Abstract
It has been suggested that transcription factor binding is temporally dynamic, and that changes in binding determine transcriptional output. Nonetheless, this model is based on relatively few examples in which transcription factor binding has been assayed at multiple developmental stages. The essential transcription factor Grainy head (Grh) is conserved from fungi to humans, and controls epithelial development and barrier formation in numerous tissues. Drosophila melanogaster, which possess a single grainy head (grh) gene, provide an excellent system to study this conserved factor. To determine whether temporally distinct binding events allow Grh to control cell fate specification in different tissue types, we used a combination of ChIP-seq and RNA-seq to elucidate the gene regulatory network controlled by Grh during four stages of embryonic development (spanning stages 5–17) and in larval tissue. Contrary to expectations, we discovered that Grh remains bound to at least 1146 genomic loci over days of development. In contrast to this stable DNA occupancy, the subset of genes whose expression is regulated by Grh varies. Grh transitions from functioning primarily as a transcriptional repressor early in development to functioning predominantly as an activator later. Our data reveal that Grh binds to target genes well before the Grh-dependent transcriptional program commences, suggesting it sets the stage for subsequent recruitment of additional factors that execute stage-specific Grh functions.
Keywords: transcription, Drosophila, pioneer factor, epithelial cell fate
TO understand how developmental processes are controlled, and how, when perturbed, they can lead to disease, it is crucial to determine the mechanisms by which transcription factors interact with the DNA to regulate gene expression. Broadly expressed transcription factors can regulate multiple, distinct developmental processes, but whether they do so through context-specific DNA-binding events, or through activities subsequent to DNA binding, remains an open question. Based on the relatively limited number of studies that have elucidated transcription factor binding over multiple stages of development, it has been suggested that functional binding events are temporally dynamic (Jakobsen et al. 2007; Zinzen et al. 2009; Wilczynski and Furlong 2010; Spitz and Furlong 2012; Yanez-Cuna et al. 2012; Slattery et al. 2013, 2014). These changes in binding site occupancy by sequence-specific transcription factors are regulated largely through alterations in chromatin structure that modulate the accessible regions of the genome (Kaplan et al. 2011; Li et al. 2011). Nonetheless, factors that act at the top of gene regulatory networks may have pioneering activity, marking cis-regulatory regions, and remaining bound to DNA in multiple developmental contexts (Spitz and Furlong 2012; Iwafuchi-Doi and Zaret 2014; Slattery et al. 2014). To begin to explore the mechanisms by which widely expressed transcription factors can regulate a variety of developmental processes, we focused on the deeply conserved transcription factor Grainy head (Grh), which is a master regulator of epithelial cell fate.
Epithelial tissues are sheets of tightly bound cells that contribute to multiple structures in adult and developing organisms, including the epidermis and lining of the digestive tract, blood vessels, lungs, and ducts. The Grh-family of proteins is an essential regulator of epithelial morphogenesis in metazoans ranging from worms to humans (Wang and Samakovlis 2012). There are three Grh family members in mammals, GRHL1, GRHL2 and GRHL3, which are necessary for neural tube closure during normal vertebrate development, and for wound healing following injury (Ting et al. 2005a,b; Gustavsson et al. 2008; Rifat et al. 2010). In Drosophila melanogaster, where Grh was first identified, the Grh family is represented by a single grh gene (Bray et al. 1988, 1989; Bray and Kafatos 1991). Similarly to its mammalian homologs, the grh gene in Drosophila is essential for embryonic development and wound healing (Bray and Kafatos 1991; Mace et al. 2005). Further highlighting the vast degree of conservation among Grh-family members, Grh proteins from worms to flies to humans bind to a shared sequence motif through a DNA-binding domain that is unique to the related Grh and CP2 protein families (Venkatesan et al. 2003). In Drosophila, Grh has been implicated in a large number of processes in addition to wound healing, including tracheal tube formation and neural stem cell differentiation (Hemphala et al. 2003; Cenci and Gould 2005; Narasimha et al. 2008; Baumgardt et al. 2009). For most of these processes, however, the direct transcriptional targets of Grh remain unknown. Thus, Drosophila Grh provides a powerful system from which to elucidate whether a broadly expressed, master regulator of differentiation influences a large number of diverse processes through temporally dynamic DNA binding, or through a regulated activity subsequent to DNA binding.
By focusing on a deeply conserved, regulator of cell fate, we also provide insight into how epithelial cells are specified in a diversity of organisms, and how misregulation can lead to disease. Morphogenic processes during embryonic development require that cells transition between epithelial and mesenchymal cell fates (Lim and Thiery 2012). During this transition, epithelial cells lose their differentiated characteristics, such as cell–cell adhesion and cell polarity, and gain the properties of mesenchymal cells, including motility, invasiveness, and resistance to apoptosis (Hay 1995; Mani et al. 2008). This epithelial–mesenchymal transition (EMT) is reversible, and, following morphogenesis, epithelial cell fates can be re-established through a mesenchymal-epithelial transition (MET) (Polyak and Weinberg 2009; Tsai and Yang 2013). Similar processes have been implicated in cancer metastasis, where transformed cells spread from a primary tumor in an EMT-like process, and relocate to additional sites in the body by reverting back to a mesenchymal cell fate (Kalluri and Weinberg 2009; Thiery et al. 2009). Misexpression of each of the mammalian Grh-family members has been associated with a range of epithelium-derived cancers (Mlacki et al. 2015). Given the function of Grh-family members in driving epithelial cell fate, it was predicted that these proteins might function as tumor suppressors by suppressing the EMT and metastasis. Indeed, Grh-family members have been found to function as tumor suppressors in a number of epithelium-derived cancers (Darido et al. 2011; Cieply et al. 2012, 2013; Werner et al. 2013; Fabian et al. 2014; Mlacki et al. 2014; Torres-Reyes et al. 2014). Surprisingly, in some cancers overexpression of the Grh-family member GRHL2 has been associated with a poor prognosis, suggesting that, in addition to its tumor suppressor activity, GRHL2 may function as an oncogene in specific cell types or at specific stages of development (Werner et al. 2013; Xiang et al. 2013; Butz et al. 2014; Quan et al. 2015). Thus, defining the factors and mechanisms that drive epithelial cell fate specification is critical for understanding both normal development and cancer progression.
To test whether temporally dynamic binding events mediate stage-specific Grh function, and to provide mechanistic insight into the function of the Grh-family of proteins in development and disease, we took advantage of the single grh gene in Drosophila to identify the transcriptional targets that drive cell-fate specification at multiple stages of development. Given the large number of processes regulated by Grh, we expected the genomic loci occupied by Grh would change dramatically over embryonic development, in accordance with the changing role in development. Unexpectedly, using chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) at multiple time points over embryonic development and in larval imaginal tissues, we demonstrated that Grh-binding sites remain largely unchanged. However, despite this stable binding profile, the Grh-dependent transcriptional profile changes. Specifically, the maternally supplied Grh acts predominantly as a transcriptional repressor, while zygotic Grh both activates and represses gene targets in the embryo. We propose that Grh regulates epithelial fate specification by marking promoters of genes required for epithelial cell fates, and that additional transcription factors or protein interaction partners affect the subsequent changes in gene expression.
Materials and Methods
Fly stocks
All stocks were grown on molasses food at 25°. Germline clones were produced as described in Harrison et al. (2010) using the FLP-FRT system (Chou and Perrimon 1996). To identify embryos mutant for zygotic grh, grhIM or grhB37 were balanced over CyO, sChFP, and embryos were scored for the absence of red fluorescence. To generate integrated reporters for hgo expression, 526 bp of the promoter region, including the transcription start site (TSS), were cloned upstream of lacZ. A potential Grh-binding sequence located 95 bp upstream of the TSS, GACCAGTT, was mutated to CATTCTT. Both wild-type and mutant transgenes were inserted into ZH-86Fb using ΦC31 integration.
Antibody generation and purification
The C-terminal antibody used for calling the ChIP peaks was described in Harrison et al. (2010). An N-terminal antibody recognizing the first 441 amino acids of Grh was used to confirm ChIP peaks. To generate these antibodies, rabbits were immunized by Josman, LLC, with GST fused to amino acids 1–441 of Grh, and purified against the same portion of the protein fused to maltose binding protein (MBP). Demonstrating specificity for Grh, these antibodies recognize a band of the correct predicted molecular weight by immunoblot on S2 cell extract only following transfection of an expression plasmid for Grh-PH. Furthermore, similar to other anti-Grh antibodies (Harrison et al. 2010), these antibodies recognize a doublet by immunoblot on 0–6 hr embryo extract (Supplemental Material, Figure S1)
Chromatin immunoprecipitation and sequencing
ChIP experiments were done with Oregon R D. melanogaster embryos collected at four different time points after egg laying: 2–3, 5–6, 11–12, and 15–16 hr. Embryos were fixed in formaldehyde, chromatin was isolated, and immunoprecipitations were performed as described in Li et al. (2008). Antibodies (described above) raised against either the N-terminus or C-terminus of Grh were used for immunoprecipitation. For each time point, single replicate immunoprecipitations using each antibody were compared (Figure S2) to confirm the successful identification of Grh-binding sites.
