Abstract
Regulator of G protein signaling (RGS) proteins negatively regulate receptor-mediated second messenger responses by enhancing the GTPase activity of Gα subunits. We describe a receptor-specific role for an RGS protein at the level of an individual brain neuron. RGS9-2 and Gβ5 mRNA and protein complexes were detected in striatal cholinergic and γ-aminobutyric acidergic neurons. Dialysis of cholinergic neurons with RGS9 constructs enhanced basal Ca2+ channel currents and reduced D2 dopamine receptor modulation of Cav2.2 channels. These constructs did not alter M2 muscarinic receptor modulation of Cav2.2 currents in the same neuron. The noncatalytic DEP-GGL domain of RGS9 antagonized endogenous RGS9-2 activity, enhancing D2 receptor modulation of Ca2+ currents. In vitro, RGS9 constructs accelerated GTPase activity, in agreement with electrophysiological measurements, and did so more effectively at Go than Gi. These results implicate RGS9-2 as a specific regulator of dopamine receptor-mediated signaling in the striatum and identify a role for GAP activity modulation by the DEP-GGL domain.
Keywords: calcium, GTPase activating protein, receptor-specific, basal ganglia, indirect pathway
Regulators of G protein signaling (RGS) are a diverse family of proteins identified by the presence of a 120-aa domain termed the RGS box. In cell lines or in purified in vitro assays, most RGS proteins enhance the GTPase activity of heterotrimeric G protein α subunits and thereby accelerate the deactivation of receptor-initiated second messenger responses. Many RGS proteins also contain one or more putative protein-protein interaction domains. These noncatalytic domains have been suggested to regulate catalytic activity, signal transduction pathway specificity, and/or subcellular targeting of RGS proteins.
One subfamily of RGS proteins (RGS6, -7, -9, and -11) all share homologous DEP (Dishevelled, Egl-10, Pleckstrin), GGL (G protein Gamma subunit Like), and RGS domains. The DEP domain of the retinal isoform of RGS9 (RGS9-1) has been shown to confer localization to a retinal membrane protein termed R9AP, and this localization has been shown to be necessary for proper RGS9-1 function in the retina (1). Several investigators have demonstrated that the GGL domain interacts with the Gβ5 subunit (2-6). In vitro, Gβ5 binding to RGS6, -7, or -11 increases the GAP specificity of these RGS proteins for Gαo (2, 3). In addition, Gβ5 binding to either RGS7 or RGS9 enhances RGS-mediated acceleration of G protein gated inwardly rectifying K+ (Kir3) channel activation and deactivation kinetics in an oocyte expression system. This enhancement may result from Gβ5-mediated increased stability of the RGS protein or enhanced GAP activity (7).
Despite the functional similarities among RGS6, -7, -9, and -11 in heterologous expression systems or when analyzed in vitro, each of these RGS proteins is likely to play a unique role in the central nervous system because they are differentially localized within the brain (8, 9). In contrast to the more ubiquitous localization of RGS6, -7, and -11, mRNA for the short splice variant of RGS9 (RGS9-1) is localized primarily to the retina and pineal (10, 11), whereas the long splice variant (RGS9-2) has been identified primarily in the striatum, nucleus accumbens, and olfactory tubercle with limited expression in the hypothalamus and amygdala (8, 12, 13). The dense and discrete expression of RGS9-2 within areas of the basal ganglia that are rich in dopaminergic innervation and the unique domain of RGS9-2 found only in these regions suggest that RGS9-2 might play a role in modulating dopaminergic receptor-mediated signaling cascades. Indeed, knockout and overexpression of RGS9-2 in mice have recently been used to show that altering levels of RGS9-2 affects dopamine D2 receptor-mediated locomotor and rewarding responses to cocaine (14).
Here, we sought to study the in vivo role of RGS9-2 at the cellular level in the striatum. Single-cell RT-PCR and immunoprecipitation techniques revealed that RGS9-2 is expressed both in medium spiny neurons and large, cholinergic interneurons. Although these interneurons represent only a small percentage of all striatal neurons, they are a well characterized, homogenous population that expresses both D2 dopamine and M2 muscarinic receptors, both of which are potently coupled to Cav2.2 Ca2+ channels through a membrane-delimited G protein signaling pathway (15, 16). On the other hand, medium spiny neurons are heterogeneous with respect to their expression of dopamine receptors and the linkage of these receptors to ion channels (17-21). As a consequence, our initial efforts at characterizing the role of RGS9-2 focused on D2/M2 receptor signaling in cholinergic interneurons. Through the introduction of various RGS9 constructs through a patch pipette in these cells, we were able to show that the RGS domain of RGS9-2 modulates the D2 receptor-mediated inhibition of Cav2.2 channels, and that this modulation was blocked by the introduction of exogenous DEP-GGL domains. This modulation was specific to D2 receptors, because RGS9 did not modulate the M2 muscarinic receptor linkage to Cav2.2 Ca2+ channels in the same cell.
Experimental Procedures
Cell Isolation. Striatal neurons from rats (>3 weeks old) were acutely dissociated by using described methods (15).
Electrophysiology. Whole cell recordings from acutely isolated rat striatal neurons were obtained as described (15).
Statistical Procedures. Data analysis was performed with systat (Version 5.2, SPSS, Chicago). Sample statistics are given as means ± standard errors. Box plots were used for graphic presentation of the data because of the small sample sizes.
