Abstract
The numerous processes that damage DNA are counterbalanced by a complex network of repair pathways that, collectively, can mend diverse types of damage. Insights into these pathways have come from studies in many different organisms, including Drosophila melanogaster. Indeed, the first ideas about chromosome and gene repair grew out of Drosophila research on the properties of mutations produced by ionizing radiation and mustard gas. Numerous methods have been developed to take advantage of Drosophila genetic tools to elucidate repair processes in whole animals, organs, tissues, and cells. These studies have led to the discovery of key DNA repair pathways, including synthesis-dependent strand annealing, and DNA polymerase theta-mediated end joining. Drosophila appear to utilize other major repair pathways as well, such as base excision repair, nucleotide excision repair, mismatch repair, and interstrand crosslink repair. In a surprising number of cases, however, DNA repair genes whose products play important roles in these pathways in other organisms are missing from the Drosophila genome, raising interesting questions for continued investigations.
Keywords: FlyBook, DNA damage, DNA repair, recombination
THE foundations of DNA repair research in Drosophila lie in studies of mutagenesis, initially to induce mutations to help elucidate foundational genetic principles, and later to investigate mechanisms of mutagenesis and molecular properties of mutations produced by different treatments. The first section of this chapter reviews studies of mutagens and mutagenesis in Drosophila. Next, approaches that have been used to study DNA repair processes in Drosophila are outlined, with emphasis on genetic approaches and fly-specific modifications of approaches used in other organisms. DNA repair genes in Drosophila are covered in the third section, and then the fourth section reviews what is known about various repair pathways, based on experimental studies and the gene content of the genome.
Mutagens and Mutagenesis
Ionizing radiation and chromosome breaks
In 1927, Hermann J. Muller published the seminal paper “Artificial transmutation of the gene,” in which he presented conclusive evidence that X-rays cause mutation (Muller 1927). Muller noted that, “In addition to the gene mutations, it was found that there is also caused by X-ray treatment a high proportion of rearrangements in the linear order of the genes.” It was presumed that X-rays somehow break chromosomes, but there was disagreement as to whether translocations resulted from the erroneous rejoining of two independent breaks, or whether a single break could lead to an illegitimate crossover (Painter and Muller 1929). In his classic address at the VIth International Congress on Genetics in 1932, Muller pointed out that the dose-response curves would be different if two breaks were involved vs. a single break (Muller 1932). Over the next decade a number of researchers published data supporting, at least at high doses, the “3/2 power rule,” consistent with two independent breaks preceding translocation formation (reviewed in Muller 1940). Implicit in this model was the notion that chromosome breaks rejoin through some chromosome repair mechanism.
Although the two-break model seemed to explain the origin of chromosome rearrangements, Friesen (1933) and Patterson and Suche (1934) showed that X-rays can also induce crossing over in male Drosophila, which had been shown to not have meiotic crossovers (Morgan 1912). Because most crossover chromosomes were homozygous viable, whereas a large fraction of translocations induced by X-rays were homozygous lethal, Patterson and Suche argued that different phenomena were involved and that perhaps these crossovers were similar to meiotic crossovers in females. Indeed, it is now thought that mitotic crossovers, like meiotic crossovers, result from repair of DNA double-strand breaks (DSBs) through homologous recombination (HR), whereas chromosome rearrangements often arise when two DSBs are repaired through an end-joining (EJ) pathway (see the section DNA Repair Mutants and Genes).
Chemical mutagenesis and base damage
The first report of a chemical mutagen was published in 1946, when Auerbach described her results from treatment of Drosophila with mustard gas (Auerbach and Robson 1944). One key difference noted between X-rays and mustard gas (and other chemical mutagens) was the ratio of chromosome rearrangements to “gene mutations,” with X-rays producing more of the former, and chemicals more of the latter. Auerbach also pointed out the ability of chemicals to produce “premutations,” wherein the progeny of males treated with mustard gas were frequently mosaic for induced mutations (Auerbach 1946). These observations, together with studies of ultraviolet light-induced mutations in bacteria, were major forces in the development of the idea of DNA repair as both a source of mutations, and a means to prevent mutation (reviewed in Auerbach 1978).
DNA Repair Assays Used in Drosophila
Mutagen sensitivity assay
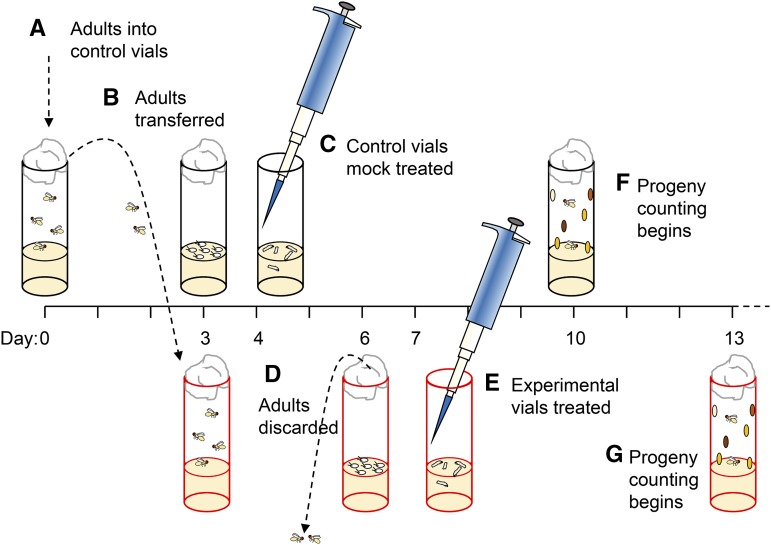
Perhaps the simplest, and most commonly used, DNA repair assay in Drosophila is the test for hypersensitivity to DNA damaging agents (Figure 1). Conceptually similar to replica plating of bacteria and fungi, this assay was first used by Smith (1973) in a screen for mutants that exhibited hypersensitivity to the alkylating agent methyl methanesulfonate (MMS). A cross that will yield both mutant and control progeny is set up in vials. After 2–4 days, the parents are transferred to another vial, from which they are discarded 2–3 days later. One vial is treated with a damaging agent, and one is left untreated or mock treated. Treatment is typically by addition of an aqueous or solvent solution containing the agent being tested, or by exposure to ionizing radiation in a gamma irradiator or a machine that generates X-rays. Ultraviolet (UV) light presents a special problem because it does not penetrate the medium. In this case, larvae may be collected on plates and spread in a “monolayer” for exposure, then transferred to vials. Treatment is done while larvae are feeding—a time when there is rapid proliferation of imaginal tissues. DNA damage leads to elevated cell death, especially in repair-defective mutants. Although there is substantial compensatory proliferation in imaginal tissues (e.g., Jaklevic and Su 2004), extensive cell death results in organismal death during pupal development.
Figure 1.
Mutagen sensitivity test. (A) On day 0, adults for a cross that generates mutant flies and sibling controls (usually heterozygous) are put into vials. (B) On day 3, the adults are transferred to new vials. The first set of vials is kept as a mock treated [(C) day 4, after eggs have hatched and larvae are feeding] or untreated control; the second set (brood two) is the treatment group. (D) On day 6, adults are removed from the brood two vials. (E) Second-brood vials are treated on day 7. (F, G) Adult progeny are counted, typically for 5–7 days after eclosion begins. The ratio of mutant to nonmutant is determined in each control vial (RC), and each corresponding treated vial (RT). Relative survival is expressed as RT/RC, with each vial being a separate biological replicate. The total number of flies in each treated vial, divided by the total number in the corresponding control vial, gives a measure of overall survival after treatment (there may be a difference in number of progeny between broods, even without treatment; some vials could be left untreated to determine the effects of brood-to-brood variation).
Adults are counted after emergence, and sensitivity is expressed as “relative survival,” calculated as the ratio of mutant to control with normalization to the untreated vial. Alternatively, an absolute measure of survival can be obtained by counting number of pupae from which adults eclose as a fraction of all pupae generated (provided they survive to pupariation). An estimate of the effects of the treatment on survival of the control class can be obtained by comparing the number of control progeny in the untreated vials to the number in the treated vials; however, there are often differences between the first and second broods, even without treatment.
A major weakness of this assay is that, for chemical mutagens, it is difficult to know the true dosage. Typically, some amount of a solution is applied to the surface of the food in a vial. The final concentration will therefore depend on the amount of food in the vial, the degree to which the agent diffuses to an even concentration (since this will not be immediate, concentrations in food ingested at early time points will be greater than those at later time points), the rate of breakdown of the agent, and other factors, some of which may be dependent on the makeup of the food. Thus, it is not possible to compare absolute sensitivity levels between different laboratories, or even at different times in the same laboratory. Nonetheless, general comparisons of the types of agents to which different mutants are hypersensitive have been useful. When interpreting such comparisons, it should be kept in mind that most agents produce multiple types of damage that may have different effects at different cell cycle stages; thus, it may not be possible to interpret sensitivity data as revealing defects in specific DNA repair pathways.
Another weakness is that the control class is often made up of siblings that are heterozygous for the repair mutation and a balancer chromosome. Although most DNA repair mutations are fully recessive, exceptions have been noted (see, for example, mei-41 in Boyd et al. 1976). Also, some balancers may have different sensitivities than others.
Because this assay measures survival in a whole organism, metabolic processes such as detoxification contribute to the outcome. Thomas et al. (2013) encountered an example of this while studying sensitivity of Brca2 mutants to camptothecin—an inhibitor of topoisomerase I. They found that one mutant was significantly more sensitive than the other, even though both carried amorphic alleles of Brca2. They traced the increased sensitivity to a common polymorphism in Cyp6d2, which encodes a cytochrome P450, and concluded that different alleles have different abilities to detoxify camptothecin and related compounds.
Genetic DNA repair assays
Several genetic assays of specific DNA repair processes have been developed. The earliest of these were for mitotic recombination, using twin spots of yellow and singed tissue in females heterozygous for y and sn mutations, as in Stern’s discovery of mitotic crossing over (Stern 1936). Somatic mitotic recombination has been assessed in flies heterozygous for mutations that affect body color, shape or color of bristles and hairs, and eye color. One of the most frequently used markers is multiple wing hairs (mwh)—a cell-autonomous marker that results in three to four hairs per cell in the adult wing, rather than the normal one hair per cell. This phenotype is easy to score under a microscope, as wings are flat and contain about 50,000 cells each. The position of mwh on distal 3L makes it possible to detect mitotic crossovers that occur along most of the length of the arm. Other types of loss of heterozygosity, such as gene conversion, deletion, and spontaneous mutation, can also be detected (Baker et al. 1978). It is not generally possible to distinguish between these different sources since molecular analyses cannot easily be conducted.
Mitotic crossing over has also been measured in the male germline. Because there are no meiotic crossovers in Drosophila males, crossovers induced by treatment, or that are elevated in mutant backgrounds, are readily detected; however, it is difficult to quantify the fold increase because spontaneous germline crossovers occur at too low a frequency to measure accurately. An advantage of this assay is that it is possible to recover the two reciprocal products of a single mitotic crossover in different progeny, and conduct molecular assays that may give insight into mechanism (e.g., Lafave et al. 2014).
A number of assays for DSB repair have been developed. In these assays, breaks are induced by excision of transposable elements, expression of a site-specific endonuclease such as I-SceI, or expression of a zinc finger nuclease or TALEN. There are several advantages to doing such assays in Drosophila. First, homologous chromosomes are paired in somatic cells and premeiotic germ cells of Dipteran insects (Stevens 1908; Metz 1916); consequently, DSB repair pathways often use the homologous chromosome as a template for DSB repair (Rong and Golic 2003). The development of site-specific transgene integration methods (Groth et al. 2004) has made it possible to engineer paired transgenes at allelic sites. This allows one to introduce a DSB in one transgene, and have the other available as a repair template at the same position on the homologous chromosome.
Cytological assays
Analyses of chromosome structure have been used for many years to assess effects of DNA damaging agents, and of spontaneous damage in mutants. Gatti and colleagues perfected techniques for analyzing squashes of chromosomes from neuroblast cells in larval brains (Gatti et al. 1974a,b, 1975; Gatti 1979). These are large, proliferative cells that are amenable to studies of mitotic chromosome structure. In addition to aneuploidy, chromosome abnormalities such as breaks (on one or both chromatids) can be observed. Gatti (1982) has also adapted a method to detecting sister chromatid exchanges (SCEs) in Drosophila neuroblasts. This method involves labeling DNA with the base analog bromodeoxyuridine (BrdU). From a dose-response curve, Gatti concluded that most SCEs in wild-type cells are induced by the BrdU labeling that is used to detect them.
Larval imaginal discs are attractive tissues in which to conduct cytological studies of repair because they are only two cell layers thick. Unfortunately, this advantage is offset by the small size of the cells. Earlier studies looked at whole-cell or whole-nucleus effects of DNA damage, such as phosphorylation of histone H2AV (the Drosophila ortholog of mammalian H2AX) (Madigan et al. 2002; Chiolo et al. 2011; Lake et al. 2013; Alexander et al. 2015), and apoptosis (e.g., Gorski et al. 2004; Jaklevic and Su 2004; Trowbridge et al. 2007). More recently, Janssen et al. (2016) performed live imaging of DNA repair focus formation imaginal disc cells after introduction of a single DSB.
