Abstract
During nervous system development, neurons and their progenitors migrate to their final destinations. In Caenorhabditis elegans, the bilateral Q neuroblasts and their descendants migrate long distances in opposite directions, despite being born in the same posterior region. QR on the right migrates anteriorly and generates the AQR neuron positioned near the head, and QL on the left migrates posteriorly, giving rise to the PQR neuron positioned near the tail. In a screen for genes required for AQR and PQR migration, we identified an allele of nfm-1, which encodes a molecule similar to vertebrate NF2/Merlin, an important tumor suppressor in humans. Mutations in NF2 lead to neurofibromatosis type II, characterized by benign tumors of glial tissues. Here we demonstrate that in C. elegans, nfm-1 is required for the ability of Q cells and their descendants to extend protrusions and to migrate, but is not required for direction of migration. Using a combination of mosaic analysis and cell-specific expression, we show that NFM-1 is required nonautonomously, possibly in muscles, to promote Q lineage migrations. We also show a genetic interaction between nfm-1 and the C. elegans Slit homolog slt-1, which encodes a conserved secreted guidance cue. Our results suggest that NFM-1 might be involved in the generation of an extracellular cue that promotes Q neuroblast protrusion and migration that acts with or in parallel to SLT-1. In vertebrates, NF2 and Slit2 interact in axon pathfinding, suggesting a conserved interaction of NF2 and Slit2 in regulating migratory events.
Keywords: Neuronal migration, Merlin, NFM-1, Q cells, C. elegans
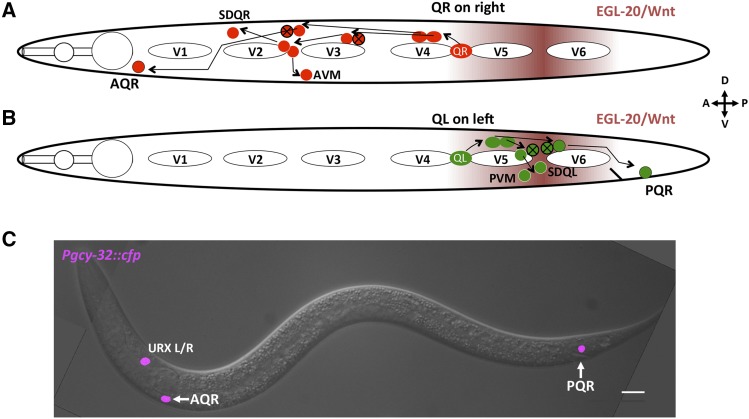
A critical process in nervous system development is the directed migration of neurons to precise destinations. Directed migration is a complex process that requires integration of extracellular cues into cytoskeletal changes, which guide the cell to a specific location. In Cenorhabditis elegans, the Q neuroblasts are an established system to study directed cell migrations (Middelkoop and Korswagen 2014). The Q neuroblasts on the right (QR) and left (QL) are born in the posterior region of the worm yet migrate in opposite directions (Sulston and Horvitz 1977; Salser and Kenyon 1992; Salser et al. 1993). Each undergoes an identical pattern of cell division and cell death to produce three neurons each: SDQL, PVM, and PQR from QL, and SDQR, AVM, and AQR from QR (Figure 1).
Figure 1.
Migration of QR and QL descendants. (A and B) Diagrams representing the migration and cell division pattern of QR on the right side (A) and QL on the left side (B) in the L1 animal, showing birthplace of the Q neuroblasts and approximate locations of the Q descendants. Maroon shading represents the posteriorly derived EGL-20/Wnt signal. White ovals are hypodermal seam cells V1–V6. Circles with black X indicate cells that undergo programmed cell death after cell division. Dorsal is up, anterior to the left. (C) Merged DIC and fluorescent micrograph showing location of Q descendants AQR and PQR in an adult wild-type animal. Pgcy-32::cfp is expressed in AQR, PQR, and URXL/R. Bar, 10 μM.
QL is born on the left side of the animal and migrates posteriorly over the seam cell V5 before dividing (Honigberg and Kenyon 2000; Chapman et al. 2008). During this initial migration, QL detects a posteriorly derived EGL-20/Wnt signal, which through canonical Wnt signaling initiates transcription of mab-5/Hox (Salser and Kenyon 1992). MAB-5 drives further posterior migration of the QL lineage, resulting in the QL.a descendant PQR migrating to the tail near the anus and posterior phasmid ganglion.
QR is born on the right side of the animal and migrates anteriorly over the seam cell V4 and away from the EGL-20/Wnt signal (Salser et al. 1993; Harris et al. 1996; Salser and Kenyon 1996). QR does not initiate mab-5 expression in response to Wnt and continues to migrate anteriorly. After division, QR.a undergoes an identical pattern of cell divisions and cell death as QL.a while migrating anteriorly, with AQR completing migration near the posterior pharyngeal bulb in the head (Figure 1) (Maloof et al. 1999; Whangbo and Kenyon 1999). Initial Q migrations are controlled autonomously by the receptor molecules UNC-40/DCC and PTP-3/LAR (Honigberg and Kenyon 2000; Sundararajan and Lundquist 2012) and nonautonomously by the Fat-like cadherin CDH-4 (Sundararajan et al. 2014). Later Q-descendant migrations are controlled by Wnt signaling (Whangbo and Kenyon 1999; Zinovyeva and Forrester 2005; Zinovyeva et al. 2008; Harterink et al. 2011), which appears to not be involved in initial migration (Josephson et al. 2016a), and by the transmembrane receptor MIG-13 in parallel with SDN-1/Syndecan (Wang et al. 2013; Sundararajan et al. 2015).
In this work, a forward genetic screen was used to identify additional molecules that regulate Q migrations. This screen identified an allele of nfm-1, which encodes a C. elegans neurofibromatosis type II (NF2)/Merlin molecule. NF2 acts as a tumor suppressor in humans, and mutations in the gene lead to development of neurofibromatosis type II (Gusella et al. 1996; Gutmann et al. 1997), a disease of benign schwannomas. NF2/Merlin is involved in signaling pathways involving hippo, mTOR, and PI3K-Akt (Zhao et al. 2007; Striedinger et al. 2008; James et al. 2009; Okada et al. 2009). Additionally, NF2 is involved in nervous system maintenance, corpus callosum development, and axon guidance (Schulz et al. 2013, 2014; Lavado et al. 2014). In corpus callosum development, NF2 inhibits the hippo pathway component Yap. In NF2 mutants, this inhibition is relieved, resulting in increased expression of Slit2, a secreted axon guidance cue that prevents midline crossing. This leads to defects in midline crossing of axons in the callosum (Lavado et al. 2014).
Here we report that nfm-1 mutants display AQR and PQR migration defects. Mosaic analysis and expression studies indicated that NFM-1 does not act in the Q cells themselves but rather nonautonomously, and possibly in muscle. Finally, we show a genetic interaction between NFM-1 and the secreted guidance cue SLT-1 in AQR migration. In vertebrates, Slit1 and Slit2 are required for guidance of many axons, acting through the Robo family receptors (Nguyen Ba-Charvet et al. 1999; Piper et al. 2000; Bagri et al. 2002; Unni et al. 2012; Kim et al. 2014). The Slit–Robo guidance pathway is conserved in C. elegans, where SLT-1 acts as a guidance cue for several neurons through SAX-3/Robo (Hao et al. 2001; Chang et al. 2006; Quinn et al. 2006; Xu and Quinn 2012). In general, detection of extracellular guidance cues such as Slit cause cytoskeletal changes that result in directed migration of cells and axonal growth cones, most typically repulsion. We show that slt-1 mutations enhance AQR migration defects of nfm-1 mutations, and that sax-3 mutants display defects in AQR and PQR migration. In sum, results presented here are consistent with a model in which NFM-1 regulates AQR and PQR migration by controlling the production of an extracellular cue that might act with SLT-1 or in parallel to SLT-1 to promote Q migrations.
