Abstract
The Arp2/3 complex, a highly conserved nucleator of F-actin polymerization, plays a key role in the regulation of actin dynamics eukaryotic cells. In animal cells and yeasts, Wiskott-Aldrich Syndrome protein (WASP)/suppressor of cAMP receptor (Scar)/WASP family verprolin homologous (WAVE) family proteins activate the Arp2/3 complex in response to localized cues. Like other eukaryotes, plants have an Arp2/3 complex, which has recently been shown to play an important role in F-actin organization and cell morphogenesis. However, no activators of the Arp2/3 complex have been identified in plants, which lack obvious homologs of WASP/Scar/WAVE family proteins. Here, we identify a family of Scar/WAVE-related plant Arp2/3 activators. Like Scar/WAVE proteins, four proteins identified in Arabidopsis thaliana (AtSCAR1 to AtSCAR4) and one in maize (ZmSCAR1) have a C-terminal WASP homology 2 (WH2)/acidic (WA)-verprolin homology/cofilin homology/acidic (VCA)-like domain, which we show can activate the bovine Arp2/3 complex. At their N termini, AtSCAR1 to ATSCAR4, along with a fifth protein lacking a VCA/WA-like domain at its C terminus (At4g18600), are related to the N-terminal Scar homology domains of Scar/WAVE family proteins. Analysis of gene expression patterns suggests functional redundancy among members of the AtSCAR family. Full-length AtSCAR1 and ATSCAR3 proteins and their Scar homology domains bind in vitro to AtBRICK 1 (AtBRK1), the Arabidopsis homolog of HSPC300, a WAVE-binding protein recently identified as a component of a complex implicated in the regulation of Scar/WAVE activity. Thus, AtSCAR proteins are likely to function in association with AtBRK1, and perhaps other Arabidopsis homologs of WAVE complex components, to regulate activation of the Arp2,3 complex in vivo.
Keywords: BRICK1, HSPC300, WAVE complex
Spatial and temporal regulation of F-actin polymerization is critical for a variety of processes in plant cells, as in other eukaryotic cells. During the past several years, work on yeast and animal cells has established a key role for the Arp2/3 complex in the spatial and temporal regulation of actin polymerization (1). Found in all eukaryotes including plants, the Arp2/3 complex nucleates polymerization of actin filaments, primarily as branches on the sides of preexisting filaments. However, efficient nucleation of actin polymerization by the Arp2/3 complex depends on the presence of an Arp2/3 activator. Although many Arp2/3 activators have been identified, the most intensively studied are members of the Wiskott-Aldrich Syndrome protein (WASP)/suppressor of cAMP receptor (Scar)/WASP family verprolin homologous (WAVE) family (2). WASP/Scar/WAVE proteins have a C-terminal domain consisting of a G-actin-binding WASP homology 2 (WH2) motif followed by an Arp2/3-binding acidic motif (A), separated by a conserved linker region (C) (3). Binding of this “WA” (WH2/acidic) or “VCA” (verprolin homology/cofilin homology/acidic) domain to the Arp2/3 complex promotes a conformational change in the complex that leads to its activation by bringing G-actin into close proximity with Arp2 and Arp3 (1). Indeed, the C-terminal VCA domain of WASP/Scar/WAVE is sufficient for activation of the Arp2/3 complex, whereas the remainder of the protein functions in the regulation of its activity (1, 3).
The fully sequenced genome of Arabidopsis thaliana contains genes encoding putative orthologs of all seven Arp2/3 complex components (4, 5). Recently, a role for the Arabidopsis Arp2/3 complex in F-actin-dependent processes has been demonstrated through the analysis of plants with mutations in genes encoding four different components of the complex (5-9). The most conspicuous feature of all Arp2/3 mutant phenotypes is a defect in the expansion of trichomes (epidermal hairs), producing abnormally shaped hairs similar to those resulting from treatment with the actin-disrupting drug cytochalasin D. As would be expected, the cell expansion defect in Arp2/3 mutants is associated with alterations in the organization and density of F-actin filaments (5-9).
Initial surveys of plant genome sequences did not reveal genes encoding VCA or WH2 domain-containing proteins, leading to the idea that plants may not have Arp2/3 activators of the WASP/Scar/WAVE family. However, recent work has identified plant homologs of three components of a mammalian WAVE-containing complex isolated from brain and HeLa cell extracts, which functions to regulate the localization and activity of WAVE/Scar in vitro and in vivo (10-12). The first of these to be identified, BRK1, is a homolog of human HSPC300. In brk1 mutants of maize, failure of lobes to form along the margins of expanding leaf epidermal cells is associated with the loss of local enrichments of cortical F-actin that are found at sites of lobe outgrowth in wild-type cells (13). More recently, the PIROGI and GNARLED genes of Arabidopsis have been shown to encode plant homologs of Pir121/SRA1 and Nap125, respectively (AtSRA-1 and AtNAP1; refs. 14-17). Arabidopsis pirogi and gnarled mutants have trichome F-actin alterations and trichome morphology defects that are nearly identical to those of Arp2/3 complex mutants. Together these findings strongly suggest that AtBRK1, AtSRA-1 and AtNAP1 function in association with a Scar/WAVE-like protein to activate the plant Arp2/3 complex. Here, we identify a family of Scar/WAVE-related proteins found in both Arabidopsis and maize that interact with BRK1 and activate the bovine Arp2/3 complex in vitro.