Single-end, 100 bp reads were sequenced at the UW Biotechnology Center DNA Sequencing Facility using an Illumina HiSeq 2000. Using the Galaxy platform (https://usegalaxy.org), reads were examined for quality, trimmed by sliding window, and filtered for low quality reads using the FASTQ groomer (Galaxy tool version 1.0.4). Reads were mapped to the BDGP R5 D. melanogaster genome using Bowtie (Bowtie for Illumina Galaxy Tool Version 1.1.2), with default settings, and allowing for two mismatches in the 28 bp seed (Hoskins et al. 2007; Langmead et al. 2009; Blankenberg et al. 2010). Because the BDGP R6 D. melanogaster genome was released during our analysis of these data, sequence reads were additionally mapped to the new version using Bowtie2 (Galaxy Tool Version 2.2.6.2), and mapping data are available along with our other sequencing data through the GEO accession number GSE83305 (Langmead et al. 2009; Langmead and Salzberg 2012). Mapped sequences were normalized to input controls, and peaks were called using MACS (Galaxy Tool version 1.0.1), with a P-value of <1e−6, MFOLD enrichment of five, and using 1000, 2500, and 5000 bp regions to calculate the maximum lambda as local lambda (Zhang et al. 2008). The MFOLD enrichment value was chosen so that model peaks could be calculated using the same settings for each ChIP-seq dataset; the sizes of regions used to find local lambda were chosen to account for the more compact Drosophila genome compared to the default settings tuned to the mouse genome (default: 1000, 5000, and 10,000 bp). A P-value of 1e−6 was chosen (compared to the default setting of 1e−5) to increase confidence in the validity of the called peaks. Peaks were further processed into subpeaks using the PeakSplitter function of the PeakAnalyzer program (Salmon-Divon et al. 2010). Genome tracks were visualized using the UCSC Genome Browser. Genomic annotations, including assigning peaks to the nearest genes and assigning genomic locations, were performed with the Bioconductor R package ChIPseeker (Bioconductor version 3.1, ChIPseeker version 1.4.7) using the default settings and the BDGP R5 Drosophila genome through the TxDb.Dmelanogaster.UCSC.dm3.ensGene package (version 3.1.2) (Huber et al. 2015; Yu et al. 2015). Peak to gene assignments were done using the default settings of the annotatePeak() function of the ChIPseeker R package, which assigns each peak to the closest TSS region (upstream or downstream). After gene assignment, each peak was further annotated to one of seven genomic classes: 5′UTR, 3′UTR, exon, intron, downstream, or intergenic, prioritized in the order listed in case of overlapping annotations. Data for GFP-Grh binding were from GSE62558 (Potier et al. 2014).
RNA-seq and analysis
RNA-seq experiments were done on embryos depleted of either zygotically expressed or maternally contributed Grh (Figure S3). Siblings with functional Grh for both the zygotically and maternally depleted sets were collected and stored in Trizol supplemented with 150 µg/ml glycogen. RNA was extracted, and cDNA libraries were prepared using Truseq RNA sample prep kit (Illumina). For the 2–3 hr after egg laying (AEL) and 11–12 hr AEL time points, two replicates of each mutant were sequenced. For the 15–16 hr AEL time point, a single replicate of each mutant was sequenced.
The cDNA 100 bp single-end reads were sequenced at the UW Biotechnology Center DNA Sequencing Facility using an Illumina HiSeq 2000. Using the Galaxy platform, reads were examined for quality, trimmed, and filtered as described above. The reads were then mapped to the BDGP R5 D. melanogaster genome using the gapped-read TopHat mapper (Galaxy Tool version 0.9; Tophat version 2.0.14) with default settings (Hoskins et al. 2007; Blankenberg et al. 2010; Kim et al. 2013). SeqMonk was used to compare expression of transcripts between mutants (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). Cufflinks (Galaxy Tool version 2.2.1.0; Cufflinks version 2.2.1) was used with default settings for transcript assembly. The resulting assembled transcripts were compared using Cuffdiff (Galaxy Tool version 2.2.1.2; Cuffdiff version 2.2.1) to identify genes that change significantly (P-value < 0.05) in expression using the mutants as replicates for the 11–12 hr AEL and 15–16 hr AEL datasets, and using biological replicates for the 2–3 hr AEL dataset (Trapnell et al. 2010). For the 2–3 hr AEL time point, only genes significantly misexpressed in both mutant backgrounds were used for further analysis. For the 11–12 hr AEL time point, only genes that were significantly misexpressed in both replicates were used for further analysis.
Motif enrichment
Motifs were identified using the online MEME-ChIP tool (version 4.11.1) in the MEME Suite of analysis tools (Bailey et al. 2009) and HOMER (Heinz et al. 2010). Sequences representing peaks identified in ChIP-seq experiments were entered as FASTA files. The program identified motifs relatively enriched in the data set compared to shuffled sequences, and also compared these motifs with a Drosophila database of known motifs combined from the OnTheFly_2014, Fly Factor Survey, FLYREG, iDMMPMM, and DMMPMM databases (Bergman et al. 2005; Kulakovskii and Makeev 2009; Kulakovskiy et al. 2009; Zhu et al. 2011; Shazman et al. 2014). Motifs are given with an e-value, calculated as the P-value of Fisher’s Exact Test of motif enrichment multiplied by the number of candidate motifs.
Gene ontology annotations
Gene ontology (GO) annotation was performed using the online GO Consortium tool (http://geneontology.org/), which uses the PANTHER classification system (Ashburner et al. 2000). Lists of gene names were entered searching for enrichment in biological processes using a Bonferroni correction. The data were collected using the PANTHER over-representation test release 20160321 with the 2016-03-25 GO ontology database release.
Quantitative RT-PCR
RNA was extracted from staged embryos (as described above), and cDNA was synthesized using Superscript III (Life Technologies). Primers designed to span exon junctions were used to perform the qPCR in triplicate for each target using GoTaq qPCR Master Mix (Promega). Samples were analyzed in triplicate for each of three or more biological replicates, and fold change was calculated using the ΔΔCt method. Primers are listed in Table S1.
Cell culture and luciferase assays
Promoter regions from hgo, Spn42De, CG13059, and CG17290, including potential upstream Grh-binding sites identified by ChIP-seq, were cloned into pGL3-Basic (Promega) to drive Firefly luciferase expression. Potential Grh-binding sites identified by similarity to the canonical binding sequence were mutated. Drosophila Schneider 2 (S2) cells were cultured in Schneider’s Media (Life Technologies) supplemented with 10% FBS (Omega Scientific) and 1% antibiotic/antimycotic (Life Technologies). Transient transfections were performed in triplicate with 900 ng wild-type or mutant reporter plasmid, 200 ng Grh-expression plasmid, and 100 ng of actin-renilla plasmid using Effectene Transfection Reagent (Qiagen). Fold activation was calculated relative to luciferase reads from controls transfected with 200 ng of empty expression vector in place of the Grh-expression plasmid. Luciferase assays were performed on cell lysate using a Dual Luciferase assay kit (Promega).
In situ hybridizations
Flies were allowed to lay for 5 hr. After aging for 12 hr, embryos were collected, fixed, and then hybridized with dioxygenin-UTP labeled RNA probes generated from plasmids containing lacZ cDNA. Stained embryos were mounted in 70% glycerol and imaged on a Leica M165 FC stereo microscope.
Data availability
The genomic data in this work was deposited in the Gene Expression Omnibus: accession number GSE83305. Strains and plasmids are available upon request.
Results
Grh binds to thousands of loci during embryonic development
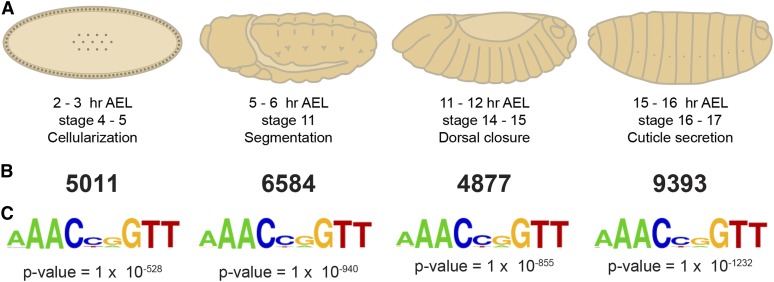
To map the gene regulatory network controlled by Grh, and how it changes over embryonic development, we performed ChIP-seq at four time points spanning the initial 16 hr of embryonic development: 2–3 hr AEL, 5–6 hr AEL, 11–12 hr AEL, and 15–16 hr AEL (Figure 1A). Essential processes occur at these time points, including cellularization (stage 4–5, 2–3 hr AEL), segment boundary formation (stage 11, 5–6 hr AEL), dorsal closure (stage 14–15, 11–12 hr AEL), and cuticle formation (stage 16–17, 15–16 hr AEL). Notably, Grh has been shown to be important for both dorsal closure and cuticle formation, with grh mutant embryos dying from cuticle defects during stage 17 (Bray and Kafatos 1991; Narasimha et al. 2008).
