Abstract
Mutations in the isocitrate dehydrogenase gene IDH1 are common in lower-grade glioma where they result in the production of 2-hydroxyglutarate (2HG), disrupted patterns of histone methylation and gliomagenesis. IDH1 mutations also co-segregate with mutations in the ATRX gene and the TERT promoter, suggesting that IDH mutation may drive the creation or selection of telomere-stabilizing events as part of immortalization/transformation process. To determine if and how this may occur, we investigated the phenotype of pRb/p53-deficient human astrocytes engineered with IDH1 wild-type (WT) or R132H mutant (IDH1mut) genes as they progressed through their lifespan. IDH1mut expression promoted 2HG production and altered histone methylation within 20 population doublings (PD), but had no effect on telomerase expression or telomere length. Accordingly, cells expressing either IDH1 WT or IDH1mut entered a telomere-induced crisis at PD 70. In contrast, only IDH1mut cells emerged from crisis, grew indefinitely in culture and formed colonies in soft agar and tumors in vivo. Clonal populations of post-crisis IDH1mut cells displayed shared genetic alterations, but no mutations in ATRX or the TERT promoter were detected. Instead, these cells reactivated telomerase and stabilized their telomeres in association with increased histone lysine methylation (H3K4me3) and c-Myc/Max binding at the TERT promoter. Overall, these results show that while IDH1mut does not create or select for ATRX or TERT promoter mutations, it can indirectly reactivate TERT, and in doing so contribute to astrocytic immortalization and transformation.
Keywords: IDH1, hTERT, transformation, 2HG, glioma
Introduction
Gliomas make up 80% of all malignant brain tumors, and remain among the most fatal of human tumors (1). Although gliomas have historically been divided into histologic sub-types based purely on morphologic resemblance to non-neoplastic counterparts, recent molecular classification schemes have defined five groups that may provide better diagnostic reproducibility (2, 3). The genetic alterations noted in these groups contribute to tumor development, although the exact mechanisms by which this occurs are not fully defined.
A particularly important mutation common to all lower grade (WHO grade II or III) adult gliomas and secondary glioblastomas is mutation in the IDH1 gene (4, 5). IDH1 encodes cytosolic isocitrate dehydrogenase 1 (IDH1WT), a metabolic enzyme critical for the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG)(6). IDH mutation, most typically at arginine-132, results in the creation of a neomorphic IDH1 protein (IDH1mut) with the unique capacity to generate 2HG (7). High levels of 2HG compete with α-KG and limit the function of α-KG-dependent enzymes, including those that control histone methylation (8–9). IDH1 mutation and 2HG production therefore result in alterations in patterns of histone methylation, DNA cytosine methylation, and gene expression. These alterations were shown in an in vitro model of gliomagenesis to be sufficient to transform immortalized cells and drive glioma formation (10). On the basis of the improved understanding of the role of IDH1 mutation in cancer, IDH1mut has become a critical target for drug development (11).
Although IDH1mut is an important driver of gliomagenesis, it is unclear if or how it contributes to the earliest stages of the process in which cells escape growth arrest caused by telomeric shortening. In tumors derived from stem or stem-like cells, endogenous telomerase expression in the cell of origin may postpone or eliminate the growth limitation caused by telomere shortening (12). In most gliomas, however, IDH mutation is associated with mutually exclusive mutations in either ATRX or the TERT promoter (13). ATRX mutations occur in IDH1 mutant lower grade astrocytomas, and ATRX-mutant tumors appear to resolve telomere shortening by a telomerase-independent, alternative lengthening of telomere (ALT) mechanism that predominantly uses recombination to generate heterogeneous telomeres up to 50 kb in length (14–16). In contrast, mutations in the TERT promoter occur primarily, but not exclusively, in IDH1 mutant oligodendroglioma (13). In these tumors, GABP binds and selectively reactivates the mutant TERT promoter, leading to telomerase expression and telomere stabilization (17). In both cases, however, it appears that glioma driven by IDH1mut require resolution of telomere dysfunction somewhere along their tumorigenic development.
Frequent mutation of ATRX or the TERT promoter in IDH mutant glioma suggest that resolution of telomere dysfunction is important in these tumors, but whether IDH1mut itself plays a role in the process remains unknown. In multi-step models of tumorigenesis, resolution of telomere dysfunction represents a distinct step, separable from the genetic alterations that drive the transformation process (18). As an example, in both in vitro and in vivo systems, mutant G12V H-Ras does not contribute to the genesis or selection of events that resolve telomeric dysfunction, but can drive the final stages of astrocytic transformation in cells immortalized by expression of exogenous hTERT (19, 20). The same may be true for IDH1mut, which in similar systems transforms p53-/pRb-deficient, hTERT-immortalized astrocytes (10). The requirement for exogenous hTERT in IDH1mut -mediated cellular transformation, however, has not been formally examined. Furthermore, the tight linkage between IDH mutations and mutations of genes and promoters involved in telomere regulation suggests that IDH mutation, in addition to driving cellular transformation, may also contribute to the genesis or selection of events that resolve telomeric dysfunction, and in so doing may contribute to both immortalization and transformation.
To address this possibility, we used a well-described in vitro gliomagenesis model (19, 21, 22) in which IDH1mut alters histone methylation and gene expression (8, 9), and drives the transformation of hTERT-expressing immortalized astrocytes (10). Using this system, we asked if IDH1mut, unlike other oncogenes, was capable of transforming non-hTERT-expressing precursor cells, and if this process involved ATRX and/or TERT promoter mutations associated with telomere stabilization in lower-grade glioma.
Materials and Methods
Cell culture and creation of cell lines
The generation and culturing of normal human astrocytes (NHAs) expressing E6E7, E6E7hTERT, E6E7 plus mutant G12V H-Ras (HRasV12), E6E7hTERT plus HRasV12 and E6E7hTERT plus IDH1mut has been described (10, 19). To generate p53-/pRb-deficient astrocytes expressing IDH1 WT or IDH1mut, NHAs expressing E6E7 were infected with lentivirus encoding GFP only or GFP with either IDH1 WT or IDH1mut. GFP-positive cells were isolated by fluorescent-activated cell sorting (FACS) and verified by Western blot for expression of the given target protein (10, 23). Following sorting, cells (except for NHA) were expanded and separated into 10 plates which were independently passaged 1:4, with each passage considered as two PDs. In vitro transformation was determined by a soft agar assay (19).
