV infected C57Bl/6 mice displayed very little viral antigen at 3 dpi that was present almost exclusively within Kupffer cells. Viral antigen increased slightly by 5 dpi to include endothelial cells. The distribution and number of EBOV-positive cells was similar in MAVS−/− mice infected with either WT- or MA-EBOV at 3 dpi, and included endothelial cells, Kupffer cells and some hepatocytes (Figure 3). By 5 dpi, EBOV-positive cells had greatly increased, the majority of which were hepatocytes. However, only MA-EBOV-infected MAVS−/− mice had VP40 in nearly every hepatocyte in the tissue section. Together, these results provide histopathologic support for the DEG data by suggesting that early differences between WT- and MA-EBOV are MAVS-dependent. However, late control of WT-EBOV evident by 5 dpi is MAVS-independent.
RLR signaling controls expression of ISGs and inflammatory responses to EBOV infection
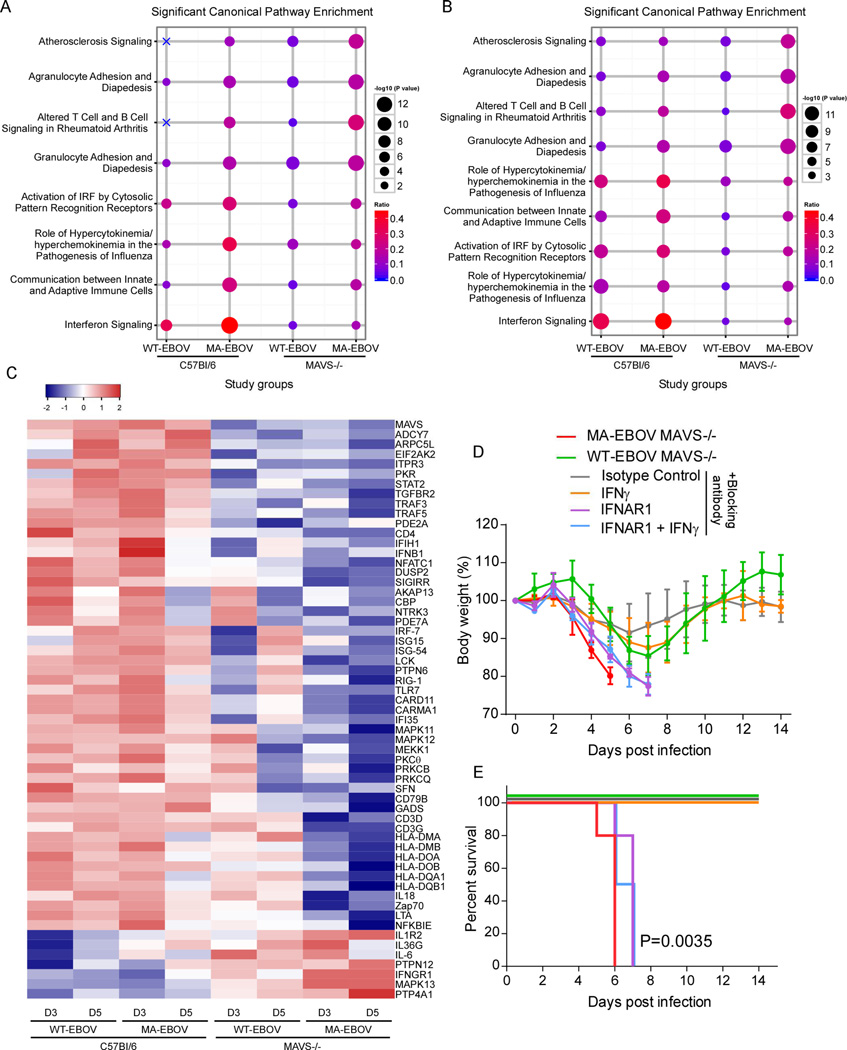
To further understand the role of RLR signaling in host resistance to EBOV, canonical pathway enrichment analysis was performed using IPA of DEG in the spleen at both 3 dpi (Figure 4A) and 5 dpi (Figure 4B). Infection of C57Bl/6 mice with either WT- or MA-EBOV induced gene expression associated with innate immune responses, including pathways of IFN signaling, activation of IRF by cytosolic PRRs, cytokine responses, and communication between innate and adaptive immune cells. These responses were generally higher in C57Bl/6 mice infected with MA-EBOV at 3 dpi, presumably due to the greater replication of this virus compared to WT-EBOV.
Figure 4. Canonical pathway analysis of DEGs, see also Figure S3.
Canonical pathway enrichment analysis of DEGs in spleen in all infection conditions on (A) day 3 and (B) day 5 p.i. Canonical pathways with the top five enrichment scores (given by the −log10 P value) in at least one gene list are shown (based on Fisher's exact test, performed with IPA software). The size of each circle is proportional to the −log10 P value, and the color gradient represents the relative frequency of DEG compared to the total number of genes associated with each pathway. X's indicate that the respective pathway had no enrichment for the DEG list of interest. (C) Hierarchical clustering of DEG derived from the canonical pathways in Figure S3A between spleen from WT and KO infected mouse strains. Gene expression is shown as log10 (ratio) of EBOV-infected to strain-matched mock-infected mice. 60 DEGs whose expression was significantly induced over strain-matched mock-infected mice are depicted in the heat map (1.5 fold; P≤0.01). Clustering was performed using Pearson correlation. (D–E) MAVS−/− mice were administered neutralizing antibody to IFNα/β, IFNγ or both one day prior to infection with WT-EBOV and again at 2–3 dpi. Mice treated with anti-IFNγ antibody alone did not succumb to infection and so were administered a third dose of antibody or isotype control at 6 dpi to no effect. The percent change in body weight (mean ± SD) and the survival curves are shown for these mice (N=4–5 per group). P value of Kaplan-Meier survival curve analysis is indicated.