Figure 1.
Grh binds to thousands of loci during embryonic development. (A) ChIP-seq was performed on chromatin from four time points encompassing the indicated key developmental events in embryogenesis. (B) Numbers of peaks identified for each time point. (C) The most highly enriched sequence motif identified by HOMER for peaks from each time point corresponds to the canonical Grh-binding site. P-values are shown below.
We identified thousands of peaks for each developmental time point (5011 peaks at 2–3 hr AE; 6584 peaks at 5–6 hr AEL; 4877 peaks at 11–12 hr AEL; 9393 peaks at 15–16 hr AEL) (Figure 1B and Table S2). For each set of peaks, the canonical Grh-binding site was the most highly enriched motif (Figure 1C). Highly overlapping peaks were identified from ChIP-seq data using an additional Grh antibody raised against a different region of the protein (Figure S2). We identified peaks corresponding to Grh-binding sites that had previously been identified either by DNase I protection assays or ChIP-qPCR, including in the regulatory regions of Dopa decarboxylase (Ddc), engrailed (en), coracle (cora), Cad96Ca (stitcher), decapentaplegic (dpp), Abdominal B (Abd-B), Sex lethal (Sxl), scute (sc), and pale (ple) (Figure 2, A–C and Figure S4) (Bray et al. 1988; Dynlacht et al. 1989; Huang et al. 1995; Blastyak et al. 2006; Narasimha et al. 2008; Pearson et al. 2009; Wang et al. 2009; Harrison et al. 2010; Brown and Kassis 2013). Together, these data demonstrate that we successfully identified in vivo Grh-binding sites during embryonic development.
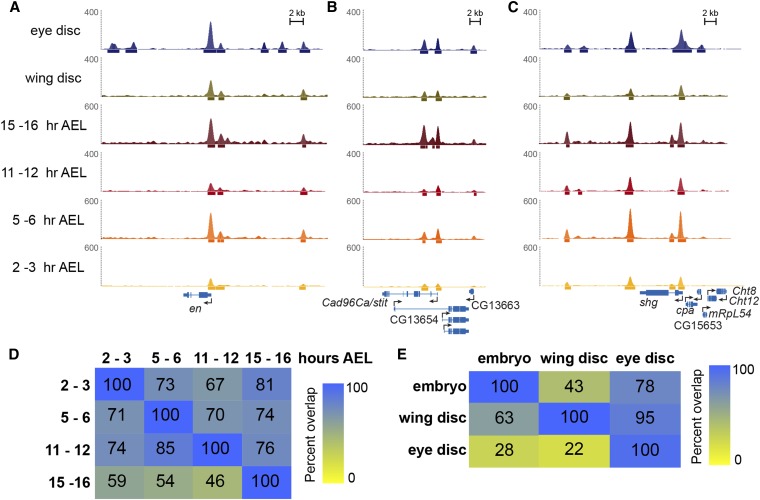
Figure 2.
Grh binding is largely invariant over three days of development. (A–C) Snapshots of Grh ChIP-seq from eye disc, wing disc, 15–16 hr embryos, 11–12 hr embryos, 5–6 hr embryos, and 2–3 hr embryos (listed from top to bottom) for three genes shown below (A) engrailed (en), (B) Cad96Ca, and (C) shotgun (shg). Eye disc data are from Potier et al. (2014). (D) Overlap among ChIP peaks identified at each embryonic time point. Numbers are the percent of the peaks from the time point indicated by the row that overlap the peaks from the time point indicated by the column. (E) Overlap of ChIP peaks shared among all four embryonic time points (embryo) and the peaks identified in wing or eye imaginal discs. Numbers are the percent of the peaks from the tissue indicated by the row that overlap the peaks from the tissue indicated by the column.
Grh-binding sites are largely invariant over development
Immediately evident from our ChIP-seq data was the high degree of overlap between the peaks from the four different time points (Figure 2, A–C). We determined the percentage of peaks that overlap among our four data sets, and found that ∼50–80% of peaks are shared between individual pairs of time points (Figure 2D). Of these, 2768 peaks were bound at all four embryonic stages. Thus, 55% of the Grh peaks identified in 2–3 hr embryos are bound throughout the first 16 hr of embryonic development, and only 7% of peaks were unique to the 2–3 hr time point. By contrast, another transcription factor, Twist, remains bound to only 29% of its binding sites through the first 8 hr of development, and 47% of the binding sites are unique to the 2–4 hr time point (Zinzen et al. 2009; Yanez-Cuna et al. 2012). Because of the lower number of peaks found in the three earlier time points as compared to the 15–16 hr time point, the percentage of peaks from the 15–16 hr embryos overlapping peaks from earlier time points was lower than the overlap for other stages (Figure 2D). A majority (82%; 2227 of 2768) of shared peaks were in the top 5000 peaks identified in 15–16 hr embryos, suggesting that the larger number of peaks identified at this time point was due to either a more successful ChIP assay from this sample, to additional lower strength binding sites, or to the presence of cell-type specific binding sites unique to this developmental time point. Furthermore, relative peak heights among these shared embryonic peaks is similar between the time points (Figure S5). Thus, the highest occupancy Grh-binding sites remain largely invariant over much of embryonic development, and these sites are likely enriched for the functional binding events (Fisher et al. 2012).
Because Grh is expressed in larvae as well as embryos, we sought to determine whether these Grh-binding sites remain occupied in larval tissues. For this purpose, we performed ChIP-seq on third instar larval wing imaginal discs, and identified 2192 peaks, the majority of which (63%; 1381 peaks) were also bound throughout embryonic development (Figure 2E).
We then compared our data to a published dataset that used a transgenic GFP-tagged version of Grh to identify binding sites in the eye imaginal disc (Potier et al. 2014). More than three quarters (78%) of the Grh-binding peaks we identified as constitutively bound throughout embryogenesis were identified in the eye imaginal disc (Figure 2E). However, only ∼¼ of the peaks identified in the eye disc overlapped with peaks we found in the embryos or wing disc. Motif searches for sequences enriched in the eye-disc peaks that did not overlap with our embryonic peaks failed to identify enrichment for the canonical Grh motif. This suggests that many of these peaks may be the result of nonspecific binding by either the anti-GFP antibody or the transgenic, tagged Grh. These results clearly indicate that thousands of Grh-binding sites are occupied throughout embryonic development, and 1146 of these loci remain bound in third instar larvae.
Grh binds to accessible chromatin in the promoter-proximal regions of embryos and larvae
Previous reports had suggested that Grh binding overlaps with regions of open chromatin in the eye (Potier et al. 2014). Given that we identified thousands of Grh-binding sites that were bound in both in the imaginal discs and in the embryo, we explored whether the correlation with accessible chromatin was a general feature of Grh binding. For this purpose, we took advantage of published FAIRE-seq data from embryos approximately stage-matched with our ChIP-seq data sets to determine whether the bound embryonic peaks overlapped with open chromatin identified at the same developmental stage (McKay and Lieb 2013). Indeed, we found that ∼½ of all Grh peaks overlapped with regions of open chromatin (47% for 2–3 hr embryos, 57% for 5–6 hr embryos, 46% for 15–16 hr embryos). Thus, Grh binding overlaps with open chromatin both in the embryo and in the imaginal disc. When we performed similar analysis focusing on the constitutively bound embryonic peaks, we identified an even stronger overlap between Grh binding sites and accessible chromatin regions (57% for 2–3 hr embryos, 62% for 5–6 hr embryos, 60% for 15–16 hr embryos).
Because we had demonstrated that many of these embryonic-bound loci remain bound in the larval imaginal discs, we investigated whether the embryonic-bound sites were correlated with chromatin accessibility in the larval imaginal tissues. When we compared our class of constitutive embryonic Grh-binding sites with FAIRE data from wing imaginal discs, we found that 76% of the embryonic Grh peaks overlap with chromatin accessible regions in the wing disc (Figure 3A). In addition there was a positive correlation (r = 0.23, P < 3 × 10−34) between average ChIP-seq peak height for Grh in the embryo and chromatin accessibility in the wing disc as measured by FAIRE (Figure S6). This suggests that DNA binding by Grh in the embryo is predictive of regions that will become accessible in the larvae. Overall, these Grh-bound open chromatin regions constitute 16% of the total number of accessible regions in the wing disc as determined by FAIRE.