Protein extraction and immunoblot analyses
Cells were lysed in RIPA lysis buffer (Life Technologies) supplemented with 1x PhosStop and protease inhibitor cocktail (Roche). Protein (30 μg) was used for Western blot analysis using primary antibodies against IDH1 (Dianova), IDH1R132H (Dianova), H-Ras (F235, Santa Cruz Biotechnology), p53 (Cell Signaling), HPV16E7 (Santa Cruz Biotechnology), ATRX (Cell Signaling), β-actin (Cell Signaling), H3K4me3, H3K9me2, H3K9me3, H3K27me3 (all Active Motif), or Histone H3 (Abcam) and the appropriate horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotech). Antibody binding was detected using ECL reagents (Amersham).
Analysis of 2-HG production
2HG levels were determined by magnetic resonance spectroscopic imaging (MRSI)(23).
Fluorescence in situ hybridization (FISH)
Chromosomal copy number was determined by single-color FISH analysis of fixed E6E7 IDH1mut pre- or post-crisis polyclonal cells using centromere enumerating probes (CEP) specific for chromosomes 3, 10, and 11 (SpectrumGreen- or SpectrumOrange-labelled CEP3, 10, and 11, Abbott)(24). Telomere-specific FISH was performed using a Telomere FISH Kit/Cy3 (DAKO)(25). ALT-positive cases were defined by large, ultra-bright telomere repeat DNA aggregates in ≥1% of the cells assayed.
Exome alignment, mutation identification, and construction of phylogenetic trees
DNA was isolated from clonal populations of E6E7 IDH1mut post-crisis populations, and paired-end sequencing data from exome capture libraries (Nimblegen SeqCap EZ Exome v3) were aligned to the reference human genome (build hg19) with the Burrows-Wheeler Aligner. Single nucleotide variants were called as previously described (26). Phylogenetic trees were constructed inferring ancestral relationships by clonal ordering (27–29).
Methylation analysis
DNA methylation analysis was performed using an Infinium MethylationEPIC BeadChip (850K) with methylation beta values for each probe and sample calculated and normalized using the ‘noob’ function in the minfi data package in R computing language. A sample was considered G-CIMP positive based on the criteria defined in (30). G-CIMP was also assessed by unsupervised hierarchical clustering using data (IDAT files,10/29/2015 download) for 647 low grade glioma and GBM cases with 450K array, copy number array and somatic mutation information. TCGA samples with >0.5% of probes with p-value >0.05 were excluded. Probes present on both the 450K and EPIC array were retained (277,562). The most variable 0.5% of probes (30) using only TCGA samples (1,332 total) were included with the cohort of samples to perform two-way unsupervised hierarchical clustering using Euclidean distance and Ward linkage.
Analysis of TERT mRNA expression and telomerase activity
Total RNA was extracted (RNeasy MiniKit, Qiagen), treated with DNase I, reverse transcribed (SuperScript II Kit, Invitrogen) and amplified (initial denaturation of 95° C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 20 s, RotorGene, Qiagen) in triplicate using forward (5′-ACTGGCTGATGAGTGTGTACGTCGT-3′) and reverse (5′-ACCCTCTTCAAGTGCTGTCTGATTCC-3′) primers. TERT mRNA was quantified by real-time PCR and normalized to the amount of β-actin mRNA. Telomerase activity was measured using TRAPeze® Kit (Millipore).
Terminal restriction fragment analysis
Genomic DNA was digested (HinfI, AluI, HaeIII, RsaI, HhaI, MspI, NEB) and resolved on a 0.7% agarose gel. The denatured and dried gel was hybridized with 32P-labeled oligonucleotide C-rich telomeric probe (31), then exposed to a PhosphorImager screen.
Mutational screening of the TERT promoter and TERT promoter assays
Genomic DNA was isolated from glioblastoma and anaplastic astrocytoma samples exhibiting nt 228 or nt 250 TERT promoter mutations, respectively (3), or from each of 9–10 cell lines derived by expansion of single cells from polyclonal populations of E6E7 IDH1mut pre- or post-crisis cells. The TERT promoter region was assessed for mutations by Sanger sequencing, which has a limit of detection of roughly 15–20% (32). TERT promoter activity was determined by cloning the TERT core promoter into the promoterless pGL4.10 luc2 Firefly luciferase vector. 72 hrs after co-transfection with a control Renilla expression construct, Firefly luciferase activity was measured and normalized to Renilla activity as described (17).
Tumorigenicity assays
Immunodeficient mice (nu/nu; Charles River) were injected intracranially with 5×105 of E6E7/empty, E6E7 IDH1 WT, or E6E7 IDH1mut cells (N = 10). Animals were monitored until they developed signs of neurological deficit, at which time they were sacrificed.
Chromatin immunoprecipitation (ChIP analysis)
E6E7 IDH1mut pre-or post-crisis cells were lysed and the chromatin was digested to mononucleosomes with micrococcal nuclease. Immunoprecipitation was performed using an IgG negative control or monoclonal antibodies (Cell Signaling) specific for H3K4me3 or the c-Myc binding partner Max. Enrichment at the TERT promoter was determined by qPCR using primers specific for the TERT+47 region. TERT-3 or ZNF333 probes were used as negative controls for non-promoter or promoter-like regions of known closed chromatin, respectively (17). Three replicate PCR reactions were carried out for each sample.
Statistical Analyses
Data are reported as mean ± standard error of at least three experiments. When two groups were compared, the unpaired Student’s t test was applied (P-value). When multiple groups were evaluated, the one-way ANOVA test with post hoc Turkey-Kramer multiple comparisons test was used. P<0.05 was considered statistically significant.