In the absence of MAVS, several pathways were enriched leading to or involving the inflammatory response, including granulocyte and agranulocyte adhesion and diapedesis, atherosclerosis signaling, and altered T and B cell signaling in rheumatoid arthritis (Figure 4A and B). These pathways were also enriched in the spleen of C57Bl/6 mice infected with MA-EBOV at 5 dpi. However, inflammatory pathway enrichment was significantly higher in MA-EBOV-infected MAVS−/− mice as indicated by a higher activation z-score (Figure 4B). These results are consistent with increased virus replication in the absence of RLR signaling driving deleterious inflammatory responses. However, it is also possible that RLR signaling in specific cell types may be required more directly to adequately control inflammatory responses to infection.
To identify individual genes whose expression was regulated by RLR signaling, comparative analysis of activation z scores of canonical pathways of all infection conditions was performed in IPA. In C57Bl/6 mice, upregulated genes were involved in antiviral innate immune responses and T cell responses, including activation of IRF by cytosolic receptors, TLR signaling, role of NFAT, CD28 signaling in T helper cells, PKCθ signaling in T lymphocytes, NFκB signaling and protein kinase A signaling (Figure S3A). Sixty individual genes in these pathways were significantly altered by MAVS deletion, which included a failure to upregulate canonical ISG expression (including IFNβ, PKR, STAT2, IRF7, ISG15, ISG54, IFI35, RIG-I, IFIH1 [Mda5]) in mice infected with either WT-EBOV or MA-EBOV (Figure 4C). However, ISG expression was markedly lower in MA-EBOV-infected MAVS−/− mice that eventually succumb to infection than following WT-EBOV infection that was not lethal, suggesting that mouse adaptation of EBOV also involves altered virus interactions with MAVS-independent responses. In contrast, a number of ISGs associated with inflammation including IL-6, the IL-1 receptor (IL1R2), and NFκB were significantly elevated in the absence of MAVS (Figure 4C), confirmed by both IL-6 and IL-1β protein in the serum of infected mice (Figure S3B and S3C). Thus, RLR signaling has a dominant role in positive regulation of intrinsic antiviral responses to EBOV infection, but may also negatively regulate inflammatory responses, either directly or indirectly through the control of virus replication.
To further understand why MAVS−/− mice survive infection with WT-EBOV, we examined the promotor region of DEGs between WT-EBOV and MA-EBOV in the absence of MAVS. The majority of these genes have STAT1 or IRF-1 binding elements within the 1500 nucleotides upstream of the +1 start site (Figure S3D), suggesting that they are ISGs that could be regulated by type I or type II IFN. Although IFNα expression following infection was dependent on MAVS, production of IFNβ was only partially MAVS-dependent (Figure 1). Alternatively, IFNγ has demonstrated anti-EBOV activity (Rhein et al., 2015), and promotes expression of MHC molecules that were a major class of DEG. Thus, mouse survival was examined in the presence of neutralizing antibodies to either IFNAR1, IFNγ or both (Figure 4D and 4E). MAVS−/− mice treated with anti-IFNAR antibody succumbed to WT-EBOV infection within a day of untreated mice infected with MA-EBOV whereas anti-IFNγ treated mice survived. Thus, MAVS-independent production of IFNα/β, but not IFNγ, was responsible for survival of WT-EBOV-infected MAVS−/− mice. Taken together, this work demonstrates that differences between MA- and WT-EBOV include two separable events: 1) greater suppression of MAVS-dependent responses by MA-EBOV, and 2) increased resistance of MA-EBOV to the antiviral effects of MAVS-independent IFN-I.
RLR signaling determines susceptibility to EBOV infection by promoting macrophage/monocyte recruitment and activation
To determine the cellular basis of host susceptibility, we first examined serum cytokines, which revealed two patterns of expression. First, cytokines associated with resistance to disease were expressed at relatively high levels in EBOV-infected C57Bl/6 mice at both 3 and 5 dpi, and included GM-CSF, RANTES, IL-10, IFNγ, IL-12p70 and IL-12p40 (Figure S4A). These cytokines were also expressed at high levels in MAVS−/− mice infected with WT-EBOV but only at 5 dpi with the exception of GMCSF and IL-12p70 that were MAVS-dependent. Second, cytokines associated with lethal infection exhibited highly elevated levels in MA-EBOV-infected MAVS−/− mice at 3dpi followed by a sharp drop in expression by 5 dpi (Figure S4B). These included KC (the mouse homolog of human IL-8), G-CSF, MCP-1, IL-5, and IL-1α. A high early inflammatory response is also associated with lethal infection in humans (Prescott et al., 2017). MIP1α, MIP1β and TNFα had no obvious association with disease phenotype (Figure S4C). These cytokine profiles raise the possibility that early monocyte and T-cell responses are important for resistance, whereas strong early cytokine responses associated with activation of epithelial cells, endothelial cells, neutrophils or eosinophils may be associated with a lethal outcome in mice.
