Abstract
Despite the high incidence of oncogenic mutations in PIK3CA, the gene encoding the catalytic subunit of phosphoinositide 3-kinase (PI3K), PI3K inhibitors have yielded little clinical benefit for breast cancer patients. Recent epidemiological studies have suggested a therapeutic benefit from aspirin intake in cancers harboring oncogenic PIK3CA. Here we show that mutant PIK3CA-expressing breast cancer cells have greater sensitivity to aspirin-mediated growth suppression than their wild-type counterparts. Aspirin decreased viability and anchorage-independent growth of mutant PIK3CA breast cancer cells independently of its effects on cyclooxygenase-2 (COX-2) and nuclear factor-kappa B (NF-κB). We ascribed the effects of aspirin to AMP-activated protein kinase (AMPK) activation, mammalian target of rapamycin complex 1 (mTORC1) inhibition, and autophagy induction. In vivo, oncogenic PIK3CA-driven mouse mammary tumors treated daily with aspirin resulted in decreased tumor growth kinetics, while combination therapy of aspirin and a PI3K inhibitor further attenuated tumor growth. Our study supports evaluation of aspirin and PI3K pathway inhibitors as combination therapy for targeting breast cancer.
Keywords: Breast cancer, aspirin, PI 3-Kinase, mTORC1, AMPK
Introduction
The phosphoinositide 3-kinase (PI3K) signaling pathway plays a critical role in cell growth, survival, motility, and metabolism (1). Deregulated PI3K signaling is observed in numerous human pathophysiologies, including cancer. In breast cancer, somatic mutations in genes that encode proteins that activate, terminate or transduce PI3K signaling are highly prevalent. Specifically, somatic mutations in PIK3CA, the gene encoding the catalytic subunit p110α, occur with a frequency of approximately 40% across all breast cancer molecular subtypes (2). The two most frequent mutations comprise single amino acid substitutions in two hotspot regions, His1047Arg and Gln545Lys (2). Expression of either of these PIK3CA mutants leads to elevated PI3K activity, downstream AKT activation, oncogenic transformation of mammary epithelial cells and formation of heterogeneous mammary tumors in vivo (3,4). Similarly, the lipid phosphatase, PTEN, which terminates PI3K signaling, is one of the most frequently mutated tumor suppressors in human cancers. Mutation or loss of at least one copy of PTEN occurs in approximately 50% of breast cancer patients, leading to hyperactivation of PI3K/AKT signaling (5). In addition, amplification and mutation of AKT genes have been identified in breast cancer, albeit with lower frequencies (6).
Given the frequency with which the PI3K/PTEN/AKT pathway is mutated in breast cancer, numerous small molecule inhibitors have been developed as targeted therapy and are under clinical evaluation. These include pan- and p110 isoform-specific inhibitors, compounds that inhibit both PI3K and the downstream effector mTOR, and also pan-AKT inhibitors. To date, most of these inhibitors have shown limited efficacy in clinical trials due to dose-limiting toxicities as well as the emergence of drug resistance. However, it is likely that use of combination therapies that target both PI3K/PTEN/AKT and other key survival pathways may result in better therapeutic responses.
Aspirin (acetylsalicylic acid) is one of the most widely used non-steroidal anti-inflammatory drugs (NSAIDs). Its medicinal use for the treatment of pain, fever and inflammatory ailment dates back to the time of Hippocrates (7). Aspirin is also widely used as an antiplatelet drug for the prevention of heart attacks and strokes (8). Recently, results from a number of observational and randomized clinical trials have suggested that regular use of aspirin reduces the risk of development and/or progression of several cancers, including breast cancer (9,10). Although the effect of aspirin on breast cancer incidence remains poorly understood, recent observations from the Nurses Health Study indicate that aspirin use is associated with a reduced risk of breast cancer distant recurrence and death (11). Additional independent observational studies have shown that aspirin use is associated with a significant improvement in survival for patients with mutant PIK3CA colorectal cancer but not for those with wild-type PIK3CA tumors (12,13). Despite these observations, the molecular basis underlying the benefit of aspirin use in mutant PIK3CA cancers remains undefined.
Here we evaluate the efficacy of aspirin either as a single agent, or in combination with PI3K inhibitors, in PI3K-driven breast cancer. We also investigate the mechanism by which aspirin may elicit a therapeutic effect in this disease.
Materials and Methods
Antibodies
Anti-p110α (#4249), anti-phospho-Akt Ser473 (#4060), anti-phospho-Akt Thr308 (#2965), anti-Akt (#4691), anti-phospho-Pras40 Thr246 (#2997), anti-Pras40 (#2691), anti-phospho-GSK3β Ser9 (#9336), anti-GSK3β (#9315), anti-βactin (#4970), anti-phospho-IKKα/β Ser176/180 (#2697), anti-phospho-IκBα Ser32/36 (#9246), anti-IκBα (#9247), anti-phospho NF-Kappa-B p65 Ser536 (#3033), anti-NF-Kappa-B p65 (#8242), anti-AMPKα (#2532), anti-phospho-AMPKα Thr172 (#2535), anti-ACC (#3676), anti-phospho-ACC Ser79 (#3661), anti-S6K (#2708), anti-phospho-S6K Thr389 (#9205), anti-S6 (#2217), anti-phospho-S6 Ser240/244 (#5364), anti-4EBP1 (#9452), anti-phosho-4EBP1 Ser65 (#9451), and anti-TSC2 (#3990) were purchased from Cell Signaling Technologies. Laminin V (#Z0097) and Ki67 (#M7240) were purchased from Dako. Horseradish peroxidase-conjugated anti-rabbit and anti-mouse immunoglobulin antibodies were purchased from Chemicon.