Materials and Methods
Nematode strains and genetics
C. elegans were grown under standard conditions at 20° on nematode growth media (NGM) plates (Sulston and Brenner 1974). N2 Bristol was the wild-type strain. Alleles used include LG III: nfm-1(ok754), nfm-1(lq132) and LG X: sax-3(ky123), slt-1(ok255), slt-1(eh15). The Pslt-1::gfp transgene kyIs174 was used (Hao et al. 2001). nfm-1(ok754) was maintained as a balanced heterozygote over the hT2 balancer (nfm-1(ok754)/hT2). Standard gonadal injection was used to create the following extrachromosomal arrays: lqEx773[nfm-1::gfp fosmid (5 ng/μl), Pgcy::32::yfp (50 ng/μl)], lqEx782 [Pnfm-1::gfp (10 ng/μl), Pgcy-32::cfp (25 ng/μl)], lqEx1065 and lqEx1066 [Pslt-1::nfm-1(+)::gfp (25 ng/μl), Pgcy-32::yfp (25 ng/μl)]; lqEx1064, lqEx1073, and lqEx1086 [Pmyo-3::nfm-1(+)::gfp (25 ng/μl), Pscm::gfp (25 ng/μl)]. Ultraviolet trimethylpsoralen (UV/TMP) techniques (Mello and Fire 1995) were used to integrate extrachromosomal arrays to generate the following transgenes: LGII: lqIs244 [lqEx737, Pgcy-32::cfp (25 ng/μl)], unknown chromosomal location lqIs247 [lqEx773, nfm-1::gfp], lqIs274 [lqEx834, Pegl-17::myr-mCherry (20 ng/μl) Pegl-l7::mCherry::his-24 (20 ng/μl)]. The nfm-1::gfp fosmid with gfp fused to the end of the nfm-1A isoform was obtained from the TransgeneOme project, clone 7039520022144752 D02 (Sarov et al. 2006). nfm-1(ok754) was maintained as a heterozygote over the hT2 balancer because homozygous ok754 animals arrest during larval stages, but positions of AQR and PQR could be scored in these arrested animals. Genotypes with M+ had maternal contribution from the hT2 balancer.
Forward genetic screen for AQR and PQR migration defects
Late L4 hermaphrodites were treated with ethyl methanesulfonate (EMS) mutagen as previously described (Sulston and Hodgkin 1988). These animals were allowed to self-fertilize, and three F1 hermaphrodites were placed on plates (three/plate). The F2 progeny of these hermaphrodites were screened using a fluorescence dissecting microscope, and animals with AQR and PQR migration defects were selected. Only one new mutant per F1 plate was retained to ensure independent events. This screen identified lq132. The genome of the lq132-bearing strain LE3406 was sequenced, and variants were annotated using the Cloudmap protocol (without Hawaiian strain mapping) (Minevich et al. 2012). lq132 was outcrossed to N2 at least three times.
Scoring Q-cell and AQR/PQR AQR migration
To score Q-cell protrusions, animals with expression of GFP in the Q neuroblasts from the egl-17 promoter (ayIs9) were synchronized to 1–2.5 hr posthatching (Chapman et al. 2008; Sundararajan and Lundquist 2012). Briefly, gravid adults were allowed to lay eggs overnight. Plates were washed with M9 buffer, and eggs remained attached to plate. Hatched larvae were collected every half hour using M9 washes and placed onto clean NGM plates for later imaging. Protrusion length was quantified from front of cell body to leading edge of protrusions in ImageJ, with significance determined by a t-test. AQR migrates to a position near post-deirid ganglia in the region of the posterior pharyngeal bulb, and PQR migrates posteriorly to a position near the phasmid ganglia posterior to the anus. We used a method as described previously to score AQR and PQR position using Pgcy-32 to drive expression of fluorescent proteins (Shakir et al. 2006; Chapman et al. 2008). Five positions in the anterior–posterior axis of the animal were used to score AQR and PQR position. Position 1 was the wild-type position of AQR and is the region around the posterior pharyngeal bulb. Neurons anterior to the posterior pharyngeal bulb were not observed. Position 2 was posterior to position 1, but anterior to the vulva. Position 3 was the region around the vulva, position 4 was the birthplace of Q cells, and position 5 was posterior to the anus, the wild-type position of PQR (see Figure 2D). A Leica DM550 equipped with YFP, CFP, GFP, and mCherry filters, was used to acquire all micrographs, and for visualization of A/PQR for scoring. Micrographs were acquired using a Qimaging Retiga camera. Significances of difference were determined using Fisher’s exact test.
Figure 2.
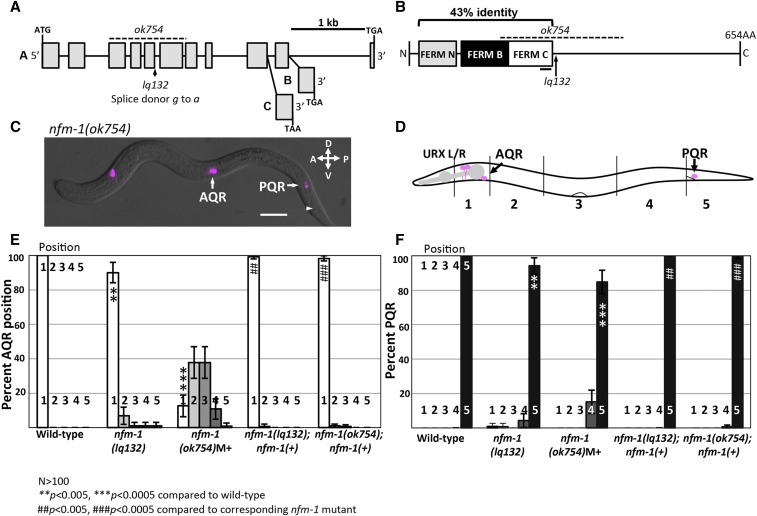
Position of Q descendants AQR and PQR in nfm-1 mutants. (A) Diagram of the nfm-1 locus and alleles used. The ok754 deletion (dashed line) and lq132 splice site mutation (arrow) are noted. The alternate 3′ exon use in the three nfm-1 isoforms A–C are shown (from WormBase). (B) NFM-1 isoform A domain structure and allele locations are shown. The FERM domain lobes N (gray), B (black), and C (white) are shown. The black bar under FERM C represents predicted actin-binding motif. The dashed line is ok754 in-frame deletion, and lq132 splice donor mutation location is marked by an arrow. NF2 and NFM-1 show 43% identity throughout the FERM N, B, and C regions. (C) Merged DIC and fluorescent micrograph of an nfm-1(ok754) arrested larval mutant animal. Both AQR and PQR failed to migrate (PQR wild-type position noted by arrowhead). Bar, 10 μM. (D) Diagram of scoring positions in an L4 animal used in E and F, with wild-type locations of AQR and PQR shown as magenta circles. (E and F) Chart showing percent of AQR (E) or PQR (F) in positions 1–5 in different genotypes as shown in D. All animals unless otherwise noted were scored using lqIs58 (Pgcy-32::cfp). M+ indicates animals were scored from heterozygous mother and have wild-type maternal contribution of nfm-1. nfm-1(+) animals harbor the array containing the nfm-1::gfp fosmid. Asterisks indicate degree of significance of difference from wild-type (N > 100; * P < 0.05, ** P < 0.005, *** P < 0.0005, Fisher’s exact test). Pound signs indicate, for that position, a significant rescue of corresponding nfm-1 mutant (N > 100; # P < 0.05, ## P < 0.005, ### P < 0.0005, Fisher’s exact test). Error bars represent two times the SE of the proportion in each direction.
Mosaic analysis
Mosaic analysis was conducted as previously dscribed (Chapman et al. 2008; Sundararajan et al. 2014) and involved generating a rescuing extrachromosomal array carrying nfm-1(+), and an independent marker of AQR and PQR position. The positions of AQR and PQR were determined in mosaics in which the rescuing extrachromosomal array was lost in AQR and/or PQR.