Studies of DNA repair in other model organisms have made extensive use of cytological methods to detect repair proteins, which often form foci at sites of damage. This requires an antibody to the protein of interest or expression of fluorescent fusion proteins. Development of reagents for such studies has lagged behind in Drosophila, but some tools are available. Most notably, several antibodies to phosphorylated H2AV have been described, and these have been used to quantify DSBs in various tissues and in cultured cells (Madigan et al. 2002; Chiolo et al. 2011; Lake et al. 2013; Alexander et al. 2015). Antibodies to Rad51 and Rad54 have also been described (Alexiadis et al. 2004; Brough et al. 2008). Chiolo and colleagues expressed a number of fluorescently tagged DNA repair proteins, including Mus304 (ATRIP) and Mus101 (TopBP1), in cultured cells (Chiolo et al. 2011; Ryu et al. 2015). Janssen et al. (2016) used eYFP-tagged MU2 (the ortholog of MDC1) to visualize DSB repair foci in imaginal disc cells.
Other assays
The comet assay, also called single-cell gel electrophoresis, is widely used in studies of DNA repair (Olive and Banath 2006). Cells are embedded in agarose on a microscope slide, lysed, and then subjected to electrophoresis. DNA moves out of the cell to form a “comet tail,” with the intensity and length of the tail being proportional to the number of chromosome breaks, either as a direct result of damage, or generated during repair processes. Gaivão et al. (2014) have adapted this method for use in Drosophila. Importantly, their method can be used on flies and tissues, thus bringing all the power of Drosophila genetics to this approach.
RNA interference (RNAi) has been used by numerous Drosophila researchers for experiments in vivo, and in cultured cell lines, and many reagents are available (reviewed in Mohr 2014). Some studies have incorporated RNAi knockdown of DNA repair genes in vivo (e.g., Marek and Bale 2006), and in cells (e.g., Chiolo et al. 2011). Ravi et al. (2009) conducted an RNAi screen in Kc cells to identify genes required for resistance to MMS. Based on the 307 genes identified, and the 13 different pathways in which they function, these authors constructed an “MMS survival network.”
Cell extracts have been used in some Drosophila repair studies. When extracts are made from embryos or other tissues, genetic manipulations can be applied (e.g., Bhui-Kaur et al. 1998). Some researchers have done “in vivo extract” studies by injecting repair substrates into embryos, then recovering the products for molecular analysis. Examples include repair of DSBs after excision of a P element from a plasmid (O’Brochta et al. 1991; Beall and Rio 1996), or after introduction of linear plasmid and repair template (Ducau et al. 2000).
DNA Repair Mutants and Genes
Genetic studies have led to the identification of many genes involved in DNA repair and other DNA damage responses. Smith (1973) described a screen for X-linked mutations that conferred hypersensitivity to MMS, recovering a mutation he named mutS. Additional screens led to identification of dozens of genes that, when mutated, caused hypersensitivity to DNA damaging agents. A systematic nomenclature was adopted in which each gene is given the name musCNN, where C is the chromosome number (1 for the X chromosome), and NN is a two-digit ascending number. Thus, musS became mus101, and a larger screen for X-linked mutations by Boyd and colleagues (1976) identified mus102 through mus106. The set of mus genes now goes to mus115, mus219, and mus327. Some of these were found to be allelic to previously identified genes so some numbers are no longer in use (Table 1). Unfortunately, the nomenclature of the mei genes is the not the same as for the mus genes, as the former are mei-[D]N, where [D] is an optional letter that describes the source [e.g., mei-P22 was induced by P element mobilization, and mei-S332 was isolated from flies caught at a winery on Via Saleria outside Rome (Sandler et al. 1968; Sekelsky et al. 1999)] and N is a 1–4 digit number that may be meaningful only to the originator (e.g., mei-41 was the 41st vial screened by Baker and Carpenter 1972).
Table 1. Drosophila mus genes that have been cloned.
| mus | Genea | Human | Notes | Cloning Reference |
|---|---|---|---|---|
| mus101 | TOPBP1 | Essential; allelic to fs(1)K451 | Yamamoto et al. (2000) | |
| mus103, mus104 | mei-41 | ATR | mei-41 | Hari et al. (1995) |
| mus110 | mei-9 | ERCC4 | Human gene also known as XPF | Sekelsky et al. (1995) |
| mus201 | ERCC5 | Human gene also known as XPG | Sekelsky et al. (2000b) | |
| mus205 | REV3L | Encodes DNA Polζ | Eeken et al. (2001) | |
| mus209 | PCNA | PCNA | Essential | Henderson et al. (1994) |
| mus210 | Xpc | XPC | Sekelsky et al. (2000b) | |
| mus301 | HELQ | Allelic to spn-C. | McCaffrey et al. (2006) | |
| mus304 | ATRIP | Brodsky et al. (2000) | ||
| mus308 | POLQ | Encodes DNA Polθ | Harris et al. (1996) | |
| mus309 | Blm | BLM | Kusano et al. (2001) | |
| mus312 | SLX4 | Yıldız et al. (2002) | ||
| mus322 | Snm1 | DCLRE1A | Human gene also known as SNM1 and PSO1 | Laurençon et al. (2004) |
| mus324 | Gen | GEN1 | Andersen et al. (2011) |
Name of gene, if a different name is used in Flybase.
As the time of writing, only 14 of the 58 genes that came out of mutagen sensitivity genes have been identified molecularly (Table 1; this includes genes that have been renamed, such as mei-41 and PCNA, but not genes that were identified through other means and then found to cause a mus phenotype when mutated). It is likely that most, or all, of the mus genes encode orthologs of DNA repair proteins identified in other organisms; unfortunately, many are so poorly mapped it is not possible to use the genome sequence to make conjectures about which proteins they may encode.
Since the sequencing of the genome, numerous DNA repair genes have been identified by similarity between predicted protein and DNA repair factors discovered in other species; however, there are many cases in which genes that are critical for certain repair processes in other model organisms are not found in Drosophila. Several such cases were pointed out in an analysis of the original draft genome sequence of D. melanogaster, but the incomplete nature of the sequence and gaps in the assembly made it impossible to definitively demonstrate the absence of these genes (Sekelsky et al. 2000a). At present, the combination of more complete sequencing and annotation of the D. melanogaster genome, and the addition of more than a dozen other Drosophila species’ genomes makes it possible to conclude with high confidence that some DNA repair genes are missing, or have diverged beyond the ability of typical alignment search algorithms to detect. The addition of genomic sequences from dozens of other insects allows one to make inferences about when in the evolutionary history of Drosophila different genes were lost (see below).
DNA Repair Pathways in Drosophila
Base excision repair
Base excision repair (BER) removes bases that are damaged or inappropriate (reviewed in Friedberg 1996). BER begins with removal of the target base by a DNA glycosylase to create an apurinic/apyrimidinic (AP) site. Some glycosylases can catalyze nicking 3′ to the AP site, and some also 5′ to the AP site to produce a 1 nt gap. Alternatively, the AP site is nicked by an AP endonuclease (APE). DNA polymerase β (Polβ) then replaces a single nucleotide (short-patch BER), or another polymerase replaces a short stretch of nucleotides (long-patch BER).
The Drosophila genome encodes several glycosylases that, based on the activities of their human orthologs and/or biochemical studies, are predicted to be able to excise a wide variety of problematic bases (Table 2). One notable absence is an ortholog of the major uracil DNA glycosylase UNG. This enzyme is missing from Diptera and Lepidoptera, along with some Coleoptera and Hymenoptera. Muha et al. (2012) hypothesized that the lack of a UNG enzyme, together with the absence of expression of deoxyuracil triphosphatase (dUTPase) in some tissues, would lead to high levels of incorporation of U into larval DNA. They presented evidence that there are indeed high levels of uracil in nonimaginal larval tissues, and that this is correctly interpreted during replication and transcription. Drosophila has a novel protein, Uracil-DNA Degrading Factor (UDE), found only in holometabolous insects (those that undergo complete metamorphosis, including the orders Diptera, Coleoptera, Lepidoptera, and Hymenoptera), which has been shown to degrade uracil-containing DNA (Bekesi et al. 2007).
Table 2. Glycosylases in humans and Drosophila.
| Human | Drosophila | Substratesa |
|---|---|---|
| UNG | — | U |
| SMUG1 | CG5825 | U and modified U |
| TDG | Thd1b | T and U mispaired with G |
| MBD4 | MBD-R2c | T and U mispaired with G |
| OGG1 | Ogg1 | 8-oxoG, FapyG |
| RPS3 | RpS3 | |
| NTH1 | CG9272 | FapyG, hoC, hoU, Tg, urea |
| NEIL1, 2, 3 | — | Similar to NTH1 |
| MYH | — | A:8-oxoG |
| MPG | — | 3-MeA, hypoxanthine |
FapyG, 2,6-diamino-4-oxo-5-formamidopyrimidine; fU, fluorouracil; hmU, 5-(hydroxymethyl)uracil; hoU, 5-hydroxyuracil; Tg, thymine glycol; 3-meA, 3-methyl-adenine.
The largest predicted isoform of human TDG is 452 residues, but it is larger in insects—four times as large in Schizophora. It appears to have been lost from the Muscoidea superfamily as well as the Nematocera suborder (mosquitoes).
MBD4 has an N-terminal methyl-CpG binding domain (MeCP), and a C-terminal endonuclease III domain. Proteins with homology to the endo III domain, but lacking a MeCP domain are found in several arthropods in which and a MeCP domain orthologous to MBD4 cannot be found. In Holometabola (at least) the apparent ortholog of the MBD4 MeCP domain is in the middle of a large protein that has an N-terminal THAP domain, followed by a Tudor domain, the MeCP domain, and then a PhD finger domain. No endonuclease domain is found, suggesting this may not be a glycosylase.
A common product of exposure to reactive oxygen species or ionizing radiation is 7,8-dihydro-8-oxoguanine (8-oxoG). In yeast and human cells, Ogg1 glycosylase excises 8-oxoG. Drosophila has an Ogg1 enzyme that efficiently removes 8-oxoG from DNA (Dherin et al. 2000). In addition, studies of Drosophila ribosomal protein S3 revealed that it also has robust glycosylase activity on 8-oxoG, as well as AP lyase activity (Wilson et al. 1994). Subsequent studies with human S3 found that it has much weaker glycosylase activity, and is unable to do the second nicking reaction, but a single amino acid substitution to match the Drosophila sequence confers these properties (Hegde et al. 2001).
Humans have two APE genes, APEX1 and APEX2. The ortholog of APEX2 is missing in many insects (including all Holometabola), but is present in noninsect arthropods. The Drosophila ortholog of APEX1 is named Rrp1 for recombination repair protein 1. Properties of this protein have been characterized in vitro (Sander et al. 1991), but genetic studies have not been reported.
Drosophila lacks a DNA Polβ ortholog (Sekelsky et al. 2000a). Most insects lack other X family DNA polymerases (Polλ, Polμ, and terminal deoxynucleotidyl transferase), but Polβ is missing only from Diptera (and possibly a few other species, though these may be sequencing gaps). This suggests that Diptera use only long-patch BER, but this assumption has not been tested.
Nucleotide excision repair
The nucleotide excision repair (NER) pathway is best known for excising damage that distorts the helix, including bulky base adducts and products of UV irradiation (primarily cyclobutane pyrimidine dimers and 6-4-photoproducts) (reviewed in Friedberg 1996). Drosophila has all of the components of the central NER pathway discovered in mammalian cells. Genes encoding the orthologs of XPC (mus210), XPG (mus201), and XPF (mei-9), were identified in screens for mutants hypersensitive to the alkylating agent MMS (Boyd et al. 1976, 1981; Sekelsky et al. 2000b), suggesting that NER is an important mechanism for removing alkylated bases as well. Vogel and colleagues used 18 different mutagens (mostly alkylating agents) to induce mutations in the vermillion gene; they sequenced >600 mutations, producing a catalog of the types of changes induced by each agent (Nivard et al. 1999). Their studies included using mei-9 and mus201 mutants to explore the effects of loss of NER on mutation avoidance (e.g., Nivard et al. 1993). One conclusion is that AP sites are important substrates for NER, suggesting that NER can function after removal of an alkylated base by a glycosylase. Subsequent studies in other organisms reached similar conclusions (reviewed in Friedberg 1996).
In many eukaryotes, the template strand of transcribed regions is repaired more rapidly than the rest of the genome through a process termed transcription-coupled NER (TC-NER). Biochemical studies failed to find evidence for TC-NER in Drosophila (De Cock et al. 1992). Consistent with this finding, two specialized proteins that mediate TC-NER, ERCC6 and ERCC8, are both missing from Drosophila (Sekelsky et al. 2000a). ERCC6 is missing only from Diptera (and some nonarthropod phyla), whereas ERCC8 cannot be found in any Holometabola. This suggests that ERCC8 has (or has acquired) at least one important function outside of TC-NER, one that is independent of ERCC6.