Results
IDH1mut expression alone drives the immortalization and transformation of p53/pRb-deficient astrocytes
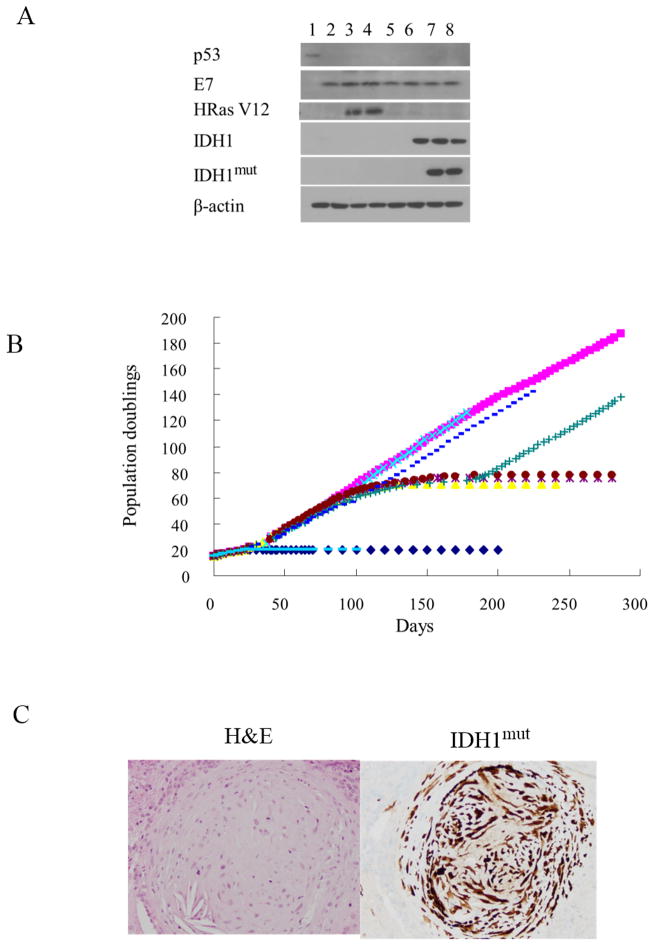
To begin to understand the role IDH1 mutation plays in immortalization and gliomagenesis, NHA rendered p53- and pRb-deficient by expression of HPV16 E6 and E7 were infected with a lentiviral construct encoding only GFP, or constructs encoding GFP and either hTERT, IDH1WT, IDH1mut, or oncogenic HRasV12. Western blot analysis of FACS-sorted, GFP-positive cells showed that cells infected with constructs encoding E6 and E7 expressed E7 and were deficient in p53 relative to blank vector control NHA (Fig. 1A). Exogenous expression of mutant H-Ras or various forms of IDH in appropriate cells was also apparent (lanes 3, 4 and lanes 6–8, respectively). Only cells infected with constructs encoding IDH1mut exhibited expression of IDH1mut using an IDH1mut -specific antibody (lanes 7, 8). Furthermore, only the E6E7 IDH1mut cells contained 2HG, and at levels that were comparable (17.9 fmol/cell) to those noted in IDH1mut -expressing human tumors (33).
Figure 1.
(A, B) Western blot validation and growth of cell populations over time: NHA (lane 1, blue diamond), NHA E6E7 hTERT (lane 2, pink square), NHA E6E7 H-Ras V12 (lane 3, yellow triangle), NHA E6E7 H-Ras V12 hTERT (lane 4, light blue x), NHA E6E7 empty vector (lane 5, purple *), NHA E6E7 IDH1WT (lane 6, brown dot), NHA E6E7 IDH1mut (lane 7, green cross), NHA E6E7 IDH1mut hTERT (lane 8, blue dash), NHA IDH1mut (light blue dash, panel B only). (C) H&E (left) and IDH1mut staining of a representative intracranial tumor formed by E6E7 IDH1mut post-crisis cells.
Following creation and characterization, the growth of each cell group in culture was followed up to one year. The ability of the cells to grow in soft agar was also monitored as a measure of in vitro cellular transformation. Control NHA, or NHA infected with a construct encoding only IDH1mut assumed a senescent-like appearance within 10 population doublings of their initial modification (Fig. 1B). Three independent cultures of each cell group also failed to grow in soft agar (Table 1), showing that IDH1mut alone cannot immortalize or transform NHAs. Cells expressing E6 and E7, in contrast, escaped replicative senescence and exhibited an extended lifespan (Fig. 1B). Additional exogenous expression of hTERT in the E6E7 cells allowed continuous growth in culture (pink line Fig. 1B), but not in soft agar (Table 1), also consistent with previous data suggesting that expression of E6, E7, and hTERT immortalizes, but does not transform, NHA (19, 20). E6E7hTERT cells additionally expressing mutant H-Ras or IDH1mut grew in a continuous manner in culture (light blue crosses and blue dashes, respectively, Fig 1B), but also formed colonies in soft agar (Table 1), consistent with the ability of both mutant H-Ras and IDH1mut to transform telomerase-positive, p53-/pRb-deficient cells (10, 19). In the absence of hTERT, however, E6E7 cells containing a blank construct, or one encoding oncogenic mutant H-Ras or IDH1mut, entered a crisis phase at roughly 70 PD. During crisis the cell number remained relatively constant although cell death was apparent. Of ten independent E6E7, E6E7 IDH1WT or E6E7 mutant H-Ras cell cultures carried into crisis, none emerged over the ensuing 250 days, and none formed colonies in soft agar. Ten E6E7 IDH1mut cultures also entered crisis approximately 100 days after creation (green crosses, Fig 1B). Surprisingly, however, all ten escaped from crisis roughly 80 days later, grew continuously in culture, and formed colonies in soft agar (Table 1). Furthermore, when the post-crisis E6E7 IDH1mut cells were injected intracranially into mice, tumors formed within six weeks in 4 of 10 animals whereas no tumors were noted in animals injected with E6E7 IDH1WT or parental E6E7 cells. Immunohistochemical analysis showed that these tumors consisted of IDH1mut cells with a low nuclear to cytoplasmic ratio consistent with a lower-grade glioma (Fig. 1C). These results show that unlike other oncogenic insults, IDH1mut can drive both the immortalization and transformation processes in p53-/pRb-deficient astrocytes.
Table 1.
Percentage of cultures escaping from crisis and number of colonies formed in soft agar (per 106 cells plated) of populations in Figure 1.