Immunoblots. Adult Sprague-Dawley rats were decapitated, and their brains placed quickly into ice-cold PBS. Cortical and striatal tissues were dissected, flash frozen in liquid nitrogen, and stored at -70°C. Frozen tissue was resuspended in buffer A (50 mM Hepes, pH 7.5/0.2 mM EGTA/150 mM KCl/1 mM DTT/0.5 mM PMSF/10 mg/ml leupeptin/10 mg/ml pepstatin/10 mg/ml aprotinin) to a concentration of 100 mg wet weight per ml, then homogenized three times for 10 s each by using an ESGE BioHomogenizer. The samples were centrifuged in a Ti-70 rotor at 30,000 rpm for 30 min at 4°C, and the supernatants were saved as the cytosolic fraction of proteins. The pellet was resuspended in 2 ml of buffer A/1% sodium cholate, homogenized once, and incubated for 10 min at 4°C while stirring. This sample was centrifuged at 25,000 rpm for 45 min at 4°C. The supernatant from this step represents the solubilized membrane fraction. Concentrations were adjusted to 2.8 mg/ml, and 200-μl aliquots were used for immunoprecipitation experiments. Two milliliters of anti-Gb5 antiserum (CytoSignal) or 1 ml of anti-RGS9 sheep antibody (generously provided by V. Arshavsky, Harvard Medical School, Boston) was added to extracts, followed by incubation at 4°C overnight. Twenty milliliters of a 50% slurry of Protein G Sepharose resin was added followed by a 1-h incubation at 4°C while rotating. The resin was collected by centrifugation at 6,000 × g and washed four times with 100 ml of homogenization buffer/0.2% BSA. Immunoprecipitated proteins were solubilized in 15 μl of 2× Tris-Glycine SDS sample buffer (Invitrogen), boiled for 5 min, and then resolved on a 10-20% Tris-Glycine SDS/PAGE gel. Proteins were transferred to a poly(vinylidene difluoride) membrane, and immunoblots for RGS9 or Gβ5 were performed and developed by using the LumiGLO Chemiluminescent Substrate kit (Kirkegaard & Perry Laboratories) and exposed to Kodak AR film.
Single-Cell RT-PCR. Protocols followed were similar to those described in refs. 22 and 23. The thermal cycling program for substance P, enkephalin, and choline acetyltransferase was 94°C for 45 s, 58°C for 45 s, and 72°C for 70 s for 45 cycles. Cycling parameters for RGS9-2 and Gβ5 were similar, except that the annealing temperature was 54°C. Primers for substance P, enkephalin, and choline acetyltransferase have been described (23). Primers for RGS9-2 were 5′-GCCCGCCTTCCCTTCCGCCAGGCTTTC-3′ and 5′-GTCCCTTGGAGGAATCGTCAAGT-3′. Primers for Gβ5 were 5′-GAGGGAGAAATCCACGCTTGA-3′ and 5′-CCAAGAAGAAGTCTGTCGCTATGC-3′.
GTPase Assay. Single turnover GTPase reactions were performed by using Gαo or Gαi1 as the substrate as described (24).
Cloning and Expression of RGS9. The cloning and expression of the core RGS9 domain has been described (25). RGS9d* encodes residues 284-484 of bovine RGS9-1 containing mutations I363T and Q330G. The DNA was amplified by PCR using primers 5′-AAAGGATCCCTGGTGGACATCCCAACCAAG-3′ (upstream) and 5′-TTTAAGCTTATTTGGGAGGCGGCTCTTTTC-3′ (downstream), digested with BamHI and HindIII and ligated into the PQE30 vector (Qiagen, Valencia, CA) (restriction sites are underlined). Expression and purification was conducted as described for RGS9d. Rat striatal mRNA was isolated by using the Rneasy Mini kit (Qiagen) according to the manufacturer's instructions. Complementary DNA was generated by using the Advantage RT-for-PCR kit and an oligo(dT) primer according to the manufacturer's instructions. The DEP-GGL domain (residues 2-284 of rat RGS9-2) was PCR amplified by using the primers 5′-AAAAGCTAGCACGATCCGACACCAAGGCCAG-3′ (upstream) and 5′-GGTTGGATCCTCCACTTACTTGGCGTTTAAATCCCAG-3′ (downstream) and RGS9-2/pcDNA3.1 vector as a template. The amplicon was digested with NheI and BamHI and ligated into amino His6 tag vector pRSETA (Invitrogen). His6-DEP-GGL was subcloned into baculovirus transfer vector pVL1392, DEP-GGL/pRSETA, by PCR with pRSETA-specific primers 5′-AGGATCTAGACATATGCGGGGTTCTC (upstream) and 5′-CCAGCTGCAGATCTCGAGCTCGGATCC (downstream) and digestion with XbaI and BamHI. Pfu polymerase (Stratagene) was used for all PCR amplifications. Transfection of Sf9 cells with DEP-GGL/pVL1392 construct was performed by using a BaculoGold transfection kit (Pharmingen), and the resulting baculovirus was amplified as suggested by the manufacturer. For expression, Sf9 cells were grown to 2 × 106 cells per ml in 500-ml suspension cultures and coinfected with baculoviruses expressing His6-DEP-GGL and untagged mouse Gβ5 (baculovirus generously provided by Mel Simon, California Institute of Technology, Pasadena). Cultures were incubated for 3 days in a 27°C shaker and harvested by centrifugation at 1,000 × g for 10 min. A total of 2 × 109 cells were resuspended in 30 ml of lysis buffer (50 mM Hepes, pH 8/50 mM NaCl/10 mM 2-mercaptoethanol/100 M PMSF/20 g/ml leupeptin/1 g/ml aprotinin) and homogenized on ice, and lysates were clarified by centrifugation at 40,000 × g for 30 min at 4°C. Supernatants were applied to 1 ml of nickel-nitrilotriacetic acid resin (Qiagen). The resin was washed with 30 ml 20 mM Hepes, pH 8/400 mM NaCl, followed by 20 ml of 20 mM Hepes, pH 8/50 mM NaCl/10 mM imidazole, 5 ml of 20 mM Hepes, pH 8/50 mM NaCl/20 mM imidazole, and finally with 2 ml of 20 mM Hepes, pH 8/50 mM NaCl/40 mM imidazole. DEP-GGL/Gβ5 complex was eluted with 5 ml of 200 mM imidazole in 20 mM Hepes buffer, pH 8/50 mM NaCl. The purified protein was buffer exchanged into 20 mM Hepes, pH 8/50 mM NaCl/20% glycerol and protease inhibitors and stored at -20°C.
