Abstract
Stereochemical control is critical in natural product biosynthesis. For ribosomally synthesized and post-translationally modified peptides (RiPPs), the mechanism(s) by which stereoselectivity is achieved is still poorly understood. In this work, we focused on the stereoselective lanthionine synthesis in lanthipeptides, a major class of RiPPS formed by addition of Cys residues to dehydroalanine (Dha) or dehydrobutyrine (Dhb). Non-enzymatic cyclization of the small subunit of a virulent lanthipeptide, the enterococcal cytolysin, resulted in the native modified peptide as the major product, suggesting that both regioselectivity and stereoselectivity are inherent to the dehydrated peptide sequence. These results support previous computational studies that a DhxDhxXxxXxxCys motif (Dhx = Dha or Dhb; Xxx = any amino acid except Dha, Dhb, and Cys) preferentially cyclizes by attack on the Re face of Dha or Dhb. Characterization of the stereochemistry of the products formed enzymatically with substrate mutants revealed that the lanthionine synthetase actively reinforces Re face attack. These findings support the hypothesis of substrate-controlled selectivity in lanthionine synthesis but also reveal likely coevolution of substrates and lanthionine synthetases to ensure the stereoselective synthesis of lanthipeptides with defined biological activities.
Natural products and their derivatives have been key sources of pharmaceuticals over the last century.1 Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a large and rapidly expanding family of natural products,2 as the burst of available genomic information has led to the identification of many RiPP biosynthetic pathways in the past decade.3, 4 The structural diversity of RiPPs is vast as a consequence of a multitude of different post-translational modifications, such as various forms of macrocyclization, methylation, hydroxylation, decarboxylation, cyclodehydration, and halogenation.2, 5 Of all the structural elements, one particular feature that accounts significantly for diversity and complexity in other natural products, the stereochemistry, has not received much attention in research related to RiPPs. The de-emphasis of stereochemistry is in part because RiPPs are initially assembled by the ribosome with amino acid building blocks that have the L configuration. However, new stereocenters may be introduced and existing stereocenters can be modified during the post-translational modification process.6–8 For instance, the presence of 18 epimerized amino acids with the non-canonical D configuration in polytheonamide, a RiPP of 48 amino acid residues, reinforces the idea that stereocenters inherited from the amino acid building blocks of the precursor peptide can be altered in the final natural product.6 Epimerization of the backbone stereocenters have also been reported for other RiPPs.9–14 Introduction of new stereocenters on side chains of selected amino acids by post-translational modifications can further increase the structural diversity of RiPPs. Thus far, relatively little is known how the stereoselective modifications are controlled during RiPP biosynthesis.
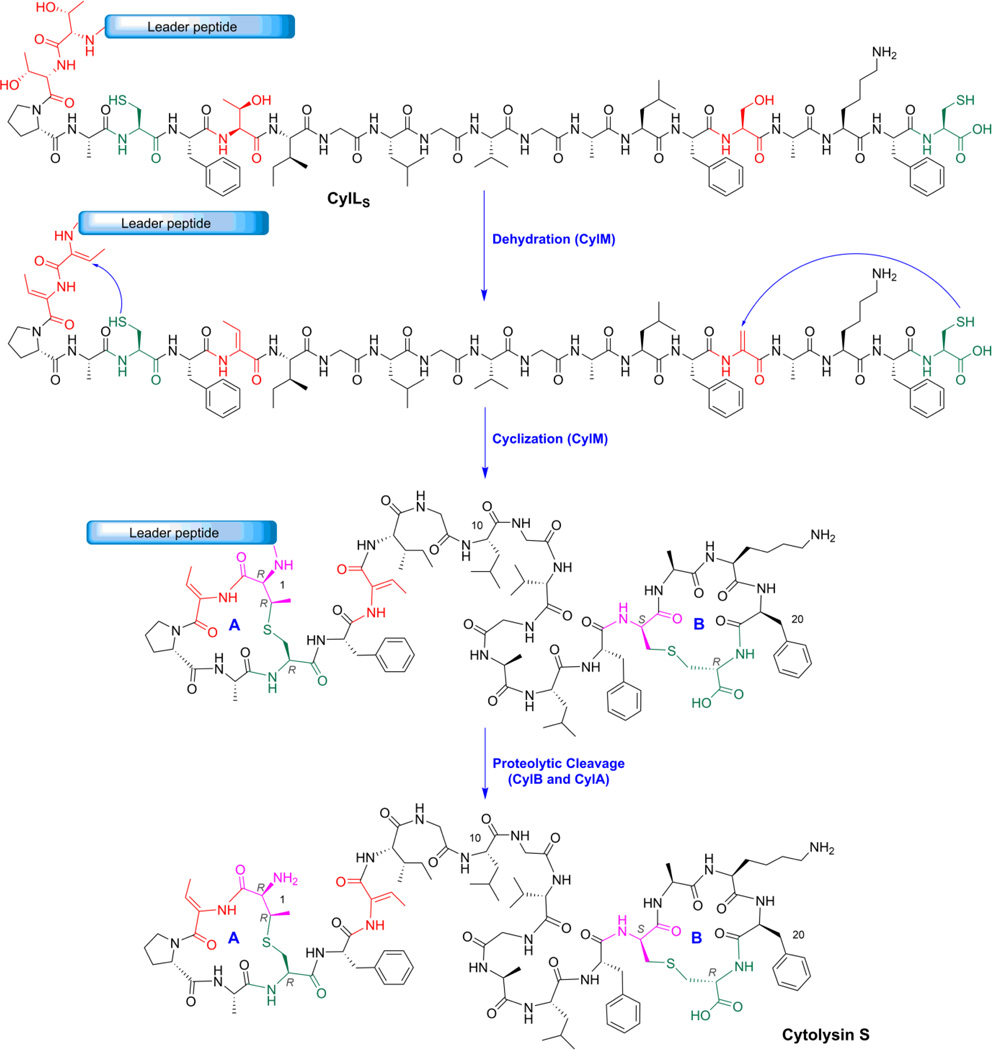
Lanthipeptides, a shorthand nomenclature for lanthionine-containing peptides, are a family of RiPPs that have been intensively studied over the past 40 years.15, 16 They are polycyclic peptides characterized by the presence of intramolecular thioether crosslinks. The biosynthesis of lanthipeptides initiates from a precursor peptide that consists of an N-terminal leader peptide and a C-terminal core peptide (Figure 1). The leader peptide is important for the recognition by the post-translational modification (PTM) machinery,17 bringing the enzyme in close proximity to the core peptide where the modifications take place,18, 19 and controlling the order of the PTMs.20 The post-translational modification of lanthipeptides involves dehydration of serine and threonine residues to dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively, and subsequent Michael-type addition of cysteine thiols onto these dehydroamino acids to form the characteristic thioether crosslinks termed lanthionine (Lan) and methyllanthionine (MeLan) (Figure 1).21 For class II lanthipeptides, both the dehydration and cyclization reactions are carried out by the same enzyme generically named LanM.22, 23
Figure 1. Schematic illustration of lanthipeptide biosynthesis using cytolysin S as an example.