| % of cultures escaped from crisis | colonies in soft asar | |
|---|---|---|
| NHA | 0 (0/3) | 0 |
| NHA IDH1mut | 0 (0/3) | 0 |
| NHA E6E7 hTERT | no crisis | 0 |
| NHA E6E7 HRasV12 | 0 (0/10) | 0 |
| NHA E6E7 HRasV12hTERT | no crisis | 1247±101 |
| NHA E6E7 empty | 0 (0/10) | 0 |
| NHA E6E7 IDH1WT | 0 (0/10) | 0 |
| NHA E6E7 IDH1mut | 100 (10/10) | 276±25 |
| NHAE6E7 IDH1mut hTERT | no crisis | 355±47 |
IDH1mut -driven alterations in histone and DNA methylation appear insufficient to drive cellular immortalization
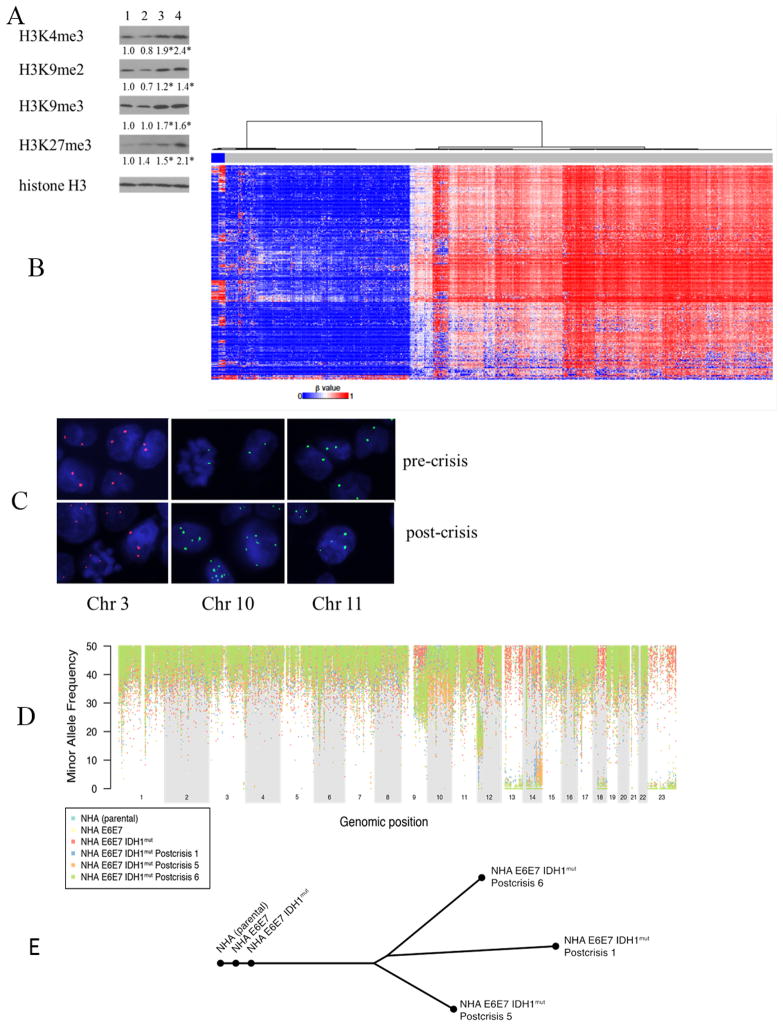
While introduction of IDH1WT had no significant effect on the levels of histone H3 modifications measured (Fig. 2A), the expression of IDH1mut caused significant increases in H3K9 dimethylation and K4, K9, and K27 trimethylation. Although these changes were detectable in the E6E7 IDH1mut cells within 40 days of IDH1mut introduction, the cells nonetheless entered into crisis as a population some 60 days later. Furthermore, when the cells emerged from crisis, the levels of H3K9 dimethylation and K4, K9, and K27 trimethylation remained elevated, but not significantly different from those in pre-crisis cells. These results suggest that while IDH1mut expression induces changes in histone modifications, these changes alone do not allow the culture as a whole to avoid crisis or become immortalized.
Figure 2.
(A) Western blot analysis of histone H3K 4,9, and 27 methylation in E6E7 empty vector (lane 1), E6E7 IDH1WT (lane 2), E6E7 IDH1mut pre-crisis (lane 3), and E6E7 IDH1mut post-crisis (lane 4) cells. Values were derived from 3 independent experiments, *, p<.05. (B) Hierarchical clustering of the methylation array data from the cell lines used in this study (far left lanes) with that of G-CIMP negative/IDH WT (blue lanes) and G-CIMP positive/IDH1mut (red lanes) TCGA tumors. (C) Fluorescence in situ hybridization of E6E7 IDH1mut pre- (top) or post (bottom) -crisis cells using probes specific for centrosomal regions on chromosome 3, 10, or 11. (D) LOH plot of NHA, E6E7, E6E7 IDH1mut pre-crisis, and three independent E6E7 IDH1mut post-crisis clonal cultures. (E) A phylogenetic tree depicts the patterns of clonal evolution of E6E7 IDH1mut post-crisis clonal populations inferred from the pattern of somatic mutations.
Because tumors driven by IDH1mut expression also frequently exhibit a hypermethylated, G-CIMP phenotype, we also determined if the G-CIMP phenotype was associated with IDH1mut –driven transformation. Although methylation array analysis showed that polyclonal and clonal populations of post-crisis E6E7 IDH1mut cells exhibited more CpG methylation than the E6E7 and E6E7 IDH1WT pre-crisis populations (right lanes under blue box at left of Figure 2B), hierarchical clustering analysis showed that all the cell lines used in this study clustered with themselves on the side of the heatmap along with G-CIMP negative/IDHWT tumors, and not with G-CIMP positive/IDH1mut glioma. Similarly, none of the post-crisis E6E7 IDH1mut cell groups met the criteria that define the G-CIMP phenotype (30). These results suggest that the cellular transformation driven by IDH1mut expression occurs independently of the G-CIMP phenotype.
p53/pRb deficient astrocytes immortalized and transformed by IDH1mut exhibit genetic alterations and divergent evolution
We also considered the possibility that the post-crisis cells emerged from a small pre-existent population of telomerase-positive cells in the pre-crisis population. If this were the case, the post-crisis E6E7 IDH1mut cells would be expected to avoid the genomic changes associated with crisis. FISH analysis using chromosome-specific centrosome probes, however, showed that unlike the diploid E6E7 IDH1mut pre-crisis cells, polyclonal populations of the post-crisis cells exhibited polysomies of multiple chromosomes (Fig. 2C). Furthermore, loss of heterozygosity of chromosmes13p and 13q, 18q, Xp and Xq, and of various areas of chromosome14 was apparent in each of three post-crisis clonal populations examined, but not in the pre-crisis population (Fig. 2D). In addition, whole exome sequencing showed that while each post-crisis clonal population contained unique genetic alterations, all clones shared 8 genetic mutations not seen in the parental cells (Table 2). These results suggest that the E6E7 IDH1mut cells arose from telomerase-deficient cells that underwent genomic alteration and divergent evolution during crisis (Fig. 2E).
Table 2.
