Abstract
The dwarf phenotype characterizes the smallest of rabbit breeds and is governed largely by the effects of a single dwarfing allele with an incompletely dominant effect on growth. Dwarf rabbits typically weigh under 1 kg and have altered craniofacial morphology. The dwarf allele is recessive lethal and dwarf homozygotes die within a few days of birth. The dwarf phenotype is expressed in heterozygous individuals and rabbits from dwarf breeds homozygous for the wild-type allele are normal, although smaller when compared to other breeds. Here, we show that the dwarf allele constitutes a ∼12.1 kb deletion overlapping the promoter region and first three exons of the HMGA2 gene leading to inactivation of this gene. HMGA2 has been frequently associated with variation in body size across species. Homozygotes for null alleles are viable in mice but not in rabbits and probably not in humans. RNA-sequencing analysis of rabbit embryos showed that very few genes (4–29 genes) were differentially expressed among the three HMGA2/dwarf genotypes, suggesting that dwarfism and inviability in rabbits are caused by modest changes in gene expression. Our results show that HMGA2 is critical for normal expression of IGF2BP2, which encodes an RNA-binding protein. Finally, we report a catalog of regions of elevated genetic differentiation between dwarf and normal-size rabbits, including LCORL-NCAPG, STC2, HOXD cluster, and IGF2BP2. Levels and patterns of genetic diversity at the LCORL-NCAPG locus further suggest that small size in dwarf breeds was enhanced by crosses with wild rabbits. Overall, our results imply that small size in dwarf rabbits results from a large effect, loss-of-function (LOF) mutation in HMGA2 combined with polygenic selection.
Keywords: whole-genome sequencing, RNA-seq, body size, IGF2BP2, mtDNA
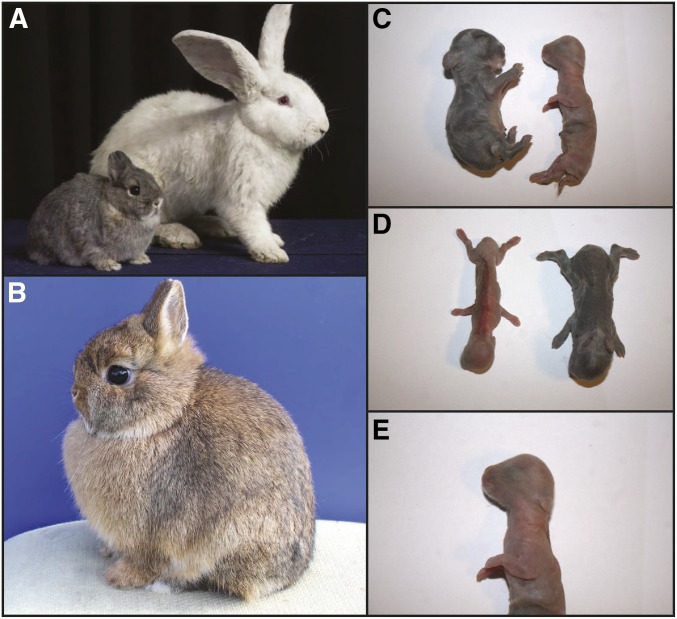
DOMESTICATION of rabbits was likely initiated after 500–600 AD in Southern France (Clutton-Brock 1999; Callou 2004; Whitman 2004). Rabbits have primarily been bred for meat production and most breeds of domestic rabbits are considerably larger (> 3 kg) compared with their wild ancestor (1.0–1.5 kg). However, more recently several dwarf breeds, such as Netherland dwarf, Holland lop, and long-haired dwarf rabbits, have been developed and these rabbits are used as pets. Most, if not all, dwarf breeds are segregating for the dwarf (dw) allele (OMIA 000299-9986, http://omia.angis.org.au/OMIA000299/9986/) first described by Greene et al. (1934) and a comprehensive summary of the phenotypic effects of this mutation is given by Robinson (1958). The dwarf allele is recessive lethal. Homozygotes (dw/dw) are smaller than litter mates and exhibit a characteristic swollen head, tiny ears, and are usually called peanuts (Figure 1). Peanuts are viable up to the time of birth but typically die within a few days of birth. Heterozygotes (Dw/dw) reach ∼2/3 of the size of wild-type litter mates (Dw/Dw) and in adulthood are typically under 1 kg in body weight, have compact and rounded bodies, a disproportionately larger head when compared to the rest of the body, small ears, and a short snout due to altered craniofacial development (Figure 1).
Figure 1.
The dwarf and wild-type phenotypes in rabbits. (A) A dwarf individual (Dw/dw) side-by-side with a normal-sized New Zealand white rabbit. (B) Dwarf individual (Dw/dw). These animals typically weigh < 1 kg, have compact and rounded bodies, short noses, a disproportionately large head when compared to rest of the body, and small ears. (C and D) Newborn dw/dw individual (peanut) side by side with a Dw/dw (dwarf) littermate. dw/dw animals are born smaller than their littermates and have a disproportionate body conformation with cone-shaped heads, small ears and limbs, and prominent eyes, which are evident at the time of birth. (E) Detail of the head of a dw/dw individual. Photo credits: (A) Sara Gutiérrez Albarran; (B–E) Javier Lopez.
Using controlled crosses, dwarfism in rabbits has been shown to be caused by a single gene in genetic linkage with Agouti (Castle and Sawin 1941). Thus, it is expected that the dwarf gene is to be found on chromosome 4 where Agouti is located. However, it is clear that the small size of dwarf rabbits is not only caused by the dwarf gene. Consistent with strong selection for small size, even those animals from dwarf breeds that are homozygous wild-type at the dwarf locus are considerably smaller than rabbits from other breeds.
The aim of the present study was to identify the causal mutation for the dwarf allele and to reveal selective sweep signals at other loci in the Netherland dwarf breed using whole-genome sequencing. We demonstrate that the causal mutation is a 12.1 kb deletion that eliminates the promoter and the first three exons of the high mobility AT-hook 2 (HMGA2) gene, resulting in inactivation of this gene. Taking advantage of this natural gene knockout, we carried out RNA-sequencing (RNA-seq) analysis in embryos to study how HMGA2 inactivation affects transcriptional regulation. We show that HMGA2 is required for the expression of IGF2BP2 and seems to be implicated in the regulation of protein-coding mitochondrial genes. We also reveal a number of putative selective sweeps when comparing the Netherland dwarf with six other breeds of normal size.
Materials and Methods
Whole-genome resequencing and SNP calling
Whole-genome resequencing data were generated for a pool of 14 unrelated dw/dw peanut individuals and a pool of 17 Dw/dw dwarf individuals collected from multiple breeders, all from the Netherland dwarf breed. Genomic DNA for dw/dw individuals was prepared from muscle tissue of newborn individuals donated by multiple breeders after death, and for Dw/dw individuals from blood extracted from live animals. Paired-end sequencing data (2 × 100 bp reads) were generated from pooled DNA using an Illumina platform. Previously published whole-genome resequencing data of pools of individuals (Carneiro et al. 2014) from six other breeds (Belgian hare, Champagne d’argent, Dutch, Flemish giant, French lop, and New Zealand white) were used in in the selective sweep scan. Reads were mapped to the reference genome assembly OryCun2 (Carneiro et al. 2014) with BWA-MEM (Li and Durbin 2009) using default parameters. Duplicate reads were flagged for subsequent analysis using Picard (http://picard.sourceforge.net).