The leader peptide is shown schematically whereas the amino acid sequence is shown for the core peptide Amino acid residues involved in post-translational modifications are highlighted in color. Sequence of the leader peptide of CylLS = MLNKENQENYSNKLELVGPSFEELSLEEMEAIQGSGDVQAE.
The formation of dehydroamino acids as intermediates allows the possible generation of one (Ser) or two (Thr) non-canonical stereocenters from the amino acid building blocks in the final Lan- or MeLan-containing products. Based on the characterization of a handful of lanthipeptide family members, it was generally assumed that the original L α-stereocenter of these residues is inverted to D during the Michael-type addition to form a (Me)Lan with DL stereochemistry (i.e. D configuration for the α carbon of the former Ser/Thr residue and L configuration for the α carbon of the former Cys residue; (2S, 6R) for Lan and (2S, 3S, 6R) for MeLan).21 This assumption was only recently challenged by the discovery of LL stereoisomers in the enterococcal cytolysin (i.e. L configuration for both α carbons; (2R, 6R) for Lan and (2R, 3R, 6R) for MeLan, Figure 1).24 The enterococcal cytolysin is produced by clinical isolates of Enterococcus faecalis and is made up of two post-translationally modified peptides named cytolysin L and S.25 Because of its tight linkage with the virulence of E. faecalis, cytolysin has been intensively studied since the 1930s with respect to its biosynthesis and biological activities.26–30 The existence of (Me)Lan residues in cytolysin that have different stereochemistry from all other lanthipeptides with previously documented structural information was unexpected, but subsequently several other lanthipeptides have been reported that contain LL-Lan or MeLan.31, 32 Sequence comparisons and mechanistic studies suggested that the LL-stereoselectivity is encoded in the substrate sequence rather than being governed by the enzyme, which is rare for naturally occurring enzymatic reactions.33 All currently known LL-(Me)Lan structures are found in class II lanthipeptides and are formed from a Dhx-Dhx-Xxx-Xxx-Cys sequence motif (Dhx = Dha or Dhb; Xxx is any amino acid except Dha/Dhb/Cys).
The hypothesis that the unusual LL stereochemistry of the (Me)Lan in cytolysin is induced by the substrate sequence has been supported by the observation that other distantly related LanM enzymes take this sequence and convert it to products with the same LL stereochemistry. However, the possibility that the cytolysin synthetase CylM also plays a role during this process has thus far not been investigated. In this work, we provide further evidence that the dehydrated cytolysin precursor peptide adopts a pre-organized conformation that facilitates the formation of the desired cyclized product even without enzymatic catalysis, reinforcing the idea of substrate-controlled reactivity and selectivity in RiPP biosynthesis. However, we also show that CylM has coevolved with its substrate to enforce the natural stereoselectivity for lanthionine synthesis in cytolysin, even when the Dhx-Dhx-Xxx-Xxx-Cys sequence motif is changed.
RESULTS AND DISCUSSION
Dehydration of CylLS in E. coli
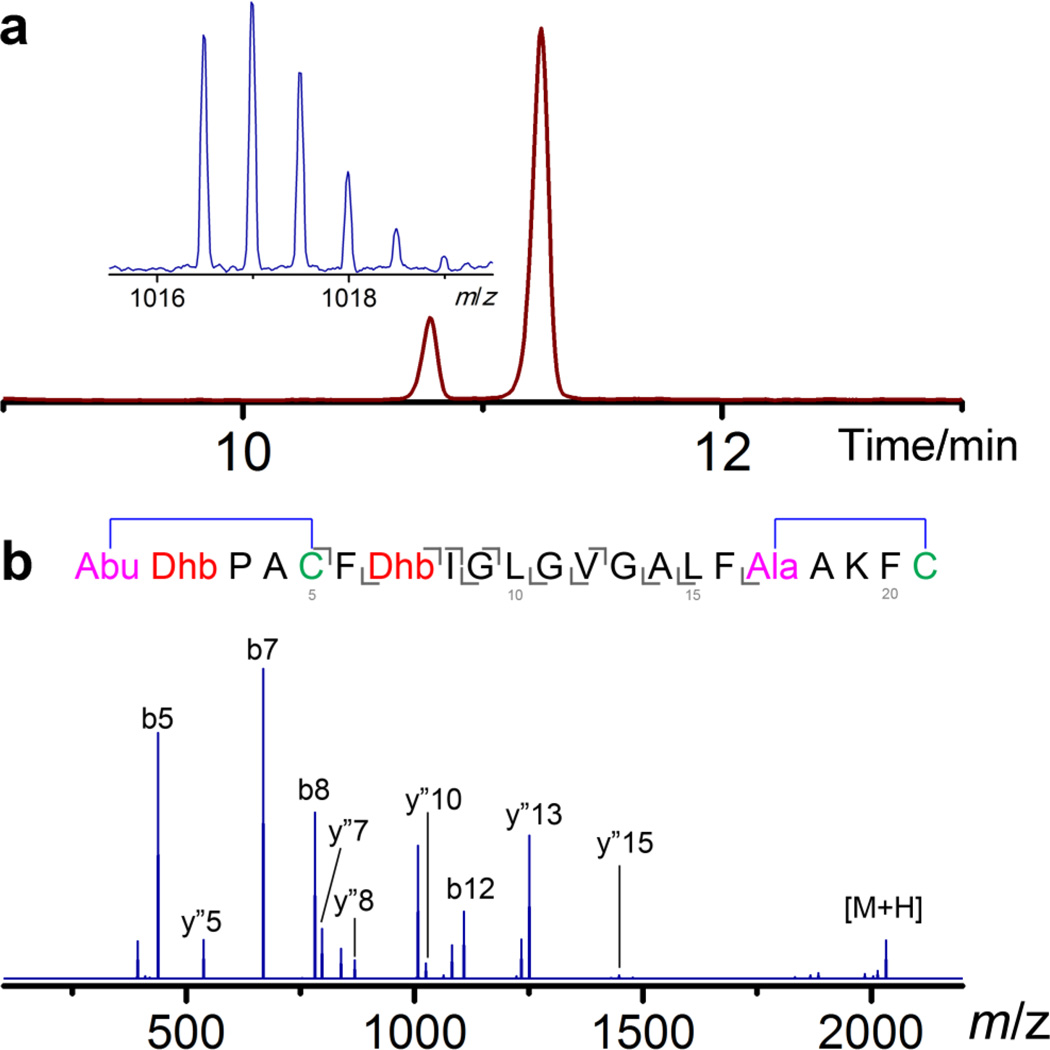
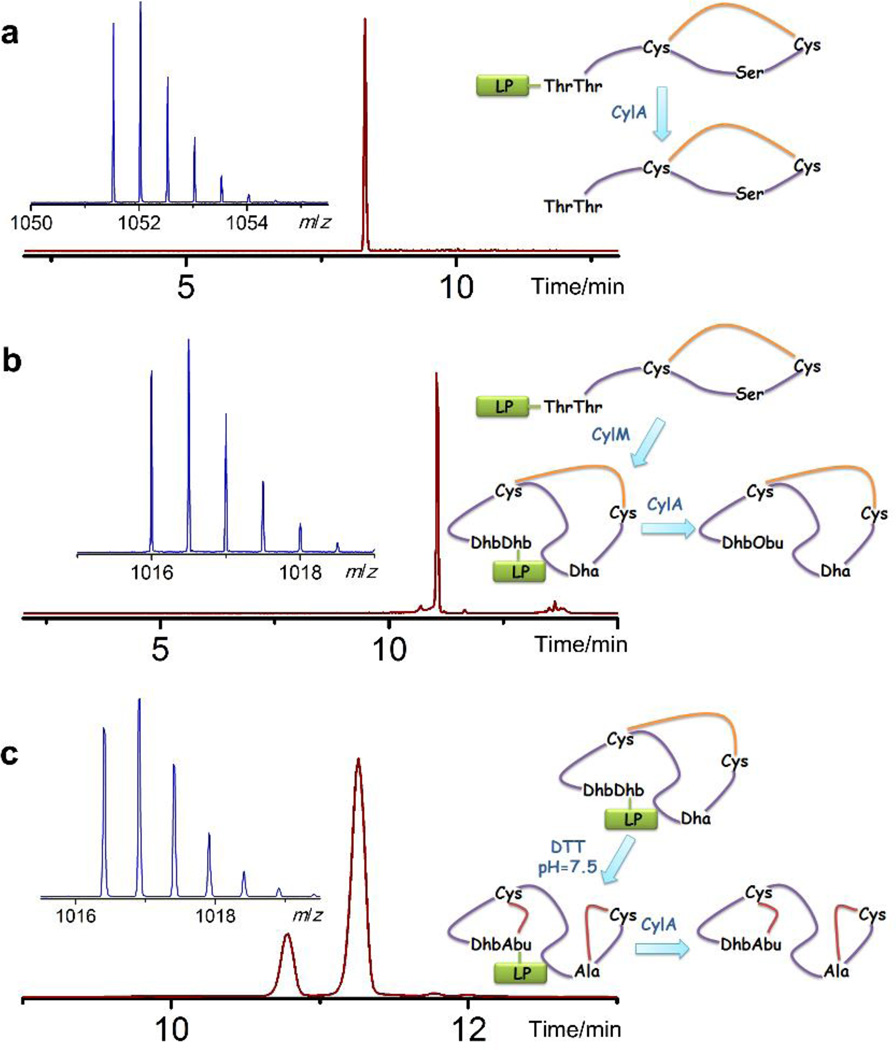
In previous work, we showed that three different lanthionine synthetases sharing limited sequence homology with CylM in their cyclase domain (<25% identity) all formed the A ring in CylLS, the precursor peptide of cytolysin S (Figure 1), with the LL stereochemistry that is found in the natural product.33 In that study, the dehydration of the CylLS peptide was executed in tandem with the cyclization reaction in E. coli, which prevents the direct investigation of the Michael-type addition process. In order to study the cyclization reaction in isolation, we first targeted obtaining dehydrated CylLS without the formation of thioether linkages. LanM proteins are bifunctional enzymes that catalyze both the