Results
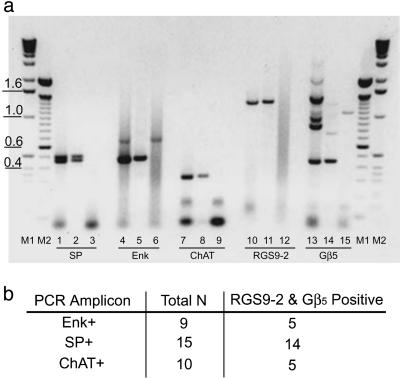
Although RGS9 and Gβ5 mRNA and/or protein have been localized at the regional level to the striatum, as well as medium spiny neurons by in situ hybridization (14), single-cell analysis of each of the striatal cell types expressing RGS9 have not been reported. Thus, our initial studies sought to determine which striatal neurons express RGS9 and to determine whether Gβ5 was coexpressed in the same neuronal populations. As shown in Fig. 1, single-cell RT-PCR analysis found both RGS9-2 and Gβ5 mRNA were identified in cholinergic interneurons within the dorsal striatum. RGS9-2 and Gβ5 were also found in both major subpopulations of medium spiny γ-aminobutyric acidergic neurons, as identified by their expression of either enkephalin or substance P. This latter finding is in agreement with the in situ hybridization studies of Rahman et al. (14).
Fig. 1.
RGS9-2 and Gβ5 mRNA are present in medium spiny neurons and in cholinergic interneurons within the striatum. (a) Representative ethidium bromide staining of RT-PCR products generated from either a single striatal neuron (lanes 2, 5, 8, 11, and 14) or a striatal cDNA-positive control preparation (lanes 1, 4, 7, 10, and 13). Lanes 3, 6, 9, 12, and 15 show the negative controls where no cDNA template was added to the PCR. Additional negative controls included the omission of reverse transcriptase (control for genomic DNA contamination of RNA samples). No PCRs were detected in these controls (data not shown). PCR fragments observed were of the expected sizes generated from rat gene-specific primers complementary to either substance P (doublet at 468 and 513 bp), enkephalin (477 bp), choline acetyltransferase (324 bp), RGS9-2 (1,300 bp), or Gβ5 (448 bp). M1 and M2 denote DNA standard markers. (b) Summary of single cell RT-PCR results indicating that RGS9-2 and Gβ5 are present in all neuronal subtypes examined. Total N is the total number of cells examined for each neuronal subtype. The number of cells positive for both RGS9-2 and Gβ5 mRNA are indicated to the right of the total.
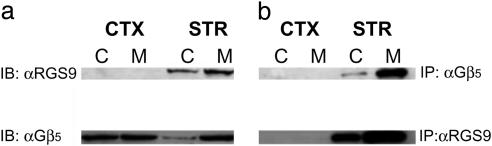
Not only were RGS9-2 and Gβ5 coexpressed in these striatal neuronal cell types, they also formed a molecular complex. RGS9-2/Gβ5 complexes were coimmunoprecipitated by anti-Gβ5 and anti-RGS9 antibodies from striatal homogenates, but not from cortical homogenates (Fig. 2b). RGS9-2/Gβ5 complexes were identified in both cytosolic and membrane compartments, with the majority of the complexes localized to the membrane. Although Gβ5 was present in the cortex, RGS9-2 protein was not detected in this tissue (Fig. 2a).
Fig. 2.
RGS9-2/Gβ5 complexes coimmunoprecipitate from the striatum. (a) Western blot showing RGS9-2 detected in striatal homogenates (STR) but not in cortical preparations (CTX), whereas Gβ5 is present in both. RGS9-2 and Gβ5 are found in both cytosolic (C) and membrane (M) extracts. (b) RGS9-2 and Gβ5 coimmunoprecipitate from both membrane and cytosolic compartments of the striatum with either RGS9 or Gβ5 antibodies, but do not coimmunoprecipitate from cortical negative controls. Data shown are representative of three identical experiments conducted. IP, immunoprecipitating antibody; IB, antibody used to probe Western blot after immunoprecipitation.