Mutations found in clonal populations transformed by expression of mutant IDH 1
| Mutations unique to: | |
| post-crisis Clone 1: | CHD5, KDR, LAMA1, SNX20, HIST1H1C, ATP6V1H, TRIM37, HIPK3, RCBTB2, DNAH11, PPARGC1A, NMB, FAM55D |
| post-crisis clone 5: | DGKK, TCF19, SLC9A6, C14orf159, L3MBTL4, OVCH1, GREB1, BAMBI |
| post-crisis clone 6: | PLD2, SLC22A7, PPP2R2A, DSCR6, ZNF782, NBEAL1, SH3RF1, COL1A2, CPOX, RARRES1, EML6, EIF2AK4, MUC16, PLA2G4C MUC17,TRIO |
| Mutations shared by two clones: | GPR19 (5 and 6) NFAFC (1 and 6) |
| Mutations shared by all clones | ATRN, YSK4, ATF7IP, SEMA6A, SDC1, TRIM29, PTPRN, UBQLNL, FGF10, ADD2 |
p53/pRb-deficient astrocytes immortalized and transformed by IDH1mut do not display ATRX mutations or the ALT phenotype
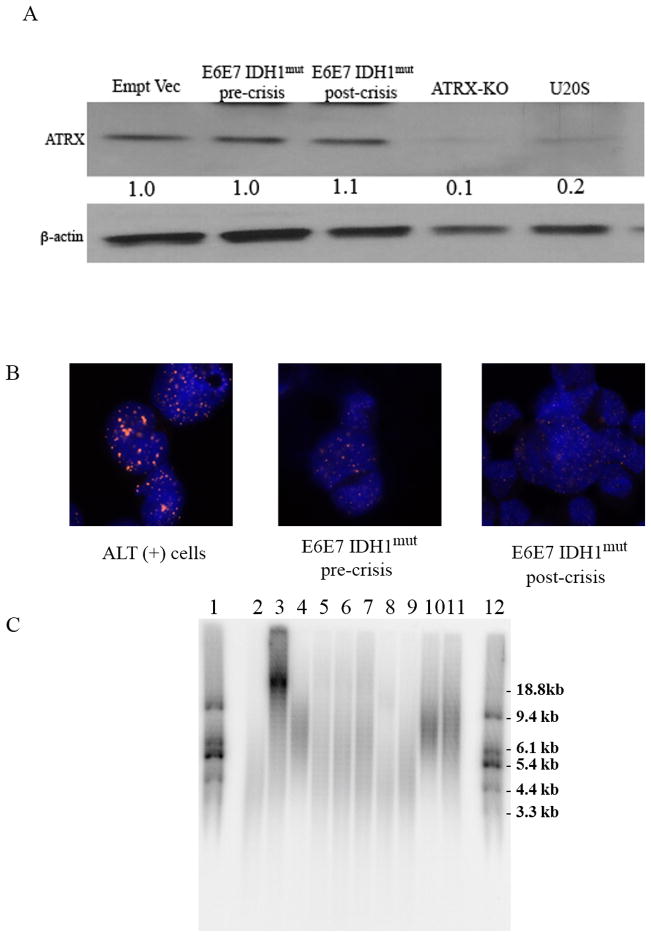
Gliomas containing IDH mutations frequently exhibit mutations in ATRX, loss of ATRX expression, and the ALT phenotype. Both the pre- and post-crisis E6E7 IDH1mut cells, however, retained ATRX expression (Fig. 3A), and no ATRX mutations were apparent in clonal post-crisis populations via whole-exome sequencing. Furthermore, while telomeric foci were readily apparent by FISH in positive control ALT+ GM847 human fibroblasts, they were not apparent in either the pre- or post-crisis E6E7 IDH1mut cells (Fig. 3B). Similarly, Southern blot analysis showed that the average telomere length in the E6E7 IDH1mut post-crisis cells was less than that in ALT+GM847, E6E7hTERT, and E6E7 IDH1mut pre-crisis cells, and only marginally greater than that in E6E7 IDH1mut cells in crisis (Fig. 3C). These results show that p53-/pRb deficient astrocytes are immortalized and transformed by IDH1mut independently of ATRX mutation and ALT.
Figure 3.
(A) Western blot analysis of ATRX and β-actin levels in E6E7 (empty vector), E6E7 IDH1mut pre-or post-crisis cells, ATRX knockout (ATRX KO) astrocytes, and U2OS cells. (B) FISH analysis of telomere aggregation using a telomere-specific probe in control ALT+ cells (GM847) and E6E7 IDH1mut pre-or post-crisis cells. (C) Southern blot analysis of average telomere length in ALT-negative, TERT+ urinary bladder cancer cells (UM-UC3, lane 2) and ALT+ (GM847, lane 3) human fibroblasts, E6E7hTERT (lane 4), E6E7 empty vector (PD38, lane 5), E6E7 IDH1WT (PD37, lane 6), pre-crisis (PD33), in crisis (PD59), and post-crisis (PD157) E6E7 IDH1mut populations (7–9, respectively), and in E6E7hTERT IDH1mut (lane 10) and U251 glioma cells (lane 11). Lanes 1 and 12, molecular weight marker.
p53/pRb-deficient astrocytes immortalized and transformed by IDH1mut reactivate telomerase in the absence of TERT promoter mutations
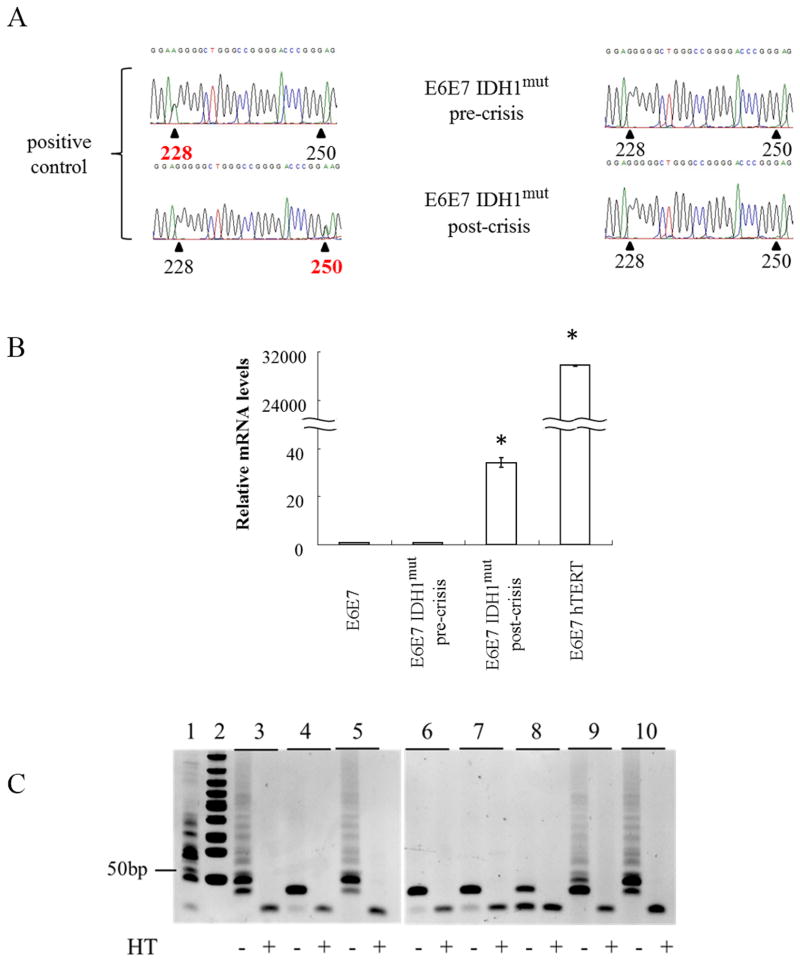
IDH1mut glioma also frequently exhibit TERT promoter mutations, which, in turn, are associated with increased TERT expression and telomerase activity (17, 34). None of nine clonal populations of E6E7 IDH1mut pre- crisis cells, or ten clonal populations of E6E7 IDH1mut post-crisis cells, however, exhibited TERT promoter mutations at nt228 or nt250, although these mutations were readily apparent in positive control glioma samples (Fig. 4A). E6E7 IDH1mut post-crisis cells did, however, have levels of TERT RNA expression that, while roughly 1% of that noted in cells containing an SV40 promoter-driven hTERT expression construct (E6E7hTERT), were still 40 times higher than the minimal TERT RNA levels noted in E6E7 and E6E7 IDH1mut pre-crisis cells (Fig. 4B). Consistent with these observations, telomerase activity was apparent in the post-crisis E6E7 IDH1mut cells (Fig 4C, lane 9), and in the positive control E6E7 and the E6E7 IDH1mut cells expressing exogenous TERT (lanes 3 and 10), but not in the E6E7 parental cells (lane 6), or in E6E7 IDH1mut cells prior to entry into crisis (lane 8). These results show that expression of IDH1mut allows a subset of p53-/pRb-deficient astrocytes to reactivate endogenous telomerase activity and to become transformed in the absence of ATRX and/or TERT promoter mutations.