A rescuing nfm-1(+) extrachromosomal array was generated using the nfm-1::gfp fosmid with a Pgcy-32::yfp marker (lqEx773). This array was crossed into nfm-1(ok754)/hT2; lqIs58 (Pgcy-32::cfp) to create the rescuing array lqEx773, referred to as nfm-1(+). Presence of the rescuing array was determined by Pgcy-32::yfp expression, and position of AQR and PQR was determined by Pgcy-32::cfp expression. nfm-1(ok754)III; nfm-1(+) animals were viable and fertile and had wild-type AQR and PQR position, indicating rescue of nfm-1(ok754). Presence of YFP in AQR or PQR indicated nfm-1(+) was present in those cells during their migrations. Pgcy-32 is also expressed in URX, and presence of YFP in the URX neurons indicates other tissues have inherited nfm-1(+). Animals that lost nfm-1(+) in AQR or PQR, and retained nfm-1(+) in the other Q descendant (PQR and AQR, respectively) and URX were scored for AQR and PQR position.
Synchronization of L1 larvae for expression analysis
L1 animals carrying Pnfm-1::gfp, the nfm-1::gfp fosmid, and Pslt-1::gfp were synchronized as described above in Scoring Q-cell and AQR/PQR AQR migration to the time of Q cell migration (3–5 hr posthatching). Pegl-17::mCherry was used as a Q-cell marker to determine overlapping expression of nfm-1 expression constructs.
Data availability
The authors state that all data necessary for confirmation of the conclusions discussed in the article are represented fully within the article.
Results
nfm-1 mutants have defective AQR and PQR migration
To identify genes required for AQR and PQR migration, a forward genetic screen was conducted (see Materials and Methods). This screen identified the new mutation lq132. The genome of the lq132-bearing strain was sequenced and variants were detected using Cloudmap (Minevich et al. 2012). The strain contained a splice donor mutation after the fifth exon in the nfm-1 gene (Figure 2A) (GTATGTGT to ATATGTGT). To determine whether nfm-1 mutation in the lq132 strain caused AQR and PQR defects, we scored AQR and PQR migration in a second allele, the existing the nfm-1(ok754) mutant generated by the C. elegans gene knockout consortium. nfm-1(ok754) is an in-frame 1042-bp deletion with breakpoints in exons 3 and 7 that removes all of exons 4–6 (Figure 2, A and B). nfm-1(ok754) homozygotes arrested as larvae, but we were able to score AQR and PQR position in arrested larvae. nfm-1(ok754) had strong AQR defects (Figure 2, C and D), with 88% of AQR failing to migrate to the head, and occasional (1%) posterior AQR migration (Figure 2, C and E). nfm-1(ok754) also had significant PQR defects, with 15% of PQR failing to migrate into the wild-type position 5, posterior to the anus (Figure 2F). To confirm that nfm-1 was the causative locus, we found that an nfm-1::gfp fosmid transgene rescued AQR and PQR defects of both lq132 and ok754 mutants (Figure 2, E and F).
nfm-1 encodes a protein similar to human NF2/Merlin, and contains Four-Point-One Ezrin Radixin and Moesin (FERM) N, B, and C domains at the N terminus (Figure 2B). Three isoforms of nfm-1 are predicted, differing at the 3′ end (WormBase) (Figure 2A). lq132 and ok754 are predicted to affect all three isoforms. The functional differences, if any, between these isoforms are not known.
The lq132 splice donor mutation occurred after the conserved FERM domain regions, and the ok754 in-frame deletion removes the entire FERM C domain, including the putative actin-binding site (Figure 2B). RNAi of nfm-1 caused embryonic lethality (Skop et al. 2004). Thus, lq132 is likely a hypomorphic mutation and retains some function. ok754 mutants have wild-type maternal contribution, which might allow the animals to bypass embryonic lethality and arrest later as larvae. It is also possible that the ok754 in-frame deletion retains some function. AQR migration defects in ok754 were significantly stronger than lq132 (P < 0.001), suggesting that ok754 is a stronger allele than lq132.
NFM-1 is required for Q-cell protrusion and migration
A Pegl-17::gfp transgene was used to inspect early Q migration (Branda and Stern 2000; Cordes et al. 2006; Josephson et al. 2016a). Between 1 and 2.5 hr after hatching, Q cells in wild-type extend robust protrusions over the neighboring seam cells in their direction of eventual migration (QR to the anterior over V4, and QL to the posterior over V5) (Figure 3, A and B) (Chapman et al. 2008). In nfm-1(ok754)M+ and nfm-1(lq132) homozygotes, Q-cell protrusions at 1–2.5 hr were significantly shorter than in wild type (Figure 3, C–F). No defects were observed in the direction of protrusion. These data indicate that NFM-1 is required for robust Q-cell protrusion. Between 3 and 3.5 hr after hatching, wild-type Q-cell bodies migrate atop the neighboring seam cells, and the first Q cell division occurs between 4 and 4.5 hr (Chapman et al. 2008). Despite reduced protrusions in nfm-1 mutants, the Q cells completed their anterior and posterior migrations before division (n > 20 for both ok754 and lq132).
Figure 3.
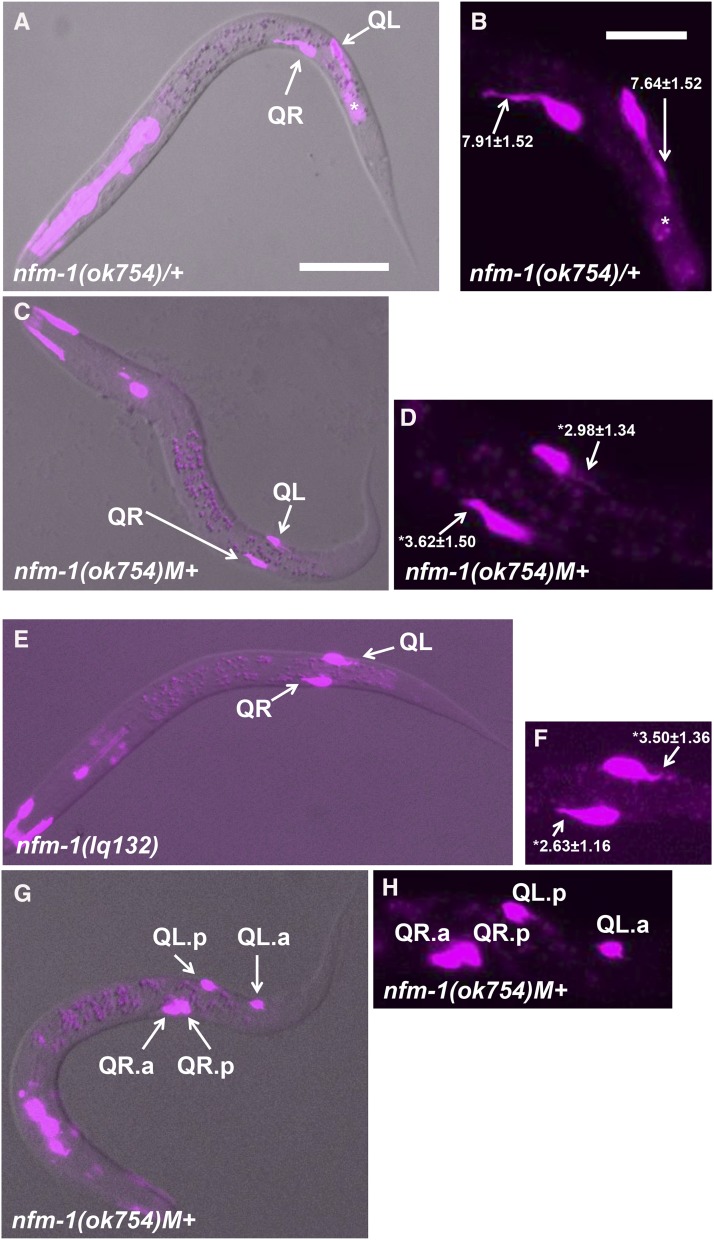
Early Q migrations in nfm-1 mutants. Fluorescent micrographs of L1 animals with ayIs9[egl-17::gfp] expression are shown (magenta). Anterior is to the left. (A, C, E, and F) Merged with a differential interference contrast image. (B, D, F, and G) Enlarged images of the migrating Q cells. The average length and SE of Q protrusions in micrometers are indicated in B, D, and F (n = 20 in all cases). The asterisks indicate statistical significance compared to wild type (t-test; P < 0.0001). (A and B). Wild-type Q cells display robust protrusions at 1–2.5 hr posthatching (arrows). This animal is a balanced nfm-1(ok754)/hT2 heterozygote with one wild-type copy of nfm-1. The bright fluorescence in the anterior is gfp expression in the pharynx associated with the hT2 balancer chromosome, and the fluorescence posterior to the Q cells is background associated with the ayIs9[Pegl-17::gfp] transgene (asterisk). (C and D). An nfm-1(ok754) homozygote with wild-type maternal contribution (M+) at 1–2.5 hr posthatching displays Q cells with shortened protrusions compared to wild-type (arrows). (E and F). An nfm-1(lq132) homozygote at 1–2.5 hr posthatching displays shortened protrusions (arrows). (G and H). An nfm-1(ok754)M+ animal at 6–7 hr posthatching. QL.p has migrated posteriorly, whereas both QR.a and QR.p have failed to migrate anteriorly and remain near their birthplace. Bars, 10 μm for A, C, E, and F and 5 μm for B, D, F, and G.