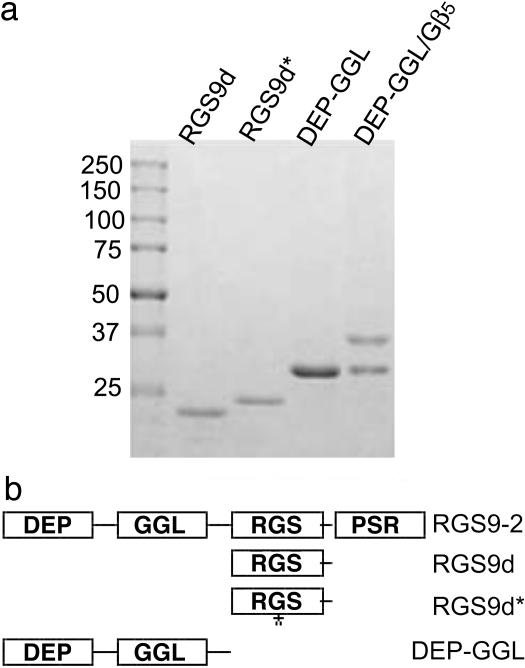
To determine the role of the catalytic and noncatalytic domains of RGS9 in regulating RGS9-2 GAP activity and function within the striatum, we purified a series of histidine-tagged RGS constructs from Escherichia coli or Sf9 insect cells for use in patch clamp experiments and in vitro GTPase activity assays. In addition to the RGS domain of RGS9 (RGS9d), we mutated RGS9 residue Ile-363 to Thr (RGS9d*). Based on the RGS4 crystal structure, Ile-363 is near one of the key residues within the RGS domain, which is believed to directly contact Gα subunits. Also, because the N-terminal domains of RGS9 have recently been shown to be necessary for localization and signaling, we purified the N-terminal DEP and GGL domains alone, and with Gβ5. As shown in Fig. 3, preparations of the RGS protein constructs were >90% pure.
Fig. 3.
Expression and purification of RGS9 constructs. (a) Coomassie-stained SDS/PAGE gel of N-terminally His6-tagged purified RGS9 constructs. Preparations were >90% pure. (b) Schematic diagram of RGS9 constructs. Domains are DEP (Dishevled/Egl10/Plextrin), GGL (G gamma-like), RGS (Regulator of G protein signaling), and PSR (proline/serine-rich).
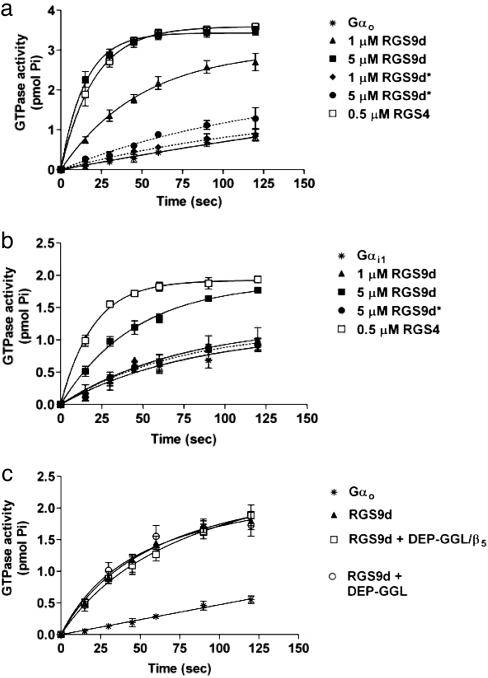
We began by testing these constructs for their ability to accelerate the intrinsic GTPase activity of Gαi and Gαo subunits in a single-turnover GTPase assay. As shown in Fig. 4 a and b, RGS9d accelerated the intrinsic GAP activity of both Gαi1 and Gαo, but it was a very poor GAP for Gαi. Addition of 5 μM RGS9d to Gαo increased the single turnover rate for GTP hydrolysis from 0.002 min-1 to 0.067 min-1 (30-fold stimulation), but it had only a 2-fold stimulation of Gαi (0.01-0.02 min-1). In contrast, RGS9d* produced only a 3-fold stimulation of Gαo GTPase activity, and RGS9d* failed to stimulate the GTPase activity of Gαi1 at concentrations as high as 10 μM. An RGS4 construct accelerated the single-turnover GTPase activity of both Gαo and Gαi1 (0.002-0.058 min-1 for Gαo and 0.01-0.04 min-1 for Gαi) suggesting that the weak ability of RGS9 to accelerate Gαi1 activity was inherent to the specific RGS9 protein sequence.
Fig. 4.
RGS9 stimulates the intrinsic GTPase activity of Gαo and Gαi1 in a single-turnover assay. (a) RGS9d accelerates Go GTPase activity (intrinsic rate constant k = 0.002 min-1) in a concentration-dependent manner (1 μM RGS9d, k = 0.021; 5 μM RGS9d, k = 0.067 min-1). Introduction of the point mutation I363T within the RGS domain near a critical Gi1 contact site (RGS9d*) eliminates the GTPase activity of this construct (1 μM RGS9d, k = 0.003; 5 μM RGS9d, k = 0.007 min-1). (b) RGS9d accelerates Gi1 GTPase activity (intrinsic rate constant k = 0.01 min-1) with lower potency than that observed for Gαo. Also similar to Gαo, RGS9d* is devoid of GAP activity toward Gαi1 (5 μM RGS9d*, k = 0.012; 1 μM RGS9d, k = 0.014; 5 μM RGS9d, k = 0.022 min-1). (c) Neither DEP-GGL nor DEP-GGL/Gβ5 directly influences the GAP activity of RGS9d toward Gαo. Neither construct affects the GTPase activity of Gαo in the absence of RGS9d.
Interestingly, neither DEP-GGL nor DEP-GGL/Gβ5 altered RGS9 GAP activity when measured in cell-free systems (Fig. 4c). This finding agreed with previous findings, which showed that the DEP domain increases the GAP activity of RGS9-1 in vivo through localization to receptor systems in the membrane rather than by directly affecting catalysis.
