Abstract
Histone methylation is a dynamic and reversible process proposed to directly impact on stem cell fate. The Jumonji (JmjC) domain–containing family of demethylases comprises 27 members that target mono-, di-, and trimethylated lysine residues of histone (or nonhistone) proteins. To evaluate their role in regulation of hematopoietic stem cell (HSC) behavior, we performed an in vivo RNAi-based functional screen and demonstrated that Jarid1b and Jhdm1f play opposing roles in regulation of HSC activity. Decrease in Jarid1b levels correlated with an in vitro expansion of HSCs with preserved long-term in vivo lymphomyeloid differentiation potential. Through RNA sequencing analysis, Jarid1b knockdown was associated with increased expression levels of several HSC regulators (Hoxa7, Hoxa9, Hoxa10, Hes1, Gata2) and reduced levels of differentiation-associated genes. shRNA against Jhdmlf, in contrast, impaired hematopoietic reconstitution of bone marrow cells. Together, our studies identified Jarid1b as a negative regulator of HSC activity and Jhdmlf as a positive regulator of HSC activity.
Introduction
Multipotent hematopoietic stem cells (HSCs) ensure sustained production of lineage-committed blood cells throughout life. The pool of long-lived HSCs is preserved due to their inherent capacity to undergo self-renewal divisions. Cell fate decisions result from integrated influences of various nuclear factors, including chromatin modifiers that regulate access of transcriptional machinery to specified genomic loci. Chromatin, consisting of intertwined DNA, histone, and nonhistone proteins, can be covalently modified by epigenetic effectors. DNA methyltransferases (DNMTs) modify carbon-5 of cytosine bases within cytosine–guanosine dinucleotides (CpGs) frequently located proximal to or in promoters.1 In mammalian cells, the mechanism and effectors of DNA demethylation are less well understood, but recent reports suggest this process likely requires the activity of the ten eleven translocation enzyme family.2,3 Post-translational modifications of histones include methylation, acetylation, phosphorylation, ubiquitination, sumoylation, or adenosine 5′-diphosphate–ribosylation (see Kouzarides4) and introduce changes in local chromatin topography that subsequently alter gene expression patterns in developmental-stage and cell context–specific manners.
Methylation of histone residues represents a classic paradigm that links epigenetics to cell fate and identity, best illustrated by the antagonistic forces of Polycomb (PcG) and Trithorax (Trx) group protein complexes on regulation of key developmental loci such as Hox gene clusters (see Mills5). In general, PcG protein repressor complexes PRC2 and PRC1 are associated with gene silencing, and Trx complexes (compass-like or MLL, mixed lineage leukemia, complexes) with gene activation. The histone methyltransferase (HMT) enhancer of zeste homolog 2 (EZH2) of the PRC2 complex catalyzes trimethylation of lysine 27 on histone H3 (H3K27me3). This covalent mark serves as a docking site for the PRC1 complex, which mono-ubiquitinates lysine 119 of histone H2A (H2AK119Ub), resulting in gene silencing. MLL complexes counteract these epigenetic marks via trimethylation of lysine 4 on histone H3 (H3K4me3) at transcription start sites, a mark associated with active gene transcription6 and recruitment of H3K27me3 demethylases UTX and JMJD3.7–9 Additional Trx-mediated modifications such as acetylation of H3K27 (H3K27Ac) and dimethylation of H3K36 (H3K36me2) further oppose the PcG-mediated gene repression.
Histone methylation status on lysine (K) or arginine (R) residues is reported to evolve through highly dynamic and finely regulated processes (see Cloos et al10). Histone demethylases (HDMs) integrate into multiunit complexes, resulting in removal of methylation marks by amine oxidation, deamination,11 or hydroxylation. The lysine-specific demethylase 1 (LSD1/KDM1A)–related HDM can demethylate mono- and dimethylated lysine residues. Jumonji C (JmjC) domain–containing HDMs (n = 27) are capable of removing all 3 lysine methylation states by an oxidative reaction requiring α-ketoglutarate and iron (Fe2+) as cofactors. Like the LSD1 family, JmjC/JARID1 proteins act as components of multi-subunit complexes, with noncatalytic domains proposed to mediate protein–protein interactions involved in regulation of demethylase activity and/or target specificity (see Secombe and Eisenman12). In addition, JMJD6 demethylates arginine residues,13 and bacterial Jumonji domain–containing AlkB protein is involved in DNA demethylation and repair,14,15 suggesting that JmjC substrates include nonhistone targets.
JmjC protein activity results in dynamic chromatin landscape changes that enable expression of distinct gene subsets required for self-renewal,16 proliferation,17 differentiation,18–20 cellular senescence,21 and cancer development.22–24 In light of these findings, an established in vivo RNAi-based screening strategy25 was undertaken, in a targeted way, to assess the impact of JmjC gene downregulation on adult primary HSC cell fate. We identify Jarid1b as a negative regulator of HSC self-renewal and progenitor cell activity, while Jhdm1f positively influences blood reconstitution. Results from these experiments and possible downstream functional networks involved are presented.
Methods
Construction of shRNA retroviral vectors
For each gene target, 3 to 5 shRNAs were designed as single-stranded oligonucleotides also incorporating miR-30 flanking arms using the RNAi Central shRNA design tool at http://cancan.cshl.edu/RNAi_central/main2.cgi and our previously established methodology.25
Mice
C57BL/6J (CD45.2+) transplant recipients and C57BL/6Ly-Pep3b (CD45.1+) congenic bone marrow donor mice were bred and manipulated in a specific pathogen-free animal facility. Experimental procedures were revised and approved by the University of Montreal Animal Ethics Committee.
Flow cytometry
Negative selection of hematopoietic lineage marker (GR-1+, B220+, Ter119+)–expressing cells (Lin−) was performed as described.25 Lin− bone marrow fraction was subsequently stained with PE-Cy7–conjugated anti-cKit, PE-Cy5–conjugated anti-Sca1 (eBioscience), PE-conjugated anti-CD150 (BioLegend), and fluorescein isothiocyanate (FITC)–conjugated anti-CD48 (BD Biosciences) antibodies, followed by isolation of HSC-enriched PE-Cy5-Sca1+/PE-Cy7-cKit+/PE-CD150+/FITC-CD48−/APC-Lin− cell population. Day E14.5d.p.c. fetal liver–derived HSCs were purified from the Lin− cell populations by isolating the fraction of PE-Cy5-Sca1+/PE-CD11b+/PE-Cy7-CD150+/FITC-CD48−/APC-Lin− cells. Cells were sorted using the FACSAria (fluorescence-activated) cell sorter (Becton-Dickinson, San Jose, CA). The frequency of Long Term Repopulating (LTR)-HSC in the sorted populations (supplemental Table 1), the proportions of transduced (green fluorescent protein [GFP+]) transplant-derived (CD45.1+) peripheral blood leukocytes,26 and the contribution of these cells to reconstitution of hematopoietic lineages25,26 were determined as described.