Figure 4.
(A) Sequence analysis of the TERT promoter in positive control DNA from nt228 or nt250 TERT promoter-mutant glioma (left) and in DNA from E6E7 IDH1mut pre- or post-crisis cells (right). (B) qPCR analysis of TERT mRNA expression in positive control E6E7hTERT cells and E6E7 IDH1mut pre- or post-crisis cells. (C) TRAP analysis of telomerase activity in untreated (−HT) and heat-inactivated (+HT) lysates from E6E7hTERT (lane 3), E6E7mut H-RasV12 (lane 4), E6E7 H-Ras hTERT (lane 5), E6E7 (lane 6), E6E7 IDH1WT (lane 7), E6E7 IDH1mut pre-crisis (lane 8), E6E7 IDH1mut post crisis (lane 9), and E6E7 IDH1mut hTERT (lane 10) cells. Lane 1, positive control lysate, lane 2, molecular weight marker. Values were derived from 3 independent experiments, *, p<.05.
Endogenous TERT reactivation in IDH1mut -expressing post-crisis cells is associated with changes in the chromatin structure and transcription factor binding at the TERT promoter
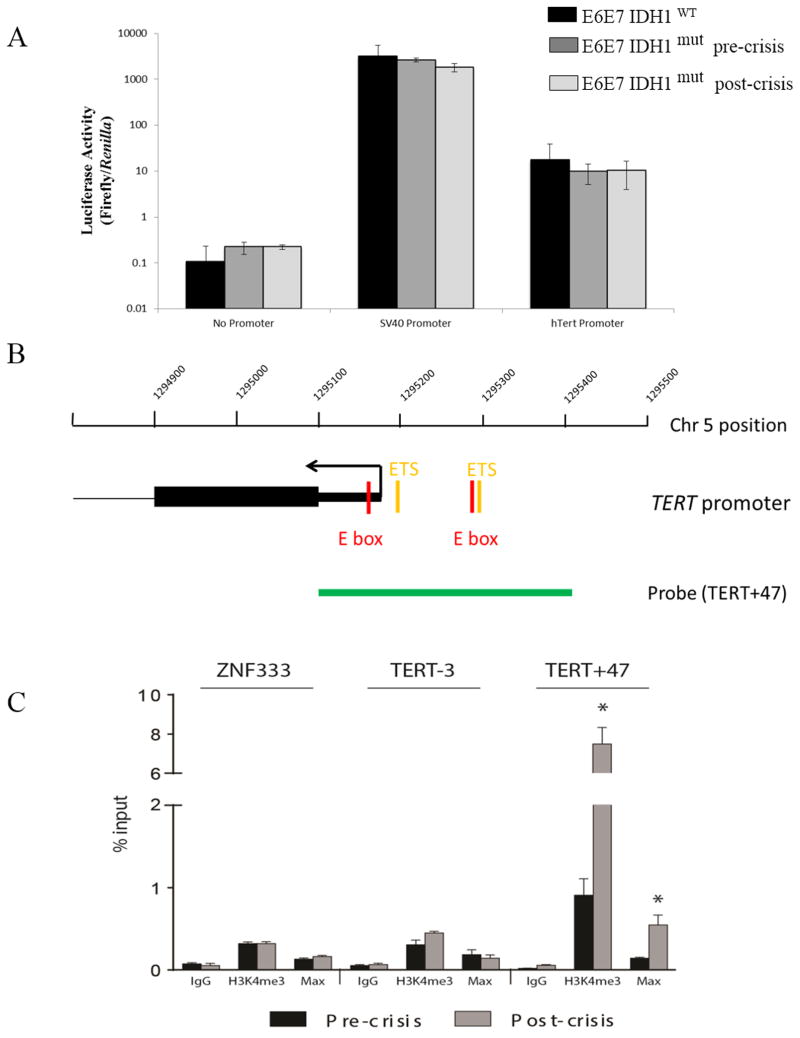
TERT promoter reactivation in IDH1mut -expressing post-crisis cells could result from increased expression or activity of factors that favor TERT expression, or from chromatin alterations at the TERT promoter that allow the binding of existing activators. To distinguish these possibilities, E6E7 IDHWT cells, and polyclonal pre- and post-crisis E6E7 IDH1mut cells were transiently co-transfected with a Renilla-encoding expression construct and a reporter construct in which Firefly luciferase expression was unpromoted (no promoter) or driven by the WT TERT promoter or a positive control SV40 promoter. Firefly luciferase activity was then measured and normalized to Renilla activity 72 hrs after transfection. Although levels of Firefly luciferase expression driven by the extrachromosomal WT TERT promoter were above background levels (TERT promoter vs no promoter, Fig. 5A), they were comparable in all cell groups examined. These results suggest that the increased endogenous telomerase expression in the post-crisis cells was related to increases in the accessibility of the endogenous TERT promoter rather than to increases in the activity of factors that favor TERT expression. To address this possibility, we used chromatin immunoprecipitation to assess levels of H3K4me3 (which recruits chromatin remodeling factors and is associated with transcriptionally active or poised genes)(35–37) and the binding of Max, whose association with c-Myc and binding to E-boxes allows expression of c-Myc target genes (including in some instances TERT)(38–42) in the TERT promoter region (probe TERT+47, Fig. 5B), and in two regions (ZNF333 and TERT-3) that lack E-boxes and are in a conformationally closed chromatin state (17). The levels of H3K4me3 and Max binding in the promoter-like and non-promoter regions covered by the ZNF333 and TERT-3 probes, respectively, were uniformly low in both the E6E7 IDH1mut pre- and post-crisis cells (Fig. 5C). Levels of both H3K4me3 and Max binding, however, were 4- to 7-fold elevated in the TERT promoter (TERT+47 probe) in post-crisis cells compared to pre-crisis cells, which in turn had levels that were not significantly different from controls. These results suggest that the passage of E6E7 IDH1mut cells through crisis allowed the emergence of cells with the endogenous TERT promoter in an H3K4me3-marked state that allowed Max binding, TERT promoter activation, telomerase expression, telomere stabilization, and ultimately cellular transformation.