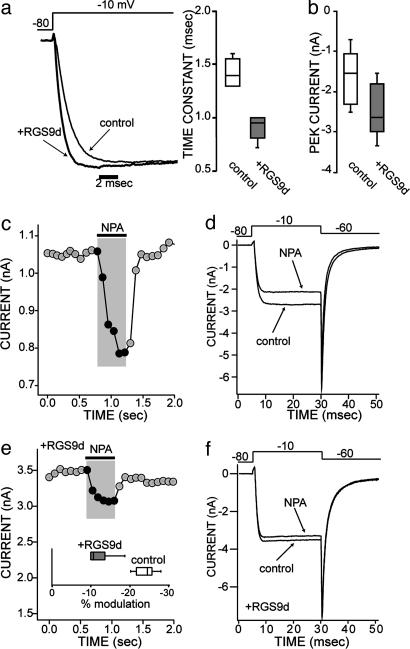
We next turned to electrophysiological approaches to determine how these constructs affected signaling in an intact neuron. Purified constructs were buffer exchanged (see Experimental Procedures) and dialyzed into cholinergic interneurons through the patch pipette. Dialysis of the core RGS domain of RGS9 (RGS9d) into the cell significantly accelerated the activation kinetics of Ca2+ channel currents and increased their amplitude (Fig. 5 a and b; n = 5, P < 0.05, Kruskal-Wallis). These alterations in current kinetics are consistent with the hypothesis that RGS9d dialysis attenuates ambient Gβγ inhibition of Cav2.2 Ca2+ channels (16, 26, 27).
Fig. 5.
The RGS domain of RGS9 (RGS9d, 10 μM) alters Ca2+ current modulation in striatal cholinergic interneurons, both in the absence of agonist (a and b) and in the presence of D2-dopaminergic receptor stimulation (c-f). In the presence of a GPCR agonist, the Ca2+ current is modulated so that the peak current is decreased (c and d). Addition of RGS9d in the pipette decreases this Ca2+ current modulation (e and f). (a and b) In the absence of agonist, addition of the RGS domain in the pipette speeds up the activation kinetics of the current (a), and the current amplitude increases (b). Patch clamp recordings showing the onset kinetics of Ca2+ currents in control conditions and with RGS9d included in the patch pipette in two typical neurons. Stimulatory step from -80 mV to -10 mV. (b) The box plot summary of the current amplitude in control conditions and in the presence of RGS9d (n = 6, P < 0.05 Kruskall-Wallis ANOVA). For smaller sized groups of data, where means do not necessarily give a good measure of central tendency, medians and ranges are given with box plots. In these plots, the central bar of the box represents the median, and the edges are the interquartiles (technically, fourths). The bars are lines drawn to the most extreme points in the sample group that are not outliers (defined as points beyond interquartile ± 1.5 interquartile range). (c) Plot of the peak current evoked by a pulse from -80 to -10 mV as a function of time; application of the D2 agonist R(-)-propylnorapomorphine (NPA, 10 μM) is indicated by the bar above the trace. (d) Individual current traces of voltage-activated Ca2+ currents after a stimulatory voltage step from -80 mV to -10 mV, then back to -60 mV. This trace shows the reduction in the current amplitude by NPA in the same cell as in a. (e) Plot of the peak current shows the NPA modulation of the Ca2+ current in one cell loaded with RGS9d. (Inset) The box plot summary of the current modulation by NPA in control conditions (n = 5) and in the presence of RGS9d (n = 6, P < 0.05 Kruskal-Wallis ANOVA). (f) Current traces corresponding to the same cell as shown in c.
Application of the D2 receptor selective agonist R(-)propylnorapomorphine (NPA) led to the inhibition of Ca2+ channel currents evoked by membrane depolarization (15). The signaling pathway mediating this modulation selectively targets Cav2.2 Ca2+ channels through a membrane-delimited Gβγ signaling cascade. NPA produced an ≈25% reduction in peak Ca2+ currents (Fig. 5 c and d), which represents a >60% reduction in Cav2.2 currents (15). RGS9d dialysis significantly blunted the modulatory influence of NPA (Fig. 5 e and f), reducing the median modulation to near 10% (n = 8, P < 0.05, Kruskal-Wallis).
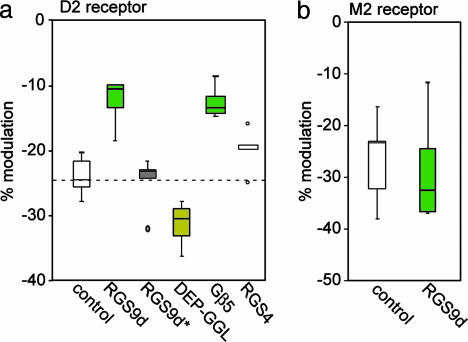
Patch-clamp recordings were repeated by using the mutant, catalytically inactive RGS9d*. Consistent with the absence of GAP activity for this construct, introduction of RGS9d* into the intracellular milieu of striatal cholinergic interneurons failed to alter D2-mediated Ca2+ current modulation (Fig. 6a; n = 5, P > 0.05, Kruskal-Wallis). These data support the contention that the RGS9d-mediated blunting of the D2 receptor modulation of Ca2+ channels resulted from its GAP activity at endogenous Gαi1 or Gαo proteins, probably Gαo (18, 28). Introduction of RGS4 did not have a significant effect on Ca2+ current modulation (n = 5, P > 0.05, Kruskal-Wallis), showing that this modulation is specific to RGS9.
Fig. 6.