HSC/progenitor cell culture, retroviral infection and transplantation
Suspension cell cultures of HSC/progenitor cell–enriched populations, generation of retrovirus-producing GP+E-86 cells, and infection of the sorted HSC/progenitor cells were performed as described.26 For validation assays, 1500 CD150+CD48− Lin− bone marrow–derived cells were introduced in coculture with retroviral producers in 96-well plates.26 After a 5-day incubation (day 0), the total cell content of each well was harvested and partitioned for transplantation and cell culture as previously reported.25 Briefly, one-eighth of the cell suspension was transplanted into sublethally irradiated congenic recipients (n =2 for each shRNA species). The remaining cells were cultured for an additional 7 days, and proportions corresponding to one-eighth of the cell input transplanted in 3 recipient mice (day 7). To compensate for the shJhdm1f-associated loss of reconstituting activity, one-fourth of the day 0 cell suspensions was transplanted in each of the 3 recipients, and no continuing suspension cultures were initiated.
Clonogenic progenitor cell assays
The total numbers and distributions of myeloid clonogenic progenitors in various cell populations recovered from the 5-day coculture with retroviral producers (day 0) or from the subsequent suspension cultures (days 5 through 7) were determined as described.25
Microphotograpy
Images of Wright-stained cytospin cell preparations were acquired using a Leica DMIRB microscope with an HCXPL FluotarL 40×/0.6 numeric aperture objective (Leica) and a Retiga EX-i camera (Q-Imaging). Images were transformed directly into TIFF files using Adobe Photoshop version 6.0 (Adobe Systems). In situ images of colonies in semisolid media were acquired using the same set up but with a HC Pl Fluotar 10×/0.30, Ph 1 lens.
Competitive repopulating unit assay
Competitive repopulation unit (CRU) assays were performed as previously described.25,26
Assessment of JmjC gene expression in HSC-enriched populations
Gene expression was assessed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) using the LightCycler 480 System (Roche, Basel, Switzerland). Reactions were performed in 384-well plates for 50 amplification cycles (95°C 10 seconds; 60°C 10 seconds; 72°C 10 seconds). Reference Taqman gene assay (Hprt) was purchased from Applied Biosystems. Primer sequences are listed in supplemental Table 2, and Δ CT (cycle threshold) values are listed in supplemental Table 3.
ChIP-chip analysis
Two-color hybridizations on NimbleGen MM8 Deluxe Promoter HX1 arrays were carried out using 34 μg of Cy5-labeled input and 34 μg of Cy3-labeled test (shLuc or shJarid1b) ChIP DNA using the NimbleGen hybridization kit as recommended by the manufacturer. Arrays were scanned at 5 μm resolution using a GenePix4000B scanner (Molecular Devices). Data extraction and peak finding analysis was done using NimbleScan 2.5 and visualized in the University of California, Santa Cruz genome browser.
RNA sequencing analysis
RNA sequencing was performed as previously described.27 RNA was extracted from 1 × 106 cells for each test (sh3Jarid1b-GFP) and control (shLuc-GFP) culture condition. Cultures were initiated with 24 000 CD150+ CD48−Lin− bone marrow cells and expanded for 4 days following infection with the respective constructs. Cells from 2 independent cultures were isolated for each condition, and only cell populations with >90% gene transfer (GT) were selected.
shRNA-mediated knockdown in NUP98-Hoxa10ΔHD-transduced HSCs
Next, 1500 purified CD150+CD48−Lin− bone marrow cells were cocultured with irradiated (40 Gy) GP+E86 cells, producing MSCV-NUP98-HOXA10HD-IRES-Puromycin (NA10HD) virus. After 4-day coculture, cells were collected and cultured for 6 days in the presence of 10 μg/mL of Puromycin, harvested, and stained with APC/Cy7-conjugated anti-CD43 (BioLegend), APC-conjugated anti-Sca1, PE/Cy7-conjugated anti-Gr1, and PE/Cy5-conjugated anti-F4/80 (eBioscience). The sorted NA10-transduced Sca1+CD43+Gr1−F4/80− cell subpopulation was cocultured with GP+E86 cells, producing shLuc, sh3Jarid1b, or sh1Moz recombinant retroviruses. After 4 days, the nonadherent cells were recovered and expanded for an additional 6 days. To assess the extent of differentiation in these cultures, cells were stained with PE/Cy7-conjugated anti-Gr1 and PE/Cy5-conjugated anti F4/80-PE/Cy5 antibodies. Data were acquired using a BD LSRII cytometer and FACSDiva version 4.1 software (BD Biosciences PharMingen) and analyzed using the FlowJo version 7.6.4 software (TreeStar).
Statistical analysis
Statistical analysis was conducted using Prism (Graphpad Software version 5) or SPSS. The 2-tailed Student t test was performed throughout the study, unless otherwise stated. Statistical significance was calculated at a 95% confidence level.
Results
JmjC gene expression in HSC-enriched populations
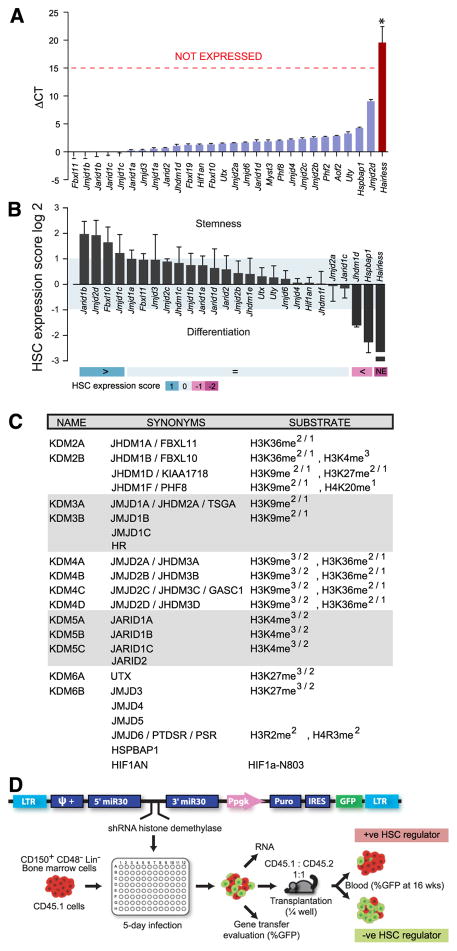
Expression profiles were obtained for all JmjC genes in sorted mouse bone marrow and fetal liver cell populations comprising >30% of functionally defined long-term–repopulating LTR-HSCs, as evaluated by CRU assays28 (supplemental Table 1). Results showed that all JmjC gene transcripts, except Hairless, were detected at relatively high levels (Figure 1A). The histone acetyl-transferase MOZ (MYST3), reported as essential for HSC expansion,29,30 was also highly expressed in this fraction and chosen as a control for the functional screen. Transcript levels of 4 JmjC genes, that is, Jarid1b, Jmjd2d, Fbxl10, and Jmjd1c, were notably increased in the LTR-HSC–enriched fractions relative to total bone marrow cells (Figure 1B). In contrast, Jhdm1d, Hspbap1, and Hairless transcript levels were notably higher in more mature cells. Genes that had greater expression in the LTR-HSC–enriched population, for example, Jarid1b, Jmjd2d, Fbxl10, and Jmjd1c, were assigned an HSC expression score of 1, while genes that that had greater expression in mature cells were attributed negative scores (Figure 1B, lower panel). Increased expression in LTR-HSCs implied an HSC biological role, and these candidates were prioritized in the functional studies.
Figure 1. JmjC gene expression in HSC/progenitor cell populations and selection for RNAi screen.