SNP calling was performed using SAMtools (Li et al. 2009) and filtered by means of the mpileup2snp implemented in VarScan2 (Koboldt et al. 2012). We produced two sets of SNPs using different stringent criteria. For the homozygosity mapping analysis (see below) we used more stringent filtering criteria as follows: (1) a minimum depth of eight reads, (2) the allele with lower frequency was observed in at least two independent reads at a frequency ≥ 0.01, (3) a minimum Phred base quality of 15, and (4) a P-value threshold for calling variants of ≤ 0.01. For the SNP functional annotation (see below) we relaxed the minimum coverage to four reads to ensure that potential causative mutations could be identified within regions of lower coverage. The latter relaxed coverage requirement was also used to call indels by means of the mpileup2indel option of VarScan2 using identical filters to the SNP calling. Allele counts for each SNP/indel were extracted for subsequent analysis.
Mapping of the dwarf mutation
To search for extended regions of homozygosity and to overcome potential genotyping errors in our SNP calls, we calculated the proportion of monomorphic SNPs in the dw/dw pool summarized in a sliding window mode. Briefly, for overlapping windows of 2000 SNPs iterated in steps of 20%, we divided the number of SNPs monomorphic in dw/dw individuals by the total number of polymorphic SNPs within domestic rabbits. We also estimated the proportion of monomorphic SNPs using only SNPs that were polymorphic in the Dw/dw pool. Since the total number of SNPs in this case was smaller, we performed the sliding window analysis in windows of 500 SNPs also iterated in steps of 20%, which resulted in an average physical size of ∼800 kb.
We also estimated FST (i.e., a measure of genetic differentiation) between the dw/dw and Dw/dw pools. To estimate FST we used the PoPoolation2 package (Kofler et al. 2011b), which corrects for possible allele sampling biases associated with pool sequencing as well as sequencing errors. We corrected for unequal sampling of alleles among positions using an unbiased estimator of FST (Karlsson et al. 2007) as implemented in the PoPoolation2 package. FST values were summarized in 1 Mb windows iterated every 200 kb. We required per position a minimum coverage of four, a maximum coverage of 30, at least two reads supporting the minor allele in polymorphic positions, and a per base Phred quality score of 20 or higher. Windows with < 30% of positions passing these quality filters were discarded.
Structural rearrangements and SNP functional annotation
The identification of structural variants was performed using Breakdancer (Chen et al. 2009) and restricted to the dw/dw pool, in which the causative mutation should be homozygous. We focused on candidates with a score ≥ 80, which were then intersected with Ensembl gene annotations (release 78). In parallel, we performed depth of coverage analysis for all breeds using nonoverlapping windows of 1000 bp. This analysis was restricted to reads with a minimum mapping quality of 10. Only windows for which at least one breed had 50% of the window covered with reads were retained.
The functional annotation of the detected variants (both SNPs and indels) was performed using the genetic variant annotation and effect prediction toolbox SNPEff (Cingolani et al. 2012). Using the extracted allele counts, we searched specifically for variants with an absolute allele frequency difference ≥ 0.85 between the dw/dw pool and breeds not carrying the dwarf allele. The Dw/dw pool was not considered in this analysis because the frequency of the causative mutation is expected to be 50% in this pool. We did not require full fixation (i.e., allele frequency difference = 1) due to potential sequencing errors or mis-phenotyping.
Diagnostic PCR test for genotyping
To genotype the deletion, we designed two pairs of PCR primers; details on the PCR reactions are given in Supplemental Material, File S1. We genotyped 19 dw/dw individuals from three different breeds (Netherland dwarf n = 14, Holland lop n = 4, and long-haired dwarf n = 1), 20 Dw/dw individuals from a single breed (Netherland dwarf), and 40 individuals belonging to six breeds that are not expected to carry the dwarf allele (Belgian hare n = 7, Champagne d’argent n = 6, Dutch n = 6, Flemish giant n = 7, French lop n = 7, and New Zealand white n = 7).
Scan for selective sweeps
To identify regions of the genome that may have been subjected to selection in the Netherland dwarf breed, we estimated levels and patterns of genetic differentiation between this breed (both pools combined) and six other breeds with normal size. FST was calculated between breeds in windows of 500 kb with 250 kb steps using the PoPoolation2 package (Kofler et al. 2011b). We applied the same filtering requirements as before for the dw/dw and Dw/dw pools FST contrast. To summarize the magnitude of allele frequency divergence, we calculated the di statistic for each window (Akey et al. 2010). This statistic provides polarized measures of differentiation for a given breed in comparisons involving multiple breeds, by standardizing locus-specific deviations for any two breeds when compared to the genome-wide average. The final statistic was obtained by summing across all pairwise combinations including the target breed. We defined candidate regions as outlier windows falling on the top 1% of the empirical distribution. The borders of these regions were extended by aggregating windows on the top 5%. Regions separated by < 500 kb were merged into a single continuous region. Nucleotide diversity (π) within each breed was calculated using an identical sliding window approach as before for FST using the PoPoolation package (Kofler et al. 2011a).
Gene expression analysis
The following experimental procedures were approved by the Ethical Committee for Animal Research of the University of Castilla la Mancha, Spain (Register number CEEA: 1012.02). Female rabbits were kept under standard conditions of housing with unrestricted access to food and water; this was done according to the European Union Directive no. 86/609/CEE. The rabbits were attended by veterinary doctors and inspected at least four times daily. Females were sedated with an intramuscular injection of a mixture of xylazine (Rompun, Bayer Co., Portugal, 8 mg/kg) and ketamine (Imalgéne 1000, Merial, Portugal, 40 mg/kg), and after killed with an injection of thiopental (Tiopental 0.5 g Braun, B. Braun, Portugal, 100 mg/kg) given intravenously in the marginal vein of the ear. After the female’s death, a laparotomy was performed, and the embryos were decapitated prior to snap-freezing in liquid nitrogen.
To choose an embryonic stage at which HMGA2 is expressed at high levels, we profiled HMGA2 expression across several developmental stages in New Zealand white rabbits (i.e., Dw/Dw) using quantitative PCR (qPCR) (File S1). We sampled 11 embryos from the following stages: three embryos at day 9.5, two embryos at day 12, and one embryo at days 15.5, 16, 18, 21, 24, and birth. For gene expression comparisons among HMGA2 genotypes, we chose to sample 10 embryos of the different genotypes with 15.5 days (see Results). Genotypes were obtained using the PCR reactions described above from DNA extracted from the tip of the left hind leg.
Before library preparation, the entire embryo was homogenized and the RNA was extracted using the Allprep DNA/RNA/miRNA Universal kit (QIAGEN, Valencia, CA). Libraries were then prepared using the TruSeq Stranded mRNA (messenger RNA) Sample Preparation Kit (Illumina), and sequenced using 125 bp paired-end reads on an Illumina instrument. We obtained an average of 39.7 million reads per individual (range 34.6–42.3) for three dw/dw, four Dw/dw, and three Dw/Dw individuals (Table S1), divided into two technical replicates. All the individuals derive from five different dwarf (Dw/dw) females crossed with a single dwarf (Dw/dw) male (Table S1).