Summary of the effects of different domains of RGS9-2 on D2 dopaminergic and M2 muscarinic modulation of Ca2+ channels. (a) Under control conditions, D2 dopaminergic stimulation causes an ≈25% decrease in Cav2.2 Ca2+ channel currents as shown in Fig. 4a. Introduction of the RGS domain of RGS9 results in a 50% decrease in D2-dopaminergic modulation of Ca2+ channels, by increasing the rate of Gi turnoff (see also Fig. 4c). RGS4 has no significant effect, demonstrating that this effect is specific to RGS9. Application of an RGS9 construct that is catalytically inactive, RGS9d*, fails to alter the D2 receptor-mediated modulation of Ca2+ currents. Introduction of the DEP/GGL domain of RGS9 (10 μM) antagonizes endogenous RGS9-2 and increases the impact of D2 receptor activation on Ca2+ channel currents. Introduction of Gβ5 decreases the ability of dopamine to modulate Ca2+ current as well as RGS9d, suggesting stabilization or recruitment of endogenous RGS9-2 to the membrane. (b) RGS9d does not modulate M2 muscarinic receptor-mediated signaling to Ca2+ channels, thereby demonstrating receptor-specific regulation by RGS9d. The box plot summary (see Fig. 5) of the current amplitude in control conditions and in the presence of RGS9d (n = 6, P < 0.05, Kruskal-Wallis ANOVA) is shown.
To determine whether the noncatalytic domains of RGS9 play a role in mediating the physiologic effects of endogenous RGS9-2, we introduced a purified DEP-GGL construct of RGS9 either alone or in a complex with Gβ5 (data not shown) into cholinergic interneurons and examined D2 receptor modulation of Ca2+ currents. Dialysis of the noncatalytic DEP-GGL domain of RGS9 enhanced the D2 receptor modulation (n = 8, P < 0.05, Kruskal-Wallis, Fig. 6a). This construct may be blocking endogenous RGS9-2 GAP activity in two ways. First, as seen with retinal RGS9-1, the DEP domain alone likely blocks endogenous RGS9-2 from reaching its intended target. Second, DEP-GGL may compete with their endogenous counterparts, thus blocking the specificity for Gαo. Dialysis with exogenous Gβ5 mimicked the effect of RGS9d, accelerating Ca2+ channel activation (n = 5, P < 0.05 Kruskal-Wallis), we believe by recruiting endogenous RGS9 to the membrane. This finding is consistent with previous work, which has shown that Gβ5 can stabilize RGS9, increasing its presence in the cell (29).
To determine whether RGS9 regulated other GPCR signaling cascades in cholinergic interneurons, we examined the M2 muscarinic receptor-mediated inhibition of Cav2.2 Ca2+ currents (15). These receptors are abundantly expressed by these neurons and also use a membrane-delimited G protein signaling cascade that targets Cav2.2 Ca2+ channels. However, there are several phenomeological differences between the M2 and D2 receptor cascades. In contrast to the D2 receptor effects, the M2 modulation is voltage-dependent and sensitive to protein kinase C inhibition, suggesting that there are key differences in the G proteins mediating the suppression of Ca2+ channel opening (30, 31). Surprisingly, RGS9d did not significantly alter M2 muscarinic receptor-mediated inhibition of Cav2.2 Ca2+ channel currents (n = 8, P > 0.05, Kruskal-Wallis) (Fig. 6b). Thus, RGS9-2 selectively regulates G proteins linked to the D2 receptor in striatal cholinergic interneurons.
Discussion
RGS proteins are key regulators of G protein lifetime, which accelerate the GTP hydrolysis that leads to signal turnoff (32-34). In the brain, many RGS proteins are expressed in a number of brain areas (8). Strikingly, one RGS protein, RGS9-2, is very highly expressed and discretely localized to the striatum, nucleus accumbens, and olfactory tubercle, all of which are integral components of the basal ganglia. This group of nuclei are involved in motor planning, drug seeking, and learning. Disruptions in striatal dopaminergic signaling are thought to underlie a variety of psychomotor disorders including drug abuse, schizophrenia, Tourette's syndrome, and Parkinson's disease.
Although RGS9 and Gβ5 mRNA and/or protein have been shown to be enriched in the striatum at the regional level, this and another recent study (14) have shown which cellular subtypes specifically express this protein. Recently, Rahman et al. (14) used double-labeling in situ hybridization to show that RGS9-2 is expressed in both the substance P/dynorphin-containing and the enkephalin-containing medium spiny neurons of the nucleus accumbens. Our single-cell RT-PCR results confirm this finding, and also show that RGS9-2 is expressed in the cholinergic interneurons of the striatum. In addition, by coimmunoprecipitation studies, we have shown that RGS9-2 and Gβ5 are part of a molecular complex in situ in the striatum. Of particular interest is the fact that RGS9 regulated D2 dopaminergic, but not M2 muscarinic, modulation of Cav2.2 Ca2+ channels in striatal cholinergic interneurons. Although most RGS proteins seem to be promiscuous in cell-free systems, this type of receptor specificity is becoming an increasingly common paradigm for RGS proteins. Receptor specificity for RGS proteins was originally demonstrated for RGS4, -1, and -16 in pancreatic acinar cells (35, 36). Recent studies In Chinese hamster ovary cells have also revealed receptor preferences for RGS proteins (37, 38). This study provides evidence of receptor specificity for RGS9 at the cellular level in native tissue. Taken together, these results imply that RGS proteins targeted for disruption by pharmacological interventions could be interrupted at very specific pathways within the cell.
This study also confirms a role for RGS9-2's noncatalytic domains in its signaling in an in vivo system. DEP domains are found in many signaling proteins, and have been shown to be involved in the localization of the retinal isoform to its membrane anchoring partner, R9AP (1). Although the striatal counterpart of R9AP has not yet been discovered, our results are consistent with this role for the DEP domain, because its introduction into the patch pipet increases D2 modulation of Ca2+ channels, presumably by blocking endogenous RGS9-2 from reaching its intended destination to speed up turn-off of the signal. These results are analogous to what Martemyanov et al. (1) have seen upon deletion of the DEP domain from RGS9-1 in mice.