(A) Transcript levels of HDM in HSC-enriched cell populations. Results show ΔCT values determined by qRT-PCR assays (with respect to endogenous Hprt expression levels, Ct~22) and represent average ± standard error of the mean (SEM) of 5 independently sorted HSC populations (bone marrow, n = 3; E14.5 d.p.c. fetal liver, n = 2). Frequencies of long-term–repopulating HSCs in these populations are shown in supplemental Table 1. (B) Comparison of HDM transcript levels detected in HSCs and total bone marrow cell populations. Relative transcript quantities (RQ) are shown in log2 scale and represent the ΔCT (HSC)/ΔCT(bone marrow) ratio determined by qRT-PCR assays (average ± SEM, n = 3). An HSC expression score was implemented based on expression levels (ΔCT) and differential expression (RQ) of individual HDM to rank gene candidates according to relative expression in HSC vs mature cells: gene not expressed in HSC, −2; less (<) expressed in HSC vs mature cells, −1; more (>) expressed in HSC, +1; equally expressed, 0. (C) List of the 23 HDM candidates tested in primary screen. HDMs subfamilies sharing similarities outside the catalytic domain are denoted by different shading. Left column, the revised current terminology; center column, synonyms; right column, proposed substrate specificity.10 Four HDM genes were excluded from the screen: Jarid1d and Uty map to chromosome Y and are thus likely not required for regulation of HSC activity; Pla3g4b belongs to the cytosolic phospholipase A2 family; Jhdm1e knockdown could not be achieved by any hairpin in 2 independent experiments. (D) Schema of shRNA retroviral vector backbone (top) and experimental outline (bottom) of the primary screen at the bottom. At 16 weeks after transplantation, an increase in the proportion of GFP+ peripheral blood leukocytes above their input levels reflects knockdown of a negative regulator of HSC activity (green box), and the inverse outcome denotes a positive HSC regulator (red box).
Functional in vivo RNAi-based primary screen
The shRNA-based screen tested 23 of the 27 known JmjC mouse genes (Figure 1C) for their potential functional role in HSC biology. shRNA Luciferase (shLuc) was used as a negative control, while shRNA Moz (shMoz) and Nup98Hoxa10-homeodomain (NA10hd) overexpression were used as controls for the loss and gain of HSC activity, respectively (data not shown for NA10hd, see Ohta et al31 and Sekulovic et al32). HDM-directed shRNA sequences (n = 112; supplemental Table 5) were subcloned into an LMP retroviral vector and assessed for their ability to modulate HSC/progenitor activity in vivo, as previously reported25 (summarized in Figure 1D). The biological impact of each shRNA was evaluated by serial sampling of peripheral blood from transplanted mice at early (3 to 4 weeks) and late (16 to 20 weeks) time points.
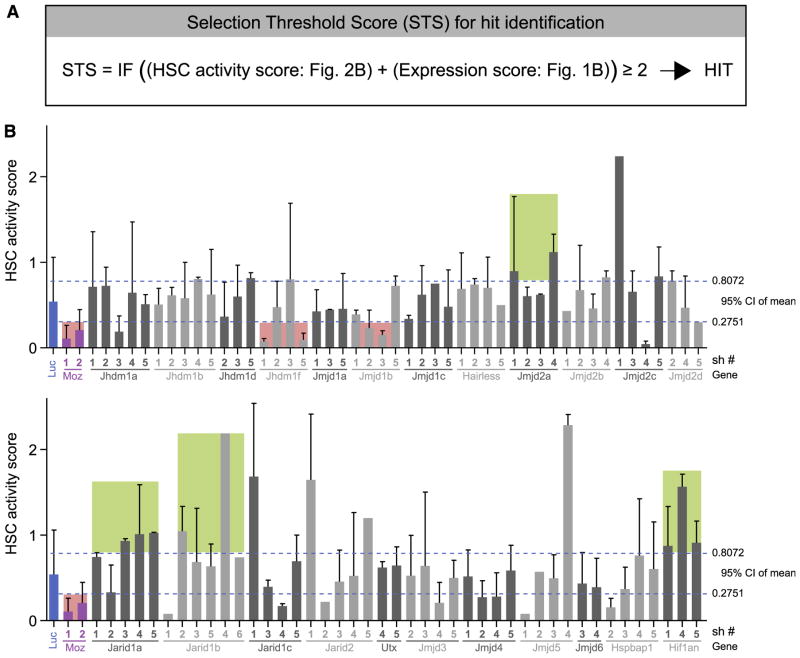
To prioritize candidates for validation and downstream functional studies, a “selection threshold scoring system” (STS), weighted for HSC-enriched genes, was established (Figure 2A and supplemental Table 3). HSC activity was determined by the long-term blood engraftment potential of the transduced donor cells (CD45.1+GFP+ cells) based on their contribution to blood leukocytes, calculated as a proportion of the total donor (CD45.1+) population, which comprises both transduced and nontransduced cells (% blood GFP= CD45.1+GFP+cells/total CD45.1+cells). The percentage of blood reconstitution of the transduced cells was then compared with the respective GT level on the day of transplantation (day 0), and this ratio (Figure 2B, y-axis) was used to document an increased (ratio >0.8072) or decreased (ratio <0.2751) contribution to the donor graft (HSC activity score). This ratio was used as GT levels for the hairpins presented in Figure 2B and ranged from 20% to 80%, with an overall average of 49% as previously reported25 (see supplemental Table 4 for complete data), and thus transduced cell expansion or attrition could be measured. Overall, 6 hits were identified: Jmjd2a, Jhdm1f, and Jmjd1b (STS of 2) and Jarid1a, Jarid1b, and Hif1an (STS of 3). Four of these genes, that is, Jarid1a, Jarid1b, Jmjd2a, and Hif1an, were identified as potential negative regulators of HSC activity (Figure 2B, green shaded areas), while Jhdm1f and Jmjd1b (Figure 2B, red area) were identified as as putative positive regulators.
Figure 2. HDM hit identification.
(A) The selection threshold score for genes was calculated from the expression score (Figure 1B) and HSC activity score or biological score (Figure 2B). Biological score represents the number of shRNAs per HDM that modulate HSC activity in recipients above or below the 95% confidence interval range established for control shLuc cells (dotted blue lines in Figure 2B). Selection threshold score of 2 and above identifies hits selected for validation experiments. (B) Contributions of GFP+ (shRNA-transduced) cells to peripheral blood reconstitution of recipients at 20 weeks after transplantation. Results are presented as proportions GFP+ cells within the transplant-derived (CD45.1+) peripheral blood cells and are normalized for the GT efficiency determined on the day of transplantation (day 0). Green shaded areas, suppressors of HSC activity; red shaded areas, enhancers of HSC activity. Raw data for GT rates and blood reconstitution levels for all recipients are provided in supplemental Table 4.