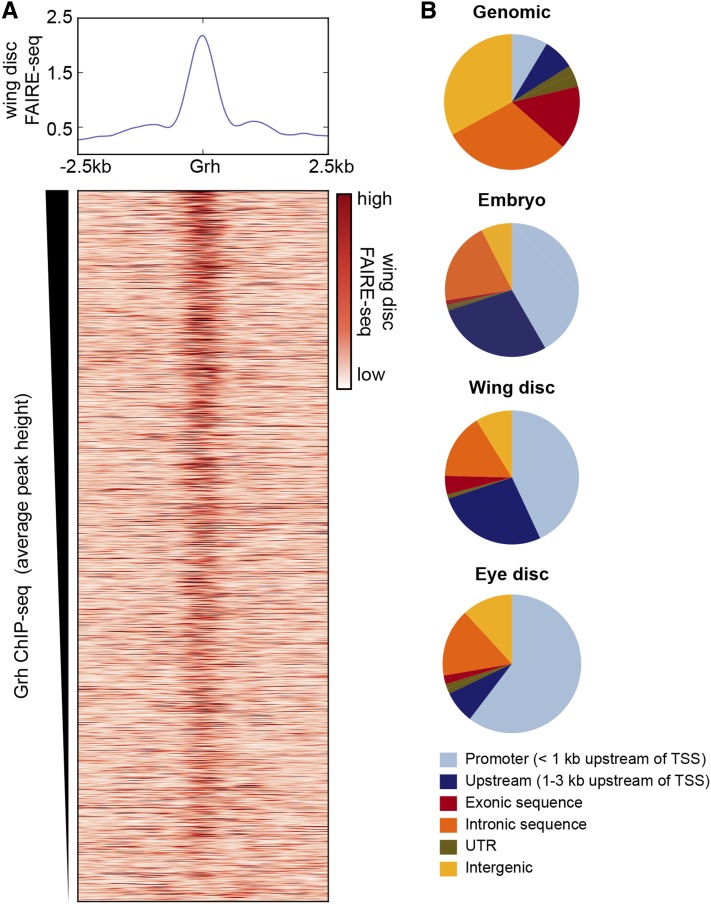
Figure 3.
Grh binding is enriched in accessible, promoter-proximal chromatin. (A) Average FAIRE-seq signal from third instar imaginal wing discs across 5 kb regions centered on the set of Grh-binding sites maintained over embryonic development (top). Heat map of FAIRE-seq signal from imaginal wing discs ranked by average Grh ChIP-seq peak height (bottom). (B) Pie chart showing the genomic distribution of promoters (<1 kb upstream of TSS), upstream regions (1–3 kb upstream of TSS), exons, introns, untranslated regions (UTR) and intergenic regions (top). Pie charts showing the distribution of Grh ChIP-seq peaks from embryos, wing discs, or eye discs (below).
Because promoters are correlated with constitutively open regions of chromatin (Thomas et al. 2011), the overlap of Grh-binding sites and open chromatin suggested that Grh might preferentially bind to promoter regions. When we analyzed the genomic distribution of our Grh-binding sites, we identified a strong enrichment to promoter-proximal regions (defined as <1 kb upstream of the TSS; Figure 3B and Figure S7). Whereas promoter-proximal regions represent only 9% of the genome, they are enriched to 30–40% of the regions bound by Grh in the embryo. Thus, Grh-binding sites are correlated with promoter-proximal regions of open chromatin that are, like the Grh-binding sites, relatively unchanged throughout days of Drosophila development.
The gene-regulatory network controlled by Grh changes over embryonic development
To determine how Grh-binding regulates gene expression, we purified mRNA from both wild-type and grh mutant embryos, and subjected them to high-throughput sequencing (RNA-seq). We used embryos stage matched with our ChIP-seq experiments to enable the discrimination of potential direct from indirect target genes. For the 2–3 hr AEL sample, we assayed embryos depleted for the maternal contribution of grh mRNA using the FLP/FRT system to generate mitotic clones in the maternal germline as previously described (Figure S3A) (Harrison et al. 2010). Embryos lacking maternal grh were identified by the absence of GFP fluorescence. For later stages, grh mutants were identified by the absence of a fluorescently marked balancer chromosome (Figure S3B). In all cases, fluorescent heterozygous siblings were used as controls. Because maternally deposited mCherry expressed from the balancer chromosome perdured in embryos until 9–10 hr AEL, we were unable to identify grh mutant embryos at 5–6 hr AEL. To control for potential background effects, we sequenced mRNA from two different grh mutants, grhIM and grhB37 (Figure 4A). Both of these mutations result in premature stop codons and are likely null alleles (Bray and Kafatos 1991; Pare et al. 2012). Data from these two mutants were highly reproducible, suggesting a minimal contribution from background mutations (Figure S8).
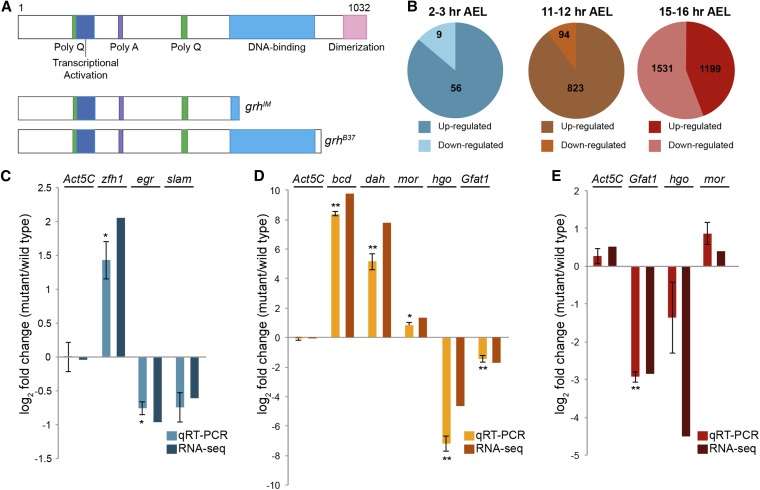
Figure 4.
Thousands of genes require Grh for expression. (A) The Grh polypeptide produced from the grh-RH isoform, and predicted peptides produced from grhIM and grhB37 mutants. DNA-binding and dimerization domains are shared among all grh splice isoforms. (B) Numbers of genes up or downregulated in grh mutants as determined by RNA-seq. Data from 2–3 hr embryos were generated by comparing embryos depleted of maternally provided grh to their heterozygous siblings. Data from 11–12 and 15–16 hr embryos were generated by comparing zygotic mutants homozygous for null mutants in grh to their heterozygous siblings. (C–E) Quantitative RT-PCR to assess gene expression levels in grhB37 mutant embryos as compared to wild type presented as log2 fold change (mutant/wild type). (C) 2–3 hr AEL. (D) 11–12 hr AEL. (E) 15–16 hr AEL. Error bars indicate the SEM for >3 biological replicates. *t-test P-value ≤0.05, **t-test P-value ≤0.005. t-test relative to Act5C control.
We identified 65 genes misexpressed when maternal grh is depleted, 917 genes misexpressed at 11–12 hr AEL in embryos lacking zygotic grh, and 2730 genes misexpressed at 15–16 hr AEL (Figure 4B and Table S3). We validated a subset of targets using quantitative PCR (Figure 4, C–E). Our data also overlapped with a previously published microarray dataset from stage 16–17 embryos, although our RNA-seq approach identified a larger number of targets (Figure S9). Misexpressed genes were then subdivided into those that were either upregulated or downregulated in the absence of Grh (Figure 4B and Table S3). The majority of the genes that changed in expression levels in the grh mutants were upregulated in the absence of Grh in both 2–3 and 11–12 hr embryos (88 and 90%, respectively). In 15–16 hr embryos, the numbers of genes upregulated and downregulated were more similar (44 and 56%, respectively).
While it has been suggested that Grh acts predominantly as a repressor early in development, Grh has largely been characterized as an activator later in development (Bray et al. 1988; Attardi et al. 1993; Uv et al. 1997; Mace et al. 2005; Narasimha et al. 2008; Pearson et al. 2009; Wang et al. 2009). Thus, over embryonic development, we might expect to see a shift from genes being directly upregulated (derepressed) in the absence of Grh, to genes being directly downregulated in its absence. We therefore combined our RNA-seq analysis with our stage-matched ChIP-seq data to determine the likely direct and indirect targets of Grh regulation. For each time point, we associated ChIP-seq peaks with the nearest gene, and identified >2000 genes with a proximal Grh-binding site for each time point, and 1525 genes with a Grh-binding site at all four embryonic time points (Table S4). To identify likely direct targets, we determined the overlap between genes with nearby Grh-binding sites and genes whose expression levels changed in the absence of Grh, allowing us to identify hundreds of potential Grh-target genes (Figure 5A and Table S5).
Figure 5.