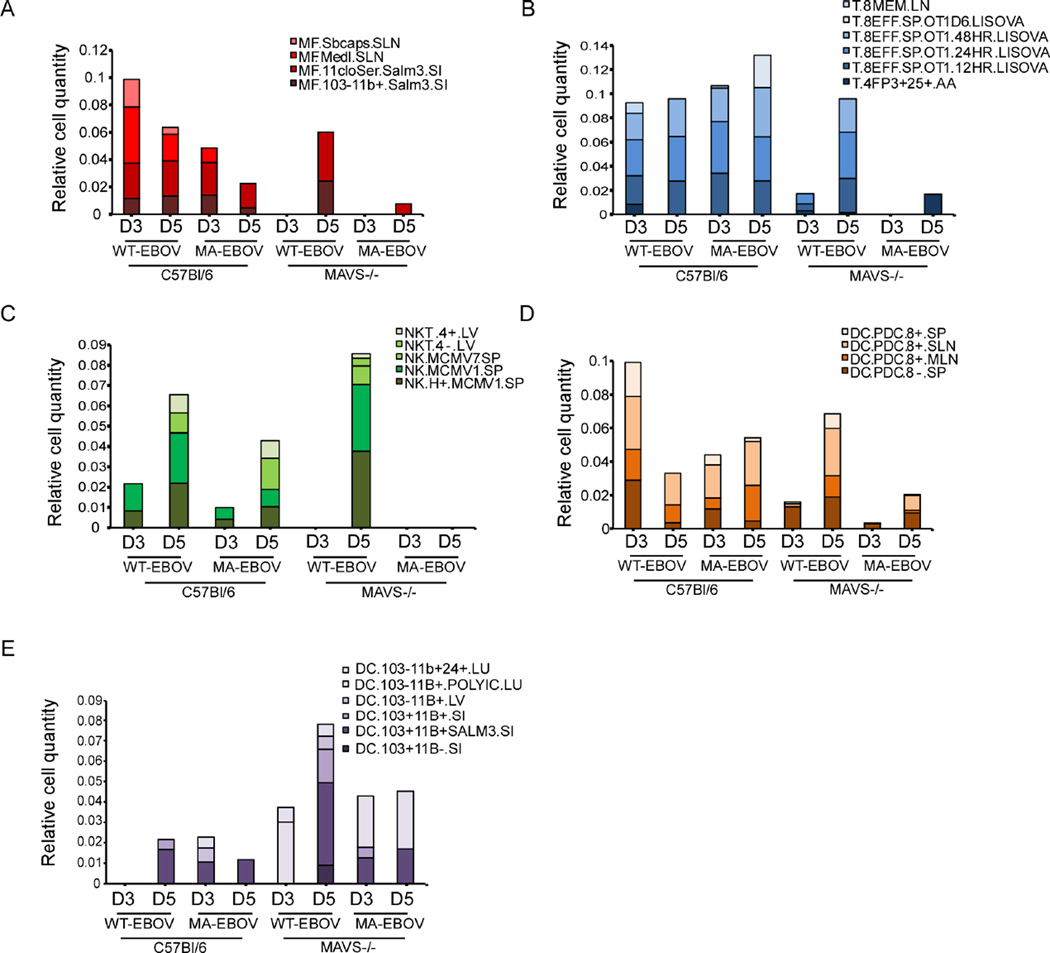
To identify immune cells that influence the outcome of EBOV infection, we used the digital cell quantifier (DCQ) algorithm for immune cell deconvolution in the spleen. The DCQ algorithm leverages cell type-specific whole-genome transcriptional expression profiles derived from 207 distinct immune cell types (ImmGen.org) to infer changes in immune cell quantities and their activation state (Heng et al., 2008). This analysis suggested early increases in macrophages (Figure 5A), and CD8 T cells (Figure 5B) following infection of C57Bl/6 mice with either WT- or MA-EBOV that were delayed in WT-EBOV-infected MAVS−/− mice and mostly absent in MA-EBOV-infected MAVS−/− mice. Similar changes in population dynamics were observed for NK T cells (Figure 5C) and plasmacytoid dendritic cells (pDCs) (Figure 5D). Thus, macrophages, CD8 T cells, NK T cells and pDCs are implicated in host resistance to EBOV infection, with transcriptional responses of these cells constituting a major source of DEG observed in the spleen of WT-EBOV-infected C57Bl/6 mice at 3 dpi (Figure 2D). These data are also consistent with serum cytokine expression profiles in Figure S4. No notable changes in T helper cells or B cells were captured throughout the course of infection (data not shown). However, the relative quantity of DCs was higher in the absence of MAVS (Figure 5E), and had an inverse relationship with macrophage recruitment. These results suggest that MAVS controls EBOV replication by promoting the early recruitment and activation of macrophages, CD8 T cells, NK T cells and pDCs, and may suppress recruitment of conventional DCs.
Figure 5. DCQ analysis of immune cell dynamics following EBOV infection and their dependence on MAVS signaling, see also Figure S4.
Immune cell dynamics in infected spleen as predicted by digital cell quantifier (DCQ). Bar graphs show the relative cell quantities for macrophages (A), CD8 T cells (B), NK/NK T cells (C), pDCs (D), and conventional DCs (E) following EBOV infection at 3 or 5 dpi. Distinct colors have been assigned to each immune cell type from the Immgen compendium. Each cell type name is followed by the tissue from which it was previously isolated, abbreviated as follows: BL, blood; BM, bone marrow; LN, lymph nodes; SI, small intestine; PC, peritoneal cavity; SLN, subcutaneous LN; and SP, spleen.