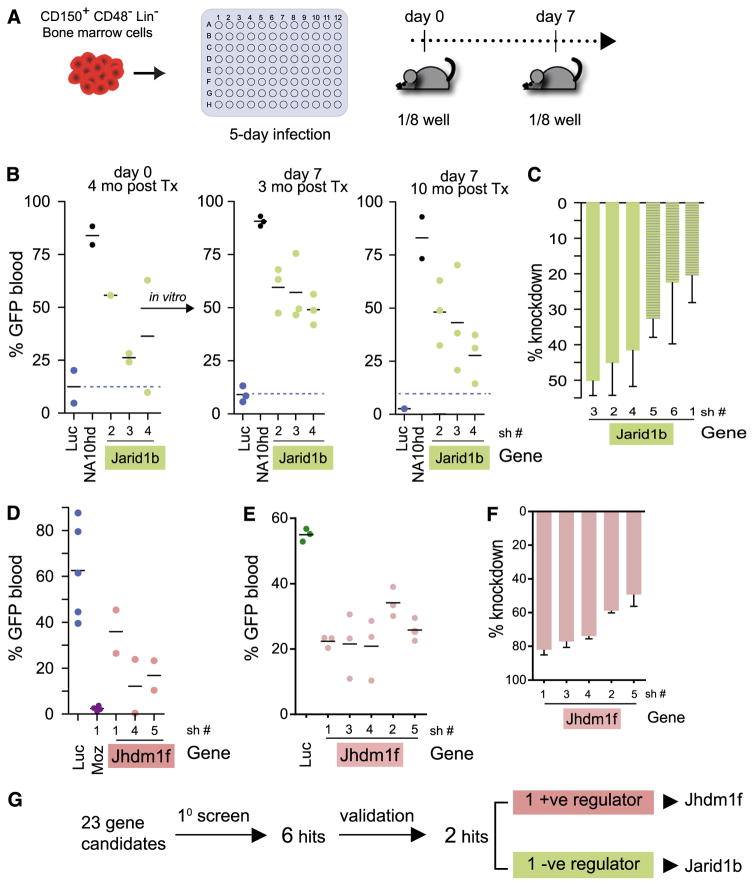
JmjC HDM knockdown validation experiments
Hit validation was further restricted to proven JmjC domain containing HDMs (HIF1AN demethylase excluded as it targets HIF1α); for technical considerations, validation of potential positive regulators was limited to Jhdm1f. The increased hematopoietic reconstitution levels observed with shRNA-mediated knockdown of the 3 remaining hits, that is, Jarid1a, Jarid1b, and Jmjd2a (Figure 2B), prompted us to examine how the transduced HSCs would behave in vitro, where control HSCs normally exhaust rapidly.26 Validation experiments were performed (Figure 3A) and a gain-of-function phenotype for Jarid1a or Jmjd2a could not be confirmed upon shRNA transduction and prolonged in vitro culture (data not shown). Conversely, reducing Jarid1b levels in HSC populations by multiple shRNA moieties (shJarid1b 2, 3, and 4) clearly conferred an in vivo competitive advantage to freshly transduced cells compared with shLuc controls (Figure 3B, left panel; n = 3 different shRNAs to Jarid1b, day 0 transplanted cells). As higher GT rates were achieved in the validation experiments (>80%), blood reconstitution levels were measured as CD45.1+GFP+ cell percentages. The positive impact of shJarid1b on HSC activity was even more noticeable for cells transplanted after 1 week of in vitro culture (Figure 3B, middle and right panels; cells transplanted after 7-day culture) to better detect HSC gain-of-function activity, as our previous studies demonstrated.26 Proportions of shJarid1b-transduced cells (GFP+) in peripheral blood remained well above those determined for shLuc controls for up to 10 months (Figure 3B). Knockdown of shJarid1b was determined for freshly transduced cells, and a >40% decrease in expression was observed for shJarid1b 2, 3, and 4 (Figure 3C). Decreased HSC activity relative to control cells was confirmed with multiple shRNA constructs against Jhdm1f (n = 5; Figure 3D–E). All 5 shRNAs against Jhdm1f had knockdown efficiencies >40% (Figure 3F). Validation experiments thus uncovered 1 negative (Jarid1b) and 1 positive (Jhdm1f) regulator of HSC activity. Further in vitro studies of shJhdm1f-transduced cells detected no significant change in proliferation or clonogenic progenitor activity of nucleated cells (supplemental Figure 1), indicating a distinct role for Jhdm1f in HSC repopulation ability. The striking impact of Jarid1b knockdown on blood reconstitution oriented research toward Jarid1b function.
Figure 3. Validation assays for identified hits.
(A) Schema of experimental design. (B) Long-term contribution of GFP+ (shLuc+, shJarid1b+, or NA10hd+) cells to peripheral blood reconstitution of recipients; GT >80% for all conditions. Left panel: recipients of day 0 cells; central and right panels, recipients of day 7 cells. NA10hd, cells engineered to overexpress NUP98Hoxa10-homeodomain fusion protein and GFP. (C) Evaluation of Jarid1b knockdown in GFP+ shJarid1b-transduced cells compared with shLuc controls. Results represent average ± SEM; n = 4). Relative transcript quantities (RQ) values determined for 6 hairpins. Bars with dashed lines correspond to shRNA constructs (sh5,6 and 1) that did not achieve significant (ie, >30%) knockdown of Jarid1b in BM HSC. (D) Long-term contribution of GFP+ (shLuc+, or shMoz+, or shJhdm1f+) cells to peripheral blood reconstitution in recipients of day 0 cells. Each recipient received a one-quarter of the transduced cell population, or twice the number of input cells transplanted for validation experiment shown in Figure 3A. (E) Short-term (3 weeks) contribution of GFP+ (shLuc+- or shJhdm1f+-transduced) cells to peripheral blood reconstitution in recipients of day 0 cells. GT = 99% for all conditions. Experiment (as in Figure 4D) was repeated to include all hairpins against Jhdm1f. (F) Evaluation of Jhmdm1f knockdown in GFP+ shJhdm1f-transduced cells compared with shLuc controls. Results represent average ± SEM (n = 3). RQ values determined for 5 hairpins. (G) Summary of screen results.
Jarid1b knockdown decreases hematopoietic cell differentiation in vitro
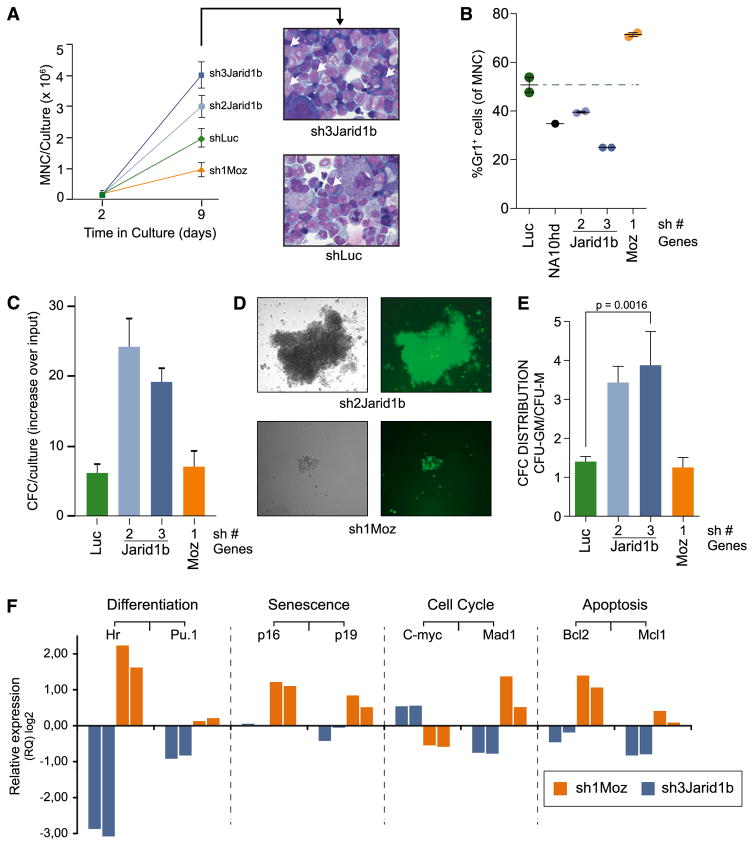
Sh2- or sh3Jarid1b-transduced primitive hematopoietic cell populations had an ~1.5- to 2-fold proliferative advantage in vitro compared with shLuc controls, while Moz knockdown decreased expansion of the transduced cells (Figure 4A, left panel). The expanded shJarid1b-transduced populations comprised high proportions of morphologically immature cells (Figure 4A, right panel) and lower percentages of differentiated (Gr1+) myeloid cells than shLuc controls (Figure 4B). Moreover, fractions of primitive (Gr1−) shJarid1b-transduced cells were comparable to those determined in response to NA10hd, a well-characterized enhancer of in vitro HSC/progenitor cell expansion, while a marked elevation in differentiation was observed for shMoz loss-of-function control (Figure 4B). During the 7-day culture period, the numbers of shJarid1b-transduced clonogenic progenitor cells (colony-forming cell [CFC]) increased ~3-fold compared with shLuc and shMoz controls (Figure 4C). Jarid1b knockdown enhanced the proliferative capacity of individual CFCs compared with shMoz controls (Figure 4D) and promoted expansion of the highly proliferative granulocyte-macrophage progenitors (Figure 4E). Although the total CFC contents of shLuc and shMoz control cultures were comparable (Figure 4C) and both cell populations differentiated into granulocytes and macrophages (Figure 4E), the antiproliferative effect of Moz knockdown was evident from the decrease in sizes of shMoz clones compared with shLuc controls (data not shown).