dehydration and cyclization reactions in the substrate peptides and these two functions are executed by two distinct domains, an N-terminal dehydratase domain and a C-terminal cyclase domain.22, 23, 34 The crystal structure of CylM reveals that each domain has its own independent active site.35 In addition, the activity of the isolated dehydratase domain of CylM has been reconstituted in vitro,35 indicating that this domain can be catalytically competent without the partner cyclase domain. Building on these results, dehydrated CylLS was obtained by co-expression in E. coli of N-terminally His6-tagged CylLS with a truncated CylM consisting of residues 1 to 625 that encompass the dehydratase domain. The peptide was then purified using immobilized metal affinity chromatography. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis illustrated a mass shift corresponding to loss of four water molecules, consistent with what was observed for fully dehydrated CylLS (Supporting Information Figure 1). To test whether the two Cys residues in CylLS obtained using this strategy were cyclized, N-ethylmaleimide (NEM) was employed as an alkylation reagent to detect free thiols in the peptide.36 Unexpectedly, dehydrated CylLS did not react with NEM under reducing conditions (Supporting Information Figure 2), suggesting the thiols of both Cys5 and Cys21 were involved in covalent linkages other than disulfide bonds. Removal of the leader peptide by CylA, the dedicated serine protease encoded in the cytolysin biosynthetic pathway,37 allowed liquid chromatography-MS (LC-MS) analysis of the modified core peptide. One major peak with a mass corresponding to cytolysin S was observed on the chromatogram and the retention time matched that of cytolysin S produced by full length CylM (Figure 2a and Supporting Information Figure 3). Notably, a minor peak with the same mass but a different retention time compared to cytolysin S was also detected on the chromatogram (Figure 2a). Electrospray ionization (ESI) MS/MS analyses of both peaks revealed a similar fragmentation pattern as native cytolysin S (Figure 2b and Supporting Information Figure 3), indicating that both products obtained in this way were cyclized and that the regioselectivity of cyclization was the same as that in the reaction catalyzed by full length CylM. We next examined the stereochemistry of (Me)Lan residues. The peptide was hydrolyzed in 6 M HCl, and individual amino acids were derivatized and subjected to gas chromatography-MS (GC-MS) analysis with a chiral stationary phase. Lan and MeLan signals were extracted from the total ion chromatogram using their characteristic fragment masses and their stereochemistry was assigned by comparing the retention time with those of synthetic standards that were derivatized using the same procedure. This examination showed that the major product contained an LL-MeLan and a DL-Lan, like the native compound, but that the minor product contains an LL-MeLan and an LL-Lan (Supporting Information Figures 4, 5 and 6). These observations were quite surprising as they indicate that regioselective cyclization of CylLS could be achieved without the cyclase domain, and suggested that dehydrated CylLS attains an organized conformation that auto-cyclizes even without the presence of a catalyst.
Figure 2. ESI MS analyses of cytolysin S peptide modified by the CylM dehydratase domain in E. coli.
a) Extracted ion chromatogram (m/z = 1017) of CylLS core peptide modified by the CylM dehydratase domain. The mass spectrum corresponding to the major peak is shown in the insert. For fully modified CylLS core peptide, calculated [M–4H2O+2H]2+: 1,016.52, monoisotopic mass; observed [M–4H2O+2H]2+: 1,016.51, monoisotopic mass. b) MS/MS analysis of the major peak on the LC chromatogram and a proposed structure deduced from the fragmentation pattern. Both MS and MS/MS results suggest that the dehydrated CylLS purified from E. coli is in its cyclized form.