To estimate differential gene expression between the three genotypes, we used the complementary (cDNA) annotation for rabbit (Ensembl release 78), which derives from a combination of empirical data across multiple tissues and gene model predictions. Mapping was performed using Bowtie2 (Langmead and Salzberg 2012) using default parameters. Following mapping, statistics for each transcript, such as number of forward and reverse reads mapped, percent covered, and read GC content, were obtained using BBMap (http://bbmap.sourceforge.net). Forward mapping read counts were extracted and used within DESeq2 (Love et al. 2014) to estimate changes in gene expression among the three genotypes. P-values were adjusted within DESeq2, using the Benjamini–Hochberg approach for multiple comparisons, to reduce the false discovery rate to < 5%. Across each of the groups, the results from both technical replicates were qualitatively similar, therefore, the results described in the main text are derived from the combined counts from both technical replicates.
IGF2 is present in the rabbit reference assembly but distributed among several unmapped scaffolds. Therefore, this gene was missed by the annotation pipelines associated with the release of the rabbit reference genome (Carneiro et al. 2014). To profile IGF2 expression, we remapped the entire set of reads to a composite transcriptome reference including a rabbit cDNA sequence for this gene (Thieme et al. 2012).
qPCR for measuring mitochondrial DNA copy number
Mitochondrial DNA (mtDNA) copy number among the three HMGA2 genotypes was estimated using a quantitative real-time PCR performed on an ABI HT7900 real-time PCR system with a 384-well block module. We used the same individuals as used for gene expression, two independent reactions using different mtDNA primers (File S1), and three replicates per individual. The seven-point standard curve method using fourfold dilution was used to estimate the PCR efficiency. The data were normalized using ACTA2 as an endogenous autosomal reference gene and relative copy numbers were calculated using the 2−ΔΔCt method. Statistical significance was evaluated using the Student’s t-test.
Data availability
Whole-genome sequencing data and RNA-seq data have been deposited in GenBank under the bioproject PRJNA354575.
Results
Mapping of the genomic region underlying dwarfism
We performed whole-genome resequencing of two pooled DNA samples of individuals homozygous (dw/dw, i.e., peanuts) or heterozygous for the dwarf allele (Dw/dw, i.e., dwarfs). All individuals belonged to the Netherland dwarf breed (Figure 1). Sequence reads were mapped to the rabbit reference genome sequence (Carneiro et al. 2014), resulting in an average effective coverage of 12.9× and 11.2×, respectively.
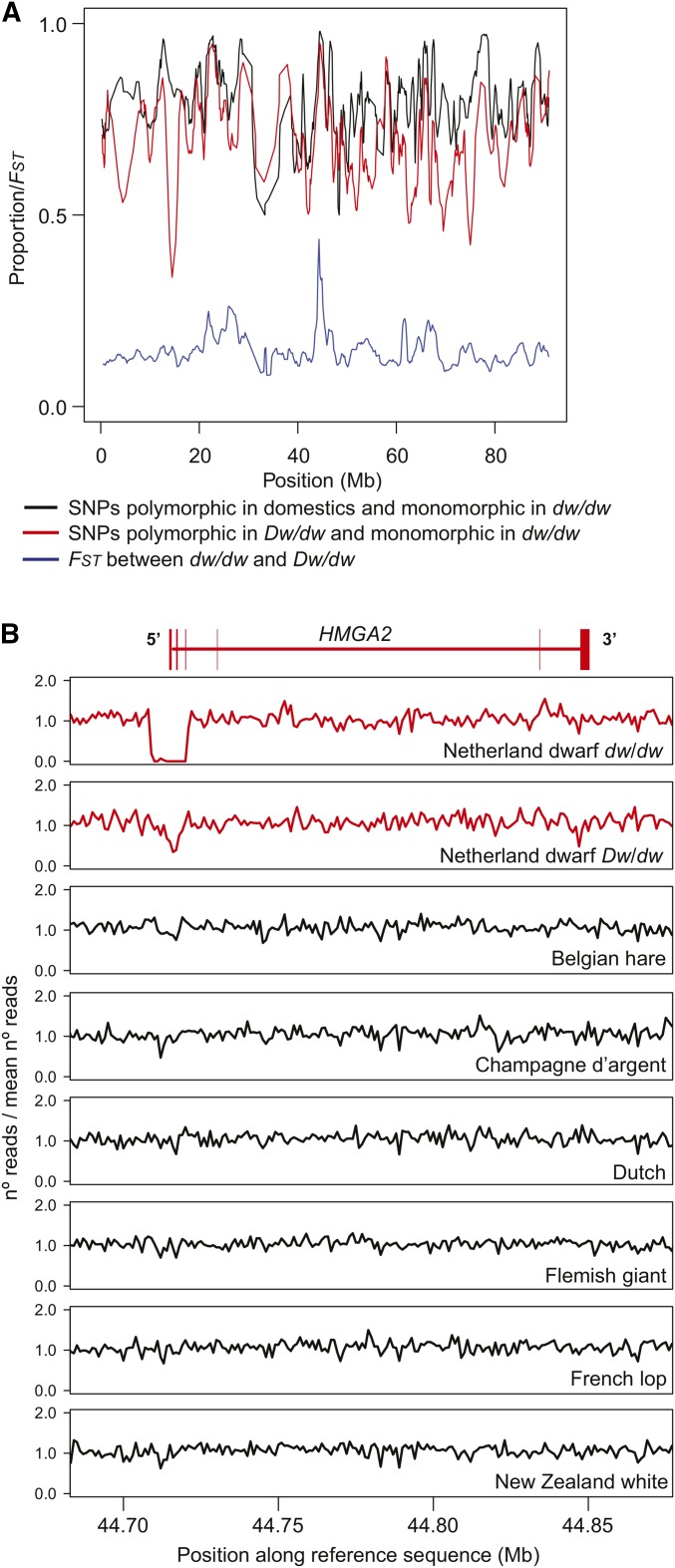
To map the specific region on chromosome 4 that was predicted to harbor the dwarf mutation based on previous linkage data (Castle and Sawin 1941), we started by applying a homozygosity mapping approach under the assumption that individuals homozygous for the dwarf allele are expected to be identical by descent for this region (Figure 2A). First, we looked for regions of high homozygosity within the dw/dw pool by calculating the proportion of monomorphic SNPs, of those previously found to be polymorphic among domestic rabbits (Carneiro et al. 2014), using a sliding window approach with a fixed number of SNPs per window (n = 2000). This proportion is expected to approach a value of one in the region associated with the phenotype. The average physical size of these windows was ∼1.06 Mb. Given that the length scale of linkage disequilibrium (LD) within domestic breeds typically decays to r2 values between 0.21 and 0.34 at genomic distances of 400 kb (Carneiro et al. 2011), this seemed an appropriate window length to avoid elevated homozygosity resulting from demography and chance events. The most extreme window in terms of the occurrence of monomorphic SNPs in dw/dw homozygotes was located at chromosome 4 (chr4):43,668,805–45,210,336 bp (Figure 2A).
Figure 2.
Mapping of the dwarf locus to a region on chromosome 4. (A) Homozygosity mapping and genetic differentiation. The red and black lines represent two statistics that summarize levels and patterns of homozygosity across chromosome 4. The red line represents the proportion of SNPs previously identified in domestic rabbits that were monomorphic in dw/dw individuals (summarized in windows of 2000 SNPs iterated every 400 SNPs); the black line represents the proportion of SNPs polymorphic in Dw/dw individuals that were monomorphic in dw/dw individuals (summarized in windows of 500 SNPs iterated every 100 SNPs). The blue line represents FST (a measure of genetic differentiation) between dw/dw and Dw/dw individuals. (B) A deletion overlapping the promoter region and the first three exons of HMGA2 associated with the dwarf allele. The lines represent the normalized number of reads mapping to nonoverlapping windows of 1000 bp across the HMGA2 region. The data for the dw/dw and Dw/dw pools are given in red at the top, while the remaining panels summarize normalized read depth for six breeds that do not carry the dwarf allele (Belgian hare, Champagne d’argent, Dutch, Flemish giant, French lop, and New Zealand white). The gene model for HMGA2 is drawn in red across the top of the figure and is based on the Ensembl annotation.