Figure 4. Jarid1b knockdown decreases hematopoietic cell differentiation in vitro.
(A) Left panel: Jarid1b knockdown increases yields of mononuclear cells in cultures initiated with shJarid-transduced HSC/progenitor cell populations (mean ± standard deviation (SD), n = 2). Right panel: Wright-stained cytospin preparations of cells on day 9 of culture, 40× magnification, white arrows indicate primitive cells. (B) shJarid1b suppresses in vitro differentiation of HSC/progenitor cell populations. Proportions of Gr1+ cells on day 9 of culture were determined by flow cytometry. Each dot represents an independent culture. (C) Jarid1b knockdown enhances in vitro expansion of myeloid CFCs. The increase in CFC numbers was calculated from MNC and CFC numbers determined on days 2 and 9 (mean ± SD, n = 4). (D) Images of the predominant colony types. shJarid1b, colony-forming unit granulocyte-macrophage (CFU-GM), high proliferative potential; shMoz, CFU-macrophage (CFU-M), low proliferation. Left panels, bright field; right panels, epifluorescence. (E) Proportions of the highly proliferative CFU-GM in cultures of shRNA-transduced cells (mean ± SD, n = 4). (F) qRT-PCR–based comparison of cell fate–associated transcript levels on day 7 (Figure 3A) shJarid1b and shMoz cells compared with shLuc controls.
qRT-PCR assays involving selected candidates revealed a marked downregulation of differentiation-associated genes Hairless (Hr) and Pu.1 in shJarid1b cells compared with shMoz control (Figure 4F). No major changes in expression levels of genes regulating senescence (Cdkn2a (p16) and Cdkn2d (p19)) or apoptosis (Bcl2, Mcl1) were detected in shJarid1b cells, while modest increases in c-Myc and decreases in Mad1 levels resembled those determined for Hoxb4-overexpressing cells (data not shown, and33). Moz knockdown was, however, clearly associated with upregulation of Hr, Cdkn2a, and Cdkn2d expression. This suggests that Jarid1b knockdown promotes in vitro HSC/progenitor expansion by suppressing differentiation, while cells remained permissive to cell cycle reentry. In contrast, reduced Moz expression enforced commitment to differentiation and senescence cell fate pathways.
Jarid1b negatively regulates HSC self-renewal
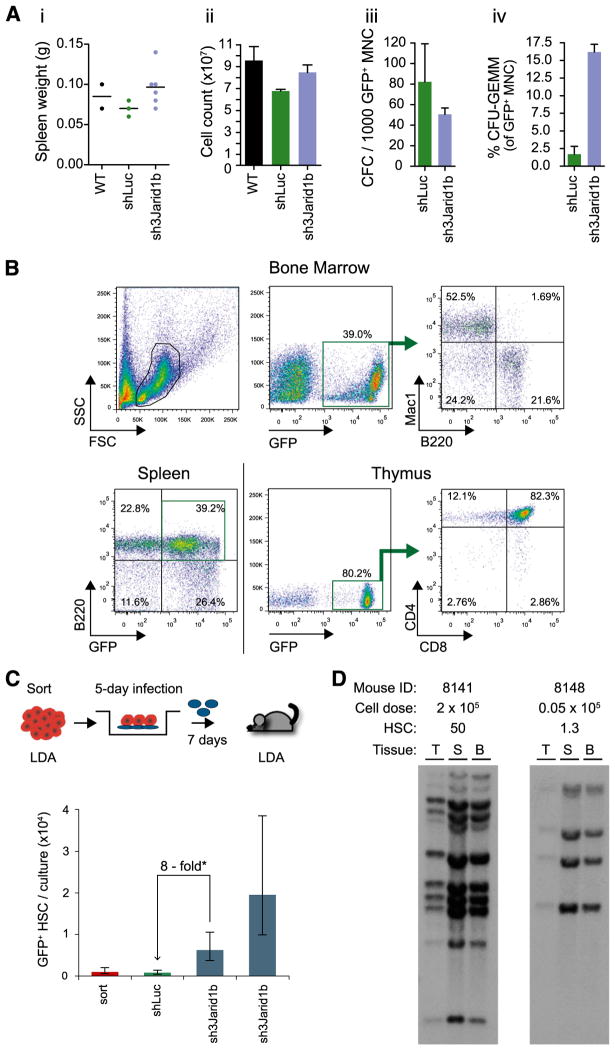
Transplant recipients of shJarid1b cells had normal numbers of total bone marrow mononuclear cells (MNCs), and no splenomegaly was observed (Figure 5A, panels i and ii). Contributions of the transduced (GFP+) shLuc and shJarid1b cells to myeloid progenitor cell compartments were comparable (Figure 5A, panel iii), but an ~8- to 10-fold increase in the frequencies of shJarid1b-multilineage progenitors (colony-forming unit-granulocyte, erythroid, macrophage, megakaryocyte [CFU-GEMM]) compared with controls was observed (Figure 5A, panel iv). This difference in immature progenitor cell content remained benign, and no hematological abnormalities could be detected during the 12-month observation period. Moreover, transplanted shJarid1b cells generated normal proportions of myeloid (Mac1+/CD11b+), B-lymphoid (B220+/CD45R+), and T-lymphoid (CD4+, CD8+) progeny (Figure 5B), suggesting that differentiation ability in vivo was not affected.
Figure 5. In vitro expanded shJarid1b-HSC retain long-term in vivo multipotency.