Chemical reagents
The IKKβ ATP competitive inhibitor, Compound A was a generous gift from the Baldwin Lab (Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill), and manufactured by Bayer Pharmaceuticals. Celecoxib (#S1261) was purchased from Selleckchem. BKM120 (#A-1108) and BYL719 (#A-1214) were purchased from Active Biochem. A769662-10mg (#ab120335) was purchased from Abcam. Aspirin (#A2093), Sodium salicylate (#A5376) and Bafilomycin A (#B1793) were purchased from Sigma Aldrich. Aspirin/salicylate was prepared as previously described (14). Briefly, aspirin was dissolved in 1M Tris-HCl (pH 7.5) to a stock concentration of 1M and final pH of 7.2. An equivalent volume of Tris-HCl (pH 7.2) was used as vehicle control.
Plasmids
JP1520-HA-PIK3CA-GFP, JP1520-HA-PIK3CA-WT (Addgene plasmid # 14570) and JP1520-HA-PIK3CA-HA-H1047R (Addgene plasmid # 14572) were generous gifts from Joan Brugge. pBABE-puro mCherry-EGFP-LC3B was a gift from Jayanta Debnath (Addgene plasmid # 22418).
RNA interference
Stable cell lines expressing COX-2 shRNA constructs were maintained in 2μg/ml puromycin. COX-2 shRNA plasmids were a kind gift from the Polyak lab (Dana Farber Cancer Center) (15). RNAi sequences of shRNA clones used in study:
COX-2 ShRNA#1 GCAGATGAAATACCAGTCTTT
COX-2 ShRNA#2 CCATTCTCCTTGAAAGGACTT
For siRNA-mediated knockdown of TSC2 and AMPKα1/α2, SMARTpool: ONTARGETplus human TSC2 siRNA (#L-003029-00-0005) and SMARTpool: ONTARGETplus human PRKAA1/AMPKα1 siRNA (L-005027-00-0005) and PRKAA2/AMPKα2 (L-005361-00-0005) were purchased from Dharmacon. Cells were transfected with each respective siRNA or control ON-TARGETplus Non-targeting control pool siRNA (Dharmacon) using Lipofectamine RNAimax transfection reagent (ThermoFischer) according to manufacturer’s protocol.
Quantitative real-time RT PCR
Total RNA was isolated using the RNeasy kit following the manufacturer’s instructions (Qiagen). Reverse transcription was performed using Quantitect Reverse transcription kit according to the manufacturer’s instructions (Qiagen). Quantitative real-time RT-PCR was performed using SYBR Green PCR Master Mix (BioRad) and the ABI Prism 7900 sequence detector (Applied Biosystems). Quantification of COX-2 mRNA expression was calculated by the ΔΔCT method with GAPDH as reference. QRT-PCR primer sequences are listed below:
COX-2 Forward 5′-TCAGCCATACAGCAAATCCTT-3′
COX-2 Reverse 5′-GTGCACTGTGTTTGGAGTGG-3′
GAPDH Forward 5′-GCAAATTCCATGGCACCGT-3′
GAPDH Reverse 5′-TCGCCCCACTTGATTTTGGAGG-3′
Cell Culture and Immunoblotting
MCF10A cells were cultured in DMEM/Ham’s F12 medium supplemented with 5% equine serum (Gibco-brl), 10μg/ml insulin (Sigma-Aldrich), 500ng/ml hydrocortisone (Sigma-Aldrich), 20ng/ml EGF (R&D Systems) and 100ng/ml cholera toxin (List Biological Labs). MCF10A cells expressing PIK3CA WT and mutants were generated and grown as described previously (16,17). Stable pools were generated by selection in 2ug/ml puromycin. SUM159-PT cells were grown in Ham’s F12 medium (Cellgro) supplemented with 5% Fetal Bovine Serum (FBS; Cyclone), 5μg/ml insulin (Sigma-Aldrich) and 1μg/ml hydrocortisone (Sigma-Aldrich). Cells used for animal studies were cultured in HUMEC medium (Cell application, 815-500) with DMEM-F12 (Cellgro, 10-090-CV) (1:1) and 5% Fetal Bovine Serum (FBS; Cyclone). MCF7, MDA-MB-468 and MDA-MB-231 cells were maintained in DMEM (Cellgro) supplemented with 10% Fetal Bovine Serum (FBS; Cyclone). MCF10A cells were provided by Joan Brugge (Harvard Medical School) and SUM159-PT cells were obtained from Kornelia Polyak (DFCI, Harvard Medical School). All other cells lines were purchased and authenticated from ATCC. All cell lines were obtained and passaged for fewer than 6 months and routinely tested for mycoplasma contamination in the years 2012–2016. For immunoblotting, cells were rinsed with PBS and lysed in RIPA buffer containing protease and phosphatase inhibitors. Lysates were resolved by SDS-PAGE and transferred by electrophoresis to nitrocellulose membrane (Bio-Rad), followed by immunoblotting.
Morphogenesis Assay
MCF10A cells were grown in three-dimensional Matrigel cultures as previously described (16). Briefly, chambers slides were coated with growth factor-reduced Matrigel (BD Biosciences) and allowed to solidify for 30 minutes. 3×103 cells suspended in assay media containing 2% Matrigel were overlayed on coated chamber slides. Assay medium contained DMEM/Ham’s F12 (Cellgro) supplemented with 2% equine serum (Cellgro), 10ug/ml insulin (Sigma-Aldrich), 500ng/ml hydrocortisone (Sigma-Aldrich), 5ng/ml EGF (R&D Systems) and 100ng/ml cholera toxin (List Biological Labs). For aspirin studies, acini were allowed to grow for 4 days followed by treatment with aspirin every 2 days. Cells were then fixed and stained with Ki67 and laminin V on day 12 as previously described (16). Phase contrast images were acquired using the Nikon Eclipse Ti microscope. Fluorescent images were acquired using Zeiss LSM 510 Meta confocal microscope.