Materials and Methods
cDNA Amplification, Cloning, and Sequence Analysis. Total RNA was isolated from clusters of unopened Arabidopsis flower buds by using TRIzol reagent (Invitrogen), and random-primed cDNA was synthesized from this RNA by using components of the RETROscript RT-PCR kit (Ambion), both according to the manufacturer's protocols. AtSCAR coding sequences were amplified from this cDNA by using the primer pairs listed in Table 1. PCR products, or clones of these PCR products isolated after ligation into pGEM-T Easy (Promega), were sequenced to determine all or part of the coding sequence of each gene. For AtSCAR1 and At4g18600, most of the coding region was determined by sequencing cDNA clones isolated from a size-selected (3-6 kb) Arabidopsis hypocotyl cDNA library (18) obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Full-length coding sequences were thus obtained from a combination of overlapping cDNA clones and RT-PCR products for AtSCAR3 (At1g29170), AtSCAR1 (At2g34150), and At4g18600, whereas EST sequences available from GenBank (AV529548, AV539904, BE520987, AV529901, and AV554877) were combined with our sequences to reconstruct full-length coding sequences for AtSCAR2 (At2g38440) and AtSCAR4 (At5g01730). Sequence alignments were created by using the clustalw alignment feature of macvector 6.5.3.
Table 1. PCR primers used for cDNA amplification.
| Primer | Product/use |
|---|---|
| AtSCAR3 (At1g29170) | |
| F: 5′-TATGCCACGGAATGTATACGGT-3′ | pSK-full-length cDNA construct |
| R: 5′-TTACGTATCACTCCATGTATCGCT-3′ | |
| F: 5′-TCAGGGAGGAGAAACAAAGTGC-3′ | pGEX-WA construct |
| R: 5′-TTACGTATCACTCCATGTATCGCT-3′ | |
| F: 5′-AAAATGCCACGGAATGTATACGG-3′ | pSK-SHD construct |
| R: 5′-TTACATTATCTCGACTCCTTGACGAA-3′ | |
| F: 5′-CCTTACCACCGCCATTTCCACCCATG-3′ | RT-PCR |
| R: 5′-TTACGTATCACTCCATGTATCGCT-3′ | |
| AtSCAR1 (At2g34150) | |
| F: 5′-CAAATGCCGCTTGTTAGGC-3′ | pSK-SHD construct |
| R: 5′-TCACTCTCGACCGCGTTGTATAGA-3′ | |
| F: 5′-CTATGATTCGTTTCACAGAGTCAATG-3′ | RT-PCR |
| R: 5′-CATGTGTCGCTATCGCTCCATGTATC-3′ | |
| AtSCAR2 (At2g38440) | |
| F: 5′-AAGCTGCTGATAAAGATGACCCTG-3′ | 5′ end of coding sequence, RT-PCR |
| R: 5′-AACGTGAGACATAGAATCCACCAAC-3′ | |
| F: 5′-GGCAATGATGGAAGAAAGGTGG-3′ | Middle segment of coding sequence |
| R: 5′-TCCTCTGGATTGGGCGGAATAG-3′ | |
| F: 5′-TTATTGGAAAGGTTCCACAT-3′ | 3′ end of coding sequence |
| R: 5′-CCAACTATCTGAATCTTCATCCTCG-3′ | |
| AtSCAR4 (At5g01730) | |
| F: 5′-GCATTGACAGAAGCATGTTGAGG-3′ | pGEX-WA construct, RT-PCR |
| R: 5′-TCACTCGCTCCAGCTATCTGAATC-3′ | |
| F: 5′-GCGTTTTGGCGGTTCGTTC-3′ | 5′ half of coding sequence |
| R: 5′-CGTCCTGTGGCACTTGTGTAACC-3′ | |
| F: 5′-AGACGAGTCATTTTCCCAGATGAC-3′ | 3′ half of coding sequence |
| R: 5′-CGCTCCAGCTATCTGAATCGTG-3′ | |
| At4g 18600 | |
| F: 5′-AGAGAGAGACAATGCCGTTGGTGAG-3′ | 5′ end of coding sequence, RT-PCR |
| R: 5′-TCAGCAGGACATAAGGACCCAG-3′ | |
| F: 5′-GCCAGGAGAGTCTATTGCCAAATATC-3′ | Internal segment of coding sequence |
| R: 5′-TTTCCCAAAGATGCACACGC-3′ | |
| F: 5′-CAAGAGTCATCAAAAGCTCGTG-3′ | Internal segment of coding sequence |
| R: 5′-CATCAGGTTCTCCTGCTGG-3′ | |
| ZmSCAR1 | |
| F: 5′-TAGGCGGTGACATTAAAAGCTTAG-3′ | pGEX-WA construct |
| R: 5′-GGTCATATATCACTCCATGTATC-3′ | |
| At UBIQUITIN | |
| F: 5′-GGTGCTAAGAAGAGGAAGAAT-3′ | RT-PCR control |
| R: 5′-CTCCTTCTTTCTGGTAAACGT-3′ |
For production of plant GST-WA fusion proteins, cDNA fragments encoding the following protein segments were amplified by using the PCR primers listed in Table 1 and cloned in frame with GST into pGEX-KG, a derivative of pGEX-4T2 (Amersham Pharmacia Biosciences) with a polyglycine linker separating GST from its fusion partner: C-terminal 73 aa of ZmSCAR1, C-terminal 80 aa of AtSCAR3, and C-terminal 89 aa of AtSCAR4. For in vitro transcription/translation experiments, full-length cDNA clones for AtSCAR1 and AtSCAR3 were assembled in pBluescript SK+; Scar homology domain (SHD)-encoding cDNA fragments of AtSCAR1 (amino acids 1-191) and AtSCAR3 (amino acids 1-196) were amplified with the primers listed in Table 1 and ligated into pBluescript SK+. To produce T7-tagged fusion proteins, cDNAs encoding full-length AtBRK1 cDNA (At2g22640), the N-terminal half of an Arabidopsis TANGLED-like protein (At3g05330, NC1) and a C-terminal fragment of a maize kinesin-like protein (unpublished, NC2), were cloned in-frame with T7 and 6his tags into pET28a, -b, or -c (Novagen).