The GGL domain of these proteins has been shown to form a complex with Gβ5. Interaction of Gβ5 with members of this RGS family leads to a selectivity for Go over Gi (3). A distinctive feature of the striatal splice variant of RGS9, RGS9-2, is the presence of a unique proline/serine rich domain of ≈200 aa (10). Although it has recently been shown that this domain shares sequence homology with the γ subunit of retinal cGMP phosphodiesterase (PDE6) and that it provides high-affinity interaction with its target G protein, probably Go (39), this domain may also impart striatal-specific functions that are unique to RGS9-2. Because we were unable to express this domain, its effects on this signaling pathway remain a target for future studies.
In summary, we have shown that RGS9-2 modulates dopamine receptor-mediated cellular responses within the striatum in a receptor-specific manner. RGS proteins (including RGS9-2) (14, 40) have been shown in a number of overexpression studies in heterologous systems to be able to regulate G protein-coupled receptor modulation of K+ and Ca2+ channels (reviewed in ref. 32). However, much less is known about physiological roles of endogenous RGS proteins in native tissues (see also refs. 41 and 42). We show that RGS9 attenuates an endogenous D2 receptor-mediated modulation of Cav2.2 Ca2+ channels. Introduction of noncatalytic domains of RGS9-2 enhances this modulation, likely by a disruption of endogenous RGS9 function. Finally, we show that, in these neurons, M2 muscarinic receptor signaling through the same Gi/o protein class to Cav2.2 Ca2+ channels is not affected by RGS9. This argues for a local, receptor-specific complex that includes RGS9-2. Because D2 receptors in cholinergic interneurons are known to control not only cellular excitability but also acetylcholine release (43), alterations in RGS9-2 function in these cells may contribute to striatal pathophysiologies with known cholinergic determinants, such as Parkinson's disease.
Acknowledgments
We thank Michelle Morrow, Sasha Ulrich, and Dr. Tatiana Tkatch for their assistance with single-cell RT-PCR measurements. This work was supported in part by National Institutes of Health (NIH) Grants EY10291 and EY06062 (to H.E.H.) and NS34696 and DA12958 (to D.J.S.), NIH Training Fellowships 5T32CA70085 and 1F32NS10955-01 (to T.M.C.-V.), and Swedish Cancer Foundation Grant 42100B99-01SAA (to A.K.S-A.).
Author contributions: M.M., D.J.S., and H.E.H. designed research; T.M.C.-V., S.H., M.M., and A.K.S.-A. performed research; T.M.C.-V., M.M., D.J.S., and H.E.H. analyzed data; and T.M.C.-V., L.R.E., M.M., D.J.S., and H.E.H. wrote the paper.
Abbreviations: RGS, regulators of G protein signaling; NPA, R(-)-propylnorapomorphine.
References
- 1.Martemyanov, K. A., Lishko, P. V., Calero, N., Keresztes, G., Sokolov, M., Strissel, K. J., Leskov, I. B., Hopp, J. A., Kolesnikov, A. V., Chen, C. K., et al. (2003) J. Neurosci. 23, 10175-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snow, B. E., Krumins, A. M., Brothers, G. M., Lee, S. F., Wall, M. A., Chung, S., Mangion, J., Arya, S., Gilman, A. G. & Siderovski, D. P. (1998) Proc. Natl. Acad. Sci. USA 95, 13307-13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posner, B. A., Gilman, A. G. & Harris, B. A. (1999) J. Biol. Chem. 274, 31087-31093. [DOI] [PubMed] [Google Scholar]
- 4.Snow, B. E., Betts, L., Mangion, J., Sondek, J. & Siderovski, D. P. (1999) Proc. Natl. Acad. Sci. USA 96, 6489-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang, J. J., Chen, H. H., Jones, P. G. & Khawaja, X. Z. (2000) J. Neurosci. Res. 60, 58-64. [DOI] [PubMed] [Google Scholar]
- 6.Zhou, J. Y., Toth, P. T. & Miller, R. J. (2003) J. Pharmacol. Exp. Ther. 305, 460-466. [DOI] [PubMed] [Google Scholar]
- 7.He, W., Lu, L., Zhang, X., El-Hodiri, H. M., Chen, C. K., Slep, K. C., Simon, M. I., Jamrich, M. & Wensel, T. G. (2000) J. Biol. Chem. 275, 37093-37100. [DOI] [PubMed] [Google Scholar]
- 8.Gold, S. J., Ni, Y. G., Dohlman, H. G. & Nestler, E. J. (1997) J. Neurosci. 17, 8024-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grafstein-Dunn, E., Young, K. H., Cockett, M. I. & Khawaja, X. Z. (2001) Brain Res. Mol. Brain Res. 88, 113-123. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, K., Howes, K. A., He, W., Bronson, J. D., Pettenati, M. J., Chen, C., Palczewski, K., Wensel, T. G. & Baehr, W. (1999) Gene 240, 23-34. [DOI] [PubMed] [Google Scholar]
- 11.He, W., Cowan, C. W. & Wensel, T. G. (1998) Neuron 20, 95-102. [DOI] [PubMed] [Google Scholar]