Mismatch repair
The mismatch repair (MMR) system corrects base–base mismatches and small insertion/deletion (indels) heterologies generated during replication and recombination (reviewed in Kunkel and Erie 2015). These distortions are recognized and bound by heterodimers of Escherichia coli MutS homologs. In humans and budding yeast, mismatches are recognized primarily by MutSα (a heterodimer of Msh2 and Msh6), whereas indels are detected primarily by MutSβ (a heterodimer of Msh2 and Msh3). Msh3 is missing from most insects and a number of other phyla. Nonetheless, both indels and mismatches are repaired efficiently in Drosophila (see Negishi 2016 for a review of MMR in Drosophila). A genetic study of spellchecker1 (spel1), which encodes the Msh2 ortholog, found elevated instability in microsatellite sequences (Flores and Engels 1999), indicating defects in repairing indels that form in these sequences during replication. Structural studies of Drosophila Msh6 that might reveal how this protein has taken on the role of Msh3 in other organisms have not been reported.
Msh dimers recruit dimers of Mlh (MutL homolog) proteins. In yeast and human cells there are four Mlh proteins (listed here as yeast/HUMAN): Mlh1/MLH1, Mlh2/PMS1, Pms1/PMS2, and Mlh3/MLH3. These make three heterodimers: MutLα (Mlh1–Pms2), MutLβ (Mlh1–Pms1), and MutLɣ (Mlh1–Mlh3). There is no ortholog of Mlh3/MLH3 in Diptera (possibly not in Hemiptera either), and no ortholog of Mlh2/PMS1 in Diptera, Lepidoptera, and Coleoptera. Drosophila therefore has the capacity to make only MutLα, but this is the major Mlh heterodimer in mismatch repair in yeast and human cells (reviewed in Kunkel and Erie 2015).
Indels and mismatches are generated frequently during meiotic recombination, since recombination is frequently, or exclusively, between homologous chromosomes and these may have abundant heterologies relative to one another. Failure to repair these indels and mismatches results in both sequences being present in a single, haploid gamete, in the two different strands of one DNA duplex. These sequences are segregated into different daughter molecules at the first postfertilization S phase, and into different daughter nuclei at the ensuing mitosis. Occurrence of such “postmeiotic segregation” (PMS) has been used to study MMR and mechanisms of recombination.
Carpenter (1982) noted a high frequency of PMS in mei-9 mutants. Using Chovnick’s purine selection method (Chovnick et al. 1970) to recover ry+ progeny of mothers heteroallelic for two mutant ry alleles, she detected PMS based on transmission of a maternally derived mutant ry allele through the germline of a phenotypically ry+ recombinant, and by staining sectioned flies for xanthine dehydrogenase activity. These findings raised the possibility that MEI-9, which was later shown to be an NER endonuclease (Sekelsky et al. 1995), plays an unanticipated role in MMR. Support for this suggestion came from in vitro studies of Bhui-Kaur et al. (1998), who found that extracts made from mei-9 mutants had MMR defects. PMS in mei-9 mutants was analyzed at sequence level by Radford et al. (2007a). Using allele-specific PCR, they detected PMS at a much lower frequency than Carpenter (1982), but argued that the difference reflected use of different ry alleles—single base pair changes rather than the small indels used by Carpenter (1982).
Further insight into the function of MEI-9 in meiotic MMR came from the analysis of Msh6 mutants (Radford et al. 2007b). PMS was frequent in these mutants, both at base–base mismatches and at indels, indicating that these are both substrates for Drosophila MutSα. Many of the tracts had sites that appeared to have been repaired adjacent to sites that were unrepaired, suggesting the existence of a short-patch MMR system that could operate in the absence of the canonical Msh-Mlh-dependent system. This partial repair was not seen in mei-9 mutants (Radford et al. 2007a), so these latter authors proposed that short-patch MMR was really NER, as had been suggested for Schizosaccharomyces pombe (Fleck et al. 1999). This hypothesis was confirmed by Crown et al. (2014), who showed that repair of mismatches and indels generated during meiotic recombination was ablated in Xpc; Msh6 double mutants.
MMR proteins have other roles in recombination. One of these is in inhibiting recombination between highly diverged sequences. Using a transgene assay in which DSBs are made with I-SceI, and divergent or identical templates are provided downstream on the same transgene, Do and Larocque (2015) showed that this function exists in Drosophila, and that it requires Msh6.
Repair of interstrand crosslinks
Interstrand crosslinks (ICLs) in DNA are one of the most toxic types of DNA damage, since both strands are affected, blocking replication and transcription. The strong cytotoxic effects of crosslinking agents, such as cisplatin and related compounds, have been used extensively as chemotherapeutic agents. Reactive aldehydes produced by metabolism of alcohol and compounds commonly found in food are thought to be an important endogenous source of ICLs (Langevin et al. 2011). Repair of ICLs is not well understood, but is thought to occur through any of several different pathways, the best-studied being the Fanconi anemia (FA) pathway (reviewed in Lopez-Martinez et al. 2016).
The FA pathway begins with recognition and binding of the damage by the FA core complex. In human cells, the core complex has at least 11 proteins. Although most of these appear to be absent from most insects, many are poorly conserved at the sequence level, and it is possible that some may be identified through more intensive searches. The one exception is Fancm, which has orthologs throughout eukaryotes, and is related to the archaeal Hef protein (reviewed in Whitby 2010). FANCM plays an important role in recognizing ICLs at blocked replication forks, and recruiting other FA core complex proteins. Consistent with this function, Drosophila Fancm mutants are hypersensitive to agents that produce ICLs (Kuo et al. 2014). As in fungi, however, Fancm also has roles outside of the FA pathway, including a role in synthesis-dependent strand annealing (SDSA, described below).
A major function of the FA core complex is to promote ubiquitylation of FANCD2–FANCI by the FANCL E3 ubiquitin ligase. Drosophila has orthologs of all three proteins. RNAi knockdown of Drosophila Fancl or FancD2 results in phenotypes reflecting defects in repairing ICLs (Marek and Bale 2006). Ubiquitylation of FANCD2–FANCI is thought to recruit downstream processing factors, including a set of HR proteins. Several of these are also conserved in Drosophila, and the corresponding genes have mutant phenotypes suggestive of defects in ICL repair. Examples include Mus312 (called SLX4/FANCP in humans) (Boyd et al. 1981; Yıldız et al. 2002), and Mei-9 (XPF/FANCQ) (Yıldız et al. 2002).
Several additional mus mutants show hypersensitivity to crosslinking agents, but limited hypersensitivity to MMS (Boyd et al. 1981; Laurençon et al. 2004). The best-studied is mus308, which encodes DNA polymerase theta (Polθ) (Harris et al. 1996). Polθ has received attention in recent years because of its role in alternative end-joining (alt-EJ) pathways (see below). The finding that some cells deficient in the FA pathway have apparent defects in alt-EJ has led to the suggestion that the FA pathway may sometimes invoke DSB repair by alt-EJ instead of homologous recombination (Lundberg et al. 2001).
Another gene implicated in ICL repair is snm1 (originally named mus322, it was renamed when it was found to be orthologous to budding yeast SNM1 [sensitive to nitrogen mustard 1]; (Laurençon et al. 2004). Snm1 is a nuclease in the same family as the Artemis protein that functions in canonical nonhomologous end joining (NHEJ). Since Saccharomyces cerevisiae snm1 mutants have defects in ICL repair but the FA pathway is not found in fungi, it seems likely that Snm1 functions in an FA-independent ICL repair pathway.
DSB repair by HR
Repair of double-strand breaks (DSBs) has been studied extensively in a number of organisms, including Drosophila. The two general strategies are HR and EJ, with multiple forms of each type of repair. Studies in Drosophila have made important contributions to understanding several aspects of DSB repair (reviewed in Holsclaw et al. 2016).
Initial processing events:
Commitment to HR begins with resection of 5′ ends to leave 3′-ended single-stranded DNA. Resection is a complex process that involves a short-resection phase requiring the MRN (Mre11–Rad50–Nbs1) complex and CtIP/Sae2, followed by a long-resection phase that can be catalyzed by either Exo1, or the combination of Dna2 and Blm helicase (reviewed in Symington 2016). Orthologs of all of these proteins are found in Drosophila (Table 3). The ortholog of CtIP is more difficult to recognize, but Uanschou et al. (2007) suggested CG5872 as a probable candidate. Chiolo et al. (2011) reported that simultaneous depletion of CG5872, Tosca (Exo1), and Blm in Kc cells reduced formation of ATRIP (Mus304) foci, consistent with resection being reduced or absent.
Table 3. Genes whose products participate in resection and strand exchange.
| Human | Drosophila | Comments |
|---|---|---|
| RBBP8 | CG5872 | Encodes CtIP |
| BRCA1 | — | BRCA1 lost in Diptera and some other clades |
| MRE11 | mre11 | |
| RAD50 | rad50 | |
| NBS1 | nbs | |
| EXOI | tos | tos is the gene symbol for tosca |
| DNA2 | CG2990 | |
| BLM | Blm | Drosophila gene formerly named mus309 |
| RAD51 | spn-A | |
| RAD51B | — | Missing in most arthropods and in nematodes |
| RAD51C | spn-D | |
| RAD51D | Rad51D | Drosophila gene formerly named Rad51C |
| XRCC2 | Xrcc2 | Drosophila gene formerly named Rad51D |
| XRCC3 | spn-B | |
| SWM1 | Swm1 | Second open reading frame on mRNA encodes Rad1 |
| RAD54L | okr | |
| RAD54B | — | Lost several times in insects |
The breast cancer 1 (BRCA1) protein has multiple functions during HR, but the earliest is thought to be in regulating resection (reviewed in Prakash et al. 2015). BRCA1 is not found in Dipteran insects, and appears to have been lost independently in Hemiptera, Acari (ticks and mites), and some nonarthropod species. It is unknown how these species have replaced or eliminated this function.
Resection is followed by a homology search and strand exchange—a process catalyzed by Rad51, and aided by a number of accessory proteins. The Drosophila ortholog of Rad51 is encoded by the spindle-A (spn-A) gene (Staeva-Vieira et al. 2003), so-named for the aberrant shape of eggs laid by mutant females. This results from defects in both anterior-posterior and dorsal-ventral axis determination during eggshell patterning, which make the eggs look like spinning wheel spindles. These axis defects arise when a meiotic DSB repair checkpoint persists because repair is delayed due the absence of strand exchange activity, thereby uncoupling signaling between the oocyte nucleus and the follicle cells that secrete the eggshell (Ghabrial and Schüpbach 1999).
Formation and stability of Rad51 filaments on single-stranded DNA, and the homology search and strand exchange process, require several Rad51 paralogs (reviewed in Suwaki et al. 2011). Mammals have five paralogs: RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3. The Drosophila orthologs of RAD51C and XRCC3 are Spn-D and Spn-B, respectively, because spn-D and spn-B mutants have oocyte phenotypes similar to those of spn-A mutants. Notably, mammalian RAD51C and XRCC3 function as a heterodimer (“CX3”). A second paralog complex is BCDX2. Drosophila has orthologs of RAD51D and XRCC2. The genes encoding these were previously misnamed Rad51C and Rad51D, respectively, but the names were revised to Rad51D and Xrcc2 in 2016. Neither has been studied genetically, though it has been noted that Rad51D may correspond to rad201 (Radford and Sekelsky 2004). RAD51B orthologs appear to be missing from most of the ecdysozoa (arthropods, nematodes, and tardigrades), perhaps offering an opportunity to understand how the BCDX2 complex might function in the absence of Rad51B.
In addition to the Rad51 paralogs, the Shu complex helps to promote Rad51 activities (reviewed in Martino and Bernstein 2016). This complex is named for the yeast Shu2 protein, but the human ortholog is named SWS1 because of the protein’s SWIM domain. Drosophila has an ortholog of SWS1. Although this has not been studied genetically or biochemically, Godin et al. (2015) noted a strong signature of coevolution between Sws1, Rad51C, and Rad51D [unfortunately, they did not specify whether this was for the true orthologs of RAD51C (Spn-D) and RAD51D (formerly Rad51C), or the proteins previously misnamed as these (Rad51D and Xrcc2)]. A curious observation about Swm1 is that it is encoded on a bicistronic transcript, with the other open reading frame encoding Rad1, a protein that functions in DNA damage checkpoint signaling. The functional significance of this arrangement, if any, is unknown, but it appears to be conserved at least as far away as Hymenoptera. In some species, these genes are annotated as two different transcripts, but there are no empirical data that address whether Swm1 and Rad1 are encoded on overlapping transcripts or, as in Drosophila (based on cDNA sequencing), on a single transcript.