Non-enzymatic cyclization of dehydrated CylLS
The unexpected observation that dehydrated CylLS was completely cyclized even in the absence of the cyclization domain may have several explanations. First, it is possible that the dehydratase domain may catalyze the cyclization. Second, another protein in E. coli may have promoted the cyclization, and third, the cyclization may have occurred non-enzymatically. To investigate the latter, most probable possibility, the cyclization reaction of dehydrated CylLS was conducted in vitro. The two cysteines in CylLS were first protected as an intramolecular disulfide (Figure 3a), and the oxidized CylLS was supplied to CylM. CylM accepted oxidized CylLS as substrate and eliminated four water molecules (Figures 3b and Supporting Information Figure 7), albeit with a lower efficiency compared to linear reduced CylLS; as no free thiols were available, cyclization could not take place. The 4-fold dehydrated and disulfide-containing CylLS peptide was then purified by reversed phase high performance liquid chromatography (RP-HPLC) (Supporting Information Figure 8). Subsequent incubation with dithiothreitol at pH 7.5 released the two Cys thiols and allowed non-enzymatic cyclization to proceed. Free thiols could no longer be detected after 12 hours of incubation at room temperature (Supporting Information Figure 9). Treatment of the peptide with the peptidase CylA and analysis of the core peptide using LC-MS again revealed a major peak on the LC chromatogram corresponding to cytolysin S as well as a minor peak with the same mass (Figure 3c). MS/MS analyses of both products demonstrated the formation of the correct ring topology (Supporting Information Figure 10). Thus, the non-enzymatic in vitro results are in good agreement with the in vivo observations with CylM-1-625, suggesting that the dehydrated CylLS produced by the dehydratase domain of CylM cyclizes non-enzymatically into two diastereomers with the correct ring topology.
Figure 3. Non-enzymatic cyclization of cytolysin S peptide in vitro.
a) ESI LC-MS analysis of the linear core peptide of CylLS in its oxidized (disulfide) form. Extracted ion chromatogram (m/z = 1052) of the LC trace for oxidized CylLS core peptide is shown in dark red and the corresponding mass spectrum in the insert. No peak corresponding to reduced CylLS core peptide was detected in the LC trace. Oxidized CylLS core peptide with a disulfide linkage, calculated [M+2H]2+: 1,051.53, monoisotopic mass; observed [M+2H]2+: 1,051.53, monoisotopic mass. b) ESI LC-MS analysis of oxidized and dehydrated core peptide of CylLS. Extracted ion chromatogram (m/z = 1016.5) of the LC trace is shown with the corresponding mass spectrum inserted. Dhb1 was hydrolyzed to a ketone after leader peptide removal, resulting in a mass increase of 1 Da. Disulfide containing and dehydrated CylLS core peptide (Dhb hydrolyzed), calculated [M–4H2O–NH+O+2H]2+: 1,016.00, monoisotopic mass; observed [M–4H2O–NH+O+2H]2+: 1,016.00, monoisotopic mass. c) ESI LC-MS analysis of non-enzymatically cyclized CylLS core peptide. Extracted ion chromatogram (m/z = 1017) of the LC trace is shown. The mass spectrum corresponding to the major peak is shown in the insert. For fully cyclized cytolysin S, calculated [M–4H2O+2H]2+: 1,016.52, monoisotopic mass; observed [M–4H2O+2H]2+: 1,016.40, monoisotopic mass. Obu: 2-oxobutyrate, LP: leader peptide. Yellow line represents disulfide.
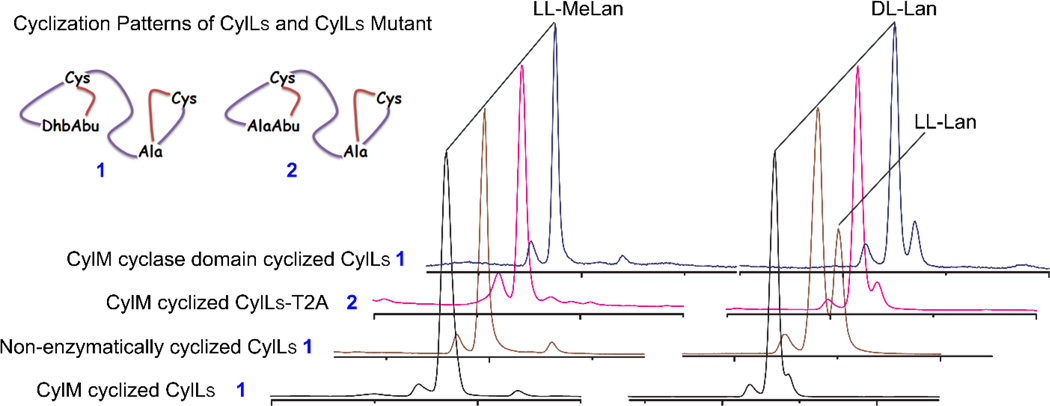
These observations at first glance raise the question whether the CylM cyclase domain is required for cytolysin S synthesis. However, both in vitro and in vivo non-enzymatic cyclization resulted in a minor product corresponding to a diastereomer of cytolysin S, where the configuration of the Lan residue in the B ring was LL rather than DL found in native cytolysin S. This peptide is never observed in vivo or in vitro for reactions with full length CylM. Therefore, we wondered whether the CylM cyclase domain could improve the fidelity for the cyclization of the B ring in CylLS. To test this hypothesis, the CylM cyclase domain consisting of residues 626 to 993 and the CylM dehydratase domain were expressed together with CylLS and allowed to modify the peptide in trans in E. coli. Fully dehydrated and cyclized product was obtained (Supporting Information Figures 11 and 12), and indeed, the minor peak corresponding to the diastereomer of cytolysin S was considerably diminished in intensity when analyzing the core peptide by LC-MS (Supporting Information Figure 13). The LL-Lan signal on the chromatogram also returned to a basal level (Supporting Information Figure 14), resulting in a similar GC trace as what was observed for CylM-modified CylLS. Collectively, these results suggest a role of the CylM cyclase domain in CylM-catalyzed modifications with respect to the stereochemical fidelity of Lan formation.
Mutation of the Dhx-Dhx-Xxx-Xxx-Cys motif
Based on previous studies,31, 33 the enzymatic formation of the non-canonical LL stereochemistry in several lanthipeptides has been linked to a Dhx-Dhx-Xxx-Xxx-Cys motif present in the substrate peptide. The second dehydroamino acid in such a motif was shown to be important for the stereoselective cyclization of the A ring of the lanthipeptide haloduracin β (Halβ).33 To test whether the second Dhb is also important for CylM-catalyzed cyclization of the Dhb-Dhb-Pro-Ala-Cys sequence (the dehydrated precursor to the A ring of cytolysin S, Figure 1), Thr2 was substituted by Ala in CylLS. In Halβ, such a mutation resulted in an inversion of the facial selectivity of the Michael-type addition catalyzed by HalM2.33 CylLS-T2A was modified by CylM in E. coli, and the resulting peptide was purified. MALDI-TOF MS analysis indicated the desired loss of three water molecules and no free thiols were present in the peptide (Supporting Information Figures 15 and 16). The modified CylLS-T2A peptide was then hydrolyzed in acid, and the amino acids were derivatized and analyzed by GC-MS. Surprisingly, signals corresponding to LL-MeLan and DL-Lan were detected (Figure 4 and Supporting Information Figure 17), similar to what was observed for native cytolysin S. The selective formation of LL-MeLan in the A ring of CylLS-T2A by CylM can be explained in two ways: 1) the second Dhb is not required in the Dhb-Dhb-Pro-Ala-Cys sequence to form an LL-MeLan; and/or 2) the synthetase CylM has a preference for the LL stereochemistry in the A ring and catalyzes the formation of LL-MeLan even when the second Dhb is missing in the substrate motif.