After division at 4–4.5 hr, the wild-type QR daughters QR.a and QR.p extend anterior protrusions and begin anterior migration, whereas the QL daughters QL.a and QL.p remain rounded and nonpolarized and do not migrate (Josephson et al. 2016a). At 5–7.5 hr after hatching, QL.a migrates posteriorly past QL.p (Josephson et al. 2016a). In nfm-1(ok754)M+, 2/20 QR.a/p daughters failed to migrate anteriorly and stayed near their birthplace, even after QL.a had migrated posteriorly (Figure 3, G and H). This defect was not observed in the weaker nfm-1(lq132) mutant, although more subtle defects in migration might have escaped detection. Failure of QR.a/p migration might explain the strong AQR migration defects observed in nfm-1(ok754), as AQR is a descendant of QR.a. These data suggest that NFM-1 is required for Q-cell and descendant protrusion and migration. Direction of protrusion and migration was not affected in nfm-1 mutants. However, as both mutants likely retain some nfm-1 function, a role of NFM-1 in controlling direction of protrusion cannot be excluded.
nfm-1::gfp transcriptional and translational reporter expression was not apparent in Q lineages
A Pnfm-1::gfp transcriptional reporter was created by using a 2.1-kb region upstream of nfm-1 to drive expression of gfp. This 2.1-kb region was the entire upstream region between nfm-1 and the next gene anmt-2. At the time of Q migration, this construct showed expression in posterior cells near the anus, including posterior intestinal cells, the three rectal gland cells, and other unidentified cells that might be the anal sphincter muscle and the stomatointestinal muscle (Figure 4, A–C). Variable expression in the hypodermis was also observed (Figure 4, A–C), as well as in body wall muscle cells (Figure 4, D–F). Pnfm-1::gfp expression was not observed in migrating Q neuroblasts (Figure 4, A–C).
Figure 4.
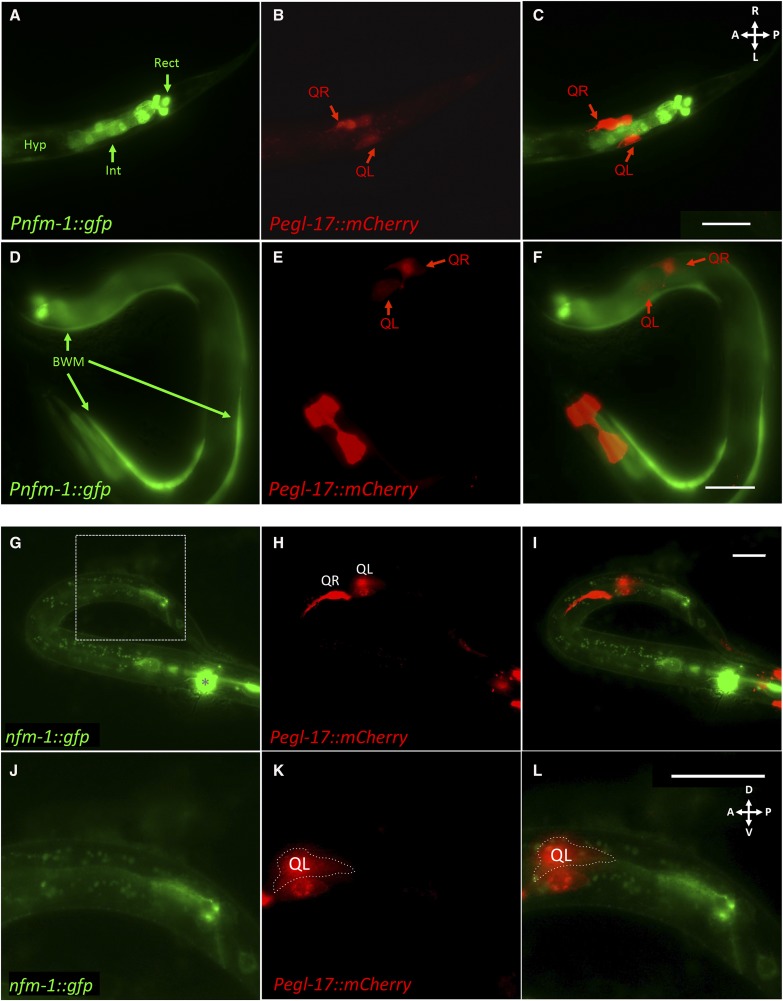
nfm-1 was not expressed in Q cells during their early migrations. (A–C) Ventral view of the posterior region of a Pnfm-1::gfp; Pegl-17::mCherry transgenic animal staged to 3–3.5 hr posthatching. (A) GFP micrograph showing expression of Pnfm-1::gfp. Expression was seen in posterior cells near the anus, including posterior intestinal cells (Int) and the three rectal gland cells (Rect). Other unidentified cells in the region were possibly the anal sphincter muscle and the stomatointestinal muscle. Variable hypodermal expression was observed along the length of the animal (Hyp). (B) An mCherry micrograph shows Q-cell-specific expression during their migrations. (C) Merged. GFP is not observed in Q cells, but is expressed in neighboring tissues and posterior cells. Bar, 10 μm for A–C. (D–F) An L1 animal 3–3.5 hr posthatching with Pnfm-1::gfp expression in body wall muscles. Bar, 20 μm for D–F. (G–L) Lateral view of a staged 3–3.5 hr posthatching L1 with full length nfm-1::gfp and Pegl-17::mCherry expression. (G) Fluorescent micrograph of GFP expression from nfm-1::gfp rescuing fosmid. Asterisk marks URX expression of Pgcy-32::yfp in the head that was not excluded by GFP filter. The dashed rectangle indicates the enlarged posterior section in J–L. (H) Pegl-17::mCherry, fluorescent micrograph showing location of early Q neuroblasts. QL is out of focus because QL and QR are on different planes, QR on the right side and QL on left side of the animal. (I) Merge of A and B. No overlap of mCherry and GFP was observed. (J–L) Enlarged posterior section of G–I. (J) Enlargement of A to show nfm-1::gfp present in posterior region near the anus. (K) Enlargement of B. QL is outlined to distinguish it from the V5 seam cell that transiently expresses Pegl-17. (L) Enlargement of C. Bar, 10 μm. In all micrographs, anterior is to the left.
Full-length NFM-1::GFP expression from the rescuing fosmid was not observed in migrating Q cells (Figure 4, G–L). NFM-1::GFP was detected in the posterior gut region. Three isoforms for nfm-1, differing at the 3′ end, are reported (WormBase). This fosmid contains the gfp tag at the end of the nfm-1A isoform and so will not report the expression of the B and C isoforms. We do not know which isoforms are required for AQR and PQR migration, but the nfm-1 promoter was not active in Q cells, and nfm-1A isoform expression was not observed in the Q cells.