Strand exchange also requires Rad54, a Swi/Snf family chromatin remodeler. Budding yeast and mammals have a paralog, but the paralog has been lost in several insect clades, including Drosophila. The gene encoding the sole Rad54 protein is named okra because mutants have the same phenotype as the spn genes (spn- names were used for genes on chromosome 3, and small vegetable names for genes on 2). Some of the other genes in this group, such as gurken (grk) and spn-E, encode proteins involved more directly in nucleus-to-follicle cell signaling rather than in DNA repair; however, spn-C encodes a helicase orthologous to HELQ (McCaffrey et al. 2006). Mutations in this gene were also identified as mus301. Although the function of this helicase is poorly understood, the similarity in oocyte phenotype suggests that it may be required for strand exchange during meiotic recombination.
Double-Holliday junction model:
In 1983, Szostak and colleagues, based on experiments with budding yeast, proposed the DSB repair model for recombination (Szostak et al. 1983). (To avoid confusion with other DSB repair models, I will refer to this as the dHJ model because of the central intermediate, the double-Holliday junction.) A key feature of this model is that each Holliday junction in the dHJ is cleaved by a Holliday junction resolvase, resulting in either crossover or noncrossover products.
Several resolvases have been identified (Table 4). The genetics and biochemistry of these enzymes is complex, as they have overlapping functions and activities (reviewed in Rass 2013). Mus81 and its noncatalytic partner Mms4/EME1 appear to be a primary resolvase in yeast and mammalian cells, but Drosophila mus81 and mms4 mutants have relatively weak phenotypes (Trowbridge et al. 2007). Conversely, Yen1/GEN1 appears be largely redundant with, and secondary to, Mus81 in yeast and mammals, but Drosophila Gen seems to play more central roles than Mus81 (Andersen et al. 2011). Slx1 is a resolvase when complexed with the scaffolding protein Slx4 (reviewed in Svendsen and Harper 2010), but the in vivo functions of this enzyme are poorly understood. In S. cerevisiae, plants, and mammals, meiotic crossovers require a resolvase that contains MutLɣ (Mlh1 and Mlh3), plus Exo1 in a noncatalytic function (Zakharyevich et al. 2010, 2012). Drosophila lacks an Mlh3 ortholog (lost multiple times, including in Diptera), but genetic data suggest that a complex containing at least MEI-9, ERCC1, and Mus312 is the meiotic HJ resolvase (reviewed in Holsclaw et al. 2016).
Table 4. Genes whose products participate in Holliday junction processing.
| Human | Fly | Comments |
|---|---|---|
| MUS81 | mus81 | |
| EME1, EME2 | mms4 | |
| SLX1 | Slx1 | |
| SLX4 | mus312 | |
| GEN1 | Gen | Gene corresponds to mus324 |
| ERCC4 | mei-9 | ERCC4 also known as XPF |
| ERCC1 | ERCC1 | |
| MEIOB | hdm | |
| BLM | Blm | |
| TOP3A | Top3α | |
| RMI1 | — | Lost in Diptera |
| RMI2 | — | Missing from most insect genomes |
An alternative to resolution is dHJ dissolution, wherein the HJs are branch migrated toward one another, and the strands are decatenated by a topoisomerase. In mammalian cells, this reaction can be catalyzed by the BTR complex, consisting of the Bloom syndrome helicase (BLM), topoisomerase 3α, RMI1, and RMI2. The S. cerevisiae orthologous complex (STR, for Sgs1, Top3, and Rmi1) seems able to perform the same process in vivo (Dayani et al. 2011). The crystal structure of human TOP3α + RMI1 shows that RMI1 provides an important function: the decatenation loop that modifies the opening and closing of TOP3α so it can promote dissolution rather than relaxation (Bocquet et al. 2014). Drosophila Blm–TOP3α can carry out the dissolution reaction in vitro (Plank et al. 2006), but Rmi1 has been lost from Schizophora. Chen et al. (2012) noted that there are insertions in the C-terminus of Drosophila Top3α that promote interaction with Blm, and suggested that, since these insertions are not found outside of Schizophora, one of them may provide the decatenation loop function.
Double mutants that lack Blm and any one of the mitotic resolvases (Mus81–Mms4, Gen, or Mus312–Slx1) are inviable, with different double mutants dying at different stages (Trowbridge et al. 2007; Andersen et al. 2009, 2011). In the case of mus81; Blm and Gen Blm, mutations in spn-A partially suppress the lethality, consistent with lethality being due to inability to process HR intermediates. Notably, Gen Blm mutants die earlier than mus81; Blm double mutants, suggesting that Gen may have a more predominant role in HR than Mus81–Mms4.
Synthesis-dependent strand annealing:
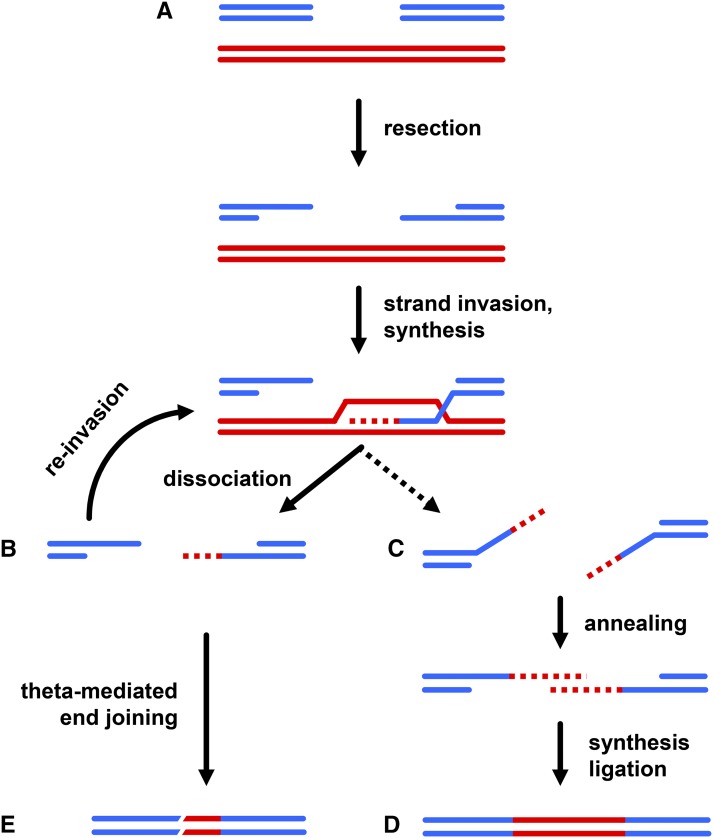
The dHJ model was the predominant model for HR for more than a decade, but many researchers now believe that a model termed synthesis-dependent strand annealing (SDSA) better reflects the most common HR pathway. The SDSA model arose out of studies of repair of double-strand gaps resulting from P element excision in Drosophila. One key observation is that repair of DSBs resulting from P element excision is rarely associated with crossing over, as would be predicted by the dHJ model (Nassif and Engels 1993). [Mobilization of P elements in the male germline does result in “male recombination,” but these arise during aberrant transposition rather than during the repair process per se (Sved et al. 1990)]. Studies with ectopic repair templates containing sequence modifications found that the two ends of the break could copy sequence from templates in different locations in the genome, and that a single end could use both an ectopic template and the sister chromatid (Nassif et al. 1994). These observations led Nassif et al. (1994) to propose a model in which the nascent strand produced by repair synthesis is displaced from the template and annealed to the second resected end; they coined the term SDSA to describe this process (Figure 2).
Figure 2.
Repair of gaps generated by P element excision. (A) Blue lines represent two strands of a DNA duplex from which a P element has excised, leaving a gap relative to the sister chromatid (red lines). Resection, Rad-51-mediated strand exchange, and repair synthesis occur as in most models of DSB repair by HR. In SDSA, the nascent strand is dissociated from the template. (B) For a gap, a single cycle of repair synthesis is unlikely to have spanned to gap to produce sequence complementary to the other end of the break (McVey et al. 2004). This end can then reinvade the template, and be extended by additional synthesis. The other end can also invade and be extended. The two ends can even invade different templates—an observation that was important in development of the SDSA model (Nassif et al. 1994). (C) Multiple such cycles of strand exchange, synthesis, and dissociation can produce single-stranded DNA that overlaps in a region of complementarity. (D) SDSA can then be completed by annealing these regions, resulting in restoration of the excised sequences. (E) In some cases, however, repair is completed by EJ, especially TMEJ (Adams et al. 2003; Chan et al. 2010). The result is partial gap filling, commonly seen as “internal deletions” in P elements.
Studies of SDSA mechanisms have been hampered by the fact that the result of SDSA, gene conversion without a crossover, usually is either invisible (if the sister chromatid or another identical template is used for repair), or is indistinguishable from dHJ dissolution or resolution that produces noncrossover products. This problem was solved through the use of specialized gap repair assays in Drosophila. Kurkulos et al. (1994) excised a P element carrying the wa allele, which has in intronic insertion of a copia retrotransposon, and found that a large fraction of the repair events had lost of all copia except a single long terminal repeat (LTR). They reasoned that the two ends of the DSB (a 14 kb gap relative to the sister chromatid) were both extended by repair synthesis off the sister chromatid, at least until each end had gone through an LTR. The LTRs from the two ends of copia then annealed via two-ended SDSA. This assay was adapted by Adams et al. (2003) to demonstrate that Blm promotes SDSA. McVey et al. (2004) subsequently presented evidence suggesting that repair of this large gap involves multiple cycles of strand exchange, synthesis, and dissociation.
Single-strand annealing:
If a DSB is made between a tandemly repeated sequence, resection may expose complementary sequences that can then anneal without strand exchange. This single-strand annealing (SSA) process was first described in budding yeast (Fishman-Lobell et al. 1992). SSA occurs efficiently in Drosophila, and has been used to reduce duplications produced by end-out gene targeting to a single copy (Rong and Golic 2003).
Depending on the extent of resection, and the length of complementarity, there may be overhangs that must be cleaved off before SSA can be completed. In yeast, this clipping is accomplished by the Rad1–Rad10 heterodimer, complexed with the scaffolding protein Slx4 and a yeast-specific protein called Saw1 (Ivanov et al. 1996; Flott et al. 2007). The orthologous nuclease (XPF–ERCC1) is also required for SSA in human cells (Al-Minawi et al. 2008). Wei and Rong (2007) screened a number of Drosophila DNA repair mutants for defects in SSA. Surprisingly, mutation of mei-9, which encodes the Rad1/XPF ortholog, or mus312, which encodes the Slx4 ortholog, did not decrease SSA significantly. Wei and Rong (2007) did find several other mutations that had an effect on SSA efficiency, the strongest identifying a gene they named ssar (SSA reducer) that maps to the distal half of 3L.
In yeast, Rad52 has an important role in promoting annealing during SSA, and other HR pathways (reviewed in Mortensen et al. 2009); however, Rad52 knockout mice have only weak repair defects, and no hypersensitivity to ionizing radiation (Rijkers et al. 1998). Thus, the absence of a Rad52 ortholog in Drosophila did not seem troubling. However, recent studies found that, in human cells, loss of RAD52 is synthetically lethal with loss of BRCA1 or BRCA2, suggesting that RAD52 is a backup for the roles of BRCA1 and BRCA2 in strand exchange (Lok et al. 2013). This begs the question of how strand exchange works in the absence of both Rad52 and BRCA1, as is the case in Diptera. This situation also seems to have arisen independently in Hemiptera, and many noninsect arthropods (but not Daphnia pulex, which orthologs of both). Several other clades, including Coleoptera and Nematoda, appear to have lost Rad52, but retained BRCA1.
DSB repair in heterochromatin:
A number of studies in yeast and mammalian cells have indicated important roles for chromatin modifications during DSB repair. Little work has been done in Drosophila on specific modifications associated with repair processes, with the exception of H2AV phosphorylation. In mammalian cells, the histone H2A paralog H2AX gets phosphorylated (termed ɣH2AX) in the vicinity of DSBs (reviewed in Dickey et al. 2009). The Drosophila counterpart, H2AV, is similarly phosphorylated at DSBs in mitotic and meiotic cells (Madigan et al. 2002; Joyce et al. 2011; Lake et al. 2013).
Chiolo et al. (2011) studied repair of DSBs generated within the heterochromatin domain of Drosophila Kc cells after exposure to X-rays. They found that these DSBs are repaired by HR, but that, after resection, the DSB is moved out of the heterochromatin domain before Rad51 filaments are assembled; within heterochromatin, Rad51 filament formation was blocked by the cohesins Smc5 and Smc6. One might expect that SSA would be a common mechanism for repair of DSBs generated in highly repetitive sequences that are common with heterochromatin, but Chiolo et al. (2011) concluded that Rad51 is required for repair of DSBs in heterochromatin.
DSB repair by EJ
In contrast to HR, EJ repair mechanisms do not make use of external homologous templates. The term NHEJ was originally used to describe the primary pathway of this type, but it is now recognized that there are several different EJ repair mechanisms.