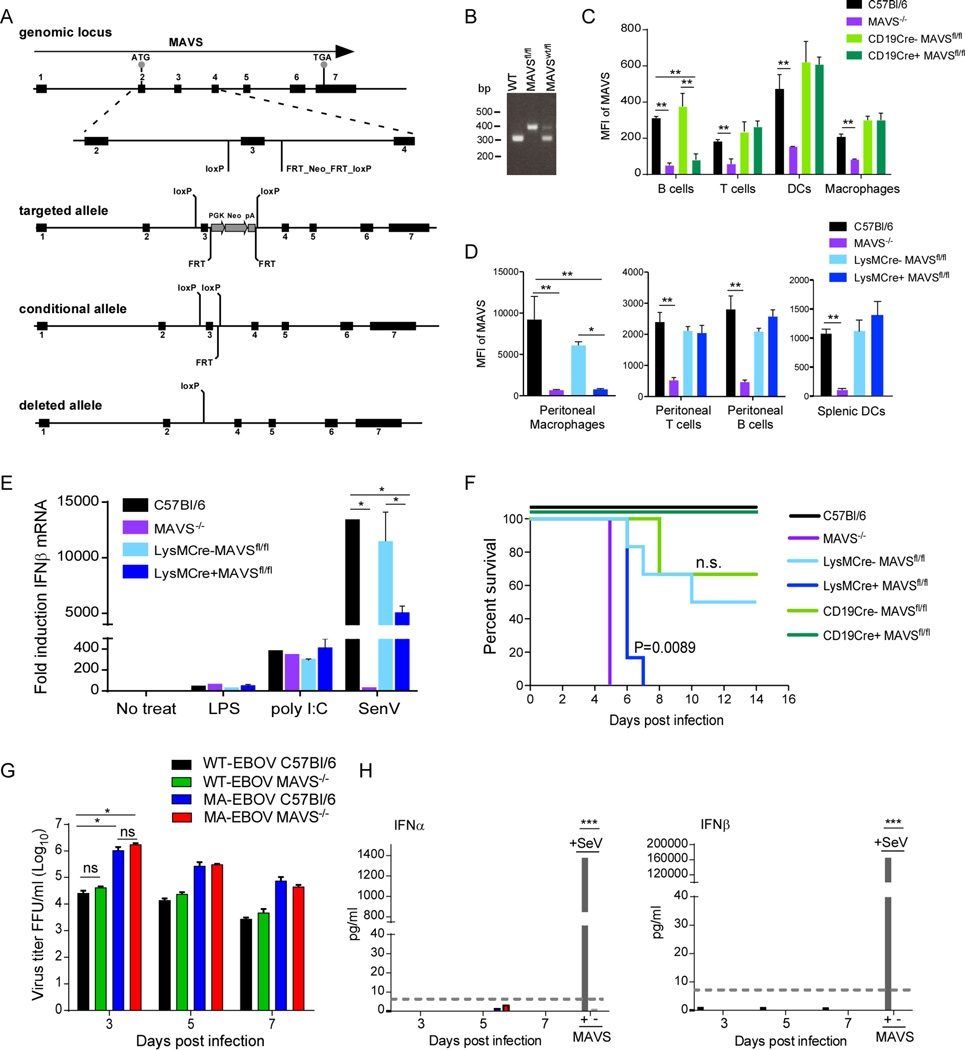
One implication from the DCQ data is that RLR signaling is specifically required in macrophages which, along with DCs, are thought to be the earliest target cells of EBOV infection (Geisbert et al., 2003). Our data also suggests that pDCs are regulated by MAVS signaling, but this is likely to be downstream of another critical cell-type since pDCs do not signal through RLRs and instead rely on TLR responses to produce high levels of IFN-I (Sun et al., 2006). To determine the predictive value of the DCQ data in EBOV infection, we generated mice with a conditional allele of the MAVS gene (Figure 6A and 6B) by flanking exon 3 with LoxP sites (MAVSfl/fl). These mice were crossed with mice expressing either the CD19-Cre or LysM-Cre transgene, specifically expressed in B cells and monocytes/macrophages, respectively, to selectively delete MAVS expression in these cell types (termed CD19Cre+MAVSfl/fl and LysMCre+MAVSfl/fl). Deletion of MAVS from B cells was used to control for any generalized effects on antigen presentation (Hoogeboom and Tolar, 2016). The status of MAVS was determined by intracellular staining for MAVS and flow cytometry (Figure 6C and 6D). In CD19Cre+MAVSfl/fl mice, MAVS was reduced to approximately a third of levels observed in C57Bl/6 B cells, but retained in T cells, DCs and macrophages (Figure 6C). In LysMCre+MAVSfl/fl mice, greater than 90% of MAVS expression was deleted from peritoneal macrophages (Figure 6D) and bone-marrow derived macrophages (BMM) (data not shown). MAVS expression was fully retained in peritoneal T cells, B cells and splenic DCs (Figure 6D). LysM-positive cells include neutrophils, but MAVS expression was not detected in neutrophils from control C57Bl/6 mice (data not shown) suggesting that potential deletion in neutrophils is not a confounding factor. To further characterize LysMCre+MAVSfl/fl mice, BMM were used in a functional assay for MAVS-dependent signaling (Figure 6E). Stimulation with Sendai virus (SeV) that is recognized by RIG-I and MDA-5 (Gitlin et al., 2010) resulted in expression of less than half of IFNβ mRNA in BMM from LysMCre+ MAVSfl/fl mice compared to that induced in BMM from LysMCre-MAVSfl/fl littermates. Induction of IFNβ mRNA expression in response to ligands for TLR4 (LPS) or TLR3 (poly I:C) was not affected in BMM from LysMCre+MAVSfl/fl or LysMCre- MAVSfl/fl mice compared to C57Bl/6 controls, confirming that RLR-independent pathways of IFN-I expression are intact in these cells.
Figure 6. MAVS expression specifically in monocytes/macrophages is required for resistance to infection with MA-EBOV.
(A) Schematic depicting the generation of the MAVS locus with the open reading frame outlined by a black arrow. Exons (1–7) are shown in black boxes. Gray circles indicate the start (ATG) and termination codons (TGA). The insert shows the locations of the loxP sites flanking exon 3 (MAVSfl/fl) in the conditional allele. Cre-recombinase removes exon 3 as depicted in the deleted allele. (B) PCR genotyping of mouse tail DNA. A single primer set was designed to distinguish between the WT (300 bp) and floxed allele (379 bp). (C) Mean fluorescence intensity (MFI) of intracellular MAVS in immune cell types from the spleen of CD19-Cre MAVSfl/fl mice as determined by flow cytometry. One of two experiments is shown. (D) MFI of intracellular MAVS in immune cell types from the peritoneum of LysM-Cre MAVSfl/fl as determined by flow cytometry. (E) IFNβ mRNA expression in BMDM stimulated with LPS, poly I:C or infected with SeV. Expression of IFNβ mRNA was determined by qRT-PCR normalized to the house-keeping gene HRPT and expressed as fold change relative to unstimulated cells. Data is represented as mean ± SD from 3 mice (D–E). (F) Survival curves of MAVS−/−, Cre-MAVSfl/fl littermate controls, LysMCre+MAVSfl/fl, or CD19Cre+MAVSfl/fl mice infected IP with 100 ffu MA-EBOV. N=3–6 per group. P value of Kaplan-Meier survival curve analysis is labeled. (G) Titers of WT-EBOV or MA-EBOV following infection of BMM derived from C57Bl/6 or MAVS−/− mice. (H) IFNα or IFNβ protein in supernatants of BMM shown in (G). SeV infection was used as a positive control for IFN-I induction. Dashed line indicates limit of detection. *P<0.05, **P<0.01, ***P<0.001, ns, not significant.