Mosaic analysis suggests a nonautonomous requirement for nfm-1 in anterior AQR migration
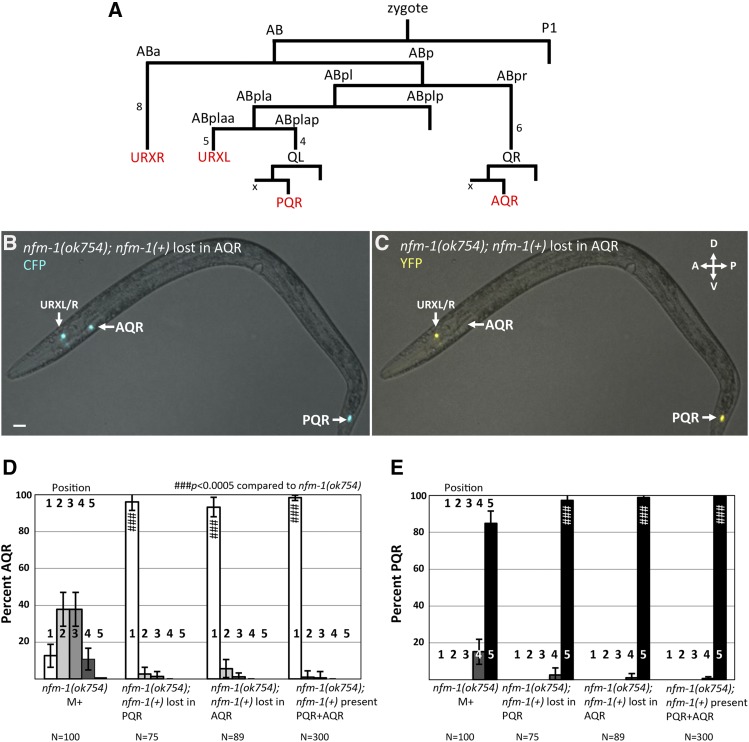
No expression of nfm-1 was observed in migrating Q neuroblasts. Genetic mosaic analysis using a rescuing nfm-1(+) extrachromosomal array was used to test whether nfm-1 was required in the Q cells themselves for proper AQR and PQR migration (see Materials and Methods). In C. elegans, extrachromosomal arrays are not stably inherited mitotically and can be lost during cell divisions, creating genetically mosaic animals. We used an established strategy to score mosaic animals that had lost an nfm-1(+) rescuing transgene in AQR or PQR lineage (see Materials and Methods and Chapman et al. 2008; Sundararajan et al. 2014). This strategy uses a stable Pgcy-32::cfp integrated transgene to visualize AQR and PQR in all animals, and an unstable array carrying the rescuing nfm-1::gfp fosmid and Pgcy-32::yfp, which we refer to as nfm-1(+). The AQR, PQR, and URX cells are derived from well-separated lineages, with URX and QR/QL lineages distinguished after the second embryonic division, and the QL and QR lineages after the third (Figure 5A), making mosaic animals with losses in specific lineages readily identifiable (Figure 5, B and C).
Figure 5.
nfm-1 mosaic analysis. (A) The abbreviated lineage of cells that express Pgcy-32 (red). Numbers next to lines indicate the number of cell divisions not shown. The X next to AQR and PQR indicates the sister of A/PQR (QL/R.aa) that undergoes programmed cell death. (B) Fluorescent micrograph taken with CFP filter of nfm-1(ok754); nfm-1(+), Pgcy-32::cfp mosaic animal with correct placement of AQR and PQR. (C) Fluorescent micrograph of the same animal from B using a YFP filter. AQR is not visible in this animal, indicating that somewhere in the AQR lineage, the nfm-1(+) transgene was lost. YFP is detected in URXL/R, and PQR indicating many tissues retained nfm-1(+). Bar, 10 μM. (D and E) Quantification of AQR (A), and PQR (B) migration as in Figure 2, with nfm-1(+) mosaic animals. nfm-1(+) represents presence of nfm-1 rescuing fosmid. nfm-1(+) rescued ok754 lethality, and animals were maintained as rescued homozygous ok754 mutants. Mosaic animals have nfm-1(+) in URX but have lost nfm-1(+) in either AQR or PQR. Pound signs indicate, for that position, a significant rescue of corresponding nfm-1 mutant (N > 100; # P < 0.05, ## P < 0.005, ### P < 0.0005, Fisher’s exact test). Error bars represent two times the SE of the proportion.
nfm-1(ok754) animals that harbored the nfm-1(+) array were viable, fertile, and were rescued for AQR and PQR migration (Figure 2, E and F). We analyzed 89 mosaic animals in which the nfm-1(+) array was lost from the AQR lineage, but retained in PQR and URX lineages as shown in Figure 5, B and C. These animals were rescued for AQR migration defects despite loss of nfm-1(+) in AQR compared to nfm-1(ok754) alone (Figure 5D), suggesting that nfm-1 is required nonautonomously for anterior AQR migration. Similarly, PQR migration defects were still rescued in 75 mosaic animals in which PQR had lost the nfm-1(+) array (Figure 5E). Loss of nfm-1(+) in AQR or PQR rescued nfm-1(ok754) defects to a similar level as in animals in which no loss occurred [nfm-1(+) in AQR and PQR] (Figure 5, D and E). It is possible that perdurance of NFM-1 protein, or array loss in the Q lineages themselves, led to nfm-1 function in the Q lineages despite loss in AQR or PQR. To account for these rare but possible events, we scored at least 70 mosaic animals. Overall, mosaic analysis suggests that nfm-1 acts nonautonomously for AQR and PQR migration, as loss of the rescuing array in AQR or PQR did not correlate with mutant phenotype.
Expression of nfm-1 in muscles rescued AQR and PQR migration defects
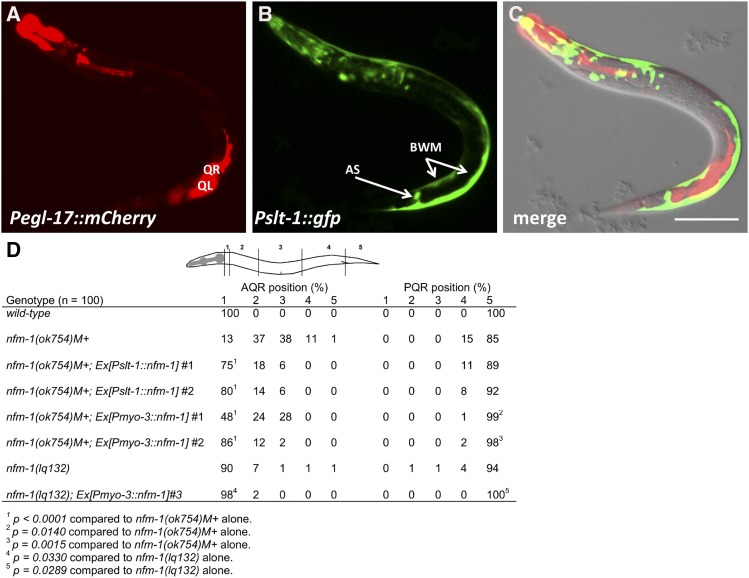
nfm-1(+) expression was driven from two promoters with expression in muscles, the slt-1 and myo-3 promoters. At the time of Q protrusion and migration, the slt-1 promoter was active in dorsal– and ventral–posterior body wall muscles (Figure 6, A–C) (Hao et al. 2001). It was also expressed in cells in the head and the anal sphincter muscle as previously reported (Figure 6, A–C) (Hao et al. 2001). Pslt-1 expression was not observed in protruding and migrating Q neuroblasts (Figure 6, A–C). The entire nfm-1A coding region fused to gfp was placed under the control of Pslt-1. Two independent lines of nfm-1(ok754)M+ animals harboring Pslt-1::nfm-1(+) still arrested as early larvae but were significantly rescued for AQR migration defects (Figure 6D). PQR defects were not significantly rescued by Pslt-1::nfm-1(+).
Figure 6.
Muscle-specific expression of nfm-1 rescues AQR and PQR defects. (A–C) Expression of Pslt-1::gfp (kyIs174) (Hao et al. 2001) and Pegl-17::mCherry in an L1 animal 3–3.5 hr posthatching. AS, anal sphincter muscle; BWM, body wall muscle. QL and QR are indicated. Bar, 20 μm for A–C. (D) Rescue of nfm-1 AQR and PQR defects by cell-specific transgenes. The positions of AQR and PQR are as described in Figure 2D. The percentage of cells in each position is indicated, with significance of differences (Fisher’s exact test). Two independent Pslt-1::nfm-1::gfp transgenes rescued nfm-1(ok754) (lqEx1065#1 and lqEx1066#2), and three independent Pmyo-3::nfm-1::gfp transgenes rescued nfm-1(ok754) (lqEx1064#1 and lqEx1086#2), and nfm-1(lq132) (lqEx1073#3).