Colony Formation in Soft Agar
For colony formation in soft agar, 5×104 cells were suspended in growth media containing aspirin or vehicle and 0.4% Noble agar. Cell suspension was plated on top of a solidified layer of 0.8% Noble agar also containing aspirin. Cells were fed with growth media containing aspirin, every 4 days. After 15 days, colonies were stained with 1mg/ml iodonitrotetrazolum chloride and quantified using Matlab software (MathWorks).
Sulforhodamine B (SRB) Assay
Cell viability was assessed using sulforhodamine B assay as previously described (18). Briefly, adherent cells were fixed with 12.5% (w/v) trichloroacetic acid for 1 hour at 4°C. Cells were then rinsed three times with water and stained with a solution of 0.5% (w/v) SRB in 1% acetic acid for at least 30 minutes at room temperature. Cells were then washed three times with 1% acetic acid and allowed to dry. SRB was dissolved in 10 mmol/l Tris (pH 10.5). Absorbance of solubilized SRB was measured at 510 nm.
Animal Studies
Female NSG (NOD.Cg-PrkdcscidIL2rgtm1Wjl/SzJ; stock no. 005557) mice aged 6 weeks, were purchased from Jackson Laboratory. All experiments were conducted in accordance with regulations of the Children’s Hospital Institutional Animal Care and Use Committee (protocol 12-11-2308R). Bilateral orthotopic injections of SUM159-PT breast cancer cells were performed by resuspending 2×10^6 cells 1:1 (vol/vol) in Matrigel (BD BioSciences) in serum-free DMEM/F12 (Cellgro). Cells were injected into the no. 4 inguinal mammary fat pads. Five mice were used for each treatment group. Tumors were measured at least once a week using a vernier caliper and tumor volume was calculated using the formula 0.5(Length × width2). Aspirin used for in vivo studies involving daily oral gavage administration was prepared as a suspension in 0.5% methyl cellulose (Sigma).
For drug combination studies, preparation of aspirin for in vivo administration was performed as previously described (19). For these experiments, mice were injected intraperitoneally with aspirin (Sigma, A2093-100G) at 100 mg/kg. BYL719 (10mg/kg) was prepared in 0.5% methyl cellulose (Sigma) and administered daily by oral gavage. A similar volume:weight ratio of vehicle (DMSO in PBS or 0.5% methyl cellulose) was administered to control animals. Tumor volumes were compared using student’s T-test with reported p-values with correction for multiple hypothesis testing using FDR (false discovery rate) method.
In addition, to compare tumor volume of the different treatment groups at a single time point we performed a tumor growth rate analysis based on the longitudinal tumor growth measurements. For this purpose, we performed a linear mixed effects model analysis (20) using R lme4 package (21). Due to the study design, we considered as fixed effects in the model, treatment type and the time points at which tumor volume was measured. To account for possible random effects due to individual mouse characteristics and side of inoculation and pre-treatment tumor volume, we also included these three variables as random effects in the model. Visual inspection of residuals plots did not reveal any obvious deviations from homoscedasticity or normality.
Results
Aspirin selectively inhibits growth of mutant PIK3CA breast cancer cells
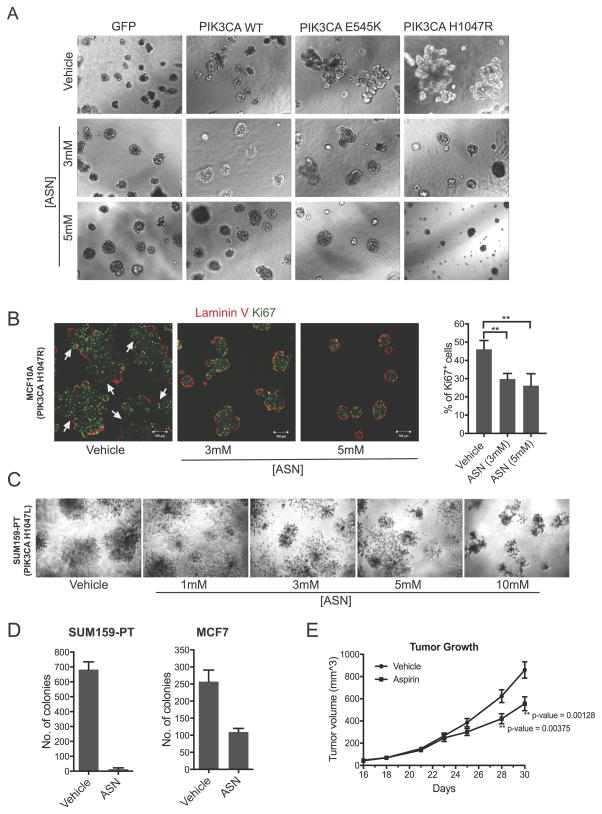
To determine whether aspirin has any effect on mutant PIK3CA breast cancer cells, we first examined the effects of aspirin on the immortalized non-tumorigenic breast epithelial cell line MCF10A stably expressing the oncogenic hotspot PIK3CA mutations H1047R or E545K. We examined cell growth in 3-dimensional Matrigel culture as a surrogate measure of in vivo tumorigenicity. In contrast to wild-type PIK3CA cells, mutant PIK3CA H1047R- and E545K-expressing cell populations both formed large multi-acinar structures (Figure 1A) and displayed irregular deposition of the basement membrane marker laminin V (Figure 1B), features of malignant. Aspirin treatment of both oncogenic mutant PIK3CA populations resulted in reversion to spheroid-like acinar structures and a localization of laminin V to the periphery of the acini, similar to that of untransformed parental MCF10A cells (Figure 1B). The aspirin concentrations (1–5mM) used were consistent with measurements of aspirin in the serum of patients treated for chronic inflammatory diseases (23–25). Aspirin treatment also resulted in a concomitant decrease in the marker Ki67, and an overall reduction in average spheroid size, supporting an anti-proliferative effect due to aspirin treatment (Figure 1B).