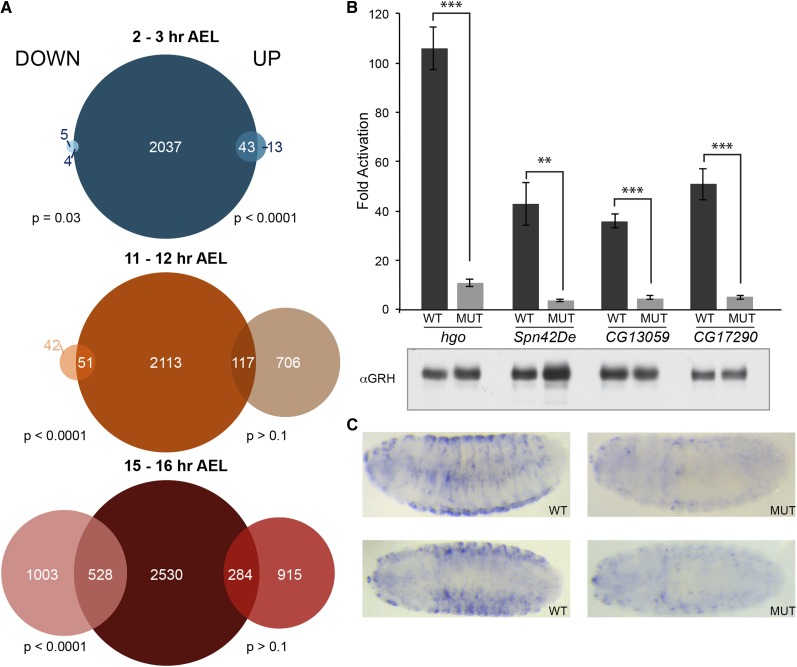
Grh transitions from a transcriptional repressor early in embryonic development to an activator and repressor later. (A) Venn diagrams for data from 2–3 hr embryos (top), 11–12 hr embryos (middle), and 15–16 hr embryos (bottom), depicting the overlap of genes downregulated (left) or upregulated (right) in the absence of Grh with genes associated with Grh-binding sites identified by ChIP-seq (center). (P-values calculated by Fisher’s exact test.) (B) Fold activation by Grh of firefly luciferase reporters driven by either wild-type (WT) promoters for hgo, Spn42De, CG13059, or CG17290, or promoters with mutations in the identified Grh-binding site (MUT). Error bars indicate SD (n = 3) ***t-test P-value <0.0002. **t-test P-value <0.002. Western blot showing Grh expression in representative assays (below). (C) In situ hybridizations against lacZ on transgenic embryos with either a wild-type hgo promoter driving lacZ (WT) or a promoter with a mutated Grh-binding site (MUT). Two examples are given for each.
Because these assays were performed on whole embryos, and Grh is expressed in only a subset of tissues in the 11–12 and 15–16 hr embryos, we wanted to determine if our analysis had successfully identified direct Grh targets. Encouragingly, when we used ImaGO to identify the tissues in which our presumptive direct targets were expressed, we found enrichment for the epidermis, pharynx, and tracheal system, tissues in which Grh is known to be expressed (Table S6) (Tomancak et al. 2007). We further confirmed that we successfully identified direct Grh targets using tissue culture. We selected four genes, homogentisate 1,2-dioxygenase (hgo), Serpin42De (Spn42De), CG13059, and CG17290, that we identified as likely direct targets and that had canonical Grh-binding sites in their promoters, underlying the identified ChIP peaks. We assayed luciferase expression levels driven by the promoters of these four genes in the presence or absence of Grh. All four genes were activated between 35- and 100-fold upon Grh expression (Figure 5B). Furthermore, this expression required binding of Grh to the promoter, as mutations in a canonical Grh-binding motif abrogated Grh-mediated activation (Figure 5B). For one of these genes, hgo, we further confirmed this Grh-mediated expression in vivo by assaying expression of a lacZ transgene driven by either the wild-type hgo promoter or one harboring mutations in the identified Grh-binding site (Figure 5C). The expression pattern of lacZ driven by the wild-type hgo promoter resembles expression of the endogenous gene as reported by the Berkeley Drosophila Genome Project (Tomancak et al. 2002, 2007; Hammonds et al. 2013). Mutations in the single canonical Grh-binding motif result in background levels of staining (Figure 5C). Together, these assays demonstrate that, by combining our ChIP-seq and RNA-seq data sets, we succeeded in identifying direct Grh targets through embryonic development.
The majority of Grh present in the 2–3 hr embryo is provided maternally (Huang et al. 1995; Harrison et al. 2010). This maternally deposited Grh was required predominantly for repressing gene expression as 43 direct targets are upregulated in the absence of Grh and only four direct targets are downregulated (Figure 5A). Of the 43 direct targets repressed by Grh, 60% (28 genes) of them were zygotically expressed (Lott et al. 2011), suggesting a role for maternally deposited Grh as a repressor in the early embryo. This role as a repressor fits with previous data (Huang et al. 1995; Liaw et al. 1995). For example, we identified engrailed, a gene that Grh has previously been shown to bind as a direct target repressed by Grh (Brown and Kassis 2013). We identified many novel targets of Drosophila Grh repression, including zinc finger homeodomain 1 (zfh1), whose mammalian ortholog, ZEB1, is similarly a target of Grh-mediated repression, and is instrumental in driving epidermal differentiation (Cieply et al. 2012, 2013; Werner et al. 2013). GO term enrichment showed that many of the genes repressed by maternal Grh were important in epithelium development, tissue morphogenesis, and central nervous system development (Table S7), processes known to require Grh. This suggests that these genes may be downstream effectors of Grh in driving these fundamental processes.
While maternally deposited Grh is not essential (Harrison et al. 2010), zygotic Grh is required for embryonic viability (Bray and Kafatos 1991). We therefore identified direct targets of Grh at 11–12 hr AEL and 15–16 hr AEL, as these target genes are likely to include those essential for proper embryonic development. At 11–12 hr AEL, only 93 genes are downregulated in the absence of Grh, but, of these 55% have associated Grh-binding sites (Figure 5A; P < 0.0001); 823 genes are upregulated at 11–12 hr AEL in the absence of zygotic Grh, but only 14% are possible direct targets (Figure 5A; P > 0.1). Despite the large number of genes upregulated in the absence of Grh, there is no statistically significant correlation between Grh-binding and upregulation in the mutants, suggesting that Grh is required predominantly to activate gene expression at this stage of development. Similarly, we identify hundreds of targets that are either activated or repressed by Grh 15–16 hr AEL, but only find a significant correlation between DNA-binding by Grh and Grh-mediated gene activation (Figure 5A). Given that we are assaying gene expression on whole embryos, there are direct targets that may have been missed if changes in gene expression occur only in specific tissue types. Nonetheless, by combining ChIP-seq and RNA-seq, we have identified hundreds of previously unknown genes that are targets of Grh regulation (Table S5). Together, our data suggest that maternally provided Grh is predominantly a repressor of gene expression, while later in development Grh functions mostly as an activator.
Gene expression levels are controlled by Grh activity, not Grh binding
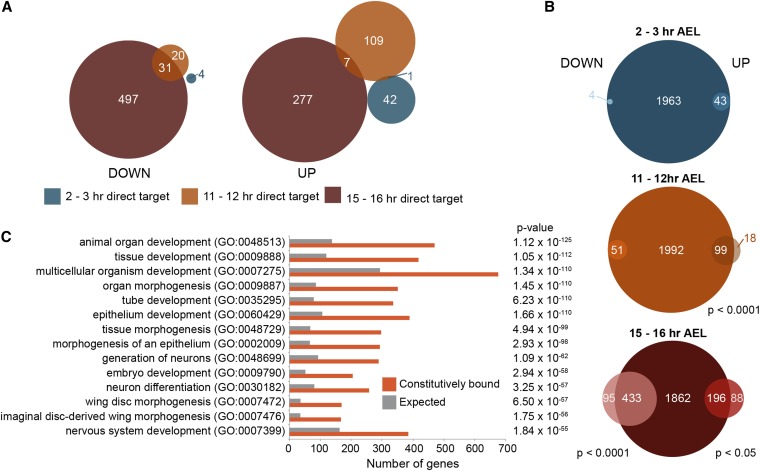
Having identified a set of likely target genes directly regulated by Grh at specific developmental stages, we assessed the degree of overlap among these targets. We found that there was only a single gene (CG18812) that was directly regulated by both maternal and zygotic Grh. Similarly, only seven genes were upregulated in the absence of Grh in both 11–12 and 15–16 hr embryos (Figure 6A). (This is equivalent to 2.5 and 5% of the 15–16 and 11–12 hr upregulated targets, respectively.) There is no significant correlation between being repressed by Grh at 11–12 hr AEL and being repressed at 15–16 hr AEL (P = 0.99). Thus, the network of genes repressed by Grh is reorganized over embryonic development; 31 genes are downregulated in 11–12 and 15–16 hr embryos (Figure 6A), which corresponds to 60% of the genes directly activated by Grh at 11–12 hr AEL. This suggests that some, but not all, of the network of genes activated by zygotic Grh are maintained through development. Similarly, there is little overlap between genes upregulated in the absence of Grh in 11–12 hr embryos and downregulated in 15–16 hr embryos (eight genes) or vice versa (six genes). Thus, it is not that Grh bound to chromatin is switching between transcriptional activities, but rather it is changing between regulating transcription and not modulating transcriptional output. Because these assays were performed on whole embryos, we cannot rule out the possibility that a subset of the genes that showed no significant change in expression levels in the grh mutants were targets of Grh regulation in only a subset of cells. Nonetheless, our data strongly suggest that Grh function and gene regulatory network is reorganized over embryonic development, with Grh activity shifting from being largely involved in transcriptional repression to activation.
Figure 6.