(A) Analysis of hematopoietic tissues in recipients of day 7 cells (Figure 3B) at 1 year after transplantation. Panel i, spleen weight; panel ii, the total numbers of bone marrow cells pooled from pelvis, femur, and tibia; panel iii, CFC frequency in the GFP+ bone marrow cell populations; panel iv, proportions of GFP+ multilineage progenitors (CFU-GEMM). Dots in first plot represent the numbers of individual mice for which all the described parameters were analyzed. (B) Contribution of day 7 (Figure 3A) shJarid 1b (GFP+) cells to reconstitution of myeloid (Mac1+), B-lymphoid (B220+), and T-lymphoid lineage (CD4+, CD8+) at 1 year after transplantation. An example of typical reconstitution observed in all recipients (n > 10) is shown. (C) Jarid1b knockdown promotes the in vitro expansion of LTR-HSCs. Upper panel, experimental outline. Lower panel, CRU numbers in freshly sorted (ie, input) and day 7 shRNA-transduced cell populations (mean ±standard error). shJarid1b CRUs were determined in 2 independent experiments (see supplemental Table 6). (D) Clonal analysis of proviral integrations in DNA isolated from hematopoietic tissues of mouse from shJarid1b cohort introduced in Figure 5C. DNA was digested with EcoRI, which cuts once within the provirus such that each DNA fragment recognized by the 32P-labeled Gfp probe represents a unique integration event. Mouse ID, the total dose of transplanted cells, and the estimated number of transplanted CRU are shown on top. T, thymus; S, spleen; BM, bone marrow.
To determine if Jarid1b knockdown favored self-renewal divisions, leading to expansion of HSC populations, equal numbers of HSC-enriched (CD150+CD48−Lin−) cell populations were cocultured with shLuc or shJarid1b retroviral producers. HSC frequencies were determined in samples immediately after cell sorting and after a cumulative 12-day ex vivo culture period (Figure 5C, panel i), using the CRU assay. CRU numbers in cell populations recovered from shLuc cultures (Figure 5C, green bar) were comparable to those determined for the input cell population (Figure 5C, red bar), suggesting no major loss or gain of HSCs during the in vitro incubation. In contrast, the CRU numbers in shJarid1b cultures increased 8- to 20-fold above shLuc control samples or sorted cells prior to shJarid1b infection (Figure 5C, blue bars), suggesting that Jarid1b knockdown promoted the in vitro maintenance/expansion of long term-repopulating HSCs. Southern blot analyses of shJarid1b revealed a common proviral integration pattern between bone marrow (mostly myeloid, erythroid and B-), thymus (mostly T-) and spleen (B- and T-) cells of individual recipients (Figure 5D), demonstrating multipotency and oligoclonal hematopoietic reconstitution of the expanded HSCs.
Impact of Jarid1b knockdown on gene expression
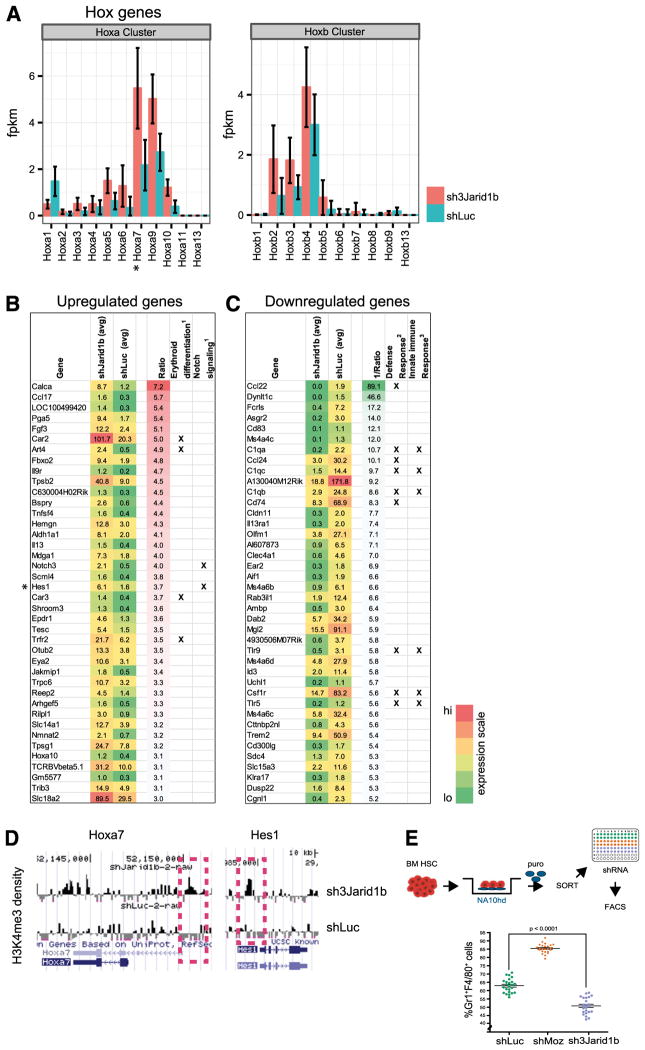
To examine downstream effects of Jarid1b knockdown, results of transcriptome next-generation sequencing (RNAseq), qRT-PCR, and ChIP-Chip assays were analyzed and compared with controls. The similarity in HSC expansion observed between Jarid1b knockdown and Hox gene overexpression31,33 and the recent report that JmjC members KDM6A (UTX) and KDM6B (JMJD3) are positive regulators of Hox gene expression21 prompted initial focus on the Hox family of transcription factors (Figure 6A). Both RNAseq and qRT-PCR assays revealed that Jarid1b knockdown induced a 2- to 5-fold increase in 5′ Hoxa gene (Hoxa5 to Hoxa11) expression levels compared with controls (Figure 6A and supplemental Figure 2). RNAseq analysis identified upregulation of genes associated with key pathways such as erythroid differentiation, notch signaling, and stem/progenitor cell function (Figure 6B and supplemental Figure 3), and down-regulation of others involved in immune responses and differentiation (Figure 6C and supplemental Figure 4). Several of the candidate downstream genes, including Hoxa7, Hes1, Hoxa10, and Hoxb2, were identified as potential direct epigenetic targets by enrichment of the associated H3K4me3 mark in 5′ loci as determined by ChIP-Chip assays (Figure 6D and supplemental Figure 5). Supporting this possibility, nonregulated genes, for example, Hoxa2, demonstrated no proximal promoter H3K4me3 enrichment following Jarid1b knockdown (supplemental Figure 5). Jarid1b knockdown also correlated with downregulation of the previously reported putative JARID1 target Cav134 (supplemental Table 7).
Figure 6. Jarid1b knockdown modulates molecular mechanisms implicated in maintenance of stemness.
(A) Quantification of Hox gene transcripts in shJarid1b cells and shLuc controls as assessed by RNA sequencing (RNAseq) analysis. For each Hox cluster (A-B) genes, FPKM (fragments per kilobase per million reads) expression values are shown for both conditions. Error bars indicate standard deviation. RNA was isolated from HSC-enriched cells in culture (4 days following retroviral infection), and only cultures with GT rates >90% were selected. For each condition, 2 biological replicates were sequenced. (B) Average FPKM and fold-change expression values of the 40 most upregulated (FPKM >1 for shJarid1b cells) and (C) downregulated (FPKM >1 for shLuc controls) genes from the RNAseq experiment described in Figure. 6A. Genes annotated to specific functions are specified by a cross mark in respective columns. Complete data for all differentially expressed genes (q value <0.05; all FPKM values included) shown in supplemental Table 7. (D) Enrichment for H3K4me3 marks (black peaks) at the Hoxa7 and Hes1 loci in shJarid1b cells. Chromatin immunoprecipitation was carried out using day 7 (Figure 5C) shJarid1b or shLuc-cells. Total H3K4me3 levels are presented in supplemental Figure 6. (E) Top panel: Experimental strategy for generation of Nup98Hoxa10-homeodomain (NA10hd) plus shRNA-overexpressing cells. Following puromycin selection, the Sca1+CD43− Gr1−F4/80− NA10hd-transduced cells were infected with shLuc-, shMoz-, and shJarid1b 1b-carrying retroviruses. Lower panel: Jarid1b knockdown suppresses differentiation of NA10hd overexpressing cells. Proportions of Gr1+F4/80+(ie, differentiated) cells in cultures were determined by flow cytometry on day 7 after shRNA transduction. Each dot represents individual culture comprising the transduced progeny of 1500 CD150+CD48−Lin− bone marrow cells1: manual curation2,3: 3.4- and 3.7-fold enrichment with false discovery rate of 4.6E-24 and 4.2E-11 in Gorilla bioinformatic tool. *Denotes high H3K4me3 densities, refer to Figure 6D.