GST Fusion Protein Preparation and Actin Polymerization Assays. Plasmids encoding GST fusions with the C-terminal WA-like regions of ZmSCAR1, AtSCAR3, and AtSCAR4 (constructed as described above) were transformed into Escherichia coli strain BL21(DE3) for protein expression. GST fusion protein induction with isopropyl β-d-thiogalactoside (IPTG) and purification on glutathione agarose (Amersham Pharmacia Biosciences) were carried out according to the manufacturer's protocols. Purified proteins were dialyzed into U buffer (50 mM Hepes, pH 7.5/100 mM KCl/3 mM MgCl2/1 mM EGTA/1 mM DTT) at 4°C; aliquots were frozen in liquid nitrogen and stored at -80°C. Protein concentrations were determined from OD280 readings by using the following extinction coefficients calculated from amino acid sequences at http://us.expasy.org/tools/protparam.html: 40920 for GST, 47890 for GST-ScarWA, and 46610 for GST-AtSCAR3WA, GST-AtSCAR4WA, and GST-ZmSCAR1. Bovine Arp2/3 complex was purified from brain extracts as described (19). Rabbit actin was purified and pyrenyl-labeled as described (20). Actin polymerization was monitored in KMET buffer (10 mM Tris, pH 7.0/50 mM KCl/1 mM MgCl2/1 mM EGTA) with 0.5 mM ATP, 1.5 μM Mg G-actin (10% pyrene-labeled), 10 nM bovine Arp2/3 complex, and GST-WA fusion (or GST) proteins. GST-ScarWA was used at 1-200 nM, and plant GST-SCARWA proteins were used at 10-1,000 nM. Polymerization was monitored by continuous pyrene fluorescence measurements (λexc = 386 nm, λem = 407 nm) by using a Cary Eclipse fluorescence spectrophotometer (Varian). Barbed end concentrations were determined as described (21). SCAR-induced Arp2/3 actin filament branches were visualized as described in ref. 22 with slight modifications. Arp2/3 complex (10 nM), 1.5 μM Mg G-actin (10% pyrene-labeled), and 25 nM or 100 nM HsScarWA or ZmSCAR1WA were incubated in KMET buffer with 0.5 mM ATP for 5 min, and with 6.6 μM AlexaFluor 488 phalloidin for 10 min at 22°C. Reactions were diluted 1/100 into fluorescence buffer (KMET with 10 mM DTT and 0.2% methylcellulose); 2 μl of this mixture was applied to 20 μg/ml polylysine-coated glass slides and then visualized by fluorescence microscopy. Branching was quantified as follows: % branching = (number of branched filaments/number of total filaments) × 100.
RT-PCR Analysis of Gene Expression. Total RNA for RT-PCR was isolated by using TRIzol Reagent (Invitrogen) from the following tissues of wild-type (Landsberg ecotype) Arabidopsis plants: flower buds (clusters of flower buds remaining after removal of open or partially open flowers), expanding siliques (after petal dehiscence but before completion of silique elongation, root tips (distal 5 mm of rapidly growing seedling roots grown aseptically on MS-agar plates), expanding cotyledons (isolated from 10-day-old seedlings), mature cotyledons (isolated from 3-week-old seedlings), expanding rosette leaves (clusters of immature rosette leaves harvested after removal of cotyledons from 3-week-old seedlings), and mature rosette leaves isolated from 5-week-old plants. One microgram of total RNA was treated with Amplification Grade DNase I (Invitrogen). First-strand cDNA was then synthesized from random decamers with the RETROscript kit (Ambion). Thirty cycles of PCR were performed by using the primers indicated in Table 1. Various dilutions of each cDNA preparation were amplified with ubiquitin-specific primers to find template dilutions resulting in equal ubiquitin product yields; such normalized dilutions were then used for amplification with AtSCAR-specific primers.
In Vitro Trancription/Translation and Binding Experiments. Full-length or SHD AtSCAR3 and AtSCAR1 cDNAs in pBluescript SK+, and T7-AtBRK1, T7-NC1, and T7-NC2 cDNAs in pET28 (generated as described above), were linearized and purified by using phenol/chloroform extraction followed by ethanol precipitation. Pairs of DNA plasmids (AtSCAR3 or AtSCAR1 plus the T7-fusion construct) were cotranscribed and cotranslated in vitro by using the TNT T7Wheat Germ Extract System (Promega) according to the manufacturer's protocols. Each 50-μl reaction contained 1 μg of each DNA template and 20 or 40 μCi (1 Ci = 37 GBq) l-[35S]methionine (Amersham Pharmacia Biosciences, 10 mCi/ml). After a 2-h incubation at 30°C, the reactions were diluted 4-fold with PBS supplemented with 1 mM DTT and 1/100 protease inhibitor mixture for plant cell and tissue extracts (Sigma). Twenty-five microliters of anti-T7 Tag antibody agarose (Novagen) was added to each reaction and incubated at 4°C for 1 h with end-over-end rotation. Beads were washed twice with 1 ml of PBS and then boiled in SDS loading buffer. Samples were electrophoresed on NuPage Novex Bistris ([bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane) gel, 4-12% (Invitrogen) by using MES SDS running buffer. After fixation and drying of gels, protein bands were detected using a PhosphorImager (Molecular Dynamics). Relative protein quantities were determined from band intensities by using iq mac 1.2 (Molecular Dynamics). Binding efficiencies were evaluated by determining the quantity of AtSCAR or AtSCAR-SHD in the immunoprecipitate relative to the quantity in the input sample per unit of bead-bound T7-tagged protein. One unit of bead-bound T7-tagged protein was arbitrarily defined as the quantity of T7-AtBRK1 bound by anti-T7 antibody agarose in Fig. 4A, lane 2. This approach was taken to account for variability in the binding efficiencies of T7-tagged proteins to anti-T7 antibody agarose. For quantitative analysis of protein binding, only the largest protein seen in input samples (corresponding to the full-length AtSCAR or AtSCAR-SHD translation product) was included in the calculation (bands with lower molecular weights seen in input samples are most likely the breakdown products of full-length proteins).