Canonical NHEJ:
Canonical NHEJ (c-NHEJ) begins with binding and bridging of the ends by DNA-dependent protein kinase (DNA-PK), which consists of the Ku heterodimer (Ku70 and Ku80) and a catalytic subunit (DNA-PKcs) (reviewed in Radhakrishnan et al. 2014). Drosophila has orthologs of Ku70 and Ku80, but DNA-PKcs was lost in Schizophora, Coleoptera, and some other insects, though it is still found in mosquitoes (Table 5). Ku70 was originally identified as inverted repeat binding protein (Irbp), because it was thought to bind specifically to P element ends (Beall et al. 1994), and was identified independently as a component of yolk protein factor 1 (YPF1), an abundant component of the embryonic yolk (Jacoby and Wensink 1996).
Table 5. Genes whose products participate in end joining repair of DSBs.
| Human | Fly | Comments |
|---|---|---|
| XRCC6 | Irbp | Encodes Ku70 |
| XRCC5 | Ku80 | Encodes Ku80 |
| PRKDC | — | Encodes DNA-PKcs. Lost in Diptera |
| DCLRE1C | — | Encodes Artemis. Lost in Diptera |
| POLK | — | Encodes Polκ. Missing in most insects |
| POLL | — | Encodes Polλ. Missing in most insects |
| DNTT | — | Encodes TdT. Missing in most insects |
| LIG4 | Lig4 | |
| XRCC4 | CG3448 | |
| NHEJ1 | CG12728 | NHEJ1 also known as XLF |
| CG32756 | Adjacent paralogs in D. melanogaster species group | |
| POLQ | mus308 | Encodes DNA polymerase θ |
If the broken ends are damaged, or there are overhangs that are not cohesive, the ends may be processed by the endonuclease Artemis, or the X family DNA polymerases Polλ, Polμ, or terminal deoxynucleotidyl transferase (TdT). Artemis is missing in Schizophora, and most insects lack Polλ, Polμ, and TdT orthologs. There is, nonetheless, strong evidence for end processing in c-NHEJ in Drosophila (e.g., Bozas et al. 2009); it is unknown what nucleases and polymerases carry out this processing.
The ligation step in c-NHEJ is accomplished by a specialized complex containing DNA ligase 4 (LigIV), XRCC4, and XLF. Gorski et al. (2003) made deletions of Lig4, and demonstrated hypersensitivity to ionizing radiation at certain developmental stages. No studies of CG3448, which encodes the ortholog of XRCC4, have been reported. D. melanogaster has two orthologs of XLF, CG12728 and CG32756, that differ only in the C-terminus. These apparently arose from a tandem duplication present only in the melanogaster species complex.
Theta-mediated EJ:
EJ that does not require Ku or LigIV has been called alt-EJ (reviewed in Rodgers and McVey 2016). In one variation, sometimes called microhomology-mediated end joining (MMEJ), short homologies (6–20 bp) near the broken ends are annealed, and the overhangs are trimmed. In other cases, there are insertions that are longer than the 1–3 bp of c-NHEJ, and these often appear to be templated from sequences near the junction; this has been called synthesis-dependent MMEJ (SD-MMEJ) (Yu and McVey 2010).
As with SDSA, Drosophila experiments employing P element excision led to a breakthrough in understanding alt-EJ pathways. The vast majority of repair after P excision normally begins with HR, perhaps in part because the 17 nt 3′ overhangs left by transposase are likely to be a poor substrate for c-NHEJ (Beall and Rio 1997; Gloor et al. 2000). However, if HR is prevented by spn-A mutation, breaks are still repaired efficiently, primarily by alt-EJ (McVey et al. 2004). Chan et al. (2010) investigated the effects of Polθ on EJ under these conditions. They found that mus308 mutants had a significant decrease in use of long microhomologies typical of MMEJ, as well as changes in nucleotide insertion. Subsequent studies revealed that human Polθ also has important roles in Ku-independent EJ, leading some to propose the name theta-mediated end joining (TMEJ) instead of alt-EJ (reviewed in Wood and Doublie 2016). Another important contribution of the Drosophila research came from the discovery that mus308 spn-A double mutants have reduced viability and fertility, indicating that TMEJ is critical when HR is compromised (Chan et al. 2010). This result was also confirmed in human mammals, when it was found that Polθ is essential and frequently upregulated in tumors in which HR is compromised (Ceccaldi et al. 2015).
Relationship between HR and EJ
Discussion of usage of HR vs. EJ typically revolves around phases of the cell cycle: EJ is thought to predominate in G1, HR in S and G2 (reviewed in Chapman et al. 2012). Studies in Drosophila have shown that there are also differences between different tissues and different developmental stages.
In the male germline, most DSBs resulting from P element excision use HR for repair (Geyer et al. 1988; Engels et al. 1990). Although gaps of at least 44 kb can be repaired by SDSA, the efficiency of completed SDSA decreases with increasing gap size (Johnson-Schlitz and Engels 2006). In cases where the gap is not completely filled, EJ (primarily TMEJ) completes repair after variable amounts of synthesis from one or both ends (Adams et al. 2003; Chan et al. 2010). It is thought that this dual usage of HR and EJ may account for the accumulation of internally deleted DNA transposable elements, contributing to their extinction in genomes (McVey et al. 2004). The signals that cause SDSA to be aborted so repair can be completed by EJ are unknown.
Although HR is used predominantly in the germline, EJ may be more predominant in somatic cells. In assays in which plasmids carrying P elements are injected into embryos, or transfected into cultured cells and then recovered after several hours, all repair appears to be by EJ (O’Brochta et al. 1991; Beall and Rio 1996; Min et al. 2004), though it is possible these plasmid-based assays do not allow HR. Even with EJ, however, there are differences between germline and somatic repair. Gloor et al. (2000) used an in vivo assay that selected P element excision events repaired by EJ to show that germline repair events retained more of the 17-nt overhang, and more sequence insertion than the somatic events (which were similar to the plasmid-based assay repair events in other studies).
There is also evidence for different usage of HR and EJ in different developmental stages. Gorski et al. (2003) found that Lig4 mutants are most sensitive to ionizing radiation in midembryonic stages of development, becoming progressively less hypersensitive as development progressed. Preston et al. (2006), using an assay in which a DSB is made by the I-SceI nuclease, found that SSA was most frequently early in male germline development, SDSA and/or dHJ in middle stages, and NHEJ in late stages (during or after meiosis). Similarly, Chan et al. (2011) expressed I-SceI from different germline promoters, and concluded that HR predominates before meiosis, whereas EJ becomes more frequent during and after meiosis.
Repair of damage encountered during replication
It is thought that problems encountered during replication are responsible for a large amount of DNA damage in proliferating cells (Liu et al. 2016). Some types of base damage can block replicative polymerases. On the lagging strand, this can result in a single-strand gap, but blocks on the leading strand can block fork progression. Two major strategies for dealing with these problems are use of translesion polymerases and fork regression. These processes, and some models of fork stalling and breakage, are discussed in this section.
Translesion synthesis:
Translesion polymerases have specialized activities that can bypass certain types of damaged bases on the template (reviewed in Waters et al. 2009). The eukaryotic translesions polymerases are Rev1, Polζ, Polκ, Polη, and Polι. Drosophila has all of these except Polκ, which appears to be missing from all insects, with the curious exception of the tephritid fruit fly Rhagoletis zephyria. Genetic studies of all but DNApol-ι have been published. The catalytic subunit of Polζ is encoded by mus205 (Eeken et al. 2001), which was originally discovered in screens for hypersensitivity to MMS (Smith et al. 1980). Kane et al. (2012) made mutations in Rev1 and DNApol-η. Rev1 mutants were hypersensitive to ionizing radiation, whereas DNApol-η mutants were hypersensitive only to UV. These authors reported defects in HR in these mutants, and in mus205 mutants, but lesion bypass studies have not been reported. Nonetheless, hypersensitivity of DNApol-η mutants to UV is consistent with the role of Polη in bypassing pyrimidine dimers (Johnson et al. 1999). Likewise, Eeken et al. (2001) argued that the mutagenicity of mus205 mutants suggests that Drosophila Polζ is involved in lesion bypass.
In yeast and human cells, the signal to switch from a replicative polymerase to a translesion polymerase involves monoubiquitylation of PCNA by the ubiquitin E3 ligase Rad18–Rad6. Curiously, Rad18 is not found in Diptera, Lepidoptera, or Coleoptera, raising the question of how the switch to translesion polymerases is regulated.
Fork regression:
When a replication block is encountered on the leading strand, the fork can undergo regression, in which the newly synthesized leading and lagging strands are annealed to one another (Branzei and Foiani 2010). This converts the three-armed fork into a four-armed Holliday junction, and minimizes the amount of ssDNA. Fork regression positions the lesion away from the junction so it can be repaired; alternatively, the leading strand can be extended, using the lagging strand as a template. In either case, reversal of the regression can restore fork structure for continued replication. In human cells, the related helicases SMARCAL1 (also called HARP), ZRANB3, and HLTF have been suggested to catalyze fork regression (Bétous et al. 2012; Weston et al. 2012; Kile et al. 2015). Neither ZRANB3 nor HLTF are found in arthropods, but orthologs of SMARCAL1 are widely distributed. However, a biochemical study of Drosophila Marcal1, done alongside human SMARCAL1, failed to detect fork migration activity in the Drosophila protein (Kassavetis and Kadonaga 2014). Thus, it is unknown whether fork regression occurs in Drosophila, and, if so, what protein(s) catalyze this process.
Several helicases in the RecQ family have been suggested to catalyze either fork regression or reversal of regressed forks (Kanagaraj et al. 2006; Ralf et al. 2006; Machwe et al. 2011). Humans have five RecQ helicases: RECQL (also called RECQ1), BLM, WRN, RECQ4, and RECQ5. Orthologs of BLM, RECQ4, and RECQ5 are found in Drosophila (Sekelsky et al. 2000a). RECQL appears to have been lost once, as it cannot be found in any of the currently sequenced genomes of species in the Brachycera suborder. WRN is more complex. Human WRN has an amino-terminal exonuclease domain, and a carboxy-terminal RecQ helicase domain; outside of Vertebrata, these domains are encoded by separate genes. Orthologs of the helicase have been lost multiple times even within Schizophora. Within the acalyptratae subsection, WRN helicase orthologs can be found in species in the Drosophila subgenus, but not those in the Sophophora subgenus (which includes D. melanogaster) or in tephritid fruit flies. In the calyptratae subsection, a WRN helicase is found in Stomoxys calcitrans (stable fly), but not Musca domestica (housefly, in the same family as Stomoxys; this may be a sequencing gap but WRN helicase is not found in either the genome or transcriptome assemblies). It is also not detectable in any of the six Glossina (tsetse fly) genome sequences, but is found in Lucillia cuprina (Australian sheep blowfly).
In contrast to this sporadic loss of the helicase domain, orthologs of the exonuclease domain are retained throughout animal species. Studies of hypomorphic mutations in WRNexo found phenotypes potentially associated with aberrant replication fork repair, including elevated mitotic recombination and sensitivity to the topoisomerase I inhibitor camptothecin (Cox et al. 2007; Saunders et al. 2008; Boubriak et al. 2009). Bolterstein et al. (2014) generated amorphic and putative nuclease-dead alleles, and reported nuclease-independent hypersensitivity to the fork-stalling agent hydroxyurea, leading these authors to propose that WRNexo functions in fork regression together with Blm.
Drosophila Blm has been studied extensively. Mutations in mus309 (the original name of the gene, used in this section for continuity) were initially recovered based on extreme hypersensitivity to a broad range of DNA damaging agents (Boyd et al. 1981). Beall and Rio (1996) reported that mus309 encoded the Ku70 NHEJ protein, based on mapping to a small deletion, and partial rescue by transgenes. This error in assignment occurred because the gene encoding Ku70 (Irbp) is only about 330 kb from mus309, and, remarkably, overexpression of Ku70 alone can partially suppress phenotypes caused by loss of Blm. Kusano et al. (2001) correctly identified the product of mus309 as Blm helicase because Df(3R)T7, which does not uncover Irbp, fails to complement mus309. Importantly, they found that both extant mutant alleles has sequences changes that affected Blm protein: mus309D2 is a nonsense mutation, and mus309D3 is a putative helicase-dead missense mutation. Kusano et al. (2001) confirmed the partial rescue of mus309 mutant phenotypes by overexpression of Ku70 that was reported previously. Interestingly, they also reported that mus309D2/Df(3R)T7 males exhibited partial sterility that was rescued by a Blm transgene, though other researchers have found normal fertility and fecundity in males heteroallelic for null alleles of mus309 (Adams et al. 2003; McVey et al. 2007). These observations illustrate the complexities inherent in studies of repair, where different pathways can compensate for one another, and in studies of Drosophila, where strain differences may influence outcome of experiments.
Genetic studies of RecQ5 and RecQ4 mutants have also been published. RecQ4 is an essential gene that is required for cell proliferation and DNA replication (Wu et al. 2008; Xu et al. 2009; Crevel et al. 2012). In contrast, RecQ5 mutants are viable and fertile, though embryos from homozygous mutant females have a small, but significantly elevated, frequency of anaphase bridges during syncytial cycles (Nakayama et al. 2009), as well as sensitivity to cisplatin (Maruyama et al. 2012)—phenotypes that suggest a role replication fork repair.