Next, we estimated the proportion of monomorphic SNPs in dw/dw, but this time for SNPs that were polymorphic in the Dw/dw individuals (Figure 2A). The rationale behind this approach is that Dw/dw are expected to carry one haplotype containing the causative mutation and one containing the wild-type allele, and are thus heterozygous for mutations that differ between these haplotypes. This approach is also expected to be less influenced by fixation of long haplotype blocks in response to genetic drift or selection specific to the Netherland dwarf breed, because it uses only positions that are polymorphic within the breed. Using a similar sliding window approach as before, the top two windows are contiguous and map to the same region (chr4:43,895,910–44,998,202 bp). However, there are a few other windows in the interval chr4:40,000,000–60,000,000 that show nearly as high a degree of homozygosity as found in the top windows.
Finally, we estimated genetic differentiation between the two pools using FST (Figure 2A). Both pools consist of individuals that belong to the same breed, and although Dw/dw individuals carry just one copy of the causative haplotype, the region containing the gene involved in dwarfism is expected to show elevated genetic differentiation between the two groups. The top five windows with elevated FST were found to be contiguous and the top window mapped again to the region on chromosome 4 revealed by homozygosity mapping (chr4:43,800,001–44,800,000 bp). Based on the annotation of the rabbit reference genome sequence (Carneiro et al. 2014) and assuming the common interval among the three statistics as the most likely candidate region (chr4:43,895,910–44,800,000 bp), we found four protein-coding genes within this interval (WIF1, LEMD3, MSRB3, and HMGA2; Table 1).
Table 1. Genes located in the region on rabbit chromosome 4 harboring the dwarf mutation.
| Gene Symbol | Name | Location (bp) |
|---|---|---|
| WIF1 | WNT inhibitory factor 1 | 43,863,790–43,948,402 |
| LEMD3 | LEM domain-containing 3 | 44,015,308–44,091,960 |
| MSRB3 | Methionine sulfoxide reductase B3 | 44,126,803–44,339,285 |
| HMGA2 | High mobility group AT-hook 2 | 44,715,271–44,850,410 |
LEM, LAP2, emerin, MAN1.
A deletion encompassing the HMGA2 coding region explains dwarfism in rabbits
To reveal the gene and causative mutation underlying the dwarf phenotype, we began by performing a detailed screen for mutations of potential functional significance (nonsynonymous, stop/gain, frameshifts, and splice site mutations). We compared the pool containing dw/dw individuals, which are supposed to be homozygous for the causative mutation, to previously reported whole-genome sequence data from pooled DNA samples from six other breeds that are not expected to carry the dwarf allele (Carneiro et al. 2014).
We failed to find any coding changes specific to dw/dw when compared to the nondwarf breeds. Although a regulatory point mutation could be causative, the failure to identify a mutation altering protein structure led us to screen our candidate region for structural changes. The software Breakdancer (Chen et al. 2009) reported two candidate variants, both deletions, within our candidate region. Sequence depth analysis revealed a sharp decrease in read count in the dw/dw pool for one of the variants, confirming the existence of a deletion (Figure 2B), but not for the other. The approximate breakpoints of the candidate deletion were estimated using soft-clipped reads (i.e., reads containing unaligned portions) from the whole-genome resequencing data obtained for dw/dw individuals. Using an individual homozygous for the dwarf allele, we then confirmed the exact breakpoints by PCR amplification followed by traditional Sanger sequencing. This deletion was ∼12.1 kb in length (chr4:44,709,089–44,721,236) and excised the promotor region and the first three exons of HMGA2, expected to cause complete gene inactivation (Figure 2B). The 5′−end of this deletion overlaps a CSINE2 element, which is the largest family of short interspersed repeat elements in the rabbit genome with an estimated copy number of ∼1,000,000. However, we did not detect any obvious sequence homology between the two breakpoint regions that may have promoted the excision of this sequence.
HMGA2 encodes for a protein that belongs to the nonhistone chromosomal high mobility group (HMG) protein family, and functions as a transcription regulator. This gene has been previously associated with body size in humans and mice (Zhou et al. 1995; Alyaqoub et al. 2012), dogs (Webster et al. 2015) (OMIA 001968-9615; http://omia.angis.org.au/OMIA001968/9615/), horses (Frischknecht et al. 2015) (OMIA 001968-9796; http://omia.angis.org.au/OMIA001968/9796/), and with beak size among Darwin’s finches (Lamichhaney et al. 2016) (OMIA 001992-48881; http://omia.angis.org.au/OMIA001992/48881/).
After confirmation of the exact breakpoints, we designed a four primer PCR test that amplified over the deletion, while amplifying an additional product for the wild-type allele with primers located within the deleted region. We genotyped a large cohort of samples (Table 2) including individuals belonging to the three genotypes and from breeds expected to carry and not carry the dwarf allele (see Materials and Methods for details). We found that all dw/dw individuals were homozygous for the deletion across three different breeds, and all individuals from nondwarf breeds were homozygous for the wild-type allele. The majority of the dwarf individuals were heterozygous (Dw/dw) as expected, but four individuals phenotyped as dwarf were homozygous wild-type (Table 2). This is likely explained by mis-phenotyping that can easily occur in rabbits of young age. Overall, these genotyping results support the causality of the deletion in HMGA2.
Table 2. Genotyping results for a 12.1 kb deletion overlapping HMGA2 at the dwarf locus.
| Group (Expected Genotype) | No. of Breeds | No. of Individuals | Genotype (Deletion) | ||
|---|---|---|---|---|---|
| del/del | del/+ | +/+ | |||
| Peanut (dw/dw) | 3a | 19 | 19 | 0 | 0 |
| Dwarf (Dw/dw) | 1b | 20 | 0 | 16 | 4 |
| Wild-type (Dw/Dw) | 6c | 40 | 0 | 0 | 40 |
del, deletion; +, wild-type.
Netherland dwarf, Holland lop, and long-haired dwarf rabbits.
Netherland dwarf.
Belgian hare, Champagne d’argent, Dutch, Flemish giant, French lop, and New Zealand white.
Differential gene expression among HMGA2 genotypes
HMGA2 functions as an architectural factor in growth regulation during embryonic development (Pfannkuche et al. 2009). Homozygosity for HMGA2-null alleles is compatible with life in mice but, as shown here, not in rabbits. To gain further insight into how HMGA2 affects transcriptional regulation, we explored differential gene expression among HMGA2 genotypes using RNA-seq (see Table S1 for details on the data set). We crossed Dw/dw heterozygotes and sampled 16-day embryos for three dw/dw, four Dw/dw, and three Dw/Dw individuals. We chose to study 15.5-day embryos after a preliminary screen for HMGA2 expression across eight developmental stages (embryonic day 9.5 to birth) using progeny of New Zealand white rabbits (Figure S1). Expression analysis of different parts of wild-type embryos revealed high HMGA2 expression during early developmental stages, from day 9.5 to day 18, and a dramatic decrease from day 21. Embryos at day 15.5 displayed the highest HMGA2 expression.