Figure 1. Aspirin inhibits growth of mutant PIK3CA breast cancer cells.
(A) Representative images of MCF10A cells expressing JP1520-GFP, PIK3CA wild-type (WT) and PIK3CA mutants E545K and H1047R, grown in 3D Matrigel.
(B) Representative images of MCF10A cells expressing mutant PIK3CA H1047R, grown in 3D Matrigel and stained for laminin V (red) and Ki67 (green). Cells were treated with 3mM and 5mM aspirin respectively, every two days, from day 4–12. Images were taken on day 12. Quantification of Ki67+ cells per condition. Ki67 was manually assessed after Z stacking immunofluorescent images with a minimum of 3 images and a total of 1000 cells per condition. (C) Defects in overall cell viability of SUM159-PT grown in 3D Matrigel. Images were taken on day 10. Cells were treated with the indicated concentration of aspirin (1–10mM) every 2 days from day 6–10. (D) Colony formation in soft agar, SUM159-PT and MCF7 cells were treated with 1.5mM aspirin every 4 days. On day 15, colonies were stained with iodonitrotetrazolum and counted using Matlab; Results show the average of technical duplicates. (E) Growth kinetics indicating average tumor volume at each time point in cohorts of mice treated with vehicle (0.5% methylcellulose) or aspirin (100mg/kg) via oral gavage (n=10 tumors/group; 5 mice each bearing bilateral tumors). Mice were treated daily with aspirin or vehicle starting at 5 days prior to orthotopic injection of SUM159-PT cells. Statistical significance at the experimental endpoints was determined using pairwise student t-test. ** denotes p-value <0.01.
A decrease in cell viability and stellate shaped acinar formation was also observed in aspirin-treated SUM159-PT breast cancer cells that harbor an endogenous oncogenic PIK3CA mutation (H1047L) (Figure 1C). Aspirin also robustly inhibited anchorage-independent growth of the mutant PIK3CA breast cancer cell lines SUM159-PT (H1047L) and MCF-7 (E545K) in soft agar (Figure 1D). Moreover, daily administration of aspirin resulted in a statistically significant decrease in tumor volume and growth kinetics in SUM159-PT orthotopic mouse xenografts (Figure 1E).
Aspirin suppresses growth of mutant PIK3CA breast cancer cells in an NF-κB and COX-2-independent manner
To explore the mechanistic basis for the growth inhibitory effects of aspirin in oncogenic PIK3CA breast cancer cells, we tested the contribution of COX-2 and IKKβ/NF-κB, two well-characterized targets of aspirin (25–29). As revealed by RT-PCR, MCF10A cells expressing PIK3CA H1047R displayed elevated levels of COX-2 mRNA (Supplementary figure 1A). Similar induction of COX-2 mRNA was observed in MDA-MB-231 breast cancer cells expressing PIK3CA H1047R (Supplementary figure 1A). Expression of mutant PIK3CA also led to elevated COX-2 protein (Supplementary figure 1B). Consistent with this observation, increased levels of PGE2α were observed in media from MCF10A cells expressing mutant PIK3CA (Supplementary figure 1C). Oncogenic PIK3CA also increased NF-κB activation as measured by phosphorylation of IKKα/β, IκB and p65 (Supplementary figure 1B), in agreement with previous studies (17). This observation coincides with the role for NF-κB as a transcriptional regulator of PTGS2 (30–32).