Figure 5.
(A) Firefly luciferase gene activity (relative to control Renilla expression) in E6E7 IDH1WT cells, and E6E7 IDH1mut pre-or post-crisis cells 72 hrs following co-transfection with constructs encoding constitutively expressed Renilla, and Firefly luciferase expression not driven (no promoter), or driven by a positive control SV40 promoter or the TERT promoter. (B) Region analyzed by chromatin immunoprecipitation analysis using antibodies specific for H3K4me3 or the c-Myc-binding partner Max. Analysis also used negative control probes (ZNF333 and TERT-3) or a probe (TERT+47, green) that spans the region of the TERT promoter containing the transcription start site (arrow), c-Myc-binding E-boxes (red), and ETS transcription factor binding sites (orange) critical for transcriptional activation. (C) H3K4me3 levels and extent of Max binding in E6E7 IDH1mut pre- or post-crisis cells as determined by chromatin immunoprecipitation analysis. *, p<.05. Values derived from 3 independent experiments.
Discussion
IDH mutation is an early event in the development of glial tumors (26). Because it is often accompanied by mutations that influence telomere function, the present study was initiated to determine whether IDH1mut expression generates or selects for events that stabilize telomeres. Our results show that while IDH1mut expression does not generate or select for ATRX or TERT promoter mutations, it contributes to the reactivation of the endogenous TERT promoter, the upregulation of telomerase activity, and the immortalization events that precede transformation.
IDH1mut as a driver of cellular immortalization and transformation
A variety of factors are important in the immortalization and transformation of NHA. Loss of p53 and pRb function allows NHA to bypass replicative senescence but does not drive the telomerase reactivation critical for immortalization and subsequent transformation (18–20). Expression of various drivers of gliomagenesis such as EGFRvIII and mutant H-Ras similarly fails to reactivate telomerase but, at least in the case of mutant H-Ras, can transform p53-/pRb-deficient cells already immortalized by hTERT expression (19). Consistent with these studies, there was no indication in the present studies that IDH1mut expression induced telomerase activity even 40 days following introduction. Accordingly, cells expressing IDH1mut entered into crisis like other telomerase-negative cultures. In contrast to other oncogenic insults, however, expression of IDH1mut resulted in the emergence of clonally-related, telomerase-positive tumorigenic populations with chromosomal abnormalities consistent with passage through crisis. As such, mutant IDH is unique among oncogenic drivers studied in that it drives both TERT reactivation and the subsequent events required for transformation of p53/pRb-deficient NHA.
IDH1mut -driven mechanisms of telomerase reactivation
If IDH1mut expression allows for an indirect activation of telomerase and resolution of telomere-induced crisis, how does this occur? In lower grade astrocytomas, mutations in IDH and TP53 are early events and are strongly associated with ATRX mutations and the ALT phenotype. Genomic instability initiated by p53 loss (43) could conceivably lead to the generation and selection of cells with mutations in IDH and/or ATRX. In the present study, however, loss of p53 function did not select for cells with IDH mutations. Similarly, although loss of p53 function in combination with the introduction of IDH1mut led to the generation and selection of clonal populations with common genetic alterations, these alterations did not include ATRX mutations. The present system relies on E6-mediated elimination of p53 and as such may not accurately mimic the effects (and possible gain of function effects) of mutant p53 found in most lower-grade astrocytoma (44, 45). It may also be possible that loss of ATRX is a prerequisite for the development of the ALT phenotype, and that in the absence of ATRX mutations, alternative means of resolving telomeric crisis are favored. Similarly, while IDH mutation is closely associated with TERT promoter mutation and TERT reactivation in oligodendroglioma, the introduction of IDH1mut in the present system did not lead to the generation or selection of TERT promoter mutations, but rather to a relatively rare and delayed reactivation of the endogenous WT TERT promoter. TERT promoter reactivation was temporally unrelated to the global changes in the histone modifications, but was associated with an increase in H3K4me3 and transcription factor binding in the TERT promoter similar to that recently noted in the transcriptionally reactivated mutant TERT promoter in GBM cells (17). These results suggest that IDH1mut -driven changes in TERT promoter chromatin structure, rather than in the expression of factors that drive TERT expression, occur in a small percentage of cells that subsequently emerge from crisis. As such the process differs significantly from that noted in NHA induced to become pluripotent by the expression of a mixture of transcription factors, and which reactivate telomerase in a population-wide manner over a shorter (3 week) period of time (46). Although IDH1mut -associated increases in 2HG could alter the activities of the αKG-dependent enzymes that regulate H3K4me3 levels and ultimately transcription factor accessibility in the TERT promoter, the explanation for IDH1mut -driven telomerase reactivation is likely more complex as telomerase reactivation appears to be a rare event. IDH1mut-induced hypermethylation of CCCTC-binding factor-binding sites near the TERT promoter may also contribute to loss of insulation and aberrant TERT promoter activation (47, 48), although post-crisis cells do not exhibit G-CIMP, and the role methylation in control of TERT promoter activation is controversial (49, 50). The mutations which arose during crisis and are shared by all post-crisis clonal populations may also provide clues to pathways that retain appropriate control of telomerase expression.