Study of epistasis between Jarid1b and Nup98Hoxa10-homeodomain
To directly examine potential gene interaction of Jarid1b and Hoxa overexpression, HSC/progenitor cell populations were first transduced with Nup98Hoxa10-homeodomain (NA10hd) and the HSC-enriched populations (Sca1+CD43+ Gr1−F4/80−; Keith Humphries, personal communication) subsequently transduced with shJarid1b, shMoz, or control shLuc. The proportions of mature GR1+F4/80+cell fractions in each condition were evaluated as a measure of differentiation (Figure 6E, panel i). Results demonstrate that the combined effects of NA10hd overexpression and Jarid1b knockdown were additive, resulting in significant suppression of in vitro differentiation (P < .0001; Student t test) below the levels determined for control NA10hd+shLuc cells (Figure 6E, panel ii). In contrast, shMoz targeted knockdown overrode the maturation arrest imposed by NA10hd overexpression and enforced differentiation above the levels determined for controls (Figure 6E, panel ii).
Together, the data presented support a role for Jarid1b in regulating key loci implicated in HSC cell fate and identify Jarid1b as a negative regulator of in vitro HSC expansion.
Discussion
Following the established pipeline strategy from HSC isolation and infection to in vivo functional assessment of hematopoietic reconstitution, the presented RNAi screen highlighted Jarid1b as a negative and Jhmd1f as a positive modulator of HSC activity. Due to defined inclusion criteria, other HDMs should not be excluded as potential HSC modifiers. Akin to Hox gene overexpression, differentiation was restrained in shJarid1b-transduced HSC cultures, as evidenced by more primitive cell morphology, reduced granulocytic maturation, and greater expansion of clonogenic progenitors relative to controls. After transplantation, ex vivo expanded HSCs were able to resume normal lympho-myeloid differentiation in recipient mice, in the absence of lineage skewing or hematological abnormalities, for up to 1 year. Logarithmic ex vivo expansion of shJarid1b-transduced HSCs was demonstrated by CRU assays indicating that Jarid1b modulation influences stem cell fate decisions. Oligoclonal origin of the repopulating HSC pool and inherent multipotency of shJarid1b-transduced parental stem cells was shown by proviral insertion patterns in long-term recipient mice. Mechanistically, the competitive advantage conferred to shJarid1b-transduced HSCs could be attributed in part to the selective upregulation of 5′ Hoxa genes. Segmental transcription of this chromosomal region is well documented35 and particularly targeted by epigenetic regulators such as MLL or its derived fusion oncoproteins both in normal and leukemic stem cells.36 Interestingly, the 5′ Hoxa cluster is also targeted by the fusion oncoprotein NUP98-JARID1A in a mouse model of myeloid leukemia.37
In agreement with the proposed substrate specificity of JARID1B for H3K4me3,34 we noted enrichment for this epigenetic mark on 5′ Hoxa genes, indicating that Jarid1b contributes to the negative regulation of Hoxa gene expression that, when relieved following knockdown, leads in part to increased HSC activity. Enhancement of NA10hd-induced maturation arrest in the presence of reduced Jarid1b levels argues for regulation of additional cell fate determinants to account for the HSC phenotype seen. Comprehensive transcriptome analysis by RNAseq identified additional downstream genes, some of which are associated with key hematopoietic or stem cell–associated pathways. Recent reports further support a role for Jarid1b in transcriptional regulation of cell fate associated genes. PU.1 induction of transcription factor EGR2 was reported to recruit Jarid1b to the miR-17–92 promoter site, resulting in H3H4 demethylation and transcriptional silencing of the cluster required for monocyte maturation in a mouse model.19 Schmitz et al recently reported that Jarid1b depletion prevents neuronal differentiation of ESCs by indirectly preventing H3K4 demethylation and silencing of pluripotency and germ cell–associated gene loci.20 Similarly, H3K4me2/1 monoamine oxidase LSD1 was also deemed essential for proper hematopoietic progenitor differentiation.38
We demonstrate that transcript levels of Jarid1b are increased in HSCs, suggesting either a role in preventing unrestricted self-renewal divisions or in enabling downregulation of cell fate–associated genes upon lineage commitment. Interestingly, 1q32 anomalies, which include Jarid1b, are common genetic mutations found in cells of chronic myeloid leukemia patients during disease progression, which is characterized by a block in myeloid differentiation.39,40 This observation supports the hypothesis that low Jarid1b levels maintain stem cell fate that, combined with BCR-ABL–induced proliferation, could result in development of overt leukemia. In our studies, transplant recipients of shJarid1b-transduced cells never developed leukemia. However, all Jarid1b hairpins tested achieved similar gene knockdown (<50%), and more drastic outcomes following complete Jarid1b depletion cannot be ruled out. Two Jarid1b null alleles, one (exon 1 deletion) embryonic lethal41 and the other (exon 6 deletion) no gross abnormalities,20 have recently been described. Precise elucidation of the role Jarid1b gene dosage plays in HSCs and leukemia development will thus likely require analyses of HSC-specific Jarid1b deletion.
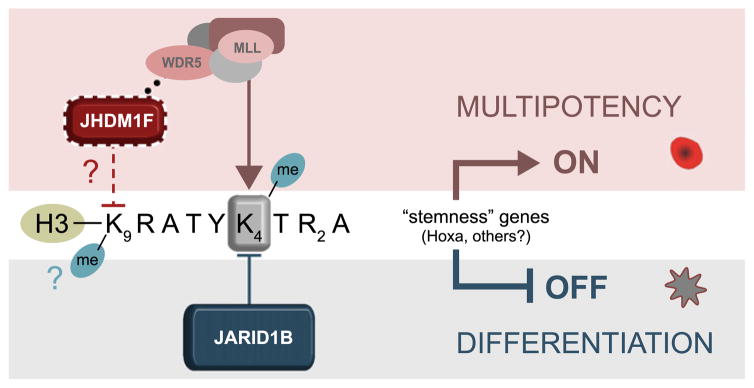
Overall, the data presented support a cellular and developmental stage–specific effect of Jarid1b levels in modulation of HSC cell fate. To sustain stem cell fate, integrated influences from various epigenetic effectors promote an active state of transcription at multipotency loci (Figure 7, upper panel). Activity of chromatin modifiers, including JARID1B, should culminate in maintenance of the H3K4me3 epigenetic mark on these loci and exclusion of repressive marks, such as H3K9me3 or H3K27me3. Jhdm1f/PHF8 has recently been identified as a key regulator of ATRA response in acute promyelocytic leukemia cells.42 Whether JHDM1F activity influences the methylation status of H3K9 or normal HSC fate decisions remains to be explored. In contrast, removal of the active H3K4me3 epigenetic mark, potentially by JARID1B, could repress transcription of “stemness” genes and favor lineage commitment (Figure 7, lower panel). Other JmjC candidates (eg, Hif1an) may also be involved in this process, underscoring the importance of further characterization of these enzymes in the context of HSC regulation.