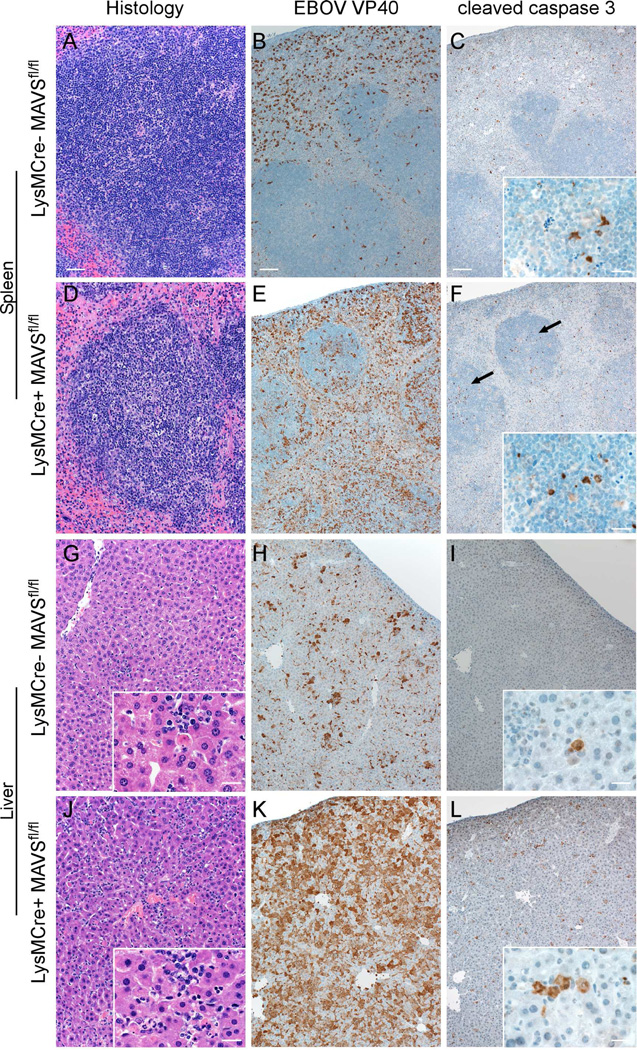
Infection of LysMCre+MAVSfl/fl mice with MA-EBOV, but not CD19Cre+MAVSfl/fl mice, resulted in 100% susceptibility by 6–7 dpi (Figure 6F). Infection of LysMCre- MAVSfl/fl or CD19Cre-MAVSfl/fl littermates showed partial susceptibility, supporting previous findings that disease associated with MA-EBOV is dependent on genetic background (Rasmussen et al., 2014). At 3dpi, both LysMCre- and LysMCre+ mice developed a mild hepatitis composed of multifocal foci of neutrophils and macrophages that occasionally surrounded a single necrotic hepatocyte (data not shown). By 5 dpi, LysMCre- mice had moderate numbers of hepatocytes, endothelial cells and Kupffer cells positive for EBOV VP40 (Figure 7). In contrast, EBOV antigen was present in nearly every hepatocyte in the tissue section in LysMCre+ mice and thus were similar to infected mice with a systemic deficiency of MAVS. Differences in the splenic lesions between LysMCre- and LysMCre+ mice were also significant (Figure 7). At 5dpi, LysMCre+ mice had moderate lymphocytic necrosis of the white pulp whereas LysMCre- mice displayed very little loss of lymphocytes. Large numbers of macrophages and DCs positive for EBOV antigen within the splenic red and white pulp of LysMCre+ mice while LysMCre- mice had considerably less viral antigen within the spleen. Also of note was an increase in cleaved caspase 3 in both the liver and splenic follicles of LysMCre+ mice (as compared to LysMCre- mice, C57Bl/6 mice or MAVS−/− mice), suggesting that the absence of MAVS specifically in myeloid cells resulted in increased apoptosis in both liver and spleen (Figure 7). Importantly, these results support the DCQ data and confirm that MAVS signaling in macrophages/monocytes has a dominant role in host resistance to EBOV.
Figure 7. Pathologic changes, EBOV VP40 antigen and active caspase 3 in the liver and spleen of MA-EBOV infected LysMCre- or LysMCre+ MAVSfl/fl mice at 5 dpi.
Depletion of MAVS in LysM+ myeloid cells resulted in increased depletion of lymphocytes in the spleen (A,D) and increased necrosis of hepatocytes in the liver (G,J). These pathological changes were associated with increased VP40 antigen staining (B,E,H,K) and increased incidence of cleaved caspase 3 (C,F,I,L) in both spleen and lover. Arrows indicate increased cleaved caspase 3 in splenic follicles. Scale bar = 100µm or 20µm (insets).
To determine whether MAVS signaling in macrophages directly controls virus replication, primary macrophages from C57Bl/6 and MAVS−/− mice were infected with WT- or MA-EBOV. As expected, MA-EBOV replicated to higher titers in mouse macrophages compared to WT-EBOV. However, the absence of MAVS did not result in increased infectious titers of either WT- or MA-EBOV. This finding was surprising given the differences in virus titers in the spleen and liver of C57Bl/6 and MAVS−/− mice (Figure 1). The expectation is that C57Bl/6 macrophages would produce higher levels of IFN-I than MAVS-deficient macrophages and release lower levels of infectious virus. However, no IFN-I was made by EBOV-infected macrophages (Figure 6H) in agreement with published work (Albarino et al., 2015). The major implication from this data is that in vivo, MAVS signaling in macrophages/monocytes may not directly control virus replication. Instead, MAVS appears to have an IFN-I-independent function in the regulation of an additional cell type(s) responsible for production of IFN-I that ultimately controls EBOV replication in tissues.