Relevance of the findings to gliomagenesis and therapy
Data from the present studies show that while the resolution of telomeric crisis is a prolonged and rate-limiting step in the IDH1mut -driven process of in vitro gliomagenesis, it can occur independently of ATRX and TERT promoter mutations. Lineage tracing of IDH mutant glioma suggests that IDH mutations precede TERT promoter and ATRX mutations, although the temporal distance between these events cannot be determined (26). It is tempting to speculate that the genesis of IDH1mut glioma may therefore also include a prolonged period of time related to resolution of telomeric dysfunction. If this is true, the unraveling of the relevant telomere maintenance mechanism may provide new therapeutic options to delay or eliminate the progression of lower-grade glioma. Furthermore, although the resolution of telomeric issues in most IDH1mut glioma involves the genesis and selection of cells with ATRX or TERT promoter mutations, a small subset of IDH1mut glioma (roughly 5–10% of IDH1mut astrocytomas and mixed oligoastrocytomas)(13) lack both ATRX and TERT promoter mutations. These tumors may reactivate the endogenous WT TERT promoter in a manner similar to that described in the present study and as such the pathways described here may be of particular importance in understanding the genesis and improving the therapy of these and other IDH1mut glioma.
Supplementary Material
Acknowledgments
Grant Support: This work was supported in part by National Institutes of Health Grants CA172845-03 (to SMR and ROP), CA171610-03 (to ROP), the Loglio Research Project (to KMW, JJP, JFC, SMR, and ROP), and The Kristian Gerhard Jebsen Foundation and The Norwegian Cancer Society (to T-CJ)
TTC was supported by the Damon Runyon Cancer Research Foundation (DRG-2168-13).
Footnotes
There are no relationships that could be construed as resulting in an actual, potential, or perceived conflict of interest with regard to this manuscript.
References
- 1.CBTRUS Statistical Report: Primary Brain and Central. Nervous System Tumors Diagnosed in the United States 2007–2011. 2014;16(Supplement 4):iv, 10. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372:2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckel-Passow JE, La
- 4.chance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372:2499–508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in glioma. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–74. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of the human cytosolic NADP-dependent isocitrate dehydrogenase from pig heart. J Biol Chem. 2004;279:33946–57. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 8.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH1 mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–78. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohba S, Mukherjee J, See WL, Pieper RO. Mutant IDH1-driven cellular transformation increases RAD51-mediated homologous recombination and temozolomide resistance. Cancer Res. 2014;74:4836–44. doi: 10.1158/0008-5472.CAN-14-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–17. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 13.Gunes C, Rudolph L. The role of telomeres in stem cells and cancer. Cell. 2013;152:390–93. doi: 10.1016/j.cell.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–26. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425–8. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: Models, mechanisms and implications. Nat Rev Genet. 2010;11:319–30. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 17.Heaphy CM, Subhawong AP, Hong SM, Goggins MG, Montgomery EA, Gabrielson E, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608–15. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell RJ, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015;348:1036–9. doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–68. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 20.Sonoda Y, Ozawa T, Hirose Y, Aldape KD, McMahon M, Berger MS, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61:4956–60. [PubMed] [Google Scholar]
- 21.Rich JN, Guo C, McLendon RE, Bigner DD, Wang X-F, Counter CM. A Genetically Tractable Model of Human Glioma Formation. Cancer Res. 2001;61:3556–62. [PubMed] [Google Scholar]
- 22.Sonoda Y, Ozawa T, Aldape KD, Berger MS, Deen DF, Pieper RO. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001;61:6674–6678. [PubMed] [Google Scholar]
- 23.Sonoda Y, Kanamori M, Deen DF, Cheng S-Y, Berger MS, Pieper RO. Over-expression of VEGF isoforms drives oxygenation and growth, but not progression to glioblastoma multiforme in a human model of gliomagenesis. Cancer Res. 2003;63:1962–68. [PubMed] [Google Scholar]
- 24.Chaumeil MM, Larson PE, Yoshihara HA, Danforth OM, Vigneron DB, Nelson SJ, et al. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun. 2013;4:2429–33. doi: 10.1038/ncomms3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips JJ, Aranda D, Ellison DW, Judkins AR, Croul SE, Brat DJ, et al. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol. 2013;23:565–73. doi: 10.1111/bpa.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abedalthagafi M, Phillips JJ, Kim GE, Mueller S, Haas-Kogan DA, Marshall RE, Croul SE, et al. The alternative lengthening of telomere phenotype is significantly associated with loss of ATRX expression in high-grade pediatric and adult astrocytomas: a multi-institutional study of 214 astrocytomas. Mod Path. 2013;26:1425–32. doi: 10.1038/modpathol.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazor T, Pankov A, Johnson BE, Hong C, Hamilton EG, Bell RJ, et al. DNA Methylation and Somatic Mutations Converge on the Cell Cycle and Define Similar Evolutionary Histories in Brain Tumors. Cancer Cell. 2015:301–17. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–28. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 31.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbert BS, Shay JW, Wright WE. Analysis of telomeres and telomerase. Current Protoc Cell Biol. 2003:18.6.1–18.6.20. doi: 10.1002/0471143030.cb1806s20. [DOI] [PubMed] [Google Scholar]
- 33.Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, et al. Comparison of Sanger Sequencing, Pyrosequencing, and Melting Curve Analysis for the Detection of KRAS Mutations. J Mol Diagn. 2010;12:425–32. doi: 10.2353/jmoldx.2010.090188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2- hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–59. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–36. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–98. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–45. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 40.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 41.McMurray HR, McCance DJ. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J Virol. 2003;77:9852–61. doi: 10.1128/JVI.77.18.9852-9861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu K-J, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. Direct activation of TERT transcription by c-MYC. Nature Genetics. 1999;21:220–24. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 43.Ho JSL, Ma W, Mao DYL, Benchimol S. p53-Dependent Transcriptional Repression of c-myc Is Required for G1 Cell Cycle Arrest. Mol Cell Biol. 2005;25:7423–31. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukasawa K, Wiener F, Vande Woude GF, Mai S. P53 Genomic instability and apoptosis are frequent in p53 deficient young mice. Oncogene. 1997;15:1295–1302. doi: 10.1038/sj.onc.1201482. [DOI] [PubMed] [Google Scholar]
- 45.Hanel W, Moll UM. Links between mutant p53 and genomic instability. J Cell Biochem. 2012;113:433–9. doi: 10.1002/jcb.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frankel RH, Bayona W, Koslow M, Newcomb EW. p53 Mutations in Human Malignant Gliomas: Comparison of Loss of Heterozygosity with Mutation Frequency. Cancer Res. 1992;52:1427–32. [PubMed] [Google Scholar]
- 47.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Pluripotent tert activation Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 48.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–14. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renaud S, Loukiniov D, Bosman FT, Lobanenkov V, Benhattar J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nuc Acids Res. 2005;33:6850–60. doi: 10.1093/nar/gki989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. 2013;14:534–42. doi: 10.1016/S1470-2045(13)70110-4. [DOI] [PubMed] [Google Scholar]
- 51.Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J. Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer. 2002;101:335–41. doi: 10.1002/ijc.10593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.