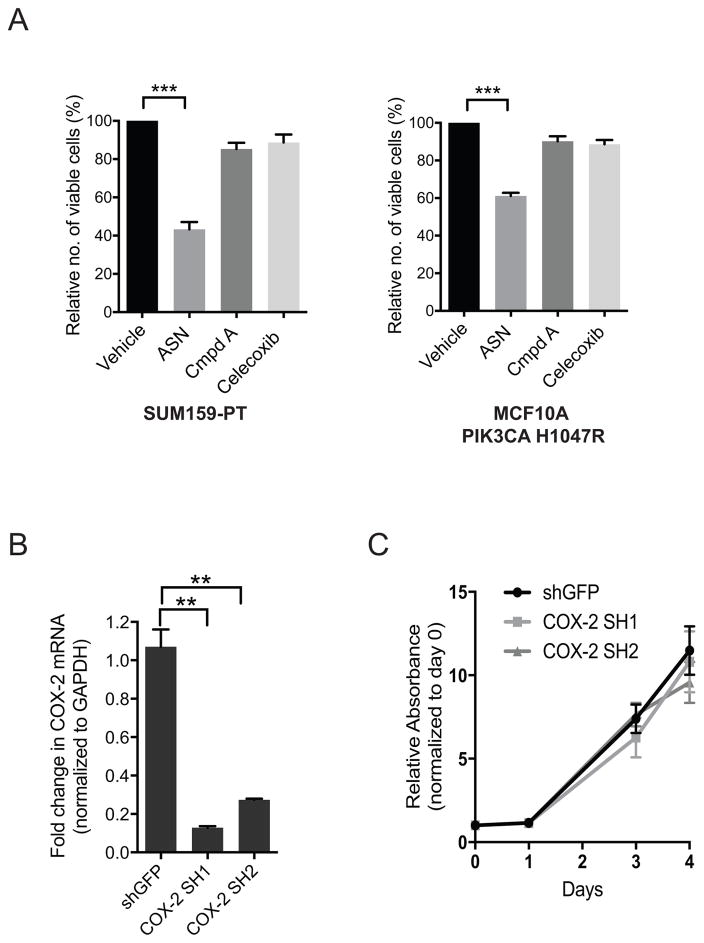
In light of these data, we investigated whether the growth inhibitory effects of aspirin on mutant PIK3CA breast cancer cells is attributable to NF-κB and/or COX-2 activity. However, we observed that treatment of mutant PIK3CA MCF10A cells or SUM159-PT breast cancer cells with Compound A (an IKKβ ATP-competitive inhibitor) (33), or Celecoxib (a COX-2 specific inhibitor), had no effect on cell growth (Figure 2A). This finding is consistent with previous work showing that attenuation of canonical NF-κB signaling in MCF10A cells expressing mutant PIK3CA E545K or H1047R with either the IKKβ ATP-competitive inhibitor BAY-65-1942, or by ectopic expression of an IκBα super-repressor does not affect monolayer growth or colony formation in soft agar (17). Furthermore, shRNA-mediated suppression of COX-2 in SUM159-PT (Figure 2B) cells did not affect cell proliferation (Figure 2C). Collectively, these findings suggested that the inhibitory effects of aspirin on proliferation of breast cancer cells expressing oncogenic PIK3CA are unlikely to be mediated exclusively by induction of NF-κB or COX-2.
Figure 2. Aspirin induces growth suppression of mutant PIK3CA breast cancer cells in an NF-κB and COX-2-independent manner.
(A) Sulforhodamine cell viability, SUM159-PT cells and MCF10A cells expressing mutant PIK3CA H1047R were treated with: 3mM aspirin (ASN), Compound A (Cmpd A) (2.5μM - MCF10A, 5μM - SUM159-PT), and 10μM Celecoxib, for 48hrs. Relative number of viable cells was determined by normalizing to vehicle (Tris-HCl, pH 7.2) control cells. Each bar represents four independent biological replicates ± the SEM. Statistical significance was determined using paired Student’s t-test, ***p<0.001. (B) QRT-PCR validation of shRNA-mediated suppression of COX-2 in SUM159-PT cells. Transcript levels were normalized to GAPDH. Each bar represents average of three technical replicates ± the SD. Statistical significance was determined using paired Student’s t-test, **p<0.01. (C) Proliferation assay, relative absorbance was determined by normalizing to day 0. Each bar represents three independent biological replicates ± the SEM. Values are not statistically siginificant at any of the indicated timepoints. Statistical significance at each timepoint was verified using paired Student’s t-test.
Aspirin activates AMPK and decreases mTORC1 signaling in mutant PIK3CA cells
Recently, aspirin was shown to target intracellular energy homeostasis and metabolism through direct activation of adenosine monophosphate-activated protein kinase (AMPK) and subsequent inhibition of mammalian target of rapamycin (mTOR) signaling (34,35). AMPK and mTORC1 are major sensors of nutrient and growth factor signaling that regulate cell proliferation and size (36,37). Thus, we investigated the contribution of AMPK and mTORC1 signaling to the growth defects induced by aspirin in mutant PIK3CA cells.
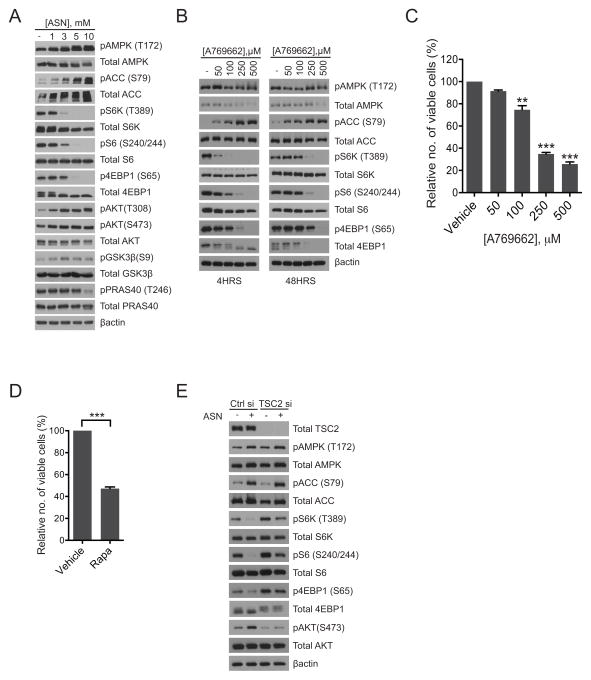
Expression of mutant PIK3CA E545K and H1047R in MCF10A cells resulted in attenuation of AMPK activity relative to cells expressing GFP vector control, or wild-type PIK3CA, as measured by phosphorylation of AMPK (pT172) and its substrate Acetyl-CoA carboxylase (ACC) (pS79). This was concomitant with increased mTORC1 signaling, as analyzed by phosphorylation of S6K1 (pT389), 4EBP1 (pS65) and S6 (pS240/244), all of which were blocked by the PIK3CA-specific inhibitor BYL719 (Supplementary figure 2A). As expected, oncogenic PIK3CA also increased phosphorylation of AKT (pS473) and downstream substrates PRAS40 (pT246) and GSK-3β (pS9) relative to PIK3CA wild-type cells (Supplementary figure 2A). In sum, these data demonstrate that mutant PIK3CA antagonizes AMPK signaling but positively regulates mTORC1 activity.