- 12.Thomas, E. A., Danielson, P. E. & Sutcliffe, J. G. (1998) J. Neurosci. Res. 52, 118-124. [DOI] [PubMed] [Google Scholar]
- 13.Rahman, Z., Gold, S. J., Potenza, M. N., Cowan, C. W., Ni, Y. G., He, W., Wensel, T. G. & Nestler, E. J. (1999) J. Neurosci. 19, 2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman, Z., Schwarz, J., Gold, S. J., Zachariou, V., Wein, M. N., Choi, K. H., Kovoor, A., Chen, C. K., DiLeone, R. J., Schwarz, S. C., et al. (2003) Neuron 38, 941-952. [DOI] [PubMed] [Google Scholar]
- 15.Yan, Z., Song, W. J. & Surmeier, J. (1997) J. Neurophysiol. 77, 1003-1015. [DOI] [PubMed] [Google Scholar]
- 16.Bean, B. P. (1989) Nature 340, 153-156. [DOI] [PubMed] [Google Scholar]
- 17.Gerfen, C. R. (1992) J. Neural Transm. Suppl. 36, 43-59. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Lopez, S., Tkatch, T., Perez-Garci, E., Galarraga, E., Bargas, J., Hamm, H. & Surmeier, D. J. (2000) J. Neurosci. 20, 8987-8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicola, S. M., Surmeier, J. & Malenka, R. C. (2000) Annu. Rev. Neurosci. 23, 185-215. [DOI] [PubMed] [Google Scholar]
- 20.Surmeier, D. J., Eberwine, J., Wilson, C. J., Cao, Y., Stefani, A. & Kitai, S. T. (1992) Proc. Natl. Acad. Sci. USA 89, 10178-10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surmeier, D. J., Yan, Z. & Song, W. J. (1998) Adv. Pharmacol. 42, 1020-1023. [DOI] [PubMed] [Google Scholar]
- 22.Baranauskas, G., Tkatch, T. & Surmeier, D. J. (1999) J. Neurosci. 19, 6394-6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mermelstein, P. G., Foehring, R. C., Tkatch, T., Song, W. J., Baranauskas, G. & Surmeier, D. J. (1999) J. Neurosci. 19, 7268-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman, D. M., Kozasa, T. & Gilman, A. G. (1996) J. Biol. Chem. 271, 27209-27212. [DOI] [PubMed] [Google Scholar]
- 25.Skiba, N. P., Yang, C. S., Huang, T., Bae, H. & Hamm, H. E. (1999) J. Biol. Chem. 274, 8770-8778. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, S. R. (1996) Nature 380, 255-258. [DOI] [PubMed] [Google Scholar]
- 27.Herlitze, S., Garcia, D. E., Mackie, K., Hille, B., Scheuer, T. & Catterall, W. A. (1996) Nature 380, 258-262. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, M., Spicher, K., Boulay, G., Wang, Y. & Birnbaumer, L. (2001) Proc. Natl. Acad. Sci. USA 98, 3577-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, C. K., Eversole-Cire, P., Zhang, H., Mancino, V., Chen, Y. J., He, W., Wensel, T. G. & Simon, M. I. (2003) Proc. Natl. Acad. Sci. USA 100, 6604-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swartz, K. J. (1993) Neuron 11, 305-320. [DOI] [PubMed] [Google Scholar]
- 31.Herlitze, S., Zhong, H., Scheuer, T. & Catterall, W. A. (2001) Proc. Natl. Acad. Sci. USA 98, 4699-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross, E. M. & Wilkie, T. M. (2000) Annu. Rev. Biochem. 69, 795-827. [DOI] [PubMed] [Google Scholar]
- 33.De Vries, L., Zheng, B., Fischer, T., Elenko, E. & Farquhar, M. G. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 235-271. [DOI] [PubMed] [Google Scholar]
- 34.Zhong, H. & Neubig, R. R. (2001) J. Pharmacol. Exp. Ther. 297, 837-845. [PubMed] [Google Scholar]
- 35.Xu, X., Zeng, W., Popov, S., Berman, D. M., Davignon, I., Yu, K., Yowe, D., Offermanns, S., Muallem, S. & Wilkie, T. M. (1999) J. Biol. Chem. 274, 3549-3556. [DOI] [PubMed] [Google Scholar]
- 36.Zeng, W., Xu, X., Popov, S., Mukhopadhyay, S., Chidiac, P., Swistok, J., Danho, W., Yagaloff, K. A., Fisher, S. L., Ross, E. M., et al. (1998) J. Biol. Chem. 273, 34687-34690. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein, L. S., Ramineni, S., Hague, C., Cladman, W., Chidiac, P., Levey, A. I. & Hepler, J. R. (2004) J. Biol. Chem. 279, 21248-21256. [DOI] [PubMed] [Google Scholar]
- 38.Ghavami, A., Hunt, R. A., Olsen, M. A., Zhang, J., Smith, D. L., Kalgaonkar, S., Rahman, Z. & Young, K. H. (2004) Cell. Signalling 16, 711-721. [DOI] [PubMed] [Google Scholar]
- 39.Martemyanov, K. A., Hopp, J. A. & Arshavsky, V. Y. (2003) Neuron 38, 857-862. [DOI] [PubMed] [Google Scholar]
- 40.Granneman, J. G., Zhai, Y., Zhu, Z., Bannon, M. J., Burchett, S. A., Schmidt, C. J., Andrade, R. & Cooper, J. (1998) Mol. Pharmacol. 54, 687-694. [PubMed] [Google Scholar]
- 41.Diverse-Pierluissi, M. A., Fischer, T., Jordan, J. D., Schiff, M., Ortiz, D. F., Farquhar, M. G. & De Vries, L. (1999) J. Biol. Chem. 274, 14490-14494. [DOI] [PubMed] [Google Scholar]
- 42.Jeong, S. W. & Ikeda, S. R. (2001) J. Physiol. 535, 335-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drukarch, B., Schepens, E. & Stoof, J. C. (1990) Neuroscience 37, 1-9. [DOI] [PubMed] [Google Scholar]