The myo-3 promoter is expressed in body wall muscles, the vulval muscles, and the anal sphincter muscle (Okkema et al. 1993). Two independent lines of Pmyo-3::nfm-1(+)::gfp rescued AQR and PQR defects of nfm-1(ok754)M+ (Figure 6D). These animals also grew to be sterile adults, indicating that Pmyo-3::nfm-1(+)::gfp expression partially rescued the larval arrest of nfm-1(ok754)M+ animals. One line of Pmyo-3::nfm-1(+)::gfp also rescued AQR and PQR defects of nfm-1(lq132) (Figure 6D). Expression of slt-1 (Figure 6), myo-3, and nfm-1 (Figure 4) overlap in the body wall muscles and possibly the anal sphincter muscle, suggesting that these tissues might be the cellular focus of nfm-1 activity in Q migration. Muscle expression of nfm-1 only partially rescued AQR and PQR defects, suggesting that nfm-1 could be required in other tissues for full rescue.
slt-1 mutations enhance AQR defects of nfm-1(lq132)
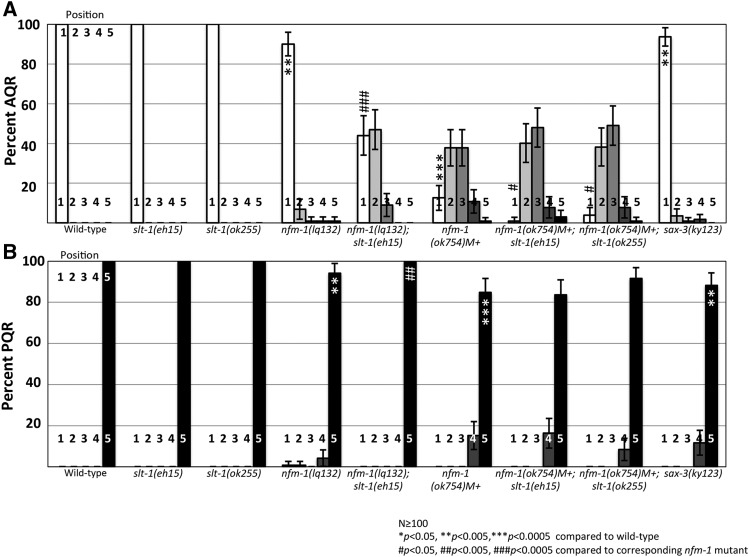
Previous studies suggested that NF2 can nonautonomously affect axon guidance in the developing mouse brain (Lavado et al. 2014). This guidance mechanism occurs through regulation of Slit2 mRNA levels, suggesting a transcriptional role of NF2 (Lavado et al. 2014). Slit2 is a secreted guidance cue for developing neurons and is detected by the Robo receptor. Because of interactions between Slit2 and NF2, we investigated the interaction of nfm-1 and the C. elegans Slit2 homolog slt-1 in Q-descendant migration. In this study we used one null allele slt-1(eh15), and one strong loss-of-function in-frame deletion allele slt-1(ok255) (Hao et al. 2001; Steimel et al. 2013). slt-1 mutations had no effect on AQR and PQR migration on their own, but enhanced AQR migration defects of nfm-1(lq132) and nfm-1(ok754) (Figure 7). slt-1 had no effect on PQR migration in double mutants. We tested the SLT-1 receptor SAX-3/Robo, and sax-3(ky123) mutants showed weak but significant defects in both AQR and PQR migration, consistent with SAX-3 promoting anterior–posterior migration of the Q lineages (Figure 7).
Figure 7.
slt-1 enhances nfm-1 AQR migration defects. (A) Percentage of AQR in each position, quantified as in Figure 2. (B) PQR migration. Asterisks indicate significant difference from wild type (N > 100; * P < 0.05, ** P < 0.005, *** P < 0.0005, Fisher’s exact test). Pound signs indicate, for that position, a significant enhancement of the corresponding nfm-1 mutant (N > 100; # P < 0.05, ## P < 0.005, ### P < 0.0005, Fisher’s exact test). Error bars represent two times the SE of the proportion.
slt-1 was not expressed in protruding and migrating Q neuroblasts (Figure 6, A–C), suggesting a likely nonautonomous effect expected of a secreted signaling molecule. Loss of NF2 in mouse led to increased levels of Slit2 expression (Lavado et al. 2014). We detected no discernible change in Pslt-1::gfp expression in nfm-1(lq132) and nfm-1(ok754)M+ animals at 1–2.5 hr posthatching, including body wall muscle and anal sphincter muscle (data not shown). However, expression of nfm-1(+) from the slt-1 promoter rescued AQR defects of nfm-1(ok754)M+ (Figure 6, D and E), suggesting that nfm-1 and slt-1 might be acting in the same cells to regulate Q migration.
Discussion
The NF2/Merlin molecule NFM-1 promotes protrusion and migration of Q cells and their descendants
Complete migration of the QR and QL descendants AQR and PQR requires the coordination of many genes (Middelkoop and Korswagen 2014). Although numerous molecules have been identified that act in the Q cells to promote migration, such as the transmembrane receptors UNC-40/DCC, PTP-3/LAR, and MIG-13 (Sundararajan and Lundquist 2012; Wang et al. 2013; Sundararajan et al. 2015), fewer have been identified that act outside the Q cells to control their migration. Of the nonautonomous genes that have been implicated in Q-descendant migration, most are secreted molecules such as Wnts (Hunter et al. 1999; Whangbo and Kenyon 1999; Korswagen 2002; Pan et al. 2006) and SPON-1/F-spondin (Josephson et al. 2016b), although the Fat-like cadherin CDH-4 has been demonstrated to nonautonomously affect Q-cell migration (Sundararajan et al. 2014).
Here we present data identifying a nonautonomous role for the FERM domain-containing molecule NFM-1, a predicted cytoplasmic protein, in promoting Q migration. NFM-1 is similar to human NF2/Merlin, the molecule affected in neurofibromatosis type II. We found that mutations in nfm-1 resulted in AQR migration defects, and to a lesser extent PQR migration defects. These defects typically manifested as incomplete migrations, suggesting that these nfm-1 mutations did not affect direction of migration along the anterior/posterior axis, but rather the migratory capacity of these cells.
We found that NFM-1 was required for robust protrusion of QR and QL in their initial migrations over V4 and V5, as well as subsequent migration of QR daughters. In no case did we observe Q protrusion or migration in the wrong direction, suggesting that NFM-1 affects the ability of Q cells to protrude and migrate, but not their direction.
Loss of NF2/Merlin function in either mouse or Drosophila results in embryonic lethality (Fehon et al. 1997; McClatchey et al. 1997). In C. elegans, nfm-1 appears to be required in embryonic development similar to other animals, as RNAi against nfm-1 is reported as embryonic lethal (Skop et al. 2004), and no null alleles of nfm-1 have been described. The two nfm-1 mutations studied here likely retain some NFM-1 function. The 5′ splice site mutant nfm-1(lq132) was viable and fertile, and the in-frame deletion allele nfm-1(ok754) caused larval arrest possibly due to wild-type maternal contribution. It is possible that complete loss of nfm-1 function results in more severe Q-migration defects, possibly even directional defects, not observed in these alleles. The nfm-1(ok754) in-frame deletion removes part of the FERMB domain and the entire FERMC domain, suggesting that these domains are important in AQR and PQR migration.