Interestingly, in breast cancer cells harboring oncogenic PIK3CA, aspirin treatment as well as its metabolite salicylate, led to activation of AMPK coincident with inhibition of mTORC1 activity, in a dose-dependent manner (Figure 3A, Supplementary figure 2B). This aspirin-induced attenuation of mTORC1 activity resulted in re-activation of AKT signaling (Figure 3A), similar to that observed with chronic Rapamycin treatment (38,39). Importantly, inhibition of COX-2 and IKKβ/NF-κB did not significantly attenuate mTORC1 activity (Supplementary figure 2B). Therefore, aspirin-induced inhibition of mTORC1 occurred independently of COX-2 and IKKβ/NF-κB signaling. Interestingly, both Celecoxib and Compound A modestly increased AMPK phosphorylation (pT172) relative to vehicle; however this did not translate into inhibition of mTORC1 signaling (Supplementary figure 2B). The mechanism that accounts for this increase remains unclear. Importantly treatment with the AMPK activator A769662 led to a dose-dependent decease in mTORC1 signaling as indicated by the decrease in phosphorylation of S6K1 (pT389), S6 (pS240/244) and 4EBP1 (pS65) (Figure 3B) relative to vehicle-treated cells, as well as a dose-dependent decrease in cell viability (Figure 3C). Interestingly, knockdown of AMPKα1/α2 partially rescued this attenuation of mTORC1 signaling upon treatment with aspirin (Supplementary figure 2C). In support of a role for mTORC1 inhibition in growth suppression, we observed that the mTORC1 inhibitor Rapamycin also decreased the growth of SUM159-PT cells (Figure 3D). Furthermore, siRNA depletion of Tuberous Sclerosis Complex 2 (TSC2) which results in an increase in phosphorylation of S6K1 (pT389), S6 (pS240/244) and 4EBP1 (pS65) (Figure 3E), partially rescued the suppression of mTORC1 signaling induced upon aspirin treatment (Figure 3E). Taken together, these mechanistic studies demonstrate that the growth inhibitory effect of aspirin can be ascribed, at least in part, to enhanced activation of AMPK and inhibition of mTORC1 signaling. They also provide a rationale for the increased sensitivity of mutant PIK3CA cells to aspirin treatment, given the importance of AMPK/mTORC1 signaling in these cells.
Figure 3. Aspirin activates AMPK and decreases mTORC1 signaling.
Immunoblot analysis of SUM159-PT cells treated with: (A) aspirin (1–10mM) for 16hrs and (B) A769662 (50–500μM) for 4hrs and 48hrs respectively. Shown are representative immunoblots from two independent experiments. (C) Sulforhodamine cell viability, SUM159-PT cells treated with increasing concentrations of A769662 and assessed after 48 hrs. (D) Sulforhodamine cell viability, SUM159-PT cells treated with 100nM Rapamycin for 48 hrs. For Sulforhodamine cell viability assays, each bar represents three independent biological replicates ± the SEM. Statistical significance was determined using paired Student’s t-test, **p<0.01, ***p<0.001. (E) Immunoblot analysis of SUM159-PT cells transfected with TSC2 SMARTpool: ONTARGETplus siRNA or control siRNA (final concentration of 20nM each) for 32 hours followed by treatment with 3mM aspirin for 16hrs. Shown are representative immunoblots from two independent experiments.
Aspirin sensitizes mutant PIK3CA breast cancer cells to PI3K inhibitors
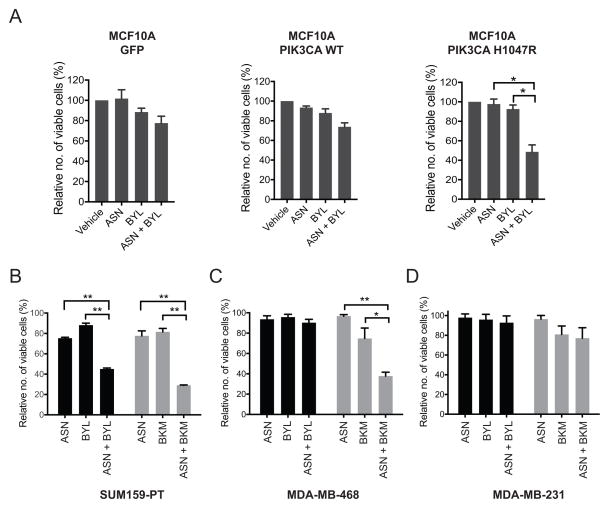
Given that aspirin treatment resulted in re-activation of AKT signaling, we hypothesized that aspirin, by modulating PI3K/mTORC1 signaling, might sensitize or prime cells for PI3K inhibition. Therefore, we investigated the ability of aspirin to attenuate the growth of PIK3CA-mutant breast cancer cell lines in combination with PI3K inhibitors. Suboptimal levels of aspirin were used in order to reveal any enhanced effects due to the combination of aspirin with various PI3K inhibitors. In contrast to GFP- or PIK3CA wild-type expressing cells, MCF10A cells expressing oncogenic PIK3CA (H1047R) displayed a significant decrease in the number of viable cells following treatment of aspirin in combination with BYL719, relative to either drug alone (Figure 4A, Supplementary figure 3). Similarly, in SUM159-PT cells (PIK3CA H1047L) enhanced growth inhibition was observed upon co-treatment with aspirin and BYL719, compared to each drug alone (Figure 4B).