In summary, studies of WRNexo, Blm, RecQ4, and RecQ5 suggest that each of these has roles in replication and/or replication fork repair, but the exact functions are not known, and even whether blocked forks under regression in Drosophila is unknown.
Fork collapse:
Some replication fork problems may lead to breakage of one or more strands, sometimes called fork collapse. In cycling cells, fork collapse results in a one-ended DSB that must be repaired in such a way as to reestablish a fork for continued replication (Branzei and Foiani 2010; Petermann and Helleday 2010). An alternative, proposed by Andersen et al. (2011) and common in Drosophila, is that replication from outside the break (from the nearest adjacent fork, or firing of a nearby dormant replication origin) converts the one-ended DSB into a two-ended DSB that can be repaired using the new sister chromatid as a template.
Drosophila offers some interesting, developmentally programmed, occurrences of fork stalling and collapse. Many larval tissues undergo endocycles, in which cells cycle through S (synthesis) and G (gap) phases with no mitosis, resulting in polyteny or polyploidy (reviewed in Lilly and Duronio 2005). The best-studied example occurs in the larval salivary glands, which can go through up to 10 S phases to produce the giant polytene chromosomes that inspired the first physical maps of the genome (Painter 1934). Gall et al. (1971) showed that pericentromeric satellite sequences are underrepresented in polytene chromosomes; genomic analyses subsequently identified a number of additional regions that are underrepresented relative to most euchromatic sequences (Belyakin et al. 2005; Nordman et al. 2011; Sher et al. 2012). This underrepresentation is due to underreplication, caused when forks encounter the SUUR (suppressor of underrepliation) protein (Belyaeva et al. 1998; Sher et al. 2012). This raises the question of what happens to forks blocked by SUUR? Glaser et al. (1992) reported evidence the forks do not build up in the transition zones between euchromatin and heterochromatin. Yarosh and Spradling (2014) analyzed whole-genome sequence from salivary glands, and found numerous deletions spanning zones of underreplication. They concluded that blocked forks break, and that different single-ended DSBs were then joined together, presumably through one of the EJ pathways.
Another interesting example comes from gene amplification. To increase production of chorion proteins during eggshell formation, follicle cells amplify genomic regions carrying these genes. This is done by repeated firing a replication origin in amplification region (Spradling and Mahowald 1980; Calvi et al. 1998). Alexander et al. (2015) investigated the fates of the resulting nested replication forks, and found evidence of DSBs. They also found that fork progression slowed in Lig4 mutants, but not in spn-A mutants, suggesting that NHEJ is involved in repairing these broken forks to allow continued elongation.
DNA damage checkpoint proteins
The presence of DNA damage can impact the cell cycle through checkpoint pathways (reviewed in Shaltiel et al. 2015). Most checkpoint genes studied in mammals and yeast are found in Drosophila, and the major pathways appear to be highly conserved (reviewed in Song 2005). These pathways are perhaps best discussed in the context of cell cycle regulation; this section focuses instead on DNA repair roles of proteins known primarily for their functions in checkpoint pathways.
Screens for hypersensitivity to DNA damaging agents identified some checkpoint genes. Most notable among these are mei-41, which encodes the ATR checkpoint kinase (Boyd et al. 1976; Hari et al. 1995), and mus304, the ortholog of ATRIP (Boyd et al. 1981; Brodsky et al. 2000). It could be argued that checkpoint defects alone can lead to hypersensitivity to DNA damage. However, Jaklevic et al. (2004) showed that mutations that affect DNA repair and the DNA damage checkpoint, or DNA repair alone, result in hypersensitivity to damage, but mutations that affect only the checkpoint, such as grp lok double mutants (these encode the orthologs of Chk1 and Chk2) do not. These authors considered MEI-41 to be in the category of proteins required in checkpoint and repair pathways. Consistent with this, LaRocque et al. (2007) found that mei-41 mutants had defects in a DSB repair assay that were more severe than those of grp lok double mutants.
The MRN complex likewise has direct roles in repair (described above), and in checkpoint signaling (reviewed in Zhang et al. 2006). However, no mutations in genes encoding MRN components (mre11, rad50, and nbs) were identified in screens for mutagen sensitivity. This is presumably because these genes are all essential due to the role of MRN in telomere capping (Bi et al. 2004; Ciapponi et al. 2004, 2006; Gorski et al. 2004). Repair functions of NBS have been studied in mutants that harbor hypomorphic or separation-of-function mutations (Mukherjee et al. 2009).
TopBP1 (topoisomerase binding protein 1) is another protein with roles in checkpoint signaling and DNA repair (reviewed in Wardlaw et al. 2014). Drosophila TopBP1 is encoded by mus101 (Yamamoto et al. 2000), one of the first mutagen-sensitivity loci to be discovered (Boyd et al. 1976). These first alleles proved to be hypomorphic, but subsequent mutations were found in screens for female sterility (Komitopoulou et al. 1983), and for temperature-sensitive lethality (Gatti et al. 1983; Smith et al. 1985).
Concluding Remarks and Unanswered Questions
As noted throughout this chapter, Drosophila is missing a number of proteins that are critical for DNA repair in other model organisms. For this reason, Drosophila has sometimes been viewed as an oddity. However, since repair pathways appear to be generally similar to those of other organisms, it is likely that continued studies of DNA repair in Drosophila will lead to new insights into repair mechanisms by revealing how repair pathways function in the absence of important components. Interesting questions include:
How is Drosophila able to tolerate large amounts of uracil in the DNA of larval tissues?
What are the consequences of lack of transcription-coupled NER, which is important in both fungi and mammals?
How does Drosophila process noncomplementary DSB ends for NHEJ, given that there are no B family polymerases, and no Artemis nuclease?
How does Drosophila accomplish Rad51 filament formation without BRCA1, Rad52, or Rad51B?
What are the signals that recruit translesion polymerases to damaged bases encountered by replication forks?
Answers are likely to include promotion of secondary pathways to primary roles, providing an avenue to better understand these pathways, and recruitment or invention of other proteins to take the place of those that are missing (see Kohl et al. 2012 for an example of this in meiotic recombination).
One area in which Drosophila has made particularly important contributions is as a model organism for studying DSB repair mechanisms and consequences, beginning with the discovery that X-rays induce chromosome rearrangements and recombination (Muller 1927; Friesen 1933; Patterson and Suche 1934), and continuing to more recent analyses of gap repair following P element excision, which resulted in the SDSA model of HR (Nassif et al. 1994), and the discovery of the key role of Polθ in Ku-independent EJ (Chan et al. 2010). Many of the details of these processes remain to be elucidated. Interestingly, a single repair event can involve both SDSA and TMEJ (Adams et al. 2003); an important question now is what causes a cell to switch from SDSA to TMEJ (Figure 2).
Understanding DSB repair mechanisms is even more important with the recent development of the bacterial CRISPR/Cas9 systems to carry out genome engineering (Cong et al. 2013; Mali et al. 2013). Empirical data from trial-and-error efforts, although based on our incomplete understanding of DSB repair pathways, have succeeded in improving and expanding the uses CRISPR/Cas9 technology to make precise genomic changes in a wide variety of organisms, both traditional models and species previously not amenable to genetic analyses (reviewed in Zhang et al. 2016). However, discussions with colleagues and observations made in my own laboratory suggest that repair events that are not easily explained by current paradigms are not infrequent. It may never be possible to explain every unusual repair event, but certainly a better understanding of the complex network of possibilities will help to further develop this technology. Experiments that take advantage of the numerous tools available to Drosophila researchers should be instrumental in this effort.
Acknowledgments
I thank members of my laboratory for comments on the manuscript. I apologize to colleagues whose work I neglected to discuss; the structure of this chapter and the choice of topics is undoubtedly biased not only by my own interests, but by my incomplete knowledge of the literature. Research in my laboratory is supported by a grant from the National Institute of General Medical Sciences under award number 5 R35 GM118127.
Footnotes
Communicating editor: T. L. Orr-Weaver
Literature Cited
- Adams M. D., McVey M., Sekelsky J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Alexander J. L., Barrasa M. I., Orr-Weaver T. L., 2015. Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair. Curr. Biol. 25: 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiadis V., Lusser A., Kadonaga J. T., 2004. A conserved N-terminal motif in Rad54 is important for chromatin remodeling and homologous strand pairing. J. Biol. Chem. 279: 27824–27829. [DOI] [PubMed] [Google Scholar]
- Al-Minawi A. Z., Saleh-Gohari N., Helleday T., 2008. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 36: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Bergstralh D. T., Kohl K. P., Larocque J. R., Moore C. B., et al. , 2009. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell 35: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Kuo H. K., Savukoski D., Brodsky M. H., Sekelsky J., 2011. Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet. 7: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach C., 1946. Chemically induced mosaicism in Drosophila melanogaster. Proc. R. Soc. Edinb. 62: 211–221. [DOI] [PubMed] [Google Scholar]
- Auerbach C., 1978. Forty years of mutation research: a pilgrim’s progress. Heredity (Edinb) 40: 177–187. [DOI] [PubMed] [Google Scholar]
- Auerbach C., Robson J. M., 1944. Production of mutations by allyl isothiocyanate. Nature 154: 81. [Google Scholar]
- Baker B. S., Carpenter A. T. C., 1972. Genetic analysis of sex chromosomal meiotic mutants in Drosophila melanogaster. Genetics 71: 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. S., Carpenter A. T., Ripoll P., 1978. The utilization during mitotic cell division of loci controlling meiotic recombination and disjunction in Drosophila melanogaster. Genetics 90: 531–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall E. L., Rio D. C., 1996. Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev. 10: 921–933. [DOI] [PubMed] [Google Scholar]
- Beall E. L., Rio D. C., 1997. Drosophila P-element transposase is a novel site-specific endonuclease. Genes Dev. 11: 2137–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall E. L., Admon A., Rio D. C., 1994. A Drosophila protein homologous to the human p70 Ku autoimmune antigen interacts with the P transposable element inverted repeats. Proc. Natl. Acad. Sci. USA 91: 12681–12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekesi A., Pukancsik M., Muha V., Zagyva I., Leveles I., et al. , 2007. A novel fruitfly protein under developmental control degrades uracil-DNA. Biochem. Biophys. Res. Commun. 355: 643–648. [DOI] [PubMed] [Google Scholar]
- Belyaeva E. S., Zhimulev I. F., Volkova E. I., Alekseyenko A. A., Moshkin Y. M., et al. , 1998. Su(UR)ES: a gene suppressing DNA underreplication in intercalary and pericentric heterochromatin of Drosophila melanogaster polytene chromosomes. Proc. Natl. Acad. Sci. USA 95: 7532–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyakin S. N., Christophides G. K., Alekseyenko A. A., Kriventseva E. V., Belyaeva E. S., et al. , 2005. Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories. Proc. Natl. Acad. Sci. USA 102: 8269–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous R., Mason A. C., Rambo R. P., Bansbach C. E., Badu-Nkansah A., et al. , 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhui-Kaur A., Goodman M. F., Tower J., 1998. DNA mismatch repair catalyzed by extracts of mitotic, postmitotic, and senescent Drosophila tissues and involvement of mei-9 gene function for full activity. Mol. Cell. Biol. 18: 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Wei S. C., Rong Y. S., 2004. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr. Biol. 14: 1348–1353. [DOI] [PubMed] [Google Scholar]
- Bocquet N., Bizard A. H., Abdulrahman W., Larsen N. B., Faty M., et al. , 2014. Structural and mechanistic insight into Holliday-junction dissolution by topoisomerase IIIalpha and RMI1. Nat. Struct. Mol. Biol. 21: 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolterstein E., Rivero R., Marquez M., McVey M., 2014. The Drosophila Werner exonuclease participates in an exonuclease-independent response to replication stress. Genetics 197: 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubriak I., Mason P. A., Clancy D. J., Dockray J., Saunders R. D., et al. , 2009. DmWRNexo is a 3′-5′ exonuclease: phenotypic and biochemical characterization of mutants of the Drosophila orthologue of human WRN exonuclease. Biogerontology 10: 267–277. [DOI] [PubMed] [Google Scholar]
- Boyd J. B., Golino M. D., Nguyen T. D., Green M. M., 1976. Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics 84: 485–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J. B., Golino M. D., Shaw K. E. S., Osgood C. J., Green M. M., 1981. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics 97: 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozas A., Beumer K. J., Trautman J. K., Carroll D., 2009. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 182: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2010. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11: 208–219. [DOI] [PubMed] [Google Scholar]
- Brodsky M. H., Sekelsky J. J., Tsang G., Hawley R. S., Rubin G. M., 2000. mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 14: 666–678. [PMC free article] [PubMed] [Google Scholar]