NFM-1 might act in muscles to promote AQR and PQR migration
As a cytoskeletal–membrane linker with a potential actin-binding domain, we hypothesized that NFM-1 might regulate actin-based membrane protrusion in migrating cells. However, no nfm-1 expression was observed in the Q cells or descendants, and a genetic mosaic analysis and cell-specific expression indicated that NFM-1 was not required in AQR or PQR for their migration. These data suggest that NFM-1 is required nonautonomously outside of the Q lineages for their migration. Pnfm-1::gfp expression was observed in the posterior region near the anus, including posterior intestine, the rectal gland cells, and potentially the anal sphincter muscle and stomatointestinal muscle. Expression was also noted in body wall muscles. Cell-specific expression of nfm-1(+) from the slt-1 and myo-3 promoters, both expressed in body wall muscles and the anal sphincter muscle, rescued AQR and PQR migration defects of nfm-1(ok754)M+. Thus, nfm-1 might be required in body wall muscles and/or the anal sphincter muscle to promote Q migrations. The Q-cell protrusions are in close proximity to the body wall muscle cells as the Q cells undergo their initial migrations. Interestingly, Pslt-1 expression of nfm-1 did not rescue PQR defects, whereas Pmyo-3 expression did. We do not understand the nature of this difference, but it is possible that AQR and PQR have differential requirements for nfm-1 expression, either different levels of expression or expression from different tissues.
nfm-1 and slt-1 interact genetically to promote anterior AQR migration
In Drosophila and mice, NF2/Merlin is known to regulate several signaling pathways, including stimulating the Hippo pathway to inhibit the Yorkie transcription cofactor (Hamaratoglu et al. 2006; Moroishi et al. 2015). In mice, loss of NF2 in neural progenitor cells results in upregulation of Yap (Lavado et al. 2014). High Yap activity leads to ectopic levels of the secreted guidance cue Slit2, which causes defects in midline axon guidance (Lavado et al. 2014). Interestingly, this is a nonautonomous role of NF2 in midline axon guidance, similar to our observation of nfm-1 in C. elegans neuronal migration. The Hippo pathway in C. elegans is poorly conserved (Hilman and Gat 2011), although C. elegans YAP-1 is similar to Yap (Iwasa et al. 2013). A role of yap-1 in AQR and PQR migration was not determined. However, nfm-1 mutants had no discernible effect on the expression of Pslt-1::gfp, suggesting that NFM-1 does not regulate SLT-1 expression, at least at the transcriptional level.
We tested the role of the single C. elegans Slit gene slt-1 in AQR/PQR migration and interaction with nfm-1. slt-1 regulates the anterior–posterior migration of the CAN neurons in embryos (Hao et al. 2001). Although no migration defects were detected in slt-1 mutants alone, they did enhance AQR migration defects of nfm-1(lq132) and nfm-1(ok754). This enhancement is consistent with NFM-1 and SLT-1 acting in parallel pathways, but since we do not know the null phenotype of NFM-1 with regard to AQR and PQR, the possibility that they act in the same pathway cannot be excluded.
Interestingly no enhancement of nfm-1 PQR migration defects was seen in slt-1; nfm-1 double mutants. This indicates that slt-1 and nfm-1 interact in AQR migration but not PQR migration. This result, together with differential rescue of AQR vs. PQR defects by cell-specific nfm-1(+) transgenes, suggests that NFM-1 might affect AQR and PQR differentially, possibly involving distinct expression levels, distinct sources of expression, or interactions with distinct genes.
sax-3/Robo mutants displayed both AQR and PQR migration defects. Possibly, SAX-3/Robo acts with SLT-1 in AQR migration, and with an unidentified ligand in PQR migration. In mice, midline axon defects are due to excess Slit2 expression in NF2 mutants. The phenotypic enhancement that we observe between slt-1 and nfm-1 suggests that these molecules are both required for AQR migration. Further studies of the interaction between nfm-1 and slt-1 will be required to understand the role of these molecules in AQR migration. However, expression of nfm-1 from the slt-1 promoter rescued AQR migration defects, suggesting that NFM-1 and SLT-1 are required in the same tissues to control AQR migration, possibly the body wall muscles and/or the anal sphincter muscles where both are expressed.
Our results are consistent with the idea that NFM-1 promotes the production of a signal or signals that regulate AQR and PQR migration. This could be SLT-1 itself, such as in vertebrates, or a molecule that acts in parallel to SLT-1. Our combined results here suggest that NFM-1 and SLT-1 might act in the body wall muscles and/or anal sphincter muscle. Of note, the Q-cell protrusions are in close proximity to body wall muscles (Figure 6), and the posterior body wall muscles are the source of SPON-1/F-spondin, which is involved in Q migrations (Josephson et al. 2016b). Possibly, the posterior body wall muscles surrounding the Q neuroblasts serve as a source for cues that promote and guide their migrations. Further studies will be required to determine whether nfm-1 can control expression of guidance cue genes in muscles or if it is involved in secretion, adhesion, or extracellular matrix function to regulate a substrate for Q neuroblast migration.
Acknowledgments
The authors thank members of the Lundquist and Ackley labs for discussion, E. Struckhoff for technical assistance, and C. Bargmann for kyIs174. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). Some next-generation sequencing was provided by the University of Kansas Genome Sequencing Core Laboratory of the Center for Molecular Analysis of Disease Pathways (NIH P20 GM103638). This work was supported by NIH grants R01 NS040945 and R21 NS070417 to E.A.L. and the Kansas Infrastructure Network of Biomedical Research Excellence (NIH P20 GM103418). M.P.J. was supported by the Madison and Lila Self Graduate Fellowship at the University of Kansas, and R.A. and M.L.N. were Kansas IdeA Netword of Biomedical Research Excellence undergraduate research scholars (NIH P20 GM103418).
Footnotes
Communicating editor: B. Goldstein
Literature Cited
- Bagri A., Marin O., Plump A. S., Mak J., Pleasure S. J., et al. , 2002. Slit proteins prevent midline crossing and determine the dorsoventral position of major axonal pathways in the mammalian forebrain. Neuron 33: 233–248. [DOI] [PubMed] [Google Scholar]
- Branda C. S., Stern M. J., 2000. Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev. Biol. 226: 137–151. [DOI] [PubMed] [Google Scholar]
- Chang C., Adler C. E., Krause M., Clark S. G., Gertler F. B., et al. , 2006. MIG-10/lamellipodin and AGE-1/PI3K promote axon guidance and outgrowth in response to slit and netrin. Curr. Biol. 16: 854–862. [DOI] [PubMed] [Google Scholar]
- Chapman J. O., Li H., Lundquist E. A., 2008. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Dev. Biol. 324: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes S., Frank C. A., Garriga G., 2006. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions. Development 133: 2747–2756. [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Oren T., LaJeunesse D. R., Melby T. E., McCartney B. M., 1997. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics 146: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella J. F., Ramesh V., MacCollin M., Jacoby L. B., 1996. Neurofibromatosis 2: loss of merlin’s protective spell. Curr. Opin. Genet. Dev. 6: 87–92. [DOI] [PubMed] [Google Scholar]
- Gutmann D. H., Giordano M. J., Fishback A. S., Guha A., 1997. Loss of merlin expression in sporadic meningiomas, ependymomas and schwannomas. Neurology 49: 267–270. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., et al. , 2006. The tumour-suppressor genes NF2/Merlin and expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8: 27–36. [DOI] [PubMed] [Google Scholar]
- Hao J. C., Yu T. W., Fujisawa K., Culotti J. G., Gengyo-Ando K., et al. , 2001. C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron 32: 25–38. [DOI] [PubMed] [Google Scholar]
- Harris J., Honigberg L., Robinson N., Kenyon C., 1996. Neuronal cell migration in C. elegans: regulation of Hox gene expression and cell position. Development 122: 3117–3131. [DOI] [PubMed] [Google Scholar]
- Harterink M., Kim D. H., Middelkoop T. C., Doan T. D., van Oudenaarden A., et al. , 2011. Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development 138: 2915–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilman D., Gat U., 2011. The evolutionary history of YAP and the hippo/YAP pathway. Mol. Biol. Evol. 28: 2403–2417. [DOI] [PubMed] [Google Scholar]
- Honigberg L., Kenyon C., 2000. Establishment of left/right asymmetry in neuroblast migration by UNC-40/DCC, UNC-73/Trio and DPY-19 proteins in C. elegans. Development 127: 4655–4668. [DOI] [PubMed] [Google Scholar]