Figure 4. Aspirin sensitizes mutant PIK3CA breast cancer cells to PI3K inhibitors.
(A) Sulforhodamine assay, MCF10A cells expressing JP1520-GFP, PIK3CA-WT and PIK3CA H1047R respectively, treated with 1.5mM aspirin and 0.5μM BYL719 for 48 hrs. Each data point represents the average of three independent biological replicates ± the SEM. (B–D) Sulforhodamine assay, SUM159-PT, MDA-MB-468 and MDA-MB-231 cells treated with 1.5mM aspirin (ASN) and 1μM BYL719 (BYL) or 1μM BKM120 (BKM) for 48hrs. Relative number of viable cells was determined by normalizing to vehicle control. Each bar represents three independent biological replicates ± the SEM. Statistical significance was determined using paired Student’s t-test, * p<0.05, ** p<0.01, ***p<0.001.
By contrast, the combination of aspirin and BYL719 showed little additive effect in MDA-MB-468 cells, a PTEN-mutant breast cancer line (Figure 4C). However, co-treatment of aspirin and BKM120, a pan class I PI3K inhibitor, showed an additive effect (Figure 4C) in these cells, consistent with the notion that PTEN-deficient tumors signal primarily through p110β (PIK3CB), and would therefore be insensitive to p110α-specific inhibitors (40). Importantly, MDA-MB-231 cells that do not harbor PI3K pathway mutations were insensitive to the combination of aspirin with either BYL719 or BKM120 (Figure 4D).
Co-treatment of aspirin and PI3K inhibitors leads to enhanced activation of AMPK, inhibition of mTORC1 and induction of autophagy
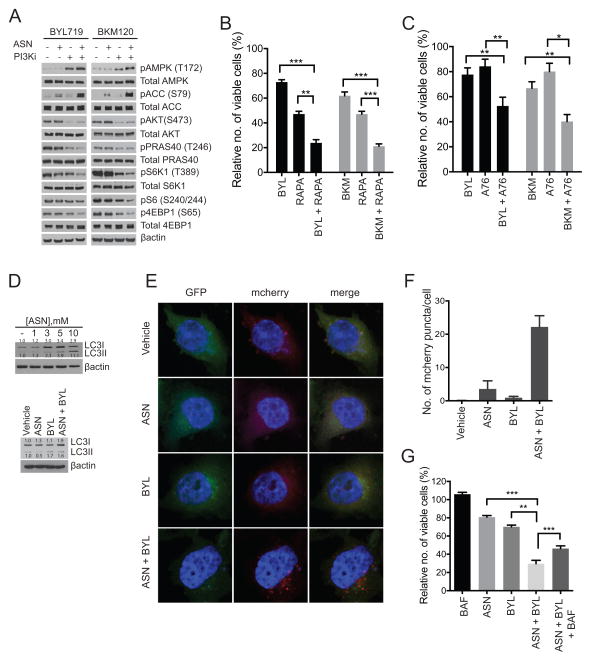
Next, we investigated the molecular basis for growth suppression of PI3K pathway mutant breast cancer cells in response to dual treatment of aspirin and PI3K inhibitors. In terms of signaling mechanisms, the combination of aspirin with either PI3K inhibitors resulted in increased phosphorylation of AMPK (pT172) and ACC (pS79) along with decreased phosphorylation of S6K1 (pT389), S6 (pS240/244) and 4EBP1 (pS65) relative to either drug alone (Figure 5A). Similar to aspirin, co-treatment of SUM159-PT cells with Rapamycin and both PI3K inhibitors (BYL719, BKM120) also led to a significant decrease in cell viability relative to either drug alone (Figure 5B). This observation also held true for the AMPK activator A769662 (Figure 5C).
Figure 5. Co-treatment of aspirin and PI3K inhibitors leads to enhanced activation of AMPK, inhibition of mTORC1 and induction of autophagy.
(A) Immunoblot analysis of SUM159-PT cells treated with 1.5mM aspirin and 1μM BYL719 for 24hrs or 1μM BKM120 for 16hrs. Representative immunoblot of three independent experiments. Sulforhodamine assay: (B) SUM159-PT cells treated with 50nM Rapamycin (RAPA) and 1μM BYL719 (BYL) or 1μM BKM120 (BKM) for 48hrs; Each bar represents three independent biological replicates ± the SEM. Statistical significance was determined using paired Student’s t-test, * p<0.05, ** p<0.01, ***p<0.001. (C) SUM159-PT cells treated with 100μM A769662 (A76) and 1μM BYL719 or 1μM BKM120 for 48hrs. Each bar represents three independent biological replicates ± the SEM. Statistical significance was determined using paired Student’s t-test, * p<0.05, ** p<0.01, ***p<0.001. (D) Immunoblot of LC3I/II in SUM159-PT cells treated with: increasing concentrations of aspirin for 48hrs (top), and 1.5mM aspirin (ASN) and 1μM BYL719 for 48hrs (bottom). Representative immunoblots from two independent experiments. (E) Immunofluorescent images of SUM159-PT cells expressing a mCherry-EGFP-LC3B reporter, treated with 1.5mM aspirin and 1μM BYL719 for 48hrs. (F) Quantification of the number of mCherry labeled puncta per cell, using Image J (n=20). (G) Sulforhodamine assay, SUM159-PT cells treated with 1.5mM aspirin, 1μM BYL719 and10nM Bafilomycin A (BAF) for 48hrs. Each bar represents three independent biological replicates ± the SEM. Relative number of viable cells was determined by normalizing to vehicle control. Statistical significance was determined using paired Student’s t-test, * p<0.05, ** p<0.01, ***p<0.001.