- Brough R., Wei D., Leulier S., Lord C. J., Rong Y. S., et al. , 2008. Functional analysis of Drosophila melanogaster BRCA2 in DNA repair. DNA Repair (Amst.) 7: 10–19. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A., Spradling A. C., 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. T. C., 1982. Mismatch repair, gene conversion, and crossing-over in two recombination-defective mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79: 5961–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Liu J. C., Amunugama R., Hajdu I., Primack B., et al. , 2015. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 518: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. H., Yu A. M., McVey M., 2010. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6: e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. S., Naujoks D. A., Huen D. S., Russell S., 2011. Insect population control by homing endonuclease-based gene drive: an evaluation in Drosophila melanogaster. Genetics 188: 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. R., Taylor M. R., Boulton S. J., 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Wu C. H., Plank J. L., Hsieh T. S., 2012. Essential functions of C terminus of Drosophila topoisomerase IIIalpha in double holliday junction dissolution. J. Biol. Chem. 287: 19346–19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S. U., Polyzos A., Costes S. V., et al. , 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovnick A., Ballantyne G. H., Baillie D. L., Holm D. G., 1970. Gene conversion in higher organisms: half-tetrad analysis of recombination within the rosy cistron of Drosophila melanogaster. Genetics 66: 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Ducau J., Flores C., Johnson-Schlitz D., et al. , 2004. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 14: 1360–1366. [DOI] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Gatti M., 2006. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics 173: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox L. S., Clancy D. J., Boubriak I., Saunders R. D., 2007. Modeling werner syndrome in Drosophila melanogaster: hyper-recombination in flies lacking WRN-like exonuclease. Ann. N. Y. Acad. Sci. 1119: 274–288. [DOI] [PubMed] [Google Scholar]
- Crevel G., Vo N., Crevel I., Hamid S., Hoa L., et al. , 2012. Drosophila RecQ4 is directly involved in both DNA replication and the response to UV damage in S2 cells. PLoS One 7: e49505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown K. N., McMahan S., Sekelsky J., 2014. Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model. PLoS Genet. 10: e1004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayani Y., Simchen G., Lichten M., 2011. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 7: e1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cock J. G., Klink E. C., Lohman P. H., Eeken J. C., 1992. Absence of strand-specific repair of cyclobutane pyrimidine dimers in active genes in Drosophila melanogaster Kc cells. Mutat. Res. 274: 85–92. [DOI] [PubMed] [Google Scholar]
- Dherin C., Dizdaroglu M., Doerflinger H., Boiteux S., Radicella J. P., 2000. Repair of oxidative DNA damage in Drosophila melanogaster: identification and characterization of dOgg1, a second DNA glycosylase activity for 8-hydroxyguanine and formamidopyrimidines. Nucleic Acids Res. 28: 4583–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey J. S., Redon C. E., Nakamura A. J., Baird B. J., Sedelnikova O. A., et al. , 2009. H2AX: functional roles and potential applications. Chromosoma 118: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do A. T., Larocque J. R., 2015. The role of Drosophila mismatch repair in suppressing recombination between diverged sequences. Sci. Rep. 5: 17601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducau J., Bregliano J. C., De La Roche Saint-Andre C., 2000. Gamma-irradiation stimulates homology-directed DNA double-strand break repair in Drosophila embryo. Mutat. Res. 460: 69–80. [DOI] [PubMed] [Google Scholar]
- Eeken J. C., Romeijn R. J., De Jong A. W., Pastink A., Lohman P. H., 2001. Isolation and genetic characterisation of the Drosophila homologue of (SCE)REV3, encoding the catalytic subunit of DNA polymerase zeta. Mutat. Res. 485: 237–253. [DOI] [PubMed] [Google Scholar]
- Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J., 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell 62: 515–525. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Rudin N., Haber J. E., 1992. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 12: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck O., Lehmann E., Schar P., Kohli J., 1999. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nat. Genet. 21: 314–317. [DOI] [PubMed] [Google Scholar]
- Flores C., Engels W., 1999. Microsatellite instability in Drosophila spellchecker1 (MutS homolog) mutants. Proc. Natl. Acad. Sci. USA 96: 2964–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S., Alabert C., Toh G. W., Toth R., Sugawara N., et al. , 2007. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell. Biol. 27: 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E. C., 1996. Cockayne syndrome - a primary defect in DNA repair, transcription, both or neither. BioEssays 18: 731–738. [DOI] [PubMed] [Google Scholar]
- Friesen H., 1933. Artificially induced crossing-over in males of Drosophila melanogaster. Science 78: 513–514. [DOI] [PubMed] [Google Scholar]
- Gaivão I., Rodríguez R., Sierra L. M., 2014. Use of the comet assay to study DNA repair in Drosophila melanogaster, pp. 397–412 in Genotoxicity and DNA Repair: A Practical Approach, edited by Sierra L. M., Gaivão I. Humana Press, New York. [Google Scholar]
- Gall J. G., Cohen E. H., Polan M. L., 1971. Repetitive DNA sequences in Drosophila. Chromosoma 33: 319–344. [DOI] [PubMed] [Google Scholar]
- Gatti M., 1979. Genetic control of chromosome breakage and rejoining in Drosophila melanogaster: spontaneous chromosome aberrations in X-linked mutants defective in DNA metabolism. Proc. Natl. Acad. Sci. USA 76: 1377–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., 1982. Sister chromatid exchanges in Drosophila, pp. 267–296 in Sister Chromatid Exchange, edited by Wolff S. John Wiley & Sons Inc, New York. [Google Scholar]
- Gatti M., Tanzarella C., Olivieri G., 1974a Analysis of the chromosome aberrations induced by X-rays in somatic cells of Drosophila melanogaster. Genetics 77: 701–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Tanzarella C., Olivieri G., 1974b Variation with sex of irradiation-induced chromosome damage in somatic cells of Drosophila melanogaster. Nature 247: 151–152. [DOI] [PubMed] [Google Scholar]
- Gatti M., Pimpinelli S., De Marco A., Tanzarella C., 1975. Chemical induction of chromosome aberrations in somatic cells of Drosophila melanogaster. Mutat. Res. 33: 201–212. [DOI] [PubMed] [Google Scholar]
- Gatti M., Smith D. A., Baker B. S., 1983. A gene controlling condensation of heterochromatin in Drosophila melanogaster. Science 221: 83–85. [DOI] [PubMed] [Google Scholar]
- Geyer P. K., Richardson K. L., Corces V. G., Green M. M., 1988. Genetic instability in Drosophila melanogaster: P-element mutagenesis by gene conversion. Proc. Natl. Acad. Sci. USA 85: 6455–6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A., Schüpbach T., 1999. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1: 354–357. [DOI] [PubMed] [Google Scholar]
- Glaser R. L., Karpen G. H., Spradling A. C., 1992. Replication forks are not found in a Drosophila minichromosome demonstrating a gradient of polytenization. Chromosoma 102: 15–19. [DOI] [PubMed] [Google Scholar]
- Gloor G. B., Moretti J., Mouyal J., Keeler K. J., 2000. Distinct P-element excision products in somatic and germline cells of Drosophila melanogaster. Genetics 155: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin S. K., Meslin C., Kabbinavar F., Bratton-Palmer D. S., Hornack C., et al. , 2015. Evolutionary and functional analysis of the invariant SWIM domain in the conserved Shu2/SWS1 protein family from Saccharomyces cerevisiae to Homo sapiens. Genetics 199: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski M. M., Eeken J. C., De Jong A. W., Klink I., Loos M., et al. , 2003. The Drosophila melanogaster DNA Ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54. Genetics 165: 1929–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski M. M., Romeijn R. J., Eeken J. C., De Jong A. W., Van Veen B. L., et al. , 2004. Disruption of Drosophila Rad50 causes pupal lethality, the accumulation of DNA double-strand breaks and the induction of apoptosis in third instar larvae. DNA Repair (Amst.) 3: 603–615. [DOI] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage φC31. Genetics 166: 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hari K. L., Santerre A., Sekelsky J., McKim K. S., Boyd J. B., et al. , 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82: 815–821. [DOI] [PubMed] [Google Scholar]
- Harris P. V., Mazina O. M., Leonhardt E. A., Case R. B., Boyd J. B., et al. , 1996. Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol. 16: 5764–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde V., Kelley M. R., Xu Y., Mian I. S., Deutsch W. A., 2001. Conversion of the bifunctional 8-oxoguanine/β-δ apurinic/apyrimidinic DNA repair activities of Drosophila ribosomal protein S3 into the human S3 monofunctional β-elimination catalyst through a single amino acid change. J. Biol. Chem. 276: 27591–27596. [DOI] [PubMed] [Google Scholar]
- Henderson D. S., Banga S. S., Grigliatti T. A., Boyd J. B., 1994. Mutagen sensitivity and suppression of position-effect variegation result from mutations in mus209, the Drosophila gene encoding PCNA. EMBO J. 13: 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsclaw J. K., Hatkevich T., Sekelsky J., 2016. Meiotic and mitotic recombination: first in flies, pp. 139–154 in Genome Stability: From Virus to Human Application, edited by Kovalchuk I., Kovalchuk O. Academic Press, San Diego, CA. [Google Scholar]
- Ivanov E. L., Sugawara N., Fishman-Lobell J., Haber J. E., 1996. Genetic requirements for the single-strand annealing pathway for double-strand break repair in Saccharomyces cerevisiae. Genetics 142: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby D. B., Wensink P. C., 1996. DNA binding specificities of YPF1, a Drosophila homolog to the DNA binding subunit of human DNA-dependent protein kinase, Ku. J. Biol. Chem. 271: 16827–16832. [DOI] [PubMed] [Google Scholar]
- Jaklevic B. R., Su T. T., 2004. Relative contribution of DNA repair, cell cycle checkpoints, and cell death to survival after DNA damage in Drosophila larvae. Curr. Biol. 14: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklevic B., Purdy A., Su T. T., 2004. Control of mitotic entry after DNA damage in Drosophila. Methods Mol. Biol. 280: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A., Breuer G. A., Brinkman E. K., Van Der Meulen A. I., Borden S. V., et al. , 2016. A single double-strand break system reveals repair dynamics and mechanisms in heterochromatin and euchromatin. Genes Dev. 30: 1645–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Prakash S., Prakash L., 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 283: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Johnson-Schlitz D. M., Engels W. R., 2006. The effect of gap length on double-strand break repair in Drosophila. Genetics 173: 2033–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Pedersen M., Tiong S., White-Brown S. K., Paul A., et al. , 2011. Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J. Cell Biol. 195: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagaraj R., Saydam N., Garcia P. L., Zheng L., Janscak P., 2006. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34: 5217–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane D. P., Shusterman M., Rong Y., McVey M., 2012. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLoS Genet. 8: e1002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Kadonaga J. T., 2014. The annealing helicase and branch migration activities of Drosophila HARP. PLoS One 9: e98173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile A. C., Chavez D. A., Bacal J., Eldirany S., Korzhnev D. M., et al. , 2015. HLTF’s ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol. Cell 58: 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komitopoulou K., Gans M., Margaritis L. H., Kafatos F. C., Masson M., 1983. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster with special attention to eggshell mutants. Genetics 105: 897–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Erie D. A., 2015. Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49: 291–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. K., McMahan S., Rota C. M., Kohl K. P., Sekelsky J., 2014. Drosophila FANCM helicase prevents spontaneous mitotic crossovers generated by the MUS81 and SLX1 nucleases. Genetics 198: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkulos M., Weinberg J. M., Roy D., Mount S. M., 1994. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics 136: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Johnson-Schlitz D. M., Engels W. R., 2001. Sterility of Drosophila with mutations in the Bloom syndrome gene—complementation by Ku70. Science 291: 2600–2602. [DOI] [PubMed] [Google Scholar]
- Lafave M. C., Andersen S. L., Stoffregen E. P., Holsclaw J. K., Kohl K. P., et al. , 2014. Sources and structures of mitotic crossovers that arise when BLM helicase is absent in Drosophila. Genetics 196: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., Holsclaw J. K., Bellendir S. P., Sekelsky J., Hawley R. S., 2013. The development of a monoclonal antibody recognizing the Drosophila melanogaster phosphorylated histone H2A variant (gamma-H2AV). G3 3: 1539–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F., Crossan G. P., Rosado I. V., Arends M. J., Patel K. J., 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475: 53–58. [DOI] [PubMed] [Google Scholar]
- Larocque J. R., Jaklevic B., Su T. T., Sekelsky J., 2007. Drosophila ATR in double-strand break repair. Genetics 175: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurençon A., Orme C. M., Peters H. K., Boulton C. L., Vladar E. K., et al. , 2004. A large-scale screen for mutagen-sensitive loci in Drosophila. Genetics 167: 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M. A., Duronio R. J., 2005. New insights into cell cycle control from the Drosophila endocycle. Oncogene 24: 2765–2775. [DOI] [PubMed] [Google Scholar]
- Liu B., Xue Q., Tang Y., Cao J., Guengerich F. P., et al. , 2016. Mechanisms of mutagenesis: DNA replication in the presence of DNA damage. Mutat. Res. Rev. Mutat. Res. 768: 53–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok B. H., Carley A. C., Tchang B., Powell S. N., 2013. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32: 3552–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Martinez D., Liang C. C., Cohn M. A., 2016. Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell. Mol. Life Sci. 73: 3097–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg R., Mavinakere M., Campbell C., 2001. Deficient DNA end joining activity in extracts from Fanconi anemia fibroblasts. J. Biol. Chem. 276: 9543–9549. [DOI] [PubMed] [Google Scholar]