Grh-bound loci in the embryo are associated with thousands of potential target genes that are likely regulated by Grh in specific tissues and at discrete developmental time points. (A) Venn diagrams demonstrating the overlap at three developmental time points (as indicated by color) of genes directly downregulated by Grh (left), or upregulated by Grh (right). (B) Venn diagrams divided by developmental time point showing the overlap of direct targets with those genes associated with Grh peaks identified in multiple embryonic time points. Downregulated genes are on the left, and upregulated genes are on the right. (Statistically significant P-values as calculated by Fisher’s exact test are shown.) A total of 2084 genes bound at 2–3 hr AEL, 2281 genes bound at 11–12 hr AEL, and 3342 genes bound at 15–16 hr AEL. Despite the fact that all genes that are direct targets in 2–3 hr AEL embryos and all genes that are upregulated in 11–12 hr AEL embryos are bound at multiple times over development, there is no statistically significant correlation between being bound by Grh at multiple time points and being a direct Grh target. This is because most of the genes bound at these two time points fall within these two classes. (C) Select GO term enrichment for the genes associated with the constitutively bound embryonic peaks (orange) vs. the expected number (gray). P-values are shown on the right. For a full list see Table S7.
The identified reorganization of the Grh-regulated network contrasts with the largely invariant Grh-chromatin occupancy we identified. Thus, we investigated whether the direct gene targets of Grh were uniquely bound by Grh during the specific developmental time point at which it is regulated, or whether Grh was bound near these target genes at multiple time points. We found that all of the targets of maternally deposited Grh in 2–3 hr embryos were bound at multiple stages of embryogenesis, and were not unique to the early embryo (Figure 6B). Similarly, in 11–12 hr embryos, all of the targets downregulated in the absence of Grh, and nearly all (85%) of the targets that are upregulated in the absence of Grh, overlapped with Grh-binding sites that are occupied at multiple time points (Figure 6B). Most of the Grh-target genes regulated in 15–16 hr embryos were bound by Grh at multiple embryonic stages (Figure 6B). Therefore, while the gene regulatory network controlled by Grh changes over development, this is not regulated at the level of Grh binding (Figure 7). Because we see a correlation between Grh peak height among constitutive Grh-binding sites across embryonic development (Figure S5), these changes in gene expression are unlikely to be explained by dramatic changes in Grh occupancy. Together, this suggests that the thousands of invariant Grh-binding sites exist proximal to genes that are regulated by Grh at distinct stages of development, or in specific tissue types.
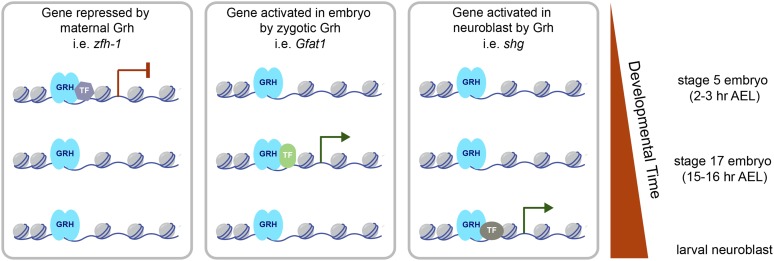
Figure 7.
Model for the role of Grh in regulating gene expression during development. Both maternally and zygotically supplied Grh bind to the regulatory regions of hundreds of genes throughout multiple stages of development. Grh likely marks cis-regulatory regions that are subsequently bound by additional transcription factors (TF) that mediate the stage-specific Grh function. Maternally supplied Grh acts predominantly as a transcriptional repressor in stage 5 embryos. Zygotically supplied Grh can function as a transcriptional repressor and activator during embryogenesis or in the larvae.
Specific, well-studied examples support the model that Grh function is regulated subsequent to DNA binding. We identified Grh binding to the regulatory regions of Fasciclin 3 and coracle throughout embryogenesis, but did not detect any changes in gene expression in grh mutants. Nonetheless, previous reports have shown that expression of both of these genes is reduced in larval imaginal disc cells lacking Grh (Narasimha et al. 2008). Thus, Grh is bound in the embryo to the regulatory regions of genes known to rely on Grh for expression in larvae. We propose that the cohort of genes associated with invariant Grh-binding sites indicates the direct targets by which Grh controls multiple aspects of development. To determine the broadly defined regulatory function of Grh, we identified enriched GO terms for the genes associated with the 2768 of Grh-binding sites occupied throughout embryonic development. These include a wide range of process that Grh is known to regulate, e.g., epithelium development, neuron differentiation, epithelium morphogenesis, and tube development (Figure 6C). Together, our data provide a list of direct target genes whose regulation by Grh likely explain the essential role of Grh in numerous developmental processes (Table S5).
Discussion
Since the initial identification of Grh in Drosophila, the Grh-family of transcription factors has been shown to have widely conserved functions in epithelial differentiation, wound healing, barrier formation, and tube morphogenesis. These roles are important in multiple different tissues in both flies and mammals (Wang and Samakovlis 2012). Nonetheless, downstream targets that affect these critical functions have been identified in only a subset of tissues. Studies of other transcription factors suggested that the varied processes regulated by Grh would be controlled by Grh binding to distinct regulatory regions at different stages of development (Jakobsen et al. 2007; Zinzen et al. 2009; Wilczynski and Furlong 2010; Spitz and Furlong 2012; Yanez-Cuna et al. 2012; Slattery et al. 2013, 2014). In contrast to this expectation, our data suggest that the Grh-binding sites remain largely unchanged over development, and that it is through occupancy of these stably bound regions that Grh regulates gene expression. We propose that Grh may function as a master regulator of epithelial cell fate specification by marking cis-regulatory regions that are bound by additional transcription factors, which provide the temporal specificity in gene expression (Figure 7). Thus, stable DNA occupancy of a transcription factor over multiple developmental time points may be a characteristic of factors that act at the top of gene regulatory networks. Future studies assaying DNA binding for multiple transcription factors across development or in multiple tissue types will be essential for determining if this is the case.
Coupling high-throughput sequencing with methods to detect transcription factor binding sites (ChIP-seq, DamID-seq, etc.) has allowed for the identification of even low occupancy binding sites. Therefore, it remains a challenge to determine which of the identified sites are functional. While it has been demonstrated that low-occupancy regions are less likely to contain regulatory information as compared to high-occupancy regions, there are multiple examples of low affinity binding sites driving specific gene expression (Fisher et al. 2012; Ramos and Barolo 2013; Crocker et al. 2015). Furthermore, binding events that are temporally dynamic over development are enriched for functional binding events, and are more predictive of activity (Jakobsen et al. 2007; Li et al. 2008, 2011; Zinzen et al. 2009; Wilczynski and Furlong 2010). Thus, there has been a focus on those DNA-binding events that change through development. Data suggest that these changes in transcription factor occupancy are likely driven by alterations in chromatin structure that allow the proteins access to the underlying DNA (MacArthur et al. 2009; Kaplan et al. 2011; Li et al. 2011). Nevertheless, these models are built around a relatively limited number of transcription factors that have been assayed at multiple developmental stages. In contrast to factors such as Biniou, Hunchback, Medea, and Twist, there are some broadly expressed proteins, like GAGA factor and Doublesex (Dsx), which bind to thousands of the same targets in multiple developmental contexts (Jakobsen et al. 2007; Wilczynski and Furlong 2010; Li et al. 2011; Spitz and Furlong 2012; Yanez-Cuna et al. 2012; Clough et al. 2014; Slattery et al. 2014). Similar to what we have demonstrated for Grh, both the male and female isoforms of Dsx bind to thousands of genes in the adult fat body, but very few are misregulated upon dsx mutation (Clough et al. 2014). Thus, factors such as Grh, GAGA factor, and Dsx are likely poising target genes, and additional factors are required for a transcriptional response.
Our data demonstrate that stage-specific activity of Grh is regulated subsequent to DNA binding. It is possible that Grh activity during development is directly regulated through post-translational modification, cofactor binding, or a combination of both. It is known that Grh is phosphorylated both in vivo and in vitro, and that the mitogen activated kinase (MAPK) pathway can regulate Grh activity (Liaw et al. 1995; Hemphala et al. 2003; Zhai et al. 2008; Wang et al. 2009; Kim and McGinnis 2011). Grh has also been found to interact with Polycomb and Trithorax group proteins, suggesting that Grh may function to recruit corepressors or coactivators to distinct loci (Tuckfield et al. 2002; Blastyak et al. 2006; Strubbe et al. 2011; Hopkin et al. 2012). It will be important to investigate the role of these regulatory mechanisms in controlling Grh activity.
Based on our data, we propose that Grh may have functions reminiscent of pioneer factors, and thus might influence chromatin accessibility. We identified an enrichment of Grh binding in the promoter-proximal regions of genes (<1 kb upstream of the TSS), which are known to be nucleosome free in many species. Further, we identified a correlation between Grh-bound regions and accessible chromatin. We find this correlation is particularly strong for those regions bound by Grh throughout embryonic development, and that the correlation extends to chromatin accessibility in the imaginal discs of the third instar larvae. Data demonstrating that the canonical Grh motif underlies species-specific changes in chromatin accessibility support a hypothesis that Grh may be a driver of this accessibility (Naval-Sanchez et al. 2015). It is interesting to note that one of the few factors shown to have remarkably consistent binding across development, GAGA factor, is instrumental in promoting chromatin accessibility (Slattery et al. 2014; Fuda et al. 2015). If Grh functions by promoting open chromatin, then its function as a transcriptional activator or repressor may be governed by the transcription factors that are present in a specific cell type to bind to these now accessible regions. Future mechanistic studies will be essential to understand what regulates Grh function in specific cell types.