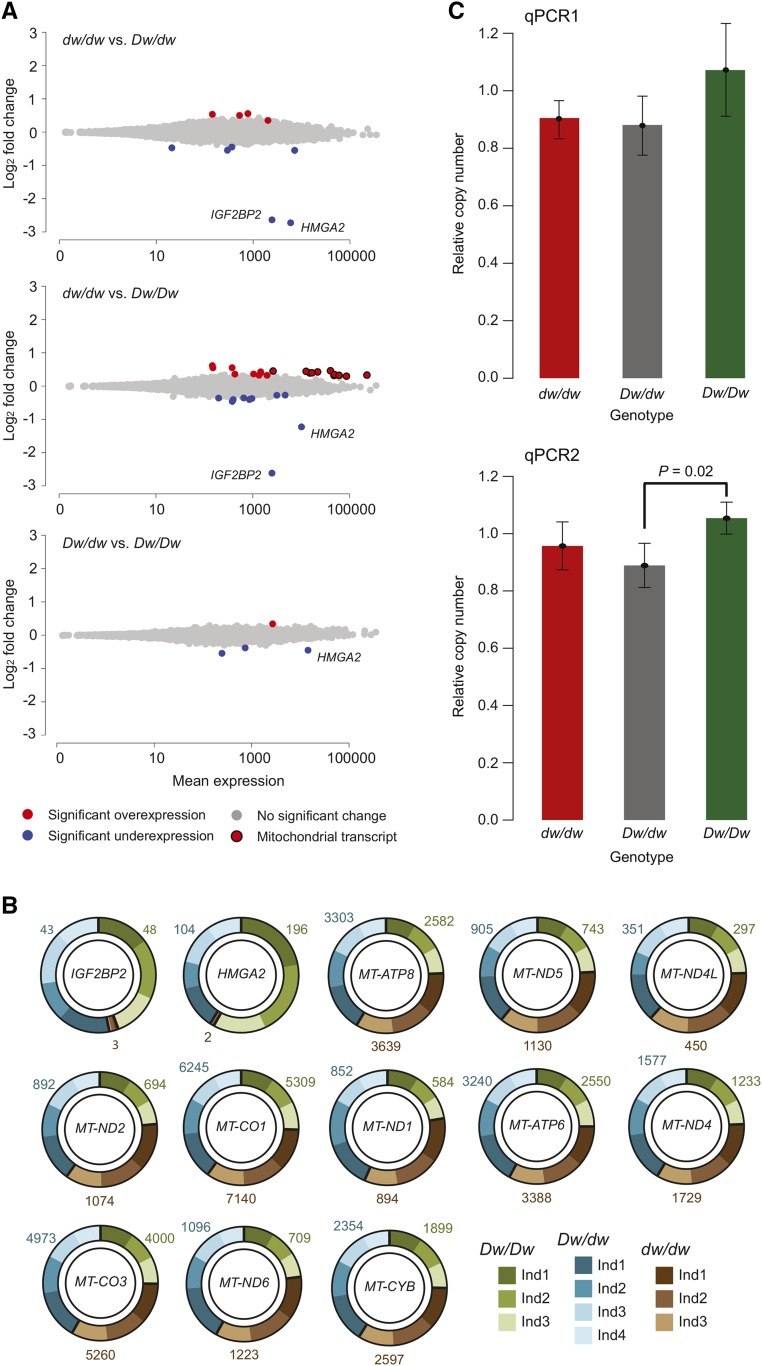
We compared gene expression among all different contrasts (summarized in File S2 and File S3). We started by comparing differential expression between dw/dw and Dw/dw individuals and found that as few as 10 transcripts were differentially expressed (Figure 3A). The two genes receiving highest statistical support for differential expression were HMGA2 and IGF2BP2, the latter encoding insulin-like growth factor 2 mRNA-binding protein 2, a member of the IGF-II mRNA-binding protein (IMP) family. The results are consistent with a complete silencing of both genes in dw/dw homozygotes (Figure 3B).
Figure 3.
Differential gene expression among the three HMGA2 genotypes and mtDNA copy number. (A) Statistically significant changes in gene expression between HMGA2 genotypes evaluated using DESeq2 (Love et al. 2014). Log fold change vs. mean expression for the 21,588 genes included in our analysis for the different contrasts (upper panel: dw/dw vs. Dw/dw; middle panel: dw/dw vs. Dw/Dw; lower panel: Dw/dw vs. Dw/Dw). Red indicates significant overexpression, blue indicates significant underexpression, and gray indicates no significant change. Mitochondrial genes are circled. The data points for HMGA2 and IGF2BP2 are indicated. Mean FPKM values for all transcripts are given in File S3. (B) Doughnut charts summarizing levels of expression for HMGA2, IGF2BP2, and protein-coding genes encoded in mtDNA. Both technical replicates have been combined. Each chart represents one gene and the different individuals are represented by different colors. Black bars separate the genotypes (green: Dw/Dw; brown: dw/dw; and cyan: Dw/dw). Numbers outside of each chart indicate the mean FPKM value for each genotype. (C) mtDNA abundance in embryos of the different HMGA2 genotypes. Barplot indicating the number of mtDNA copies relative to the autosomes for the three HMGA2 genotypes. The bars reflect the average values and SD among three dw/dw (peanut), four Dw/dw (dwarf), and three Dw/Dw (wild-type) individuals. qPCR1 and qPCR2 represent two independent reactions based on different sets of primers targeting the rabbit mitochondrial genome. FPKM, fragments per kilobase million; Ind, individual; mtDNA, mitochondrial DNA; qPCR, quantitative PCR.
There were 29 differentially expressed transcripts in the contrast between dw/dw and Dw/Dw homozygotes (Figure 3A). Again, the two most significant genes were HMGA2 and IGF2BP2 (Figure 3B). Two interesting aspects were revealed by this contrast. First, two of the genes (MSRB3 and LEMD3) showing upregulated expression in the HMGA2 knockouts are located only ∼700 kb upstream of HMGA2. Interestingly, MSRB3 together with a third gene (WIF1) in the near vicinity (Table 1) also showed upregulated expression in the dw/dw vs. Dw/dw contrast (File S2). Five other genes showing differential expression between dw/dw and Dw/Dw homozygotes mapped to chromosome 4 at larger distances (> 5 Mb). Thus, 8 out of 29 genes showing differential expression are located on this chromosome (File S2). Second, and most strikingly, 11 out of the 13 protein-coding genes encoded by the mtDNA were found to be significantly upregulated in dw/dw homozygotes (Figure 3B).
Only four genes showed differential expression between Dw/dw and Dw/Dw rabbits (Figure 3A). The most significant transcript was HMGA2, which was downregulated in Dw/dw and the only gene in common with the dw/dw vs. Dw/Dw comparison. Although the levels of expression for mtDNA genes were not significantly different in this contrast, they were intermediate between dw/dw and Dw/Dw (Figure 3B).