DISCUSSION
We examined transcriptional profiles in four infection conditions that have three distinct phenotypes of host resistance (complete resistance, partial resistance with weight loss, or lethality) to understand the role of RLR sensing following EBOV infection in vivo. These comparisons revealed kinetic changes in transcriptionally regulated antiviral and inflammatory responses in an organ-specific manner and demonstrated both IFN-I-dependent and –independent roles for MAVS signaling. Most strikingly, 90% of the differential expression of over 2500 genes in the spleen of WT-EBOV-infected mice at 3 dpi associated with complete resistance to disease was MAVS-dependent. RLR signaling orchestrated the majority of interferon and pattern-recognition associated gene expression in the spleen, thus demonstrating a dominant role of RLRs in sensing of EBOV infection and IFN-I expression in vivo.
Collectively, our findings suggest a model of early events in the control of EBOV replication and host resistance. In this model, early infection of macrophages/monocytes (and likely DCs) at the inoculation site and subsequent trafficking to lymphoid organs such as the spleen is ideally associated with production of IFNα and ISG expression dependent on RLR signaling. However, the intrinsic antiviral response of these cells may not contribute directly to early control of EBOV due to viral antagonism of RLR signaling and suppression of IFN-I production. Our data also suggest that MAVS itself may also participate in IFN-I-independent control of EBOV infection in tissues. Indeed, MAVS has IFN-I-independent roles in antiviral responses (Olagnier et al., 2014) including ISG expression (Dixit et al., 2010), inflammasome activation (Chakrabarti et al., 2015; Subramanian et al., 2013) and induction of apoptosis (Kumar et al., 2015). In the context of macrophages, MAVS signaling may also participate in cell-to-cell communication through the induction of proinflammatory mediators and promotion of cell-mediated immune responses (Lazear et al., 2013; Suthar et al., 2013). EBOV does not antagonize MAVS directly suggesting that MAVS will be available to signal in infected cells in any pathways independent of the proteins targeted by EBOV (RNA sensing and PACT, IRF3/7, TBK1, IFN-dependent JAK-STAT signaling), such as the inflammasome. In order to recover from infection even when the virus displays high resistance to direct anti-viral effects of IFN-I, MAVS-dependent signaling initiates further cascades of innate and adaptive responses that include NK and NK T cells, CD8 T cells and pDCs. The consequences of failure to induce IFN-I and ISGs include loss of protective macrophage responses, increased inflammatory cytokine production and loss of subsequent cascades of protective innate and adaptive immune cell activation. These events then enable uncontrolled virus replication that spreads to additional cell types and further drives deleterious inflammatory responses and cell death, particularly in the liver.
Despite the dominant role of RLR-MAVS in IFN-I production, this work also demonstrated that MAVS-independent production of IFN-I was sufficient to control replication of WT-EBOV, but not MA-EBOV. This was associated with greater suppression of ISG expression by MA-EBOV suggesting increased resistance of MA-EBOV to IFN-I in vivo. This finding is supported by the observation that MA-EBOV is more resistant to the antiviral effects of IFN-I in mouse macrophages in vitro (Ebihara et al., 2006). Alternatively, MA-EBOV may suppress the generation of the MAVS-independent IFN-I better than WT-EBOV in mice. The source of this IFN-I was not confirmed, but was unlikely to be infected macrophages. The main candidate from our studies is pDCs that do not signal through RLR-MAVS and instead rely on TLR7 signaling to produce large amounts of IFN-I in response to RNA viruses (Swiecki and Colonna, 2015). This may be an important mechanism of host resistance in the context of infection because the role of pDC-derived IFN-I becomes more important in protection from RNA virus infection following elimination of macrophage-derived IFN-I (Kumagai et al., 2007) such as occurs as a result of EBOV antagonism. In addition, pDCs are resistant to EBOV infection (Leung et al., 2011a) and VP35 does not inhibit IFN-I production by pDCs (Leung et al., 2011b), suggesting that these cells could participate in anti-EBOV responses.
The distinct DEG profiles distinguishing WT-EBOV from MA-EBOV infection in the spleen at 3 dpi were largely lost in the absence of MAVS. Thus, an additional major difference between virus variants was increased suppression of MAVS-dependent transcriptional responses in the spleen by MA-EBOV which was directly responsible for lack of viral control. However, the antagonist functions of viral VP35 or VP24 are not different between WT- and MA-EBOV (Ebihara et al., 2006; Reid et al., 2007), suggesting that additional mechanisms of antagonism may exist. It is also possible that virus interactions with individual ISGs that function to directly restrict virus replication in infected cells may differ between these virus variants. The ISGs with the highest differential expression in our study that might be candidates included PKR, ISG15 and ISG54, of which PKR and ISG15 are implicated as having antiviral activity against WT-EBOV (Feng et al., 2007; Malakhova and Zhang, 2008; Okumura et al., 2008). However, VP35 antagonizes PKR (Feng et al., 2007) and the single mutation in VP35 does not strongly influence virulence of MA-EBOV (Ebihara et al., 2006), suggesting that PKR does not specifically influence EBOV host-adaptation.