Because the thousands of Grh-binding sites we identified likely represent functional binding events in multiple tissue types, we suggest that the role of Grh in specific cells, such as neuroblasts, may be explained by stage-specific activity at these stably bound regions. For example, we have identified constitutively occupied Grh-bindings sites upstream of many known regulators of neuroblast differentiation, including brain tumor, numb, castor, Dichaete, klumpfuss, pointed, and Enhancer of split mγ (Homem and Knoblich 2012). We propose that Grh may regulate neuroblast differentiation in part by directly regulating some, or all, of these genes. Similarly, the role of Grh in other processes, such as tracheal development or epithelial morphogenesis, is likely mediated through direct regulation of a subset of genes identified through our studies. Thus, by defining the gene regulatory network controlled by Grh, we have identified specific targets through which Grh likely controls essential developmental processes (Table S5).
We have previously shown that Grh can compete with the transcription factor Zelda for DNA binding (Harrison et al. 2010), and others have similarly suggested that competition with additional transcription factors could regulate Grh function (Huang et al. 1995; Garcia and Stathopoulos 2011). While we found that the TAGteam binding sequence recognized by Zelda is enriched in Grh-bound peaks, and that ∼44% of Grh-bound peaks in the 2–3 hr embryo overlap with Zelda-bound peaks, this high degree of overlap is not unique to Grh. Zelda binding is predictive of where large numbers of other transcription factors bind (Harrison et al. 2011). Our data cannot rule out the possibility that competition with additional factors regulates Grh function at specific loci or in discrete tissues. Nonetheless, the large number of Grh-bound loci that remain occupied throughout much of embryonic and larval development suggest that, at many genes, Grh activity is not regulated by competition for chromatin.
The role of Grh in cancer has been linked to its function as a driver of epithelial cell fate, and Grh suppresses the EMT in a subset of cancer cell lines. A feedback loop between GRHL2 and ZEB1 has been implicated in driving the EMT in some cancers (Cieply et al. 2012, 2013; Werner et al. 2013). GRHL2 represses expression of ZEB1 by binding to its promoter, but ZEB1 can also repress GRHL2 expression (Cieply et al. 2013; Werner et al. 2013). In addition, GRHL2 activates expression of essential markers of the EMT, such as E-cadherin. We identified Grh-binding sites upstream of the Drosophila homologs of both ZEB1 and E-cadherin, zinc finger homeodomain 1 (zfh1), and shotgun (shg), respectively. Similar to what has been shown in mammalian cell culture, our data demonstrate that zfh1 is likely a target of direct repression by Grh in the early embryo. While our RNA-seq did not identify changes in shg expression in grh mutant embryos, previous reports have shown that E-cadherin levels are decreased in the larval neuroblasts of grh mutants (Narasimha et al. 2008). This supports our hypothesis that the Grh-binding sites we identified represent Grh targets in a diversity of tissues extending through larval development. Together, these data provide support for conservation of distinct Grh targets as well as the biological processes they control. Thus, the gene regulatory network we identified helps to explain the apparently contradictory data that Grh behaves as an oncogene in some cell types, and a tumor suppressor in others. Importantly, our developmental time course demonstrates that, for most genes, Grh activity is not regulated at the level of DNA binding, and that instead subsequent events control target-gene expression. These mechanistic insights suggest that the role of Grh is to mark cis-regulatory regions. Whether Grh functions as an oncogene in some tumor types, and a tumor suppressor in others, may be due to the additional transcription factors and cofactors expressed in individual cell types.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/lookup/suppl/doi:10.1534/genetics.116.195685/-/DC1.
Acknowledgments
The authors thank Catherine Fox, Andy Mehle, Sophia Sdao, and members of the Harrison laboratory for useful discussions and comments on the manuscript. Fly strains carrying the grhB37 allele were kindly provided by Sarah Bray. We would also like to thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility. This work was supported by a Basil O’Connor Starter Scholar Research Award #5-FY14-29 and Wisconsin Partnership Program New Investigator Award #2826 to M.M.H. T.K. is a member of the Israeli Center of Excellence (I-CORE) for Gene Regulation in Complex Human Disease (no. 41/11) and the Israeli Center of Excellence (I-CORE) for Chromatin and RNA in Gene Regulation (no. 1796/12). K.N.S. was supported by the National Institutes of Health (NIH) National Research Service Award T32 GM07215.
Footnotes
Communicating editor: P. K. Geyer
Literature Cited
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi L. D., Von Seggern D., Tjian R., 1993. Ectopic expression of wild-type or a dominant-negative mutant of transcription factor NTF-1 disrupts normal Drosophila development. Proc. Natl. Acad. Sci. USA 90: 10563–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., et al. , 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M., Karlsson D., Terriente J., Diaz-Benjumea F. J., Thor S., 2009. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139: 969–982. [DOI] [PubMed] [Google Scholar]
- Bergman C. M., Carlson J. W., Celniker S. E., 2005. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics 21: 1747–1749. [DOI] [PubMed] [Google Scholar]
- Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., et al. , 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26: 1783–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A., Mishra R. K., Karch F., Gyurkovics H., 2006. Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Mol. Cell. Biol. 26: 1434–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J., Kafatos F. C., 1991. Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev. 5: 1672–1683. [DOI] [PubMed] [Google Scholar]
- Bray S. J., Johnson W. A., Hirsh J., Heberlein U., Tjian R., 1988. A cis-acting element and associated binding factor required for CNS expression of the Drosophila melanogaster dopa decarboxylase gene. EMBO J. 7: 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. J., Burke B., Brown N. H., Hirsh J., 1989. Embryonic expression pattern of a family of Drosophila proteins that interact with a central nervous system regulatory element. Genes Dev. 3: 1130–1145. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Kassis J. A., 2013. Architectural and functional diversity of polycomb group response elements in Drosophila. Genetics 195: 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz H., Szabo P. M., Nofech-Mozes R., Rotondo F., Kovacs K., et al. , 2014. Integrative bioinformatics analysis reveals new prognostic biomarkers of clear cell renal cell carcinoma. Clin. Chem. 60: 1314–1326. [DOI] [PubMed] [Google Scholar]
- Cenci C., Gould A. P., 2005. Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development 132: 3835–3845. [DOI] [PubMed] [Google Scholar]
- Chou T. B., Perrimon N., 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B., Riley P., IV, Pifer P. M., Widmeyer J., Addison J. B., et al. , 2012. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res. 72: 2440–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B., Farris J., Denvir J., Ford H. L., Frisch S. M., 2013. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res. 73: 6299–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Jimenez E., Kim Y. A., Whitworth C., Neville M. C., et al. , 2014. Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Dev. Cell 31: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J., Abe N., Rinaldi L., McGregor A. P., Frankel N., et al. , 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darido C., Georgy S. R., Wilanowski T., Dworkin S., Auden A., et al. , 2011. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 20: 635–648. [DOI] [PubMed] [Google Scholar]
- Dynlacht B. D., Attardi L. D., Admon A., Freeman M., Tjian R., 1989. Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev. 3: 1677–1688. [DOI] [PubMed] [Google Scholar]
- Fabian J., Lodrini M., Oehme I., Schier M. C., Thole T. M., et al. , 2014. GRHL1 acts as tumor suppressor in neuroblastoma and is negatively regulated by MYCN and HDAC3. Cancer Res. 74: 2604–2616. [DOI] [PubMed] [Google Scholar]
- Fisher W. W., Li J. J., Hammonds A. S., Brown J. B., Pfeiffer B. D., et al. , 2012. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 109: 21330–21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda N. J., Guertin M. J., Sharma S., Danko C. G., Martins A. L., et al. , 2015. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 11: e1005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., Stathopoulos A., 2011. Lateral gene expression in Drosophila early embryos is supported by Grainyhead-mediated activation and tiers of dorsally-localized repression. PLoS One 6: e29172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson P., Copp A. J., Greene N. D., 2008. Grainyhead genes and mammalian neural tube closure. Birth Defects Res. A Clin. Mol. Teratol. 82: 728–735. [DOI] [PubMed] [Google Scholar]
- Hammonds A. S., Bristow C. A., Fisher W. W., Weiszmann R., Wu S., et al. , 2013. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 14: R140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. M., Botchan M. R., Cline T. W., 2010. Grainyhead and Zelda compete for binding to the promoters of the earliest-expressed Drosophila genes. Dev. Biol. 345: 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. M., Li X. Y., Kaplan T., Botchan M. R., Eisen M. B., 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7: e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay E. D., 1995. An overview of epithelio-mesenchymal transformation. Acta Anat. (Basel) 154: 8–20. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Bertolino E., Lin Y. C., Laslo P., et al. , 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphala J., Uv A., Cantera R., Bray S., Samakovlis C., 2003. Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development 130: 249–258. [DOI] [PubMed] [Google Scholar]
- Homem C. C., Knoblich J. A., 2012. Drosophila neuroblasts: a model for stem cell biology. Development 139: 4297–4310. [DOI] [PubMed] [Google Scholar]