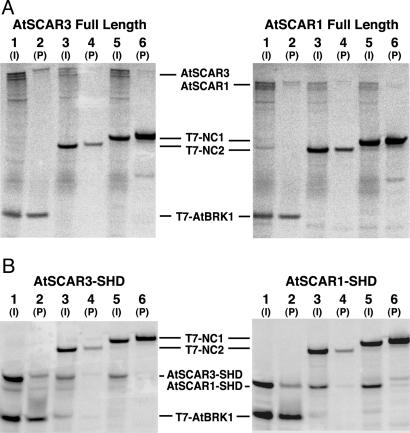
Fig. 4.
Binding interactions between AtSCAR proteins and AtBRK1 in vitro. AtSCAR3 (Left) or AtSCAR1 (Right) full-length proteins (A) or SHDs (B) were cotranscribed and translated with T7-tagged AtBRK1 (lanes 1 and 2 in each panel), T7-tagged negative control protein 2 (NC2; lanes 3 and 4), or T7-tagged negative control protein 1 (NC1; lanes 5 and 6). Inputs (I, 1/10th of total sample before anti-T7 bead incubation) are loaded in lanes 1, 3, and 5; proteins bound to anti-T7 beads (P, one-half of total sample) are loaded in lanes 2, 4, and 6.
Results
Scar/WAVE-Related Proteins Are Present in Arabidopsis and Maize. The domain organization of Scar/WAVE proteins is summarized in Fig. 1A. Like related WASP proteins, Scar/WAVE proteins have a C-terminal VCA/WA domain, which is preceded by an extensive proline-rich region characterized by the presence of several stretches of two to nine consecutive prolines. Scar/WAVE proteins differ from WASP by having an SHD at the N terminus, which is immediately followed by a short basic region (34-47% basic residues in a stretch of 29-34 aa). blast searches of Arabidopsis genome sequence revealed the presence of five predicted genes with weak homology at their N termini to the SHD of mammalian WAVE1 (26-27% identity, AtSCAR1 to AtSCAR4 and At4g18600). Although the overall level of homology is low, the plant and animal sequences are conserved at almost all (17/21) positions in the alignment shown where Scar/WAVE proteins from four distantly related animal species are identical to each other (mouse WAVE1, Drosophila melanogaster Scar, Caenorhabditis elegans Scar, Dictyostelium discoideum Scar; Fig. 1B, asterisks).
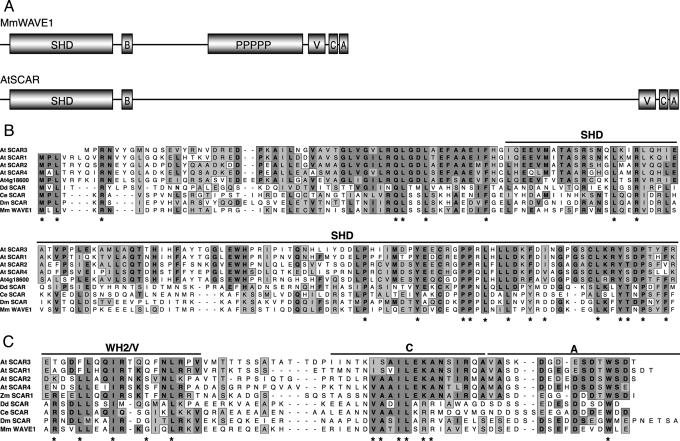
Fig. 1.
Comparison of plant and animal Scar/WAVE proteins. (A) Schematic summary of the domain organization of mammalian WAVE1 (after ref. 29) and of AtSCAR proteins. B, basic region; PPPPP, polyproline-rich region; V, verprolin homology/WH2 motif; C, cofilin homology/connector region; A, acidic region. (B) Alignment of N termini of five SHD-containing Arabidopsis proteins: AtSCAR1 (At2g34150), AtSCAR2 (At2g38440), AtSCAR3 (At1g29170), AtSCAR4 (At5g01730), and At4g18600, with N termini of Scar/WAVE proteins from D. discoideum (AAD29083), C. elegans (NP_493028), D. melanogaster (NP_609477), and Mus musculus (NP_114083). (C) Alignment of C termini of the same proteins shown in B without At4g18600, which has no VCA-like region at its C terminus, but including Zea mays SCAR1 (BM340311). In B and C, domain boundaries are shown as defined previously (29). Asterisks mark residues that are invariant in animal Scar/WAVE proteins, including several not shown here. GenBank accession numbers for sequence-verified AtSCAR full-length coding regions are AY743923-AY743927.