Additional factors that likely influence control of EBOV in mice include ISGs or other DEGs that control innate cellular responses or communication between innate and adaptive immune responses. Specifically, recruitment or activation of distinct populations of macrophages, NK cells, pDCs and CD8 T cells were delayed in the absence of MAVS following WT-EBOV infection, and were largely absent even at 5 dpi with MA-EBOV. In contrast, transcriptional signatures of conventional DCs were highest in MA-EBOV-infected MAVS−/− mice. Multiple possibilities could explain this result, although the inverse relationship between macrophage and DC populations in the spleen may suggest that differentiation of inflammatory monocytes recruited to sites of infection favors DCs over macrophages under inflammatory conditions in the absence of RLR signaling. These cells may then act as an additional source of infection or drive deleterious inflammatory responses. Alternatively, the lack of certain cell populations such as CD8 T cells may simply reflect cell death. Lethal EBOV infection in humans, NHP and mice is associated with severe loss of lymphocytes by apoptosis (Bradfute et al., 2007; Wauquier et al., 2010), which may underlie the striking lack of representation of the various cell types in MA-EBOV infected MAVS−/− mice.
In summary, the application of a systems biology approach to infection with a host-adapted variant of EBOV has revealed the critical nature of RLR signaling specifically in myeloid cells for antiviral resistance and establishment of cellular immunity. Examining the interplay between MAVS and EBOV demonstrated that host adaptation to cause disease in mice includes two separable events, increased antagonism of MAVS-dependent responses by MA-EBOV, and increased resistance of MA-EBOV to the antiviral effects of MAVS-independent IFN-I. Identification of the cellular source of this IFN-I and whether it operates similarly in other models of infection including NHPs may provide a target for therapeutics to potentiate this response in humans.
EXPERIMENTAL PROCEDURES
Biosafety and biocontainment
Experiments using infectious EBOV were performed in the BSL-4/ABSL-4 research facility at the Rocky Mountain Laboratories (RML), NIAID, NIH. All handling of infectious EBOV including sample inactivation was performed according to standard operating protocols approved by the RML Institutional Biosafety Committee.
Mice and infections
C57Bl/6J mice were purchased from the Jackson Laboratory. MAVS−/− mice on C57Bl/6J background were kindly provided by Dr. Michael Gale (University of Washington) (Suthar et al., 2013). Mice bearing a loxP-flanked MAVS allele (MAVSfl/fl) on a C57Bl/6J background were generated by Ozgene Pty Ltd (Bentley WA, Australia). A conditional allele of mouse MAVS was created by introducing through homologous recombination two loxP sites flanking exon 3 of the gene in C57Bl/6(J) embryonic stem cells, which were then used to generate chimeric mice by standard procedures. The genotypes of offspring mice were determined by PCR analysis of DNA extracted from tail biopsies using Qiagen DNeasy Tissue and Blood Kit. The following primer set was used to distinguish WT-MAVS allele (300bp) and floxed-MAVS allele (394bp) (forward primer: CTTCCTTCACCCTTGGACCTTCT and reverse primer: TGACTGGGTGTAGACTCTGTACT). To obtain conditional knockout of MAVS, homozygous MAVSfl/fl mice were crossed to mice expressing the Cre recombinase gene under control of the LysM promoter (LysMCre) or the CD19 promoter (CD19Cre), both purchased from the Jackson Laboratory (C57Bl/6J mixed with C57Bl/6N). The NIAID Animal Care and Use Committee, as part of the NIH Intramural Research Program, approved all experimental procedures.
Six to 9 week-old male mice (C57Bl/6J, MAVS−/−, CD19Cre+MAVSfl/fl, CD19Cre-MAVSfl/fl LysMCre+MAVSfl/fl, LysMCre-MAVSfl/fl) were used in all experiments. Mice were infected by IP injection with 100 ffu of either MA-ZEBOV or WT ZEBOV diluted in 0.2 ml of Dulbecco’s minimum essential medium (DMEM). Mock-infected mice were IP inoculated with DMEM alone. Mice were weighed and observed for clinical disease at least once daily and twice daily when animals showed onset of clinical signs of disease for 28 days. For survival studies, criteria for euthanasia were ≥20% weight loss generally with signs of ataxia, lethargy, bloody discharge or paralysis. To initially establish this model, we used 1, 10, 100 and 1000 infectious units of MA-EBOV in wild-type and MAVS−/− mice. This did not change the outcome of lethality from that observed with 100 ffu which resulted in the greatest difference in weight loss between MAVS−/− and C57Bl/6 mice and was therefore chosen for the experimental model.
Statistical analysis
Survival curves were analyzed by Kaplan-Meier log-rank test. Multiple comparisons were determined by one-way ANOVA with Dunnett’s multiple comparison post-test. Statistical analysis of microarray data is explained in detail in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
Thank you to Nicki Arndt of the RML Veterinary Branch for assistance with animal husbandry and genotyping, Rebecca Rosenke for immunohistochemistry, Dr. Katy Bosio for providing LysM-Cre mice, Dr. Michael Gale for providing MAVS−/− mice, and Dr. Hideki Ebihara for helpful discussions. This work was supported in part by the Division of Intramural Research, National Institutes of Health, National Institute of Allergy and Infectious Diseases, and by the Center for Research on Diagnostics and Discovery grant AI109761.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Experimental Procedures
Generation of bone marrow-derived macrophages, in vivo neutralization of IFNs, flow cytometry, quantitative RT-PCR, cytokine analysis by Bio-Plex assay and Lumikine ELISA, RNA preparation and oligonucleotide microarray processing, microarray analysis and bioinformatics, and immunohistochemistry are detailed in Supplemental Experimental Procedures.
Author contributions:
Conceptualization, MGK, ALR and SMB; Methodology, MD, SJR, AO, DPS, MGK, ALR and SMB; Investigation, SJR, AO, GLS, FF, EH, AIC, SSP and JDD; Resources, MGK and SMB; Data curation, MD, SJR, AO, JC, JMW, ALR; Writing – original draft, MD, SJR and SMB; Writing –Review and Editing, MD, SJR, MGK, ALR and SMB; Visualization, MD, SJR, AO, DPS, ALR and SMB; Supervision, MGK, ALR and SMB.
Accession Numbers:
Primary data from microarrays are available through the Gene Expression Omnibus (GEO: GSE83309).