- Hopkin A. S., Gordon W., Klein R. H., Espitia F., Daily K., et al. , 2012. GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 8: e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Kennedy C., Acevedo D., Evans-Holm M., et al. , 2007. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science 316: 1625–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. D., Dubnicoff T., Liaw G. J., Bai Y., Valentine S. A., et al. , 1995. Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev. 9: 3177–3189. [DOI] [PubMed] [Google Scholar]
- Huber W., Carey V. J., Gentleman R., Anders S., Carlson M., et al. , 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K. S., 2014. Pioneer transcription factors in cell reprogramming. Genes Dev. 28: 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen J. S., Braun M., Astorga J., Gustafson E. H., Sandmann T., et al. , 2007. Temporal ChIP-on-chip reveals Biniou as a universal regulator of the visceral muscle transcriptional network. Genes Dev. 21: 2448–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., Weinberg R. A., 2009. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T., Li X. Y., Sabo P. J., Thomas S., Stamatoyannopoulos J. A., et al. , 2011. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS Genet. 7: e1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., McGinnis W., 2011. Phosphorylation of Grainy head by ERK is essential for wound-dependent regeneration but not for development of an epidermal barrier. Proc. Natl. Acad. Sci. USA 108: 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskii I. V., Makeev V., 2009. [Integration of data obtained by different experimental methods to determine the motifs in DNA sequences recognized by transcription-regulating factors]. Biofizika 54: 965–974. [PubMed] [Google Scholar]
- Kulakovskiy I. V., Favorov A. V., Makeev V. J., 2009. Motif discovery and motif finding from genome-mapped DNase footprint data. Bioinformatics 25: 2318–2325. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., MacArthur S., Bourgon R., Nix D., Pollard D. A., et al. , 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., Thomas S., Sabo P. J., Eisen M. B., Stamatoyannopoulos J. A., et al. , 2011. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biol. 12: R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw G. J., Rudolph K. M., Huang J. D., Dubnicoff T., Courey A. J., et al. , 1995. The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev. 9: 3163–3176. [DOI] [PubMed] [Google Scholar]
- Lim J., Thiery J. P., 2012. Epithelial-mesenchymal transitions: insights from development. Development 139: 3471–3486. [DOI] [PubMed] [Google Scholar]
- Lott S. E., Villalta J. E., Schroth G. P., Luo S., Tonkin L. A., et al. , 2011. Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLoS Biol. 9: e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S., Li X. Y., Li J., Brown J. B., Chu H. C., et al. , 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace K. A., Pearson J. C., McGinnis W., 2005. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308: 381–385. [DOI] [PubMed] [Google Scholar]
- Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., et al. , 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. J., Lieb J. D., 2013. A common set of DNA regulatory elements shapes Drosophila appendages. Dev. Cell 27: 306–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlacki M., Darido C., Jane S. M., Wilanowski T., 2014. Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS One 9: e89247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlacki M., Kikulska A., Krzywinska E., Pawlak M., Wilanowski T., 2015. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp. Biol. Med. (Maywood) 240: 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha M., Uv A., Krejci A., Brown N. H., Bray S. J., 2008. Grainy head promotes expression of septate junction proteins and influences epithelial morphogenesis. J. Cell Sci. 121: 747–752. [DOI] [PubMed] [Google Scholar]
- Naval-Sanchez M., Potier D., Hulselmans G., Christiaens V., Aerts S., 2015. Identification of lineage-specific cis-regulatory modules associated with variation in transcription factor binding and chromatin activity using Ornstein-Uhlenbeck models. Mol. Biol. Evol. 32: 2441–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare A., Kim M., Juarez M. T., Brody S., McGinnis W., 2012. The functions of grainy head-like proteins in animals and fungi and the evolution of apical extracellular barriers. PLoS One 7: e36254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. C., Juarez M. T., Kim M., Drivenes O., McGinnis W., 2009. Multiple transcription factor codes activate epidermal wound-response genes in Drosophila. Proc. Natl. Acad. Sci. USA 106: 2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K., Weinberg R. A., 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9: 265–273. [DOI] [PubMed] [Google Scholar]
- Potier D., Davie K., Hulselmans G., Naval Sanchez M., Haagen L., et al. , 2014. Mapping gene regulatory networks in Drosophila eye development by large-scale transcriptome perturbations and motif inference. Cell Rep. 9: 2290–2303. [DOI] [PubMed] [Google Scholar]
- Quan Y., Xu M., Cui P., Ye M., Zhuang B., et al. , 2015. Grainyhead-like 2 promotes tumor growth and is associated with poor prognosis in colorectal cancer. J. Cancer 6: 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A. I., Barolo S., 2013. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat Y., Parekh V., Wilanowski T., Hislop N. R., Auden A., et al. , 2010. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 345: 237–245. [DOI] [PubMed] [Google Scholar]
- Salmon-Divon M., Dvinge H., Tammoja K., Bertone P., 2010. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shazman S., Lee H., Socol Y., Mann R. S., Honig B., 2014. OnTheFly: a database of Drosophila melanogaster transcription factors and their binding sites. Nucleic Acids Res. 42: D167–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Voutev R., Ma L., Negre N., White K. P., et al. , 2013. Divergent transcriptional regulatory logic at the intersection of tissue growth and developmental patterning. PLoS Genet. 9: e1003753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M., Ma L., Spokony R. F., Arthur R. K., Kheradpour P., et al. , 2014. Diverse patterns of genomic targeting by transcriptional regulators in Drosophila melanogaster. Genome Res. 24: 1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F., Furlong E. E., 2012. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13: 613–626. [DOI] [PubMed] [Google Scholar]
- Strubbe G., Popp C., Schmidt A., Pauli A., Ringrose L., et al. , 2011. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci. USA 108: 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J. P., Acloque H., Huang R. Y., Nieto M. A., 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890. [DOI] [PubMed] [Google Scholar]
- Thomas S., Li X. Y., Sabo P. J., Sandstrom R., Thurman R. E., et al. , 2011. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 12: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting S. B., Caddy J., Hislop N., Wilanowski T., Auden A., et al. , 2005a A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308: 411–413. [DOI] [PubMed] [Google Scholar]
- Ting S. B., Caddy J., Wilanowski T., Auden A., Cunningham J. M., et al. , 2005b The epidermis of grhl3-null mice displays altered lipid processing and cellular hyperproliferation. Organogenesis 2: 33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., et al. , 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3: research0088.1–88.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., et al. , 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8: R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Reyes L. A., Alvarado-Ruiz L., Pina-Sanchez P., Martinez-Silva M. G., Ramos-Solano M., et al. , 2014. Expression of transcription factor grainyhead-like 2 is diminished in cervical cancer. Int. J. Clin. Exp. Pathol. 7: 7409–7418. [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai J. H., Yang J., 2013. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 27: 2192–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckfield A., Clouston D. R., Wilanowski T. M., Zhao L. L., Cunningham J. M., et al. , 2002. Binding of the RING polycomb proteins to specific target genes in complex with the grainyhead-like family of developmental transcription factors. Mol. Cell. Biol. 22: 1936–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uv A. E., Harrison E. J., Bray S. J., 1997. Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol. Cell. Biol. 17: 6727–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan K., McManus H. R., Mello C. C., Smith T. F., Hansen U., 2003. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res. 31: 4304–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Samakovlis C., 2012. Grainy head and its target genes in epithelial morphogenesis and wound healing. Curr. Top. Dev. Biol. 98: 35–63. [DOI] [PubMed] [Google Scholar]
- Wang S., Tsarouhas V., Xylourgidis N., Sabri N., Tiklova K., et al. , 2009. The tyrosine kinase Stitcher activates Grainy head and epidermal wound healing in Drosophila. Nat. Cell Biol. 11: 890–895. [DOI] [PubMed] [Google Scholar]
- Werner S., Frey S., Riethdorf S., Schulze C., Alawi M., et al. , 2013. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J. Biol. Chem. 288: 22993–23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynski B., Furlong E. E., 2010. Dynamic CRM occupancy reflects a temporal map of developmental progression. Mol. Syst. Biol. 6: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J., Fu X., Ran W., Chen X., Hang Z., et al. , 2013. Expression and role of grainyhead-like 2 in gastric cancer. Med. Oncol. 30: 714. [DOI] [PubMed] [Google Scholar]
- Yanez-Cuna J. O., Dinh H. Q., Kvon E. Z., Shlyueva D., Stark A., 2012. Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 22: 2018–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L. G., He Q. Y., 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zhai B., Villen J., Beausoleil S. A., Mintseris J., Gygi S. P., 2008. Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. J., Christensen R. G., Kazemian M., Hull C. J., Enuameh M. S., et al. , 2011. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 39: D111–D117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen R. P., Girardot C., Gagneur J., Braun M., Furlong E. E., 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data in this work was deposited in the Gene Expression Omnibus: accession number GSE83305. Strains and plasmids are available upon request.