Sequencing of overlapping RT-PCR products and cDNA clones was carried out to confirm that all five plant genes are expressed and to determine the full coding sequence of each gene (GenBank accession numbers AY743923-AY743927). The domain organization of AtSCAR1 to AtSCAR4 is shown schematically in Fig. 1A. With molecular masses from 92 kDa for AtSCAR1 to 222 kDa for At4g18600, Arabidopsis SHD-containing proteins are much larger than animal Scar/WAVE proteins (e.g., 48-76 kDa for the four Scar/WAVE proteins shown in Fig. 1). Like Scar/WAVE proteins, immediately after the SHD-like region, each of these plant proteins has a basic region, although these are typically shorter than the basic regions found in animal Scar/WAVE proteins, consisting of a 16- to 26-aa stretch containing 36-62% basic residues. Between the basic region and the C terminus, we found no recognizable sequence homology to Scar/WAVE proteins, and there are no proline-rich regions comparable with those found in Scar/WAVE and WASP family proteins. Indeed, this central region shows no significant homology to any other proteins or functional motifs, although AtSCAR1 to AtSCAR4 do show modest sequence conservation with each other in this region. However, at their C termini, four of the SHD-containing Arabidopsis proteins identified (AtSCAR1 to AtSCAR4) do have a WA/VCA-like region, as does a maize protein identified from a partial cDNA sequence, which we have designated ZmSCAR1 (Fig. 1C). The plant WA-like sequences share 4 to 8 of the 11 residues that are invariant among animal Scar/WAVE proteins in the WH2 and C regions (marked with asterisks in Fig. 1C), with the most similar being ZmSCAR1. After the C region, the plant proteins have an acidic region of similar length and composition to those found in animal Scar/WAVE proteins, which includes the invariant tryptophan residue found very near the C terminus of all Scar/WAVE and WASP proteins (Fig. 1C). Interestingly, the largest of the five SHD-containing Arabidopsis proteins, At4g18600, lacks a WA/VCA-like region at its C terminus, but shares sequence homology near its C terminus to the central regions of the other four SHD-containing plant proteins. Because of the lack of a C-terminal WA/VCA domain, At4g18600 has not been designated as a SCAR protein.
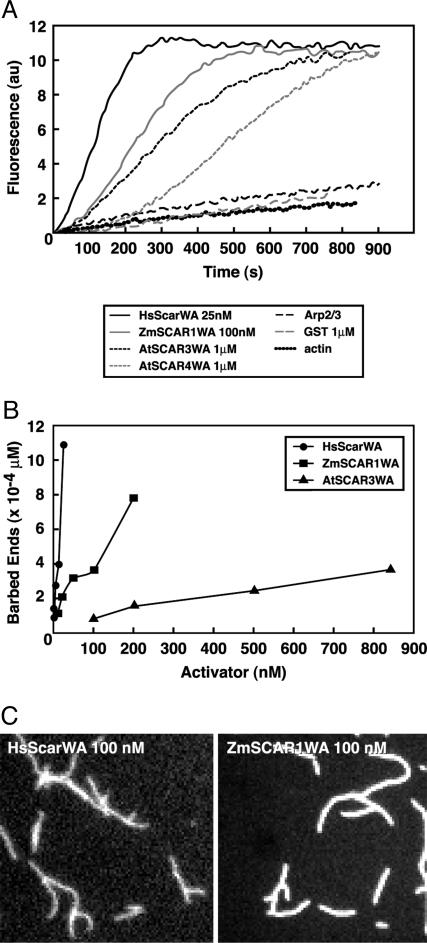
The C Termini of Plant Scar Proteins Activate the Arp2/3 Complex. Because the C-terminal WA/VCA domains of Scar/WAVE proteins are sufficient for activation of the Arp2/3 complex (23), we sought to determine whether the WA-like regions of the plant SHD-containing proteins would function similarly. Because the plant Arp2/3 complex has not been purified and biochemically characterized, we tested the ability of GST fusions with the WA-like regions of ZmSCAR1, AtSCAR3, and AtSCAR4 to activate bovine Arp2/3 complex-mediated actin polymerization by using purified proteins. Bovine Arp2/3 complex was incubated with actin (10% pyrene-labeled) and varying concentrations of GST-WA fusion proteins or GST alone as a negative control. Fig. 2A shows the kinetics of actin polymerization for each protein tested at its optimal concentration. GST-ZmSCAR1WA was the most potent Arp2/3 activator of the plant proteins tested; 100 nM GST-ZmSCAR1WA stimulates actin assembly at a rate approaching that of the postive control, 25 nM GST-HsScarWA. GST-AtSCAR3WA and GST-AtSCAR4WA were also active but less potent, stimulating significant but lower nucleation rates at 1 μM (Fig. 2 A). In Fig. 2B, barbed end concentrations (reflecting filament nucleation rates) are shown for different activator concentrations tested, demonstrating an increase in actin-nucleating activity with increasing activator concentration for GST-HsScarWA, GST-ZmSCAR1WA, and AtSCAR3WA (this calculation was not performed for GST-AtSCAR4WA because no activity was observed at concentrations below 1 μM). As expected, activation of bovine Arp2/3 by ZmSCAR1WA leads to formation of branched actin filaments, a hallmark of Arp2/3-mediated actin filament nucleation (Fig. 2C). ZmSCAR1WA induces the formation of 8% and 15% branched filaments at 25 nM and 100 nM, respectively (compared with 30% and 50% branched filaments at 25 nM and 100 nM HsScarWA).