References
- Albarino CG, Wiggleton Guerrero L, Lo MK, Nichol ST, Towner JS. Development of a reverse genetics system to generate a recombinant Ebola virus Makona expressing a green fluorescent protein. Virology. 2015;484:259–264. doi: 10.1016/j.virol.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Bradfute SB, Braun DR, Shamblin JD, Geisbert JB, Paragas J, Garrison A, Hensley LE, Geisbert TW. Lymphocyte death in a mouse model of Ebola virus infection. J. Infect. Dis. 2007;196(Suppl 2):S296–S304. doi: 10.1086/520602. [DOI] [PubMed] [Google Scholar]
- Bradfute SB, Warfield KL, Bray M. Mouse models for filovirus infections. Viruses. 2012;4:1477–1508. doi: 10.3390/v4091477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M. The role of the Type I interferon response in the resistance of mice to filovirus infection. J. Gen. Virol. 2001;82:1365–1373. doi: 10.1099/0022-1317-82-6-1365. [DOI] [PubMed] [Google Scholar]
- Chakrabarti A, Banerjee S, Franchi L, Loo YM, Gale M, Jr, Nunez G, Silverman RH. RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe. 2015;17:466–477. doi: 10.1016/j.chom.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara H, Takada A, Kobasa D, Jones S, Neumann G, Theriault S, Bray M, Feldmann H, Kawaoka Y. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog. 2006;2:e73. doi: 10.1371/journal.ppat.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 2003;163:2347–2370. doi: 10.1016/S0002-9440(10)63591-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb TR, Bray M, Geisbert TW, Steele KE, Kell WM, Davis KJ, Jaax NK. Pathogenesis of experimental Ebola Zaire virus infection in BALB/c mice. J. Comp. Pathol. 2001;125:233–242. doi: 10.1053/jcpa.2001.0502. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Benoit L, Song C, Cella M, Gilfillan S, Holtzman MJ, Colonna M. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 2010;6:e1000734. doi: 10.1371/journal.ppat.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of ebola virus. J. Virol. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TS, Painter MW Immunological Genome Project, C. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hoogeboom R, Tolar P. Molecular Mechanisms of B Cell Antigen Gathering and Endocytosis. Curr. Top. Microbiol. Immunol. 2016;393:45–63. doi: 10.1007/82_2015_476. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, Aozasa K, Kawai T, Akira S. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity. 2007;27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Kumar S, Ingle H, Mishra S, Mahla RS, Kumar A, Kawai T, Akira S, Takaoka A, Raut AA, Kumar H. IPS-1 differentially induces TRAIL, BCL2, BIRC3 and PRKCE in type I interferons-dependent and -independent anticancer activity. Cell Death Dis. 2015;6:e1758. doi: 10.1038/cddis.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Pinto AK, Ramos HJ, Vick SC, Shrestha B, Suthar MS, Gale M, Jr, Diamond MS. Pattern recognition receptor MDA5 modulates CD8+ T cell-dependent clearance of West Nile virus from the central nervous system. J. Virol. 2013;87:11401–11415. doi: 10.1128/JVI.01403-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung LW, Martinez O, Reynard O, Volchkov VE, Basler CF. Ebola virus failure to stimulate plasmacytoid dendritic cell interferon responses correlates with impaired cellular entry. J. Infect. Dis. 2011a;204(Suppl 3):S973–S977. doi: 10.1093/infdis/jir331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung LW, Park MS, Martinez O, Valmas C, Lopez CB, Basler CF. Ebolavirus VP35 suppresses IFN production from conventional but not plasmacytoid dendritic cells. Immunol. Cell Biol. 2011b;89:792–802. doi: 10.1038/icb.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi I, Amarasinghe GK, Basler CF. Filovirus pathogenesis and immune evasion: insights from Ebola virus and Marburg virus. Nat. Rev. Microbiol. 2015;13:663–676. doi: 10.1038/nrmicro3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagnier D, Scholte FE, Chiang C, Albulescu IC, Nichols C, He Z, Lin R, Snijder EJ, van Hemert MJ, Hiscott J. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J. Virol. 2014;88:4180–4194. doi: 10.1128/JVI.03114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Marzi A, Safronetz D, Feldmann H, Best SM. Immunobiology of Ebola and Lassa virus infection. Nat. Rev. Immunol. 2017 doi: 10.1038/nri.2016.138. In Press Feb 2017. [DOI] [PubMed] [Google Scholar]
- Prins KC, Delpeut S, Leung DW, Reynard O, Volchkova VA, Reid SP, Ramanan P, Cardenas WB, Amarasinghe GK, Volchkov VE, et al. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J. Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AL, Okumura A, Ferris MT, Green R, Feldmann F, Kelly SM, Scott DP, Safronetz D, Haddock E, LaCasse R, et al. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J, Bradfute S, Bray M. Filovirus infection of STAT-1 knockout mice. J. Infect. Dis. 2011;204(Suppl 3):S986–S990. doi: 10.1093/infdis/jir335. [DOI] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 2007;81:13469–13477. doi: 10.1128/JVI.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhein BA, Powers LS, Rogers K, Anantpadma M, Singh BK, Sakurai Y, Bair T, Miller-Hunt C, Sinn P, Davey RA, et al. Interferon-gamma Inhibits Ebola Virus Infection. PLoS Pathog. 2015;11:e1005263. doi: 10.1371/journal.ppat.1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian N, Natarajan K, Clatworthy MR, Wang Z, Germain RN. The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell. 2013;153:348–361. doi: 10.1016/j.cell.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Suthar MS, Brassil MM, Blahnik G, McMillan A, Ramos HJ, Proll SC, Belisle SE, Katze MG, Gale M., Jr A systems biology approach reveals that tissue tropism to West Nile virus is regulated by antiviral genes and innate immune cellular processes. PLoS Pathog. 2013;9:e1003168. doi: 10.1371/journal.ppat.1003168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. Human fatal zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl. Trop. Dis. 2010;4(10):e837. doi: 10.1371/journal.pntd.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.