Figure 4. GC-MS traces for hydrolyzed and derivatized (Me)Lan residues from CylLS and CylLS–T2A peptides that were cyclized using different set ups.
Gas chromatograms showing signals corresponding to derivatized MeLan (left panel) and Lan (right panel) from CylLS peptides cyclized by CylM (black trace)24, by the CylM cyclase domain (blue trace) or without a cyclase (brown trace), and CylLS–T2A peptide cyclized by CylM (magenta trace) are overlaid. For co-injection traces with synthetic (Me)Lan standards that were used for stereochemistry assignments, see Supporting Information Figures 4, 14 and 17.
To attempt to differentiate these two possibilities, we first expressed CylLS-T2A together with the CylM dehydratase domain in E. coli. As expected three dehydrations were observed, corresponding to fully dehydrated CylLS-T2A (Supporting Information Figure 18). However, NEM analysis indicated one of the two thiols in the peptide remained non-cyclized (Supporting Information Figure 19), different from what was observed for wild type CylLS modified by the CylM dehydratase domain and for CylLS-T2A modified by full length CylM. Removal of the leader peptide and MS analysis of the resulting mutated core peptide confirmed that it was the A ring that was not properly cyclized as hydrolysis of the N-terminal Dhb to 2-oxobutyrate after leader peptide removal was exclusively observed (Supporting Information Figure 20). The partially cyclized CylLS-T2A peptide was purified and non-enzymatic cyclization of the A ring was pursued in vitro. Unlike the facile non-enzymatic cyclization of the A ring in the wild-type sequence described above, very little ring closure was observed after 15 hours of incubation at pH 9 (Supporting Information Figure 21). Collectively, our results suggest that the cyclization of the A ring of dehydrated CylLS-T2A requires the cyclase domain of CylM, and that non-enzymatic cyclization is greatly aided by the second (underlined) Dhb in the Dhb-Dhb-Pro-Ala-Cys motif.
The inability to cyclize the A ring of CylLS-T2A non-enzymatically prevented determination whether cyclization of the Dhb-Ala-Pro-Ala-Cys sequence in dehydrated CylLS-T2A to form an LL-MeLan residue is controlled by CylM. A second question is whether this activity is unique to this enzyme. To answer the latter question, we tested whether a different LanM, the Halβ synthetase HalM2, would modify CylLS-T2A to form an LL-MeLan in the A ring. A gene encoding a chimeric peptide with the leader peptide of Halβ connected to the CylLS-T2A core peptide was constructed and co-expressed with HalM2 in E. coli. HalM2 carried out the desired three dehydrations of the CylLS-T2A core peptide (Supporting Information Figure 22). The efficiency of the cyclization reaction was evaluated using the NEM assay. Partially cyclized HalA2-CylLS-T2A peptide with one free thiol was detected almost exclusively (Supporting Information Figure 23), suggesting that HalM2 is also inefficient in catalyzing the cyclization of the A ring of dehydrated CylLS-T2A.
Previous studies have demonstrated that both enzymatic and non-enzymatic cyclization involving a Dha is much more facile than cyclization onto a Dhb.38, 39 Thus, in a last effort to achieve cyclization of the A ring of cytolysin S in the absence of two consecutive Dhx residues we generated the double mutant CylLS-T1S/T2A, which after dehydration would generate a Dha-Ala-Pro-Ala-Cys sequence. The peptide was co-expressed individually with either CylM-1-625 or HalM2 in E. coli. Both proteins carried out the desired three dehydrations and evaluation of cyclization by the NEM assay as well as tandem MS analysis also showed that both rings were formed (Supporting Information Figures 24 and 25). Hence, the increased reactivity of Dha indeed resulted in cyclization of the A ring. Subsequent acid hydrolysis of the peptide, derivatization of the amino acids, and analysis by GC-MS demonstrated that the experiment with CylM-1-625 generated predominantly DL-Lan (Supporting Information Figure 26), indicating that non-enzymatic cyclization of the Dha-Ala-Pro-Ala-Cys sequence preferentially provides the canonical stereochemistry. However, the experiment with HalM2 generated a near 1:1 ratio of DL-Lan and LL-Lan (Supporting Information Figure 27). The most likely explanation of these data is that the A ring is formed with LL stereochemistry and the B ring with DL stereochemistry, indicating that, like CylM, HalM2 catalyzes the enzymatic cyclization of the Dha-Ala-Pro-Ala-Cys sequence to provide the LL stereochemistry even in the absence of the second Dhx. Thus, both proteins impart onto the substrate a preference for attack by the thiol/thiolate onto the Re face of Dha even when non-enzymatic cyclization preferentially occurs by attack onto the Si face. Collectively, the data in this study clearly show that whereas the Dhx-Dhx-Xxx-Xxx-Cys motif indeed has an inherent preference for LL stereochemistry, the enzymes investigated further enforce this preference even without the motif.