Fig. 2.
Effect of AtSCARWA proteins on bovine Arp2/3 complex-dependent actin filament formation. (A) Nucleation activity was assessed by monitoring polymerization kinetics by using 1.5 μM actin (10% pyrene-labeled) and 10 nM bovine Arp2/3 complex. (B) Barbed end concentrations obtained at 50% polymerization with the indicated concentrations of GST-HsScarWA, GST-ZmSCAR1WA, and GST-AtSCAR3WA. (C) Like GST-HsScarWA, GST-ZmSCAR1WA promotes the formation of branched actin filaments. Actin filaments were polymerized in the presence of Arp2/3 complex and HsScarWA or ZmSCAR1WA, stained with AlexaFluor 488 phalloidin, and visualized by fluorescence microscopy.
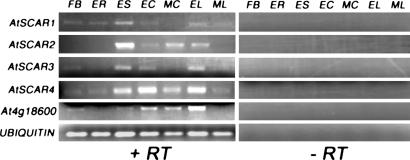
Arabidopsis Scar Genes Have Overlapping Expression Profiles. Analysis of the Arabidopsis genome sequence shows that most components of the Arp2/3 complex are encoded by single copy genes, as are three putative WAVE/Scar complex components, AtBRK1, AtSRA-1, and AtNAP1. Therefore, it is somewhat surprising that the Arabidopsis genome contains four SCAR genes. One possible explanation is that these genes have become specialized to function in different tissues, as is the case for mammalian WAVEs [WAVE1 and WAVE3 are predominantly expressed in the brain, whereas WAVE2 is widely expressed (24)]. To investigate this possibility, we analyzed the expression of AtSCAR genes in a variety of tissues by means of semiquantitative RT-PCR, emphasizing those in which cells are expanding where we could anticipate that the SCAR genes would be expressed. As illustrated in Fig. 3, this analysis revealed overlapping, although not identical, expression profiles for the AtSCAR genes and At4g18600. All five genes are expressed in expanding cotyledons, expanding rosette leaves, and expanding siliques (containing developing embryos), and these tissues are among those in which the highest expression levels were observed for all five genes. Expression of all five genes is reduced (in some cases to undetectable levels) in mature leaves compared with expanding leaves; reduced expression is also observed in mature cotyledons compared with expanding cotyledons for AtSCAR1 and AtSCAR3. Expression was detected in unopened flower buds for all genes except AtSCAR2, and in the expanding tip region of roots for AtSCAR1 and AtSCAR4. These results are consistent with a role for AtSCAR genes in cell expansion (and/or other actin-dependent processes) in multiple organs. Although there is some variation in expression patterns, results of the RT-PCR analysis are more consistent with functional redundancy among the AtSCAR genes than with tissue-specific functional specialization within this gene family.
Fig. 3.
RT-PCR analysis of the expression of AtSCAR1 to AtSCAR4 and At4g18600. Tissues analyzed (prepared as described in Materials and Methods) were flower buds (FB), expanding tip region of roots (ER), expanding siliques (ES), expanding cotyledons (EC), mature cotyledons (MC), expanding rosette leaves (EL), and mature rosette leaves (ML). RT, reverse transcriptase.
Full-Length AtSCAR Proteins and Their N-Terminal SH Domains Bind Directly to AtBRK1 in Vitro. If the Arabidopsis SCAR proteins identified function in a complex containing AtBRK1, then we could expect that they would bind directly to AtBRK1, because HSPC300 has been shown to bind directly to WAVE1 and WAVE2 (10, 11). Therefore, full-length AtSCAR3 and AtSCAR1 were produced by in vitro transcription/translation in the presence of [35S]methionine together with T7 epitope-tagged AtBRK1 or two different T7-tagged negative control proteins (NC1 and NC2). Binding of AtSCAR to T7-AtBRK1 and T7-NC proteins was then assessed after capture of the T7-tagged proteins on monoclonal anti-T7 antibody-coupled beads. As shown in Fig. 4A, full-length AtSCAR3 and AtSCAR1 bound significantly better to T7-AtBRK1 than to the T7-NC proteins. Binding efficiencies were analyzed quantitatively by calculating the proportion of input SCAR protein that bound to the beads per unit of bead-bound T7-tagged protein (see Materials and Methods for details of this analysis). Using this approach, we determined that AtSCAR3 bound ≥7-fold more efficiently to T7-AtBRK1 than to either T7-NC1 or T7-NC2, and AtSCAR1 bound ≥55-fold more efficiently to T7-AtBRK1 than to either negative control T7-tagged protein.
Other than the actin- and Arp2/3-binding WA domain, the N-terminal SHD is the only region of sequence homology between AtSCAR proteins and animal WAVE/Scar proteins. Thus, we reasoned that the SHD was likely to be the region responsible for BRK1 binding. As illustrated in Fig. 4B, using the same approach described for the full-length proteins, we found that the SHDs of AtSCAR3 and AtSCAR1 (AtSCAR3-SHD and AtSCAR1-SHD) bound ≥5-fold more efficiently to T7-tagged AtBRK1 than to the T7-tagged NC proteins.