- Machwe A., Karale R., Xu X., Liu Y., Orren D. K., 2011. The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) Holliday junctions to functional replication forks. Biochemistry 50: 6774–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan J. P., Chotkowski H. L., Glaser R. L., 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30: 3698–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek L. R., Bale A. E., 2006. Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst.) 5: 1317–1326. [DOI] [PubMed] [Google Scholar]
- Martino J., Bernstein K. A., 2016. The Shu complex is a conserved regulator of homologous recombination. FEMS Yeast Res. 16: pii: fow073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S., Ohkita N., Nakayama M., Akaboshi E., Shibata T., et al. , 2012. RecQ5 interacts with Rad51 and is involved in resistance of Drosophila to cisplatin treatment. Biol. Pharm. Bull. 35: 2017–2022. [DOI] [PubMed] [Google Scholar]
- McCaffrey R., St Johnston D., Gonzalez-Reyes A., 2006. Drosophila mus301/spindle-C encodes a helicase with an essential role in double-strand DNA break repair and meiotic progression. Genetics 174: 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Adams M. D., Staeva-Vieira E., Sekelsky J. J., 2004. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz C. W., 1916. Chromosome studies on the Diptera. II. The paired association of chromosomes in the Diptera, and its significance. J. Exp. Biol. 21: 213–279. [Google Scholar]
- Min B., Weinert B. T., Rio D. C., 2004. Interplay between Drosophila Bloom’s syndrome helicase and Ku autoantigen during nonhomologous end joining repair of P element-induced DNA breaks. Proc. Natl. Acad. Sci. USA 101: 8906–8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S. E., 2014. RNAi screening in Drosophila cells and in vivo. Methods 68: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H., 1912. Complete linkage in the second chromosome of the male of Drosophila. Science 36: 719–720. [Google Scholar]
- Mortensen U. H., Lisby M., Rothstein R., 2009. Rad52. Curr. Biol. 19: R676–R677. [DOI] [PubMed] [Google Scholar]
- Muha V., Horvath A., Bekesi A., Pukancsik M., Hodoscsek B., et al. , 2012. Uracil-containing DNA in Drosophila: stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 8: e1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Lafave M. C., Sekelsky J., 2009. DNA damage responses in Drosophila nbs mutants with reduced or altered NBS function. DNA Repair (Amst.) 8: 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1927. Artificial transmutation of the gene. Science 46: 84–87. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1932. Further studies on the nature and causes of gene mutations, pp. 213–254 in Sixth International Congress of Genetics, edited by Jones D. F. Brooklyn Botanic Garden, Ithaca, New York. [Google Scholar]
- Muller H. J., 1940. An analysis of the process of structural change in chromosomes of Drosophila. J. Genet. 40: 1–66. [Google Scholar]
- Nakayama M., Yamaguchi S., Sagisu Y., Sakurai H., Ito F., et al. , 2009. Loss of RecQ5 leads to spontaneous mitotic defects and chromosomal aberrations in Drosophila melanogaster. DNA Repair (Amst.) 8: 232–241. [DOI] [PubMed] [Google Scholar]
- Nassif N., Engels W., 1993. DNA homology requirements for mitotic gap repair in Drosophila. Proc. Natl. Acad. Sci. USA 90: 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi T., 2016. Genome stability in Drosophila: mismatch repair and genome stability, pp. 155–161 in Genome Stability: From Virus to Human Application, edited by Kovalchuk I., Kovalchuk O. Academic Press, San Diego, CA. [Google Scholar]
- Nivard M. J., Pastink A., Vogel E. W., 1993. Impact of DNA nucleotide excision repair on methyl methanesulfonate-induced mutations in Drosophila melanogaster. Carcinogenesis 14: 1585–1590. [DOI] [PubMed] [Google Scholar]
- Nivard M. J., Aguirrezabalaga I., Ballering L. A., Pastink A., Sierra L. M., et al. , 1999. Evaluation of the database on mutant frequencies and DNA sequence alterations of vermilion mutations induced in germ cells of Drosophila shows the importance of a neutral mutation detection system. Mutat. Res. 431: 39–57. [DOI] [PubMed] [Google Scholar]
- Nordman J., Li S., Eng T., Macalpine D., Orr-Weaver T. L., 2011. Developmental control of the DNA replication and transcription programs. Genome Res. 21: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brochta D. A., Gomez S. P., Handler A. M., 1991. P element excision in Drosophila melanogaster and related drosophilids. Mol. Gen. Genet. 225: 387–394. [DOI] [PubMed] [Google Scholar]
- Olive P. L., Banath J. P., 2006. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 1: 23–29. [DOI] [PubMed] [Google Scholar]
- Painter T. S., 1934. Salivary chromosomes and the attack on the gene. J. Hered. 25: 465–475. [Google Scholar]
- Painter T. S., Muller H. J., 1929. Parallel cytology and genetics of induced translocations and deletions in Drosophila. J. Hered. 20: 287–298. [Google Scholar]
- Patterson J. T., Suche M. L., 1934. Crossing over induced by X-rays in Drosophila males. Genetics 19: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E., Helleday T., 2010. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11: 683–687. [DOI] [PubMed] [Google Scholar]
- Plank J. L., Wu J., Hsieh T. S., 2006. Topoisomerase IIIa and Bloom’s helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc. Natl. Acad. Sci. USA 103: 11118–11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Zhang Y., Feng W., Jasin M., 2015. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7: a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. R., Flores C. C., Engels W. R., 2006. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., Sekelsky J., 2004. Taking Drosophila Rad51 for a SPiN. Nat. Struct. Mol. Biol. 11: 9–10. [DOI] [PubMed] [Google Scholar]
- Radford S. J., McMahan S., Blanton H. L., Sekelsky J., 2007a Heteroduplex DNA in meiotic recombination in Drosophila mei-9 mutants. Genetics 176: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. J., Sabourin M. M., McMahan S., Sekelsky J., 2007b Meiotic recombination in Drosophila Msh6 mutants yields discontinuous gene conversion tracts. Genetics 176: 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan S. K., Jette N., Lees-Miller S. P., 2014. Non-homologous end joining: emerging themes and unanswered questions. DNA Repair (Amst.) 17: 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C., Hickson I. D., Wu L., 2006. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281: 22839–22846. [DOI] [PubMed] [Google Scholar]
- Rass U., 2013. Resolving branched DNA intermediates with structure-specific nucleases during replication in eukaryotes. Chromosoma 122: 499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi D., Wiles A. M., Bhavani S., Ruan J., Leder P., et al. , 2009. A network of conserved damage survival pathways revealed by a genomic RNAi screen. PLoS Genet. 5: e1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijkers T., Van Den Ouweland J., Morolli B., Rolink A. G., Baarends W. M., et al. , 1998. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18: 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers K., McVey M., 2016. Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 231: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T., Spatola B., Delabaere L., Bowlin K., Hopp H., et al. , 2015. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 17: 1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Lowenhaupt K., Rich A., 1991. Drosophila Rrp1 protein: an apurinic endonuclease with homologous recombination activities. Proc. Natl. Acad. Sci. USA 88: 6780–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Lindsley D. L., Nicoletti B., Trippa G., 1968. Mutants affecting meiosis in natural populations of Drosophila melanogaster. Genetics 60: 525–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders R. D., Boubriak I., Clancy D. J., Cox L. S., 2008. Identification and characterization of a Drosophila ortholog of WRN exonuclease that is required to maintain genome integrity. Aging Cell 7: 418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., McKim K. S., Chin G. M., Hawley R. S., 1995. The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., McKim K. S., Messina L., French R. L., Hurley W. D., et al. , 1999. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 152: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., Brodsky M. H., Burtis K. C., 2000a DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol. 150: F31–F36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J. J., Hollis K. J., Eimerl A. I., Burtis K. C., Hawley R. S., 2000b Nucleotide excision repair endonuclease genes in Drosophila melanogaster. Mutat. Res. 459: 219–228. [DOI] [PubMed] [Google Scholar]
- Shaltiel I. A., Krenning L., Bruinsma W., Medema R. H., 2015. The same, only different – DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 128: 607–620. [DOI] [PubMed] [Google Scholar]
- Sher N., Bell G. W., Li S., Nordman J., Eng T., et al. , 2012. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 22: 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. D., 1973. Mutagen sensitivity of Drosophila melanogaster. I. Isolation and preliminary characterization of a methyl methanesulphonate-sensitive strain. Mutat. Res. 20: 215–220. [DOI] [PubMed] [Google Scholar]
- Smith P. D., Snyder R. D., Dusenbury R. L., 1980. Isolation and characterization of repair-deficient mutants of Drosophila melanogaster, pp. 170–188 in DNA Repair and Mutagenesis in Eukaryotes, edited by Generoso W. M., Shelby M. D., De Serres F. J. Plenum Press, New York. [DOI] [PubMed] [Google Scholar]
- Smith D. A., Baker B. S., Gatti M., 1985. Mutations in genes encoding essential mitotic functions in Drosophila melanogaster. Genetics 110: 647–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., 2005. Drosophila melanogaster: a model for the study of DNA damage checkpoint response. Mol. Cells 19: 167–179. [PubMed] [Google Scholar]
- Spradling A. C., Mahowald A. P., 1980. Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77: 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staeva-Vieira E., Yoo S., Lehmann R., 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern C., 1936. Somatic crossing over and segregation in Drosophila melanogaster. Genetics 21: 625–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N. M., 1908. A study of the germ cells of certain Diptera, with reference to the heterochromosomes and the phenomena of synapsis. J. Exp. Biol. 8: 359–374. [Google Scholar]
- Suwaki N., Klare K., Tarsounas M., 2011. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 22: 898–905. [DOI] [PubMed] [Google Scholar]
- Sved J. A., Eggleston W. B., Engels W. R., 1990. Germ-line and somatic recombination induced by in vitro modified P elements in Drosophila melanogaster. Genetics 124: 331–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen J. M., Harper J. W., 2010. GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev. 24: 521–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., 2016. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 51: 195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W., 1983. The double-strand-break repair model for recombination. Cell 33: 25–35. [DOI] [PubMed] [Google Scholar]
- Thomas A. M., Hui C., South A., McVey M., 2013. Common variants of Drosophila melanogaster Cyp6d2 cause camptothecin sensitivity and synergize with loss of Brca2. G3 (Bethesda) 3: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge K., McKim K. S., Brill S., Sekelsky J., 2007. Synthetic lethality in the absence of the Drosophila MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176: 1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uanschou C., Siwiec T., Pedrosa-Harand A., Kerzendorfer C., Sanchez-Moran E., et al. , 2007. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 26: 5061–5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw C. P., Carr A. M., Oliver A. W., 2014. TopBP1: a BRCT-scaffold protein functioning in multiple cellular pathways. DNA Repair (Amst.) 22: 165–174. [DOI] [PubMed] [Google Scholar]
- Waters L. S., Minesinger B. K., Wiltrout M. E., D’Souza S., Woodruff R. V., et al. , 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73: 134–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D. S., Rong Y. S., 2007. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics 177: 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston R., Peeters H., Ahel D., 2012. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 26: 1558–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M. C., 2010. The FANCM family of DNA helicases/translocases. DNA Repair (Amst.) 9: 224–236. [DOI] [PubMed] [Google Scholar]
- Wilson D. M., III, Deutsch W. A., Kelley M. R., 1994. Drosophila ribosomal protein S3 contains an activity that cleaves DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 269: 25359–25364. [PubMed] [Google Scholar]
- Wood R. D., Doublie S., 2016. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair (Amst.) 44: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Capp C., Feng L., Hsieh T. S., 2008. Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev. Biol. 323: 130–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Lei Z., Huang H., Dui W., Liang X., et al. , 2009. dRecQ4 is required for DNA synthesis and essential for cell proliferation in Drosophila. PLoS One 4: e6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R. R., Axton J. M., Yamamoto Y., Saunders R. D., Glover D. M., et al. , 2000. The Drosophila mus101 gene, which links DNA repair, replication and condensation of heterochromatin in mitosis, encodes a protein with seven BRCA1 C-terminus domains. Genetics 156: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh W., Spradling A. C., 2014. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes Dev. 28: 1840–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yıldız Ö., Majumder S., Kramer B. C., Sekelsky J., 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A. M., McVey M., 2010. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 38: 5706–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Ma Y., Tang S., Hwang P. Y., Boiteux S., et al. , 2010. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40: 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhou J., Lim C. U., 2006. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 16: 45–54. [DOI] [PubMed] [Google Scholar]
- Zhang J. H., Adikaram P., Pandey M., Genis A., Simonds W. F., 2016. Optimization of genome editing through CRISPR-Cas9 engineering. Bioengineered 7: 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]