We investigated in further detail levels and patterns of expression in other genes associated with the insulin-growth factor pathway (IGF1, IGF1R, IGF2, and IGF2R). These genes have been frequently implicated in size differences and muscle growth (Van Laere et al. 2003; Sutter 2007; Hoopes et al. 2012). However, we found no evidence for differential expression of these genes in any of the contrasts (Figure S2).
mtDNA copy number does not vary among HMGA2 genotypes
In the dw/dw individuals, we observed a consistent upregulation of many genes encoded in mtDNA (Figure 3C). Cell type composition can potentially explain gene expression differences among genotypes. Thus, it is unclear whether this finding is a direct result of mtDNA misregulation caused by the HMGA2 knockout or is explained by variation in the number of mitochondria in the sequenced sample. Therefore, we estimated mtDNA copy number for the same individuals used for the gene expression profiling using two independent qPCRs based on different sets of primers (Figure 3C). We found that copy number of mtDNA was not significantly different between most contrasts, with the exception of Dw/dw and Dw/Dw for just one of the qPCR reactions. If anything, the results were opposite to the expectation based on the gene expression pattern; dw/dw individuals tend to have slightly lower mtDNA copy numbers than Dw/Dw individuals. Another potential explanation for differential expression among genotypes is differential mapping efficiency associated with distinct mtDNA haplotypes. However, since this study is based on a within-litter comparison this explanation can be ruled out (Table S1), and gene expression is consistent among individuals within genotype class (Figure 3B). Taken together, we can conclude that mtDNA gene expression is altered as a result of the inactivation of HMGA2, suggesting that HMGA2 expression might be directly associated with mitochondrial function.
Genome-wide scan for signatures of selection in Netherland dwarfs
Netherland dwarfs that do not carry the dwarf allele are still smaller in size when compared to most breeds, and this is true for most dwarf breeds in rabbits. To identify additional genomic regions that may have been subjected to directional selection for reduced size in Netherland dwarfs, we scanned the genome of this breed for regions of increased differentiation compared with normal-sized domestic rabbits by means of the di statistic (Akey et al. 2010).
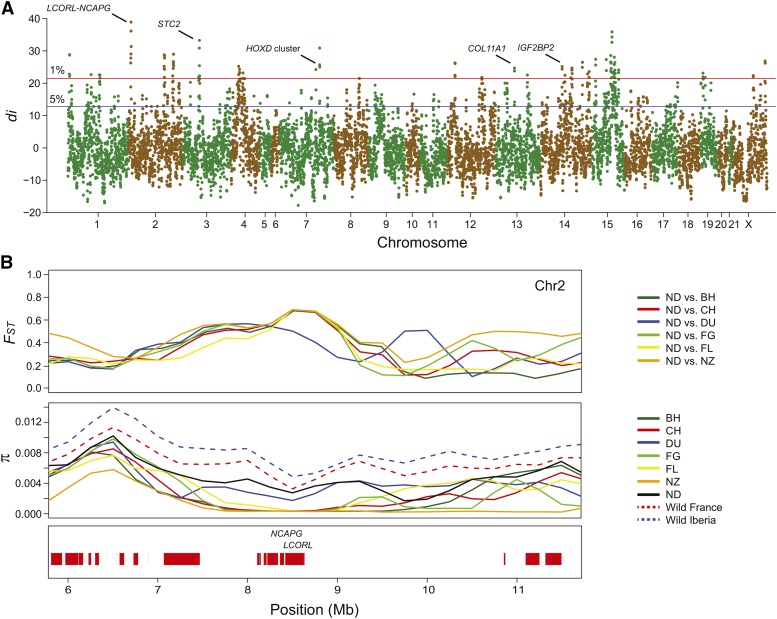
We identified 34 genomic regions with unusual levels of population differentiation between Netherland dwarf and six other breeds (Figure 4A; outlier regions and their gene content are detailed in Table S2). These regions varied largely in size (750,000–6,250,000 bp, median 1,500,000 bp), and contained 646 genes in total. Within the most differentiated regions we noticed several genes previously associated with size or skeletal features in other species including LCORL-NCAPG (e.g., Gudbjartsson et al. 2008; Rubin et al. 2012; Tetens et al. 2013; Sahana et al. 2015), STC2 (Gagliardi et al. 2005; Rimbault et al. 2013), HOXD cluster (Zakany and Duboule 2007), COL11A1 (Li et al. 1995; Annunen et al. 1999), and IGF2BP2 (Dai et al. 2015) (Figure 4A).
Figure 4.
Selective sweep screen for additional genomic regions associated with small size in Netherland dwarf rabbits. (A) Genome-wide distribution of di. Values were summarized in 500 kb windows iterated every 250 kb. The red and blue lines indicate the top 1 and 5% of the empirical distribution, respectively. Unmapped contigs were included in the analysis but are not represented in the figure. (B) FST and nucleotide diversity (π) in the LCORL-NCAPG region. Each π and FST data point is based on a sliding window analysis using 500 kb windows with 250 kb steps. Annotated protein-coding genes are drawn across the bottom of each region, and putative candidate genes are highlighted. Nucleotide diversities of wild rabbits were averaged over three populations of wild rabbits from France (Caumont, La Roque, and Villemolaque) and two populations of wild rabbits from the Iberian Peninsula (Guadalajara and Zaragoza) belonging to the Oryctolagus cuniculus cuniculus subspecies; data from Carneiro et al. (2014). BH, Belgian hare; CH, Champagne d’argent; Chr, chromosome; DU, Dutch; FG, Flemish giant; FL, French lop; ND, Netherland dwarf; NZ, New Zealand white.
In the genome-wide screen, the strongest signal overlaps the LCORL-NCAPG region (Figure 4A). In spite of the high FST values between Netherland dwarfs and other breeds, we failed to find a reduction of nucleotide diversity in Netherland dwarfs across this region (Figure 4B). In fact, Netherland dwarfs have the highest genetic diversity in this region, and most other breeds have nucleotide diversity values close to zero. With the exception of dwarf breeds, domestic rabbits are larger in size than wild rabbits, and our results for this region are consistent with a selective sweep for a growth-promoting allele at the LCORL-NCAPG locus in rabbit breeds that are large compared with wild rabbits. Given that the Netherland dwarf breed is the most recently developed breed in our data set, the increase in genetic diversity around this locus specific to Netherland dwarfs is suggestive of crosses with wild rabbits to achieve such small sizes. Historical records support such a scenario (Whitman 2004).
Discussion
The small size of Netherland dwarf rabbits and probably most, if not all, dwarf rabbits from other breeds, is due to heterozygosity for an HMGA2 LOF allele combined with polygenic selection for small size at other loci. Although genome-wide association studies in humans have shown that body size is a highly polygenic trait, it is striking how notoriously HMGA2 has been associated with variation in body size across species (Zhou et al. 1995; Alyaqoub et al. 2012; Frischknecht et al. 2015; Webster et al. 2015), including in humans (Buysse et al. 2009). We propose that HMGA2 is a major regulator of body size in mammals and probably in other vertebrates because of an important role in regulating growth combined with minimal, if any, negative pleiotropic effects on other traits. An important difference between the dwarf mutation in rabbits and the pygmy allele in mouse (Zhou et al. 1995), the two HMGA2 LOF alleles described so far, is that pygmy homozygotes are fully viable and show the pygmy phenotype whereas dwarf in rabbits is a recessive lethal (Figure 1). Human HMGA2 LOF alleles are probably also recessive lethal, since an intragenic deletion in HMGA2 is associated with short stature (Buysse et al. 2009) and no LOF homozygotes at this locus have yet been reported. Both pygmy mice and dwarf rabbits show altered craniofacial development, characterized by shortened heads (Zhou et al. 1995) (Figure 1). Interestingly, a recent study showed that HMGA2 is a major locus affecting craniofacial development (beak size) in Darwin’s finches (Lamichhaney et al. 2016), but in that case the phenotypic effect is likely caused by regulatory changes.