Discussion
Activators of the Arp2/3 complex have not been previously identified in plants. In this study, we have identified and analyzed a family of four A. thaliana proteins distantly related to Scar/WAVE (AtSCAR1 to AtSCAR4), which can activate the bovine Arp2/3 complex. At their N termini, AtSCAR1 to AtSCAR4, along with a fifth protein (At4g18600), share 26-27% identity with the N-terminal SHDs of animal Scar/WAVE proteins. The function of the SHD of Scar/WAVE is not well understood, but the SHD of WAVE2 has recently been shown to bind directly to Abi-1, a core component of the WAVE regulatory complex that is critical for complex assembly (11, 25). A family of four predicted Arabidopsis genes encoding proteins with a low level of homology to mammalian Abi-1 has recently been identified (16); the products of these genes may bind to the SHDs of AtSCAR proteins and play a role in assembly of an AtSCAR regulatory complex. Here, we have shown that the SHDs of AtSCAR1 and AtSCAR3 (as well as the corresponding full-length proteins) bind to AtBRK1, the Arabidopsis homolog of another mammalian WAVE complex component, HSPC300. Thus, it is likely that the AtSCAR proteins we have identified interact with AtBRK1 in vivo, perhaps together with other WAVE complex components, to regulate the activity and/or localization of AtSCARs. However, further work will be required to understand the functional significance of the AtBRK1-AtSCAR interaction we have observed.
As in animal Scar/WAVE proteins, a short basic region immediately follows the SHDs of all four AtSCAR proteins and At4g18600. The basic region of human WAVE2 has recently been shown to bind to PtdIns(3,4,5)P3 and mediate recruitment of WAVE2 to the plasma membrane in response to growth factor stimulation (26). Thus, the basic regions of AtSCAR proteins may also function to mediate their interaction with membranes. However, AtSCAR proteins lack the extensive proline-rich region that makes up most of the C-terminal half of both Scar/WAVE and WASP family proteins. Binding of both profilin and the SH3 domains of the adaptor proteins Grb2 and Nck to the proline-rich regions of WASP proteins facilitates their activation (1). Because profilins do play a role in regulating actin polymerization in plant cells (27, 28), it will be interesting to determine whether they play any role in the regulation of plant SCAR proteins in the absence of a proline-rich region. A particularly interesting question concerning the plant SCAR proteins is: What is the functional significance of the novel stretch of 560-1,140 aa between the basic region and the C-terminal WA-like domain, which comprises the majority of each of the AtSCAR proteins and has no counterpart in animal Scar/WAVE proteins?
An ≈65-aa region at the C terminus of AtSCAR1 to AtSCAR4, as well as a maize protein we have called ZmSCAR1, resembles the C-terminal VCA/WA region of Scar/WAVE and WASP family proteins, consisting of a WH2-like region, a conserved linker region, and a tryptophan-containing acidic region at the extreme C terminus. Very recent work has shown that the WA-like domain of AtSCAR1 has actin-binding activity (16). We have shown that all three of the WA-like domains of plant SCAR proteins tested (AtSCAR3, AtSCAR4, and ZmSCAR1) can activate the bovine Arp2/3 complex in vitro. Of these, the most potent activator is ZmSCAR1, whereas AtSCAR3 was less active and AtSCAR4 far less active. However, definitive conclusions concerning the relative activities of the WA-like domains of these proteins cannot be drawn until their nucleation abilities have been tested by using plant actin and plant Arp2/3 complex. It may be that the WA-like domain of ZmSCAR1 simply binds more efficiently to mammalian Arp2/3 complex and/or actin than WA-like domains of AtSCAR3 and AtSCAR4. Indeed, among the plant proteins tested, the presumptive G-actin binding the WH2-like region of ZmSCAR1 is the most similar to the WH2 domains of animal Scar/WAVEs. Given that the Arp2/3-binding activity of the C-terminal acidic region of WASP is used to facilitate purification of animal and yeast Arp2/3 complex (e.g., ref. 19), the acidic C termini of plant SCAR proteins may prove to be instrumental in purification of the plant Arp2/3 complex for such biochemical studies.
Nothing is yet known about the in vivo functions of plant SCAR proteins. Gene expression, protein localization, and reverse genetic analyses should all be helpful in elucidating the roles of these proteins in Arp2/3-dependent cellular processes. It will also be interesting to investigate the roles of plant homologs of mammalian WAVE complex components in regulating the activity and localization of plant SCAR proteins.
Acknowledgments
We thank John Fowler for pointing out the SHD-containing Arabidopsis proteins to us, Michael Burke for help with RT-PCR analysis, Laura Machesky (University of Birmingham, Birmingham, U.K.) for the GST-HsScarWA construct, and the Arabidopsis Biological Resource Center (Columbus, OH) for providing the 3- to 6-kb Arabidopsis cDNA library. This work was supported by National Science Foundation Grant IBN-0212724 (to L.G.S.) and Human Frontier Science Program Postdoctoral Fellowship LT00029/2000-M (to C.E.).
Author contributions: M.F., C.E., J.D., R.L., and L.G.S. designed research; M.F., C.E., J.D., S.D., M.N., and L.G.S. performed research; M.F. and L.G.S. contributed new reagents/analytical tools; M.F., C.E., J.D., S.D., M.N., and L.G.S. analyzed data; L.G.S. wrote the paper; R.L. supervised work at Harvard Medical School; and L.G.S. supervised all work at the University of California at San Diego.
Abbreviations: WASP, Wiskott-Aldrich Syndrome protein; WH2, WASP homology 2; WA, WH2/acidic; VCA, verprolin homology/cofilin homology/acidic; Scar, suppressor of cAMP receptor; SHD, Scar homology domain; WAVE, WASP family verprolin homologous; BRK1, BRICK1.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY743923-AY743927).