Ring size requirement for LL stereochemistry
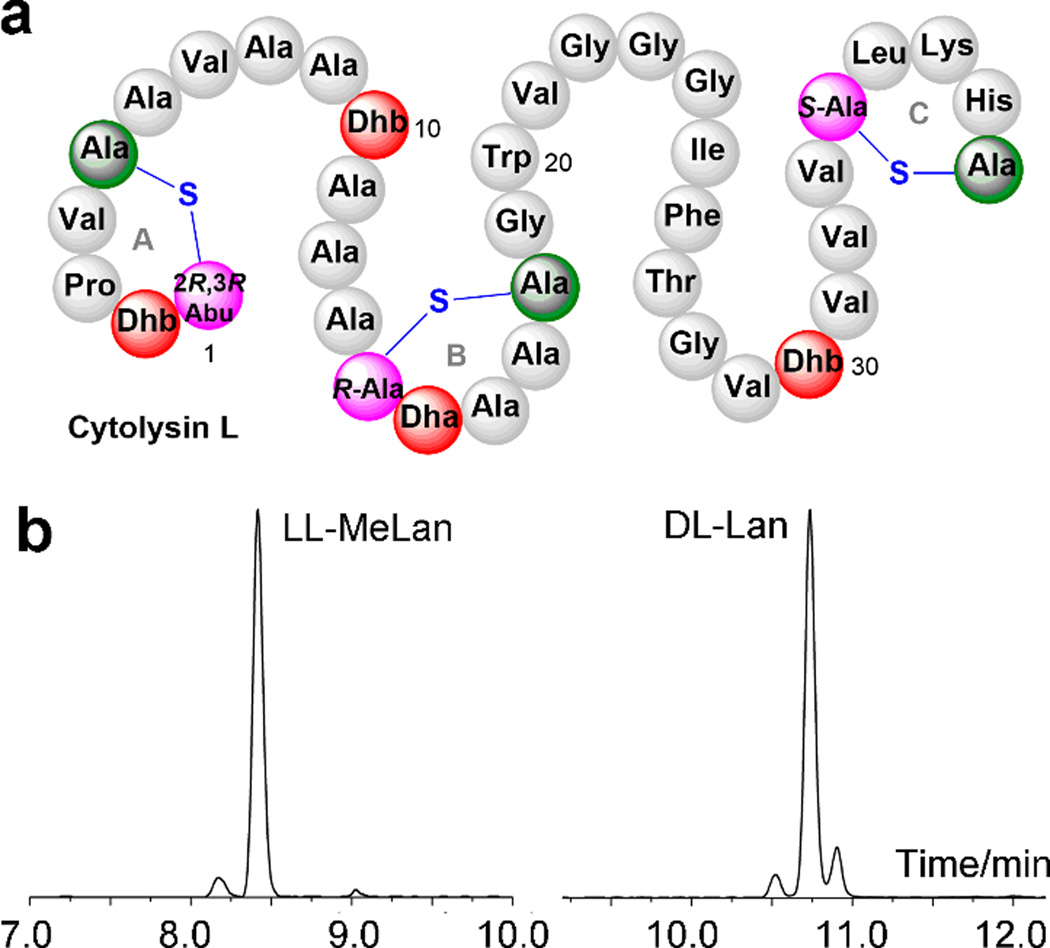
Thus far, all naturally occurring LL-(Me)Lan residues have been identified in five amino acid-rings. We therefore decided to probe whether this ring size is important for lanthionine synthetases to produce thioether links with the non-canonical configuration. The large subunit of cytolysin, cytolysin L (Figure 5a) contains two rings with LL stereochemistry, a MeLan A ring and a Lan B ring. Given the higher reactivity for Lan formation as discussed in the previous section, we therefore generated a series of mutants of the CylLL B ring with altered sizes. We first investigated ring contraction by deleting Ala16. Its precursor peptide CylLL-A16del was co-expressed with CylM in E. coli and the desired seven dehydrations were observed (Supporting Information Figure 28a). NEM analysis confirmed that the CylM-modified CylLL-A16del peptide was fully cyclized (Supporting Information Figure 28b). Acid hydrolysis of the modified peptide and derivatization of the individual amino acids were followed by analysis by GC-MS. DL stereochemistry was exclusively observed for the Lan signal (Figure 5b), suggesting that CylM cyclized the four-amino-acid B ring of cytolysin L to form a lanthionine residue with the DL configuration. As expected, a signal corresponding to LL-MeLan (derived from the A ring) was also observed on the chromatogram (Figure 5b and Supporting Information Figure 29). To construct an expanded B ring in cytolysin L, an alanine was inserted after Ser15, resulting in a Dha14-Dha15-Ala-Ala-Ala-Cys sequence. To be able to determine which dehydroamino acid was cyclized with the Cys residue in this sequence, Ser14 or Ser15 were individually mutated to Thr, which would then result in either a MeLan (cyclization with Dhb) or Lan (cyclization with Dha). CylLL-S14T-A16ins and CylLL-S15T-A16ins were co-expressed in E. coli with CylM and the desired 7-fold dehydrated peptides were produced (Supporting Information Figures 30a and 30b). Free thiols were not detected in either peptide using the NEM assay, indicating complete cyclization (Supporting Information Figure 30c). The modified peptides were then hydrolyzed, and the resulting amino acids derivatized and subjected to GC-MS analysis. For CylM-modified CylLL-S15T-A16ins, signals corresponding to LL-MeLan and DL-Lan were observed (Supporting Information Figure 31), whereas for CylM-modified CylLL-S14T-A16ins, DL-Lan and a 1:1 ratio of DL- and LL-MeLan were observed in the chromatogram (Supporting Information Figure 32). The most straightforward interpretation of these results is that the Cys reacts with the underlined dehydroamino acid in Dhx14-Dhx15-Ala-Ala-Ala-Cys. In CylLL-S15T-A16ins, this results in a DL-Lan B ring (Dhx14 = Dha), to go along with an LL-MeLan for the A ring and a DL-Lan for the C-ring. For CylLL-S14T-A16ins, this results in a DL-MeLan (Dhx14= Dhb), again along with an LL-MeLan for ring A (resulting in the 1:1 observed ratio for LL and DL-MeLan) and DL-Lan for ring C. Importantly, regardless of the mutant, the ring in such a six amino acid ring was synthesized with the canonical DL configuration. Collectively, this suggests that the ring size with five amino acids is required for CylM to catalyze the formation of the unusual LL stereochemistry, at least for the B ring of CylLL. We also note that cyclization occurred in both cases on Dhx14, rather than Dhx15 which would have given a five-amino-acid B ring as found in native cytolysin L. These observations are in accord with the aforementioned lower reactivity of a Dhx-Ala-Xxx-Xxx-Cys sequence compared to a Dhx-Dhx-Xxx-Xxx-Cys sequence. In addition, these findings suggest that two consecutive Dhx residues may generally increase the reactivity of the first Dhx, either electronically or because of conformational preferences.
Figure 5. Stereochemical characterization of CylM-modified CylLL-16Adel.
a) Structure of cytolysin L. b) GC-MS traces for hydrolyzed and derivatized (Me)Lan residues from CylM-modified CylLL-16Adel. For co-injection traces with synthetic (Me)Lan standards that were used for stereochemistry assignments, see Supporting Information Figure 29.
Discussion
In this study, we provide additional support for the hypothesis that the substrate controls in part the regioselectivity and stereoselectivity of ring formation, but we also show that the enzymes clearly play important roles as well. First, the successful non-enzymatic cyclization of dehydrated CylLS, both in E. coli and in vitro, confirmed that a substrate containing a Dhb-Dhb-Pro-Ala-Cys sequence inherently prefers formation of the LL stereochemistry as predicted computationally (Table 1).33 The in vitro auto-cyclization of dehydrated CylLS proceeded efficiently at nearly neutral pH, which was unexpected before this work because of results obtained for another group of lanthipeptides, the prochlorosins, where the non-enzymatic Michael-type addition required a higher pH to activate the thiols and was extremely slow, especially for the formation of MeLan residues.40 Indeed, we also observed very slow non-enzymatic cyclization for the MeLan A ring of dehydrated CylLS-T2A substrate. Our observations for wild type cytolysin S suggest that dehydrated CylLS can access a conformation with increased reactivity, which allows the cyclization of both rings to proceed efficiently even in the absence of an enzyme catalyst. Previous computational analysis demonstrated that in the optimal transition state structure of Re-face attack of the thiol on the alkene of Dhb en route towards the LL-MeLan A ring of cytolysin S, the negative charge of the enolate intermediate is efficiently stabilized.33 The hydrogen bonding network that stabilizes this enolate accounts significantly for the lower activation energy barrier for the Michael-type addition to yield LL stereochemistry and might be the reason why this reaction can proceed efficiently and with the same stereochemistry even without a cyclase.
Table 1.