HMGA2 does not bind a specific DNA sequence but contributes to transcriptional regulation through interaction with other DNA-binding transcription factors (Reeves 2001; Pfannkuche et al. 2009), and it has a well-established role in cancer biology (Young and Narita 2007 and references therein). Despite the abundant genetic data demonstrating the important role of HMGA2 for controlling growth, the underlying mechanism is still poorly understood. Therefore, we carried out expression analysis of whole embryos of all three genotype classes Dw/Dw, Dw/dw, and dw/dw; embryos at day 15.5 were used because HMGA2 expression was found to peak at this day. There were only three transcripts, besides HMGA2, that showed significant differential expression in the contrast between dwarf (Dw/dw) and wild-type (Dw/Dw) rabbits, and none of these three transcripts have an established role in regulating growth. This indicates that HMGA2 haploinsufficiency causes dwarfism in Dw/dw rabbits by subtle changes in gene expression, or that the effects are very tissue-specific and therefore not revealed in our whole-embryo analysis. Only a limited number of transcripts showed significant differential expression even in the comparison between the two opposite homozygotes (dw/dw vs. Dw/Dw). However, there were two striking observations in this contrast: (1) silencing of IGF2BP2 expression, and (2) mild (log2 fold change ≈ 0.5) upregulation of the transcripts for most mtDNA-encoded proteins.
Our result shows that the presence of HMGA2 is critical for the expression of IGF2BP2, consistent with a previous study showing that Igf2bp2 undergoes a large-scale downregulation in pygmy mice (Hmga2-null) (Brants et al. 2004). Similar to our study (Figure 2A), it was essentially only the Hmga2 and Igf2bp2 transcripts that were significantly downregulated in pygmy 12.5-day embryos. HMGA2 also regulates expression of Igf2bp2 in mouse myoblasts and the HMGA2-IGF2BP2 axis is an important regulator of skeletal muscle development (Li et al. 2012). IGF2BP2 is an RNA-binding protein and was first described as a protein that binds the IGF2 mRNA, and it enhances IGF2 translation (Dai et al. 2011). However, it also binds many other mRNAs and affects their translation (Li et al. 2012; Dai et al. 2015). Genome-wide association studies in humans have consistently shown that IGF2BP2 is associated with susceptibility to Type 2 Diabetes (e.g., Scott et al. 2007; Zeggini et al. 2007). Igf2bp2-deficient mice are modestly smaller, more lean, show improved glucose tolerance, higher insulin sensitivity, and have a significantly longer life span than wild-type litter mates (Dai et al. 2015). The fact that Hmga2-null (pygmy) mice show a much more drastic reduction in body size than Igf2bp2-null mice implies that the reduced Igf2bp2 expression in Hmga2-null mice and HMGA2 heterozygous (dwarf) rabbits can only partly be mediated by reduced IGF2BP2 expression. Interestingly, our screen for selective sweeps in Netherland dwarf rabbits indicated that genetic changes at the IGF2BP2 locus itself might also have contributed to small size in this breed (Figure 4A).
Eleven out of the 13 protein-coding genes in mtDNA were significantly upregulated in the HMGA2 knockout animals (Figure 3, A and B). The upregulation of mtDNA transcripts did not reflect an increased copy number of mtDNA per cell (Figure 3C), suggesting that HMGA2 has a direct or indirect role in regulating transcription of these genes. This functional role in mtDNA function is a novel finding not previously reported, but we note that the Igf2bp2-null mice show enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins, which in turn is expected to affect transcription of mtDNA genes (Dai et al. 2015).
Another interesting observation in this study was that the three genes MSRB3, LMD3, and WIF1 located upstream of HMGA2 show a mild overexpression in HMGA2 dw/dw and Dw/dw rabbits (File S2), suggesting that these four genes are coregulated to some extent. There are several possible explanations for this observation: (1) HMGA2 may be a negative regulator of these genes, (2) the 12.1 kb region deletion in HMGA2 contains regulatory elements affecting the expression of the neighboring genes, or (3) the elimination of HMGA2 expression may simply increase transcriptional activity from the promoters in the near vicinity. Gene targeting experiments will be required to evaluate these possible explanations.
A striking finding in our genome-wide screen for loci showing signatures of selection in Netherland dwarf rabbits was the enrichment of loci previously associated with variation in body size. The most prominent example is the two closely linked genes LCORL (Ligand-Dependent Nuclear Receptor Corepressor-Like), which encodes a transcription factor, and NCAPG (Non-SMC Condensin I Complex, Subunit G), which encodes a subunit of the condensin complex required for mitosis and meiosis progression. LCORL-NCAPG are, together with HMGA2, two of the loci that are most consistently associated with stature and body size variation across multiple studies and species. The LCORL-NCAP region has previously been associated with body size in human, cattle, dog, pig, and horse (Gudbjartsson et al. 2008; Pryce et al. 2011; Vaysse et al. 2011; Rubin et al. 2012; Signer-Hasler et al. 2012; Tetens et al. 2013; Sahana et al. 2015). Our results are consistent with a selective sweep at the LCORL-NCAP region related to selection for increased size in domestic rabbits, since all normal-sized rabbits included in this study are significantly larger than wild rabbits and were fixed for a single haplotype (Figure 4B). In contrast, Netherland dwarf rabbits as well as Dutch rabbits, which are well below the average size of domestic rabbits, appear to carry wild-type haplotypes at this locus. Overall, our results show that small size in dwarf rabbits results from a large effect, 12.1 kb deletion at the HMGA2 locus, combined with polygenic selection.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.196667/-/DC1.
Acknowledgments
We thank innumerous rabbit breeders and associations who contributed samples, namely Javier López Rosell, Mar Novoa Rodríguez, Carlos Moura, Francisco Teixeira, Rita Maria Oliveira Araújo, Paulo Martins, Emily Clark, Joe Rogers, Shelly Rowland, and the Portuguese and Spanish associations of dwarf rabbit breeders. We thank Bernardino Silva for help with animal breeding. The work was supported by POPH-QREN funds from the European Social Fund and Portuguese MCTES [Fundação para a Ciência e a Tecnologia Investigator, IF/00283/2014/CP1256/CT0012) and postdoc grants to M.C. (SFRH/BPD/72343/2010)]; by the European Research Council project BATESON to L.A.; by FEDER funds through the COMPETE program and Portuguese national funds through the Fundação para a Ciência e a Tecnologia (project PTDC/CVT/122943/2010); by the projects “Genomics and Evolutionary Biology” and “Genomics Applied to Genetic Resources” cofinanced by North Portugal Regional Operational Programme 2007/2013 (ON.2 – O Novo Norte) under the National Strategic Reference Framework and the European Regional Development Fund ERDF; by a European Union FP7 REGPOT grant [CIBIO-New-Gen] [286431]; and by travel grants to M.C. (COST Action TD1101).