References
- 1.Pollard, T. D. & Borisy, G. G. (2003) Cell 112, 453-465. [DOI] [PubMed] [Google Scholar]
- 2.Weaver, A. M., Young, M. E., Lee, W. L. & Cooper, J. A. (2003) Curr. Opin. Cell Biol. 15, 23-30. [DOI] [PubMed] [Google Scholar]
- 3.Welch, M. D. & Mullins, R. D. (2002) Annu. Rev. Cell Dev. Biol. 18, 247-288. [DOI] [PubMed] [Google Scholar]
- 4.Klahre, U. & Chua, N. H. (1999) Plant Mol. Biol. 41, 65-73. [DOI] [PubMed] [Google Scholar]
- 5.Li, S., Blanchoin, L., Yang, Z. & Lord, E. M. (2003) Plant Physiol. 132, 2034-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le, J., El-Assal, S. E., Basu, D., Saad, M. E. & Szymanski, D. B. (2003) Curr. Biol. 13, 1341-1347. [DOI] [PubMed] [Google Scholar]
- 7.Mathur, J., Mathur, N., Kernebeck, B. & Hulskamp, M. (2003) Plant Cell 15, 1632-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B. P. & Hulskamp, M. (2003) Development (Cambridge, U.K.) 130, 3137-3146. [DOI] [PubMed] [Google Scholar]
- 9.El-Assal, S. E., Le, J., Basu, D, Mallery, E. L. & Szymanski, D. B. (2004) Plant J. 38, 526-538. [DOI] [PubMed] [Google Scholar]
- 10.Eden, S., Rohatgi, R., Podtelejnikov, A. V., Mann, M. & Kirschner, M. W. (2002) Nature 418, 790-793. [DOI] [PubMed] [Google Scholar]
- 11.Gautreau, A., Ho, H. H., Li, J., Steen, H., Gygi, S. P. & Kirschner, M. W. (2004) Proc. Natl. Acad. Sci. USA 101, 4379-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blagg, S. L. & Insall, R. H. (2004) Nat. Cell Biol. 6, 279-281. [DOI] [PubMed] [Google Scholar]
- 13.Frank, M. J. & Smith, L. G. (2002) Curr. Biol. 12, 849-853. [DOI] [PubMed] [Google Scholar]
- 14.El-Assal, S. E., Le, J., Basu, D., Mallery, E. L. & Szymanski, D. B. (2004) Curr. Biol. 14, 1405-1409. [DOI] [PubMed] [Google Scholar]
- 15.Basu, D., El-Assal, S. E., Le, J., Mallery, E. L. & Szymanski, D. B. (2004) Development (Cambridge, U.K.) 131, 4345-4355. [DOI] [PubMed] [Google Scholar]
- 16.Deeks, M. J., Kaloriti, D., Davies, B., Malho, R. & Hussey, P. J. (2004) Curr. Biol. 14, 1410-1414. [DOI] [PubMed] [Google Scholar]
- 17.Brembu, T., Winge, P., Seem, M. & Bones, A. M. (2004) Plant Cell 16, 2335-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieber, J., Rothenberg, M., Roman, G., Feldmann, K. A. & Ecker, J. R. (1993) Cell 72, 427-441. [DOI] [PubMed] [Google Scholar]
- 19.Egile, C., Loisel, T. P., Laurent, V., Li, R., Pantaloni, D., Sansonetti, P. J. & Carlier, M. F. (1999) J. Cell Biol. 146, 1319-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardee, J. D. & Spudich, J. A. (1982) Methods Cell Biol. 24, 271-289. [DOI] [PubMed] [Google Scholar]
- 21.Li, F. & Higgs, H. N. (2003) Curr. Biol. 13, 1335-1340. [DOI] [PubMed] [Google Scholar]
- 22.Blanchoin, L., Amann, K. J., Higgs, H. N., Marchand, J. B., Kaiser, D. A. & Pollard, T. D. (2000) Nature 404, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 23.Machesky, L. M., Mullins, D. M., Higgs, H. N., Kaiser, D. A., Blanchoin, L., May, R. C., Hall, M. E. & Pollard, T. D. (1999) Proc. Natl. Acad. Sci. USA 96, 3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suetsugu, S., Miki, H. & Takenawa, T. (1999) Biochem. Biophys. Res. Commun. 260, 296-302. [DOI] [PubMed] [Google Scholar]
- 25.Innocenti, M., Zucconi, A., Disanza, A., Frittoli, E., Areces, L. B., Steffen, A., Stradal, T. E. B., Di Fiore, P. P., Carlier, M.-F. & Scita, G. (2004) Nat. Cell Biol. 6, 319-327. [DOI] [PubMed] [Google Scholar]
- 26.Oikawa, T., Yamaguchi, H., Itoh, T., Kato, M., Ijuin, T., Yamazaki, D., Suetsugu, S. & Takenawa, T. (2004) Nat. Cell Biol. 6, 420-426. [DOI] [PubMed] [Google Scholar]
- 27.Staiger, C.J., Yuan, M., Valenta, R., Shaw, P.J., Warn, R. M. & Lloyd, C. W. (1994) Curr. Biol. 4, 215-219. [DOI] [PubMed] [Google Scholar]
- 28.Ramachandran, S., Christensen, H.E., Dong, C. H., Chao-Ming, W., Cleary, A.L. & Chua, N. H. (2000) Plant Physiol. 124, 1637-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stradal, T. E. B., Rottner, K., Disanza, A., Confalonieri, S., Innocenti, M. & Scita, G. (2004) Trends Cell Biol. 14, 303-311. [DOI] [PubMed] [Google Scholar]