Product summary of CylLS, CylLS–T2A and CylLS–T1S/T2A peptides that were cyclized under different conditions.a
| Substrate | Modification enzymes | A ringb | B ring | |||
|---|---|---|---|---|---|---|
| Dehydration | Cyclization | thioether | stereochemistry | thioether | stereochemistry | |
| CylLS | CylM | CylM | MeLan | LL | Lan | DL |
| HalM2 | HalM2 | MeLan | LL | Lan | DL | |
| CylM dehydratase domain |
Non- enzymatically |
MeLan | LL | Lan | DL (~ 80%)c LL (~ 20%) |
|
| CylM dehydratase domain |
CylM cyclase domain (in trans) |
MeLan | LL | Lan | DL | |
| CylLS- T2A |
CylM | CylM | MeLan | LL | Lan | DL |
| HalM2 | HalM2 | Not formed |
N/Ad | Lan | NDe | |
| CylM dehydratase domain |
Non- enzymatically |
Not formed |
N/A | Lan | ND | |
| CylLS- T1A/T2A |
HalM2 | HalM2 | Lan | LL (major) DL (minor) |
Lan | DL |
| CylM dehydratase domain |
Non- enzymatically |
Lan | DL (major) LL (minor) |
Lan | DL | |
Ring topologies of the final products were deduced form MS/MS fragmentation patterns, whereas configurations of (Me)Lan rings were assigned using GC-MS.
Under all tested conditions, CylLS and its variants cyclized with the same ring topology as that found in the natural cytolysin S.
Ratio of products was estimated based on integration of the LC trace.
N/A: not applicable.
ND: not determined.
Second, the formation of the correct ring topology for cytolysin S by non-enzymatic cyclization provides further support for our previous hypothesis of substrate-controlled selectivity in the lanthipeptide family,36 and suggests that both regioselectivity and stereoselectivity for cytolysin S synthesis are encoded, at least in part, in the CylLS sequence.41 There are 144 possible constitutional isomeric products of the cyclization process of dehydrated CylLS, considering different ring topologies and stereochemistries of (Me)Lan residues with a fixed L configuration for the former Cys residue that does not alter during the process. However, only one major product was observed for non-enzymatically cyclized CylLS, suggesting that the conformational energy landscape of dehydrated CylLS is such that the transition states leading to the natural product efficiently outcompete the other 143 possibilities and dominate the reaction outcome. Collectively, these observations reinforce the idea of reactivity and selectivity for lanthipeptide biosynthesis in which the sequence of the substrate is important for the final outcome.
Third, we show that having two consecutive dehydroamino acids alone is not sufficient for obtaining the LL stereochemistry. Only when those amino acids are imbedded at the start of a five amino acid sequence with a Cys at the 5th position, was cyclization by anti addition on the Re face of the alkene observed. When the Dhx-Dhx sequence was present at the start of either a four or six amino acid sequence, the canonical DL stereochemistry was obtained.
Fourth, our results show that the dehydratase and cyclase activities of CylM can be independently accessed using truncated CylM proteins. Although cyclization of dehydrated CylLS could proceed non-enzymatically with similar regioselectivity and stereoselectivity as the native enzyme-synthesized product, the cyclase domain is clearly important since the stereochemical fidelity of B ring formation was improved over non-enzymatic cyclization in the presence of the C-terminal domain (Table 1). In addition to ensuring the correct stereochemistry of the B ring of CylLS, the CylM cyclase domain can overcome the inherent stereoselectivity of cyclization that is encoded in the substrate for the A ring (Table 1). The second dehydroamino acid in a Dhx-Dhx-Xxx-Xxx-Cys motif (underlined) has been proposed to be important for the formation of LL-(Me)Lan as mutation of the second Dhb to Ala in its natural substrate HalA2 resulted in an inversion of the face selectivity of cyclization during formation of the A ring of Halβ by HalM2.24 However, in this work, we show that CylM cyclizes the A ring of CylLS-T2A to form an LL-MeLan residue even when the second Dhb is replaced by Ala. To determine if such stereoselectivity is perhaps still inherent to the substrate sequence, we tried to cyclize the mutated A ring non-enzymatically, but unfortunately, the Dhb-Ala-Pro-Ala-Cys sequence was very unreactive towards non-enzymatic cyclization. We therefore employed a double mutant to obtain a more reactive Dha-Ala-Pro-Ala-Cys sequence (A ring formed from CylLS-T1S/T2A). This substrate cyclized when co-expressed with CylM dehydratase domain, which reports on non-enzymatic cyclization, and the product formed contained mostly DL-Lan. Thus, CylM generates an A ring with LL stereochemistry when the substrate inherently prefers the DL stereochemistry. Co-expression of CylLS-T1S/T2A with HalM2 also generated predominately the LL-Lan isomer for the A ring, and hence this selectivity is not specific to CylM. Collectively, these observations indicate that the stereoselectivity of CylM is more complicated than a simple preference of the substrate for a certain configuration. The outcome with the CylLS-T2A mutant is puzzling since CylM cyclized this sequence to generate the LL stereochemistry for the A ring while at the same time converting the Dha-Ala-Lys-Phe-Cys sequence of the B ring to a DL-Lan. It appears that the enzyme has evolved to provide LL stereochemistry at the N-terminus of the core peptide and DL stereochemistry at the C-terminus. The reason why CylM coevolved with its substrates to introduce the LL stereochemistry remains unknown. Both subunits of cytolysin (L and S) contain the unusual LL stereochemistry, and cytolysin is unique in the lanthipeptide family for its lytic activity against eukaryotic cells.16, 24 It is tempting to speculate that the unusual LL stereochemistry may therefore be an essential feature that accounts for the virulence activity of cytolysin. In that scenario, the LL stereochemistry probably originally arose from the inherent preference of the substrate, but during evolution the enzyme co-evolved to help enforce the LL stereochemistry such that now even when the dominant motif is removed, the enzyme still forms the LL product. Whether there is indeed a structure-activity correlation between the unusual LL stereochemistry and the lytic activity of cytolysin requires further investigation.
METHODS
Materials and experimental procedures are described in Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01 GM058822 to W.v.d.D.)