Footnotes
Communicating editor: S. F. Chenoweth
Literature Cited
- Akey J. M., Ruhe A. L., Akey D. T., Wong A. K., Connelly C. F., et al. , 2010. Tracking footprints of artificial selection in the dog genome. Proc. Natl. Acad. Sci. USA 107: 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyaqoub F., Pyatt R. E., Bailes A., Brock A., Deeg C., et al. , 2012. 12q14 microdeletion associated with HMGA2 gene disruption and growth restriction. Am. J. Med. Genet. A. 158A: 2925–2930. [DOI] [PubMed] [Google Scholar]
- Annunen S., Korkko J., Czarny M., Warman M. L., Brunner H. G., et al. , 1999. Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome, but other mutations cause overlapping Marshall/Stickler phenotypes. Am. J. Hum. Genet. 65: 974–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brants J. R., Ayoubi T. A. Y., Chada K., Marchal K., Van de Ven W. J. M., et al. , 2004. Differential regulation of the insulin-like growth factor II mRNA-binding protein genes by architectural transcription factor HMGA2. FEBS Lett. 569: 277–283. [DOI] [PubMed] [Google Scholar]
- Buysse K., Reardon W., Mehta L., Costa T., Fagerstrom C., et al. , 2009. The 12q14 microdeletion syndrome: additional patients and further evidence that HMGA2 is an important genetic determinant for human height. Eur. J. Med. Genet. 52: 101–107. [DOI] [PubMed] [Google Scholar]
- Callou C., 2004. De la garenne au clapier: etude archeozoologique du lapin en Europe occidentale. Bull. Soc. Préhist. Fr. 101: 371–372. [Google Scholar]
- Carneiro M., Afonso S., Geraldes A., Garreau H., Bolet G., et al. , 2011. The genetic structure of domestic rabbits. Mol. Biol. Evol. 28: 1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M., Rubin C. J., Di Palma F., Albert F. W., Alfoldi J., et al. , 2014. Rabbit genome analysis reveals a polygenic basis for phenotypic change during domestication. Science 345: 1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle W. E., Sawin P. B., 1941. Genetic linkage in the rabbit. Proc. Natl. Acad. Sci. USA 27: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wallis J. W., McLellan M. D., Larson D. E., Kalicki J. M., et al. , 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock J., 1999. A Natural History of Domesticated Mammals. Cambridge University Press, Cambridge. [Google Scholar]
- Dai N., Rapley J., Angel M., Yanik M. F., Blower M. D., et al. , 2011. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev. 25: 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N., Zhao L., Wrighting D., Kramer D., Majithia A., et al. , 2015. IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and other mRNAs encoding mitochondrial proteins. Cell Metab. 21: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht M., Jagannathan V., Plattet P., Neuditschko M., Signer-Hasler H., et al. , 2015. A non-synonymous HMGA2 variant decreases height in Shetland ponies and other small horses. PLoS One 10: e0140749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi A. D., Kuo E. Y., Raulic S., Wagner G. F., DiMattia G. E., 2005. Human stanniocalcin-2 exhibits potent growth-suppressive properties in transgenic mice independently of growth hormone and IGFs. Am. J. Physiol. Endocrinol. Metab. 288: E92–E105. [DOI] [PubMed] [Google Scholar]
- Greene H. S. N., Hu C. K., Brown W. H., 1934. A lethal dwarf mutation in the rabbit with stigmata of endocrine abnormality. Science 79: 487–488. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D. F., Walters G. B., Thorleifsson G., Stefansson H., Halldorsson B. V., et al. , 2008. Many sequence variants affecting diversity of adult human height. Nat. Genet. 40: 609–615. [DOI] [PubMed] [Google Scholar]
- Hoopes B. C., Rimbault M., Liebers D., Ostrander E. A., Sutter N. B., 2012. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm. Genome 23: 780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson E. K., Baranowska I., Wade C. M., Salmon Hillbertz N. H. C., Zody M. C., et al. , 2007. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 39: 1321–1328. [DOI] [PubMed] [Google Scholar]
- Koboldt D. C., Zhang Q., Larson D. E., Shen D., McLellan M. D., et al. , 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Orozco-terWengel P., De Maio N., Pandey R. V., Nolte V., et al. , 2011a PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6: e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Pandey R. V., Schlotterer C., 2011b PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27: 3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S., Han F., Berglund J., Wang C., Sällman Almén M., et al. , 2016. A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 352: 470–474. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lacerda D. A., Warman M. L., Beier D. R., Yoshioka H., et al. , 1995. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell 80: 423–430. [DOI] [PubMed] [Google Scholar]
- Li Z., Gilbert J. A., Zhang Y., Zhang M., Qiu Q., et al. , 2012. An HMGA2–IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev. Cell 23: 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannkuche K., Summer H., Li O., Hescheler J., Dröge P., 2009. The high mobility group protein HMGA2: a co-regulator of chromatin structure and pluripotency in stem cells? Stem Cell Rev. Rep. 5: 224–230. [DOI] [PubMed] [Google Scholar]
- Pryce J. E., Hayes B. J., Bolormaa S., Goddard M. E., 2011. Polymorphic regions affecting human height also control stature in cattle. Genetics 187: 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., 2001. Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277: 63–81. [DOI] [PubMed] [Google Scholar]
- Rimbault M., Beale H. C., Schoenebeck J. J., Hoopes B. C., Allen J. J., et al. , 2013. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 23: 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R., 1958. Genetic studies of the rabbit. Bibliographia Genetica 17: 229–558. [Google Scholar]
- Rubin C. J., Megens H. J., Martinez Barrio A., Maqbool K., Sayyab S., et al. , 2012. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 109: 19529–19536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahana G., Höglund J. K., Guldbrandtsen B., Lund M. S., 2015. Loci associated with adult stature also affect calf birth survival in cattle. BMC Genet. 16: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L. J., Mohlke K. L., Bonnycastle L. L., Willer C. J., Li Y., et al. , 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer-Hasler H., Flury C., Haase B., Burger D., Simianer H., et al. , 2012. A genome-wide association study reveals loci influencing height and other conformation traits in horses. PLoS One 7: e37282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter N. B., 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316: 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetens J., Widmann P., Kühn C., Thaller G., 2013. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim. Genet. 44: 467–471. [DOI] [PubMed] [Google Scholar]
- Thieme R., Schindler M., Ramin N., Fischer S., Muhleck B., et al. , 2012. Insulin growth factor adjustment in preimplantation rabbit blastocysts and uterine tissues in response to maternal type 1 diabetes. Mol. Cell. Endocrinol. 358: 96–103. [DOI] [PubMed] [Google Scholar]
- Van Laere A. S., Nguyen M., Braunschweig M., Nezer C., Collette C., et al. , 2003. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425: 832–836. [DOI] [PubMed] [Google Scholar]
- Vaysse A., Ratnakumar A., Derrien T., Axelsson E., Rosengren Pielberg G., et al. , 2011. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 7: e1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M., Kamgari N., Perloski M., Hoeppner M., Axelsson E., et al. , 2015. Linked genetic variants on chromosome 10 control ear morphology and body mass among dog breeds. BMC Genomics 16: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman B. D., 2004. Domestic Rabbits & Their Histories: Breeds of the World. Leathers Publishing, Overland Park, KS. [Google Scholar]
- Young A. R., Narita M., 2007. Oncogenic HMGA2: short or small? Genes Dev. 21: 1005–1009. [DOI] [PubMed] [Google Scholar]
- Zakany J., Duboule D., 2007. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 17: 359–366. [DOI] [PubMed] [Google Scholar]
- Zeggini E., Weedon M. N., Lindgren C. M., Frayling T. M., Elliott K. S., et al. , 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Benson K. F., Ashar H. R., Chada K., 1995. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376: 771–774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequencing data and RNA-seq data have been deposited in GenBank under the bioproject PRJNA354575.