- Hunter C. P., Harris J. M., Maloof J. N., Kenyon C., 1999. Hox gene expression in a single Caenorhabditis elegans cell is regulated by a caudal homolog and intercellular signals that inhibit wnt signaling. Development 126: 805–814. [DOI] [PubMed] [Google Scholar]
- Iwasa H., Maimaiti S., Kuroyanagi H., Kawano S., Inami K., et al. , 2013. Yes-associated protein homolog, YAP-1, is involved in the thermotolerance and aging in the nematode Caenorhabditis elegans. Exp. Cell Res. 319: 931–945. [DOI] [PubMed] [Google Scholar]
- James M. F., Han S., Polizzano C., Plotkin S. R., Manning B. D., et al. , 2009. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol. Cell. Biol. 29: 4250–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson M. P., Chai Y., Ou G., Lundquist E. A., 2016a EGL-20/Wnt and MAB-5/Hox act sequentially to inhibit anterior migration of neuroblasts in C. elegans. PLoS One 11: e0148658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson M. P., Miltner A. M., Lundquist E. A., 2016b Nonautonomous roles of MAB-5/Hox and the secreted basement membrane molecule SPON-1/F-Spondin in Caenorhabditis elegans neuronal migration. Genetics 203: 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Farmer W. T., Bjorke B., McMahon S. A., Fabre P. J., et al. , 2014. Pioneer midbrain longitudinal axons navigate using a balance of Netrin attraction and Slit repulsion. Neural Dev. 9: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen H. C., 2002. Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: variations on a common signaling theme. BioEssays 24: 801–810. [DOI] [PubMed] [Google Scholar]
- Lavado A., Ware M., Pare J., Cao X., 2014. The tumor suppressor Nf2 regulates corpus callosum development by inhibiting the transcriptional coactivator Yap. Development 141: 4182–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof J. N., Whangbo J., Harris J. M., Jongeward G. D., Kenyon C., 1999. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development 126: 37–49. [DOI] [PubMed] [Google Scholar]
- McClatchey A. I., Saotome I., Ramesh V., Gusella J. F., Jacks T., 1997. The Nf2 tumor suppressor gene product is essential for extraembryonic development immediately prior to gastrulation. Genes Dev. 11: 1253–1265. [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A., 1995. DNA transformation. Methods Cell Biol. 48: 451–482. [PubMed] [Google Scholar]
- Middelkoop T. C., Korswagen H. C., 2014. Development and migration of the C. elegans Q neuroblasts and their descendants. WormBook 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G., Park D. S., Blankenberg D., Poole R. J., Hobert O., 2012. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192: 1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T., Park H. W., Qin B., Chen Q., Meng Z., et al. , 2015. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 29: 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba-Charvet K. T., Brose K., Marillat V., Kidd T., Goodman C. S., et al. , 1999. Slit2-Mediated chemorepulsion and collapse of developing forebrain axons. Neuron 22: 463–473. [DOI] [PubMed] [Google Scholar]
- Okada M., Wang Y., Jang S. W., Tang X., Neri L. M., et al. , 2009. Akt phosphorylation of merlin enhances its binding to phosphatidylinositols and inhibits the tumor-suppressive activities of merlin. Cancer Res. 69: 4043–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P. G., Harrison S. W., Plunger V., Aryana A., Fire A., 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C. L., Howell J. E., Clark S. G., Hilliard M., Cordes S., et al. , 2006. Multiple Wnts and frizzled receptors regulate anteriorly directed cell and growth cone migrations in Caenorhabditis elegans. Dev. Cell 10: 367–377. [DOI] [PubMed] [Google Scholar]
- Piper M., Georgas K., Yamada T., Little M., 2000. Expression of the vertebrate Slit gene family and their putative receptors, the Robo genes, in the developing murine kidney. Mech. Dev. 94: 213–217. [DOI] [PubMed] [Google Scholar]
- Quinn C. C., Pfeil D. S., Chen E., Stovall E. L., Harden M. V., et al. , 2006. UNC-6/netrin and SLT-1/slit guidance cues orient axon outgrowth mediated by MIG-10/RIAM/lamellipodin. Curr. Biol. 16: 845–853. [DOI] [PubMed] [Google Scholar]
- Salser S. J., Kenyon C., 1992. Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature 355: 255–258. [DOI] [PubMed] [Google Scholar]
- Salser S. J., Kenyon C., 1996. A C. elegans Hox gene switches on, off, on and off again to regulate proliferation, differentiation and morphogenesis. Development 122: 1651–1661. [DOI] [PubMed] [Google Scholar]
- Salser S. J., Loer C. M., Kenyon C., 1993. Multiple HOM-C gene interactions specify cell fates in the nematode central nervous system. Genes Dev. 7: 1714–1724. [DOI] [PubMed] [Google Scholar]
- Sarov M., Schneider S., Pozniakovski A., Roguev A., Ernst S., et al. , 2006. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods 3: 839–844. [DOI] [PubMed] [Google Scholar]
- Schulz A., Zoch A., Morrison H., 2014. A neuronal function of the tumor suppressor protein merlin. Acta Neuropathol. Commun. 2: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz A., Baader S. L., Niwa-Kawakita M., Jung M. J., Bauer R., et al. , 2013. Merlin isoform 2 in neurofibromatosis type 2-associated polyneuropathy. Nat. Neurosci. 16: 426–433. [DOI] [PubMed] [Google Scholar]
- Shakir M. A., Gill J. S., Lundquist E. A., 2006. Interactions of UNC-34 enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics 172: 893–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop A. R., Liu H., Yates J., III, Meyer B. J., Heald R., 2004. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305: 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimel A., Suh J., Hussainkhel A., Deheshi S., Grants J. M., et al. , 2013. The C. elegans CDK8 Mediator module regulates axon guidance decisions in the ventral nerve cord and during dorsal axon navigation. Dev. Biol. 377: 385–398. [DOI] [PubMed] [Google Scholar]
- Striedinger K., VandenBerg S. R., Baia G. S., McDermott M. W., Gutmann D. H., et al. , 2008. The neurofibromatosis 2 tumor suppressor gene product, merlin, regulates human meningioma cell growth by signaling through YAP. Neoplasia 10: 1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J., 1988. Methods, pp. 587–606 in The Nematode Caenorhabditis elegans, edited by Wood W. B., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sulston J. E., Brenner S., 1974. The DNA of Caenorhabditis elegans. Genetics 77: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sundararajan L., Lundquist E. A., 2012. Transmembrane proteins UNC-40/DCC, PTP-3/LAR, and MIG-21 control anterior-posterior neuroblast migration with left-right functional asymmetry in Caenorhabditis elegans. Genetics 192: 1373–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan L., Norris M. L., Lundquist E. A., 2015. SDN-1/Syndecan acts in parallel to the transmembrane molecule MIG-13 to promote anterior neuroblast migration. G3 5: 1537–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan L., Norris M. L., Schoneich S., Ackley B. D., Lundquist E. A., 2014. The fat-like cadherin CDH-4 acts cell-non-autonomously in anterior-posterior neuroblast migration. Dev. Biol. 392: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unni D. K., Piper M., Moldrich R. X., Gobius I., Liu S., et al. , 2012. Multiple Slits regulate the development of midline glial populations and the corpus callosum. Dev. Biol. 365: 36–49. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhou F., Lv S., Yi P., Zhu Z., et al. , 2013. Transmembrane protein MIG-13 links the Wnt signaling and Hox genes to the cell polarity in neuronal migration. Proc. Natl. Acad. Sci. USA 110: 11175–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo J., Kenyon C., 1999. A Wnt signaling system that specifies two patterns of cell migration in C. elegans. Mol. Cell 4: 851–858. [DOI] [PubMed] [Google Scholar]
- Xu Y., Quinn C. C., 2012. MIG-10 functions with ABI-1 to mediate the UNC-6 and SLT-1 axon guidance signaling pathways. PLoS Genet. 8: e1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R. S., Yang Q., et al. , 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21: 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Forrester W. C., 2005. The C. elegans Frizzled CFZ-2 is required for cell migration and interacts with multiple Wnt signaling pathways. Dev. Biol. 285: 447–461. [DOI] [PubMed] [Google Scholar]
- Zinovyeva A. Y., Yamamoto Y., Sawa H., Forrester W. C., 2008. Complex network of Wnt signaling regulates neuronal migrations during Caenorhabditis elegans. Dev. Genet. 179: 1357–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirmation of the conclusions discussed in the article are represented fully within the article.