Footnotes
Supporting Information
Detailed procedures and Supporting Figures and Table. The Supporting Information is available free of charge on the ACS Publication website at DOI:
References
- 1.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs from 1981 to 2014. J. Nat. Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RE, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velásquez JE, van der Donk WA. Genome mining for ribosomally synthesized natural products. Curr. Opin. Chem. Biol. 2011;15:11–21. doi: 10.1016/j.cbpa.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega MA, van der Donk WA. New Insights into the Biosynthetic Logic of Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products. Cell Chem. Biol. 2016;23:31–44. doi: 10.1016/j.chembiol.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Nat. Prod. Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freeman MF, Gurgui C, Helf MJ, Morinaka BI, Uria AR, Oldham NJ, Sahl HG, Matsunaga S, Piel J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338:387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 7.Sit CS, McKay RT, Hill C, Ross RP, Vederas JC. The 3D structure of thuricin CD, a two-component bacteriocin with cysteine sulfur to alpha-carbon cross-links. J. Am. Chem. Soc. 2011;133:7680–7683. doi: 10.1021/ja201802f. [DOI] [PubMed] [Google Scholar]
- 8.Lohans CT, Vederas JC. Structural characterization of thioether-bridged bacteriocins. J. Antibiot. 2014;67:23–30. doi: 10.1038/ja.2013.81. [DOI] [PubMed] [Google Scholar]
- 9.Skaugen M, Nissenmeyer J, Jung G, Stevanovic S, Sletten K, Abildgaard CIM, Nes IF. In-Vivo Conversion of L-Serine to D-Alanine in a Ribosomally Synthesized Polypeptide. J. Biol. Chem. 1994;269:27183–27185. [PubMed] [Google Scholar]
- 10.Kawulka K, Sprules T, McKay RT, Mercier P, Diaper CM, Zuber P, Vederas JC. Structure of Subtilosin A, an Antimicrobial Peptide from Bacillus subtilis with Unusual Posttranslational Modifications Linking Cysteine Sulfurs to a-Carbons of Phenylalanine and Threonine. J. Am. Chem. Soc. 2003;125:4726–4727. doi: 10.1021/ja029654t. [DOI] [PubMed] [Google Scholar]
- 11.Martin NI, Sprules T, Carpenter MR, Cotter PD, Hill C, Ross RP, Vederas JC. Structural characterization of lacticin 3147, a two-peptide lantibiotic with synergistic activity. Biochemistry. 2004;43:3049–3056. doi: 10.1021/bi0362065. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, van der Donk WA. Post-translational Introduction of D-Alanine into Ribosomally Synthesized Peptides by the Dehydroalanine Reductase NpnJ. J. Am. Chem. Soc. 2015;137:12426–12429. doi: 10.1021/jacs.5b05207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo L, van der Donk WA. Discovery and Characterization of Bicereucin, an Unusual d-Amino Acid-Containing Mixed Two-Component Lantibiotic. J. Am. Chem. Soc. 2016;138:5254–5257. doi: 10.1021/jacs.6b02513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milne BF, Long PF, Starcevic A, Hranueli D, Jaspars M. Spontaneity in the patellamide biosynthetic pathway. Org. Biomol. Chem. 2006;4:631–638. doi: 10.1039/b515938e. [DOI] [PubMed] [Google Scholar]
- 15.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 16.Bierbaum G, Sahl HG. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 2009;10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 17.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2015;517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehnke J, Mann G, Bent AF, Ludewig H, Shirran S, Botting C, Lebl T, Houssen WE, Jaspars M, Naismith JH. Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat. Chem. Biol. 2015;11:558–563. doi: 10.1038/nchembio.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thibodeaux CJ, Wagoner J, Yu Y, van der Donk WA. The Leader Peptide Establishes Dehydration Order, Promotes Efficiency, and Ensures Fidelity During Lacticin 481 Biosynthesis. J. Am. Chem. Soc. 2016 doi: 10.1021/jacs.6b00163. 10.1021/jacs.1026b00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee C, Paul M, Xie L, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 2005;105:633–684. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 22.Siezen RJ, Kuipers OP, de Vos WM. Comparison of lantibiotic gene clusters and encoded proteins. Antonie van Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 23.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Lacticin 481: in vitro reconstitution of lantibiotic synthetase activity. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 24.Tang W, van der Donk WA. The sequence of the enterococcal cytolysin imparts unusual lanthionine stereochemistry. Nat. Chem. Biol. 2013;9:157–159. doi: 10.1038/nchembio.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox CR, Coburn PS, Gilmore MS. Enterococcal cytolysin: A novel two component peptide system that serves as a bacterial defense against eukaryotic and prokaryotic cells. Curr. Protein Pept. Sci. 2005;6:77–84. doi: 10.2174/1389203053027557. [DOI] [PubMed] [Google Scholar]
- 26.Todd EW. A comparative serological study of streptolysins derived from human and from animal infections, with notes on pneumococcal h ae molysin, tetanolysin and staphylococcus toxin. J. Pathol. Bateriol. 1934;39:299–321. [Google Scholar]
- 27.Ike Y, Clewell DB. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis using transposon Tn917 as an insertional mutagen. J. Bacteriol. 1984;158:777–783. doi: 10.1128/jb.158.3.777-783.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huycke MM, Spiegel CA, Gilmore MS. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis, Antimicrob. Agents Chemother. 1991;35:1626–1634. doi: 10.1128/aac.35.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, Zervos MJ. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 1993;37:2474–2477. doi: 10.1128/aac.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Tyne D, Martin MJ, Gilmore MS. Structure, function, and biology of the enterococcus faecalis cytolysin. Toxins. 2013;5:895–911. doi: 10.3390/toxins5050895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohans CT, Li JL, Vederas JC. Structure and Biosynthesis of Carnolysin, a Homologue of Enterococcal Cytolysin with d-Amino Acids. J. Am. Chem. Soc. 2014;136:13150–13153. doi: 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- 32.Zhao X, van der Donk WA. Structural Characterization and Bioactivity Analysis of the Two-Component Lantibiotic Flv System from a Ruminant Bacterium. Cell Chem. Biol. 2016;23:246–256. doi: 10.1016/j.chembiol.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang W, Jiménez-Osés G, Houk KN, van der Donk WA. Substrate control in stereoselective lanthionine biosynthesis. Nat. Chem. 2015;7:57–64. doi: 10.1038/nchem.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.You YO, van der Donk WA. Mechanistic investigations of the dehydration reaction of lacticin 481 synthetase using site-directed mutagenesis. Biochemistry. 2007;46:5991–6000. doi: 10.1021/bi602663x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong SH, Tang W, Lukk T, Yu Y, Nair SK, van der Donk WA. The enterococcal cytolysin synthetase has an unanticipated lipid kinase fold. eLife. 2015;4:e07607. doi: 10.7554/eLife.07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thibodeaux CJ, Ha T, van der Donk WA. A price to pay for relaxed substrate specificity: a comparative kinetic analysis of the class II lanthipeptide synthetases ProcM and HalM2. J. Am. Chem. Soc. 2014;136:17513–17529. doi: 10.1021/ja5089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Booth MC, Bogie CP, Sahl H-G, Siezen RJ, Hatter KL, Gilmore MS. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol. Microbiol. 1996;21:1175–1184. doi: 10.1046/j.1365-2958.1996.831449.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhou H, van der Donk WA. Biomimetic stereoselective formation of methyllanthionine. Org. Lett. 2002;4:1335–1338. doi: 10.1021/ol025629g. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Ni W, van der Donk WA. On the regioselectivity of thioether formation by lacticin 481 synthetase. Org. Lett. 2007;9:3343–3346. doi: 10.1021/ol071301h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee S, van der Donk WA. Mechanistic Studies on the Substrate-Tolerant Lanthipeptide Synthetase ProcM. J. Am. Chem. Soc. 2014;136:10450–10459. doi: 10.1021/ja504692v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Yu Y, Velásquez JE, van der Donk WA. Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18361–18366. doi: 10.1073/pnas.1210393109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.