Figure 7. Proposed model for JARID1B activity in modulation of HSC fate.
Gray shaded area: JARID1B erases the tri-methyl marks of H3K4 at stemness loci and represses activity of multipotency genes. Pink area: Decrease in JARID1B activity shifts balance in favor of histone methylases, preserving the active H3K4me3 mark at stemness loci to sustain multipotency.
Supplementary Material
Key Points.
Jarid1b knockdown promotes enhanced HSC activity.
Acknowledgments
The authors acknowledge Mélanie Fréchette and Andrea Evelyn Mejia Alfaro for their assistance with animal care and transplantation experiments and the Institute for Research in Immunology and Cancer technological platform members: Danièle Gagnè from Flow Cytometry Core Facility for help with cell sorting; Christian Charbonneau from Bio-imaging Core Facility for assistance with image acquisition and figure preparation; and Raphaëlle Lambert, Pierre Chagnon, and Simon Drouin from Genomic Core Facility for qRT-PCR and ChIP-chip experiments. This work was supported by grants from the Canadian Institute for Health Research (CIHR), the Canadian Cancer Society Research Institute and Fonds de Recherche en Santé du Québec to G.S. G.S. holds a Canada Research Chair in the Molecular Genetics of Stem Cells. S.C. is recipient of a CIHR Clinician-Scientist Fellowship Award and a Cole Foundation Transition Award. K.H. is recipient of a CIHR Post-Doctoral Fellowship Award and a Cole Foundation Award. S.B.T. is recipient of National Health and Medical Research Council of Australia, Royal Australian College of Physicians and CIHR Postdoctoral Fellowships.
Footnotes
Contribution: S.C. and G.S. established the gene candidate list; M.S., J.C., and S.C. contributed to isolation and functional assessment of highly purified HSC populations used for expression profile studies of gene candidates; S.C., K.J.H., and G.S. planned and performed the initial screen; S.C., K.J.H., and N.M. designed and performed validation experiments; S.C., N.M., K.J.H., S.B.T., J.C., E.D., J.K., and M.S. contributed to HSC isolation experiments required for initial screen, validation experiments, and subsequent experiments; S.C. and N.M. performed confirmation experiments for shJarid1b-transduced cells including cell culture, progenitor assays, and FACS; T.M. and S.C. performed RNA extraction and qRT-PCR analyses involving shJarid1b-transduced cells; S.C., N.M., and J.C. performed LDA experiments and analyses of long-term recipient mice; B.T.W. performed RNAseq studies; A.T. optimized and performed qRT-PCR assays for HoxA gene expression studies; E.D. performed southern blot analyses; J.R.L., T.M., S.C., and K.J.H. performed ChIP-chip experiments; M.S. and N.M. performed experiments involving cotransduction of NA10hd and shRNA constructs; K.H. helped in designing experimental strategy reported in Figure 6E and in writing the paper; and S.C., G.S., J.K., and A.T. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The online version of this article contains a data supplement.
References
- 1.Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33(20):e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10(10):669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 7.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Issaeva I, Zonis Y, Rozovskaia T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27(5):1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 10.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585(13):2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Secombe J, Eisenman RN. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection. Cell Cycle. 2007;6(11):1324–1328. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- 13.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 14.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419(6903):178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 15.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419(6903):174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 16.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21(20):2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayami S, Yoshimatsu M, Veerakumarasivam A, et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer. 2010;9:59. doi: 10.1186/1476-4598-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jepsen K, Solum D, Zhou T, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 19.Pospisil V, Vargova K, Kokavec J, et al. Epigenetic silencing of the oncogenic miR-17–92 cluster during PU.1-directed macrophage differentiation. EMBO J. 2011;30(21):4450–4464. doi: 10.1038/emboj.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz SU, Albert M, Malatesta M, et al. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J. 2011;30(22):4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood. 2011;117(14):3869–3880. doi: 10.1182/blood-2010-10-312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41(4):465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Y, Zhu Z, Han G, et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104(49):19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hope KJ, Cellot S, Ting SB, MacRae T, Mayotte N, Iscove NN, Sauvageau G. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell. 2010;7(1):101–113. doi: 10.1016/j.stem.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Deneault E, Cellot S, Faubert A, et al. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 2009;137(2):369–379. doi: 10.1016/j.cell.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon C, Chagraoui J, Krosl J, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26(7):651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szilvassy SJ, Humphries RK, Lansdorp PM, Eaves AC, Eaves CJ. Quantitative assay for totipotent reconstituting hematopoietic stem cells by a competitive repopulation strategy. Proc Natl Acad Sci USA. 1990;87(22):8736–8740. [Google Scholar]
- 29.Katsumoto T, Aikawa Y, Iwama A, Ueda S, Ichikawa H, Ochiya T, Kitabayashi I. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20(10):1321–1330. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas T, Corcoran LM, Gugasyan R, et al. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20(9):1175–1186. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta H, Sekulovic S, Bakovic S, et al. Near-maximal expansions of hematopoietic stem cells in culture using NUP98-HOX fusions. Exp Hematol. 2007;35(5):817–830. doi: 10.1016/j.exphem.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekulovic S, Gasparetto M, Lecault V, et al. Ontogeny stage-independent and high-level clonal expansion in vitro of mouse hematopoietic stem cells stimulated by an engineered NUP98-HOX fusion transcription factor. Blood. 2011;118(16):4366–4376. doi: 10.1182/blood-2011-04-350066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cellot S, Krosl J, Chagraoui J, Meloche S, Humphries RK, Sauvageau G. Sustained in vitro trigger of self-renewal divisions in Hoxb4hiPbx1 (10) hematopoietic stem cells. Exp Hematol. 2007;35(5):802–816. doi: 10.1016/j.exphem.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamane K, Tateishi K, Klose RJ, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25(6):801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorrance AM, Liu S, Yuan W, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006;116(10):2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang GG, Song J, Wang Z, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sprüssel A, Schulte JH, Weber S, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26(9):2039–2051. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 39.Karrman K, Sallerfors B, Lenhoff S, Fioretos T, Johansson B. Cytogenetic evolution patterns in CML post-SCT. Bone Marrow Transplant. 2007;39(3):165–171. doi: 10.1038/sj.bmt.1705560. [DOI] [PubMed] [Google Scholar]
- 40.Shah NK, Wagner J, Santos G, Griffin CA. Karyotype at relapse following allogeneic bone marrow transplantation for chronic myelogenous leukemia. Cancer Genet Cytogenet. 1992;61(2):183–192. doi: 10.1016/0165-4608(92)90084-l. [DOI] [PubMed] [Google Scholar]
- 41.Catchpole S, Spencer-Dene B, Hall D, et al. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol. 2011;38(5):1267–1277. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- 42.Arteaga MF, Mikesch JH, Qiu J, et al. The histone demethylase PHF8 governs retinoic acid response in acute promyelocytic leukemia. Cancer Cell. 2013;23(3):376–389. doi: 10.1016/j.ccr.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.