AMPK and mTORC1 are critical regulators of autophagy, a catabolic process that facilitates organelle and protein recycling and/or degradation to maintain cellular homeostasis (41). Activation of autophagy can be tumor suppressive as demonstrated by loss-of-function studies of autophagy genes such as Beclin1 (42). Early stages of autophagy involve recruitment and processing of cytosolic light chain LC3-I to membrane-associated LC3-II, while the latter stages involve maturation and fusion of the autophagosome to the lysosome (41). Given that co-treatment of aspirin and BYL719 led to activation of AMPK and suppression of mTORC1 signaling and that aspirin results in an increase in autophagy (Figure 5D), we investigated the effect of this drug combination on autophagy. In order to assess activation of autophagy we employed a mCherry-GFP-LC3B reporter system (43,44). Upon activation of autophagy, cytosolic mCherry-GFP-LC3B is first recruited to autophagosomes and can be visualized as distinct puncta. Following this, autophagosomes undergo fusion with the lysosome to form the autolysosome. Due to the acidic environment of the autolysosomes, positive pH-insensitive mCherry fluorescence is mainly observed while pH sensitive GFP signal is diminished. Thus formation of mCherry-LC3B positive puncta can be used as a surrogate marker for activation of autophagy. We observed that treatment of SUM159-PT cells with a combination of aspirin and BYL719 resulted in an increase in LC3I/II total protein (Figure 5D) as well as an increase in the number of fluorescently labeled mCherry-LC3B puncta, relative to each drug alone (Figure 5E, 5F). Together, these observations demonstrate that aspirin in combination with BYL719 results in activation of autophagy.
We next investigated whether activation of autophagy was responsible for the decrease in cell viability observed upon co-treatment with aspirin and BYL719. To address this, we treated SUM159-PT cell with the autophagy inhibitor, Bafilomycin A. This autophagy inhibitor partially rescued the decrease in cell viability induced by co-treatment of aspirin and BYL719 (Figure 5G).
Aspirin augments tumor growth suppression induced by BYL719
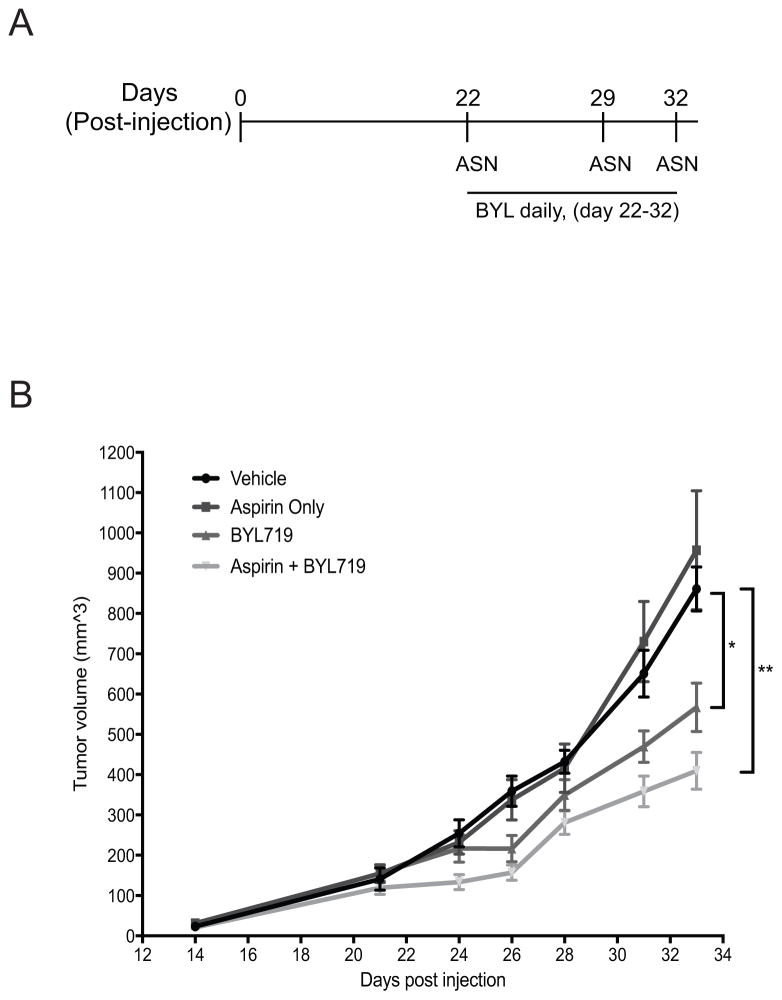
Given the pleiotropic effects of aspirin, we investigated the effects of aspirin in combination with BYL719 in SUM159-PT mouse xenografts. Tumor cells were implanted into NSG mice and allowed to grow for a period of 22 days, when average tumor volume attained ~120–160 mm3. Mice were then divided into 4 cohorts with comparable tumor size across all cohorts and treated with vehicle control, aspirin alone, BYL719 alone, or aspirin + BYL719 combination (Figure 6A). Tumor-bearing mice were treated daily with BYL719 by oral gavage while aspirin was administered by intraperitoneal injection, as indicated (Figure 6A). At the end of the experiment, tumors of mice treated with BYL719 alone showed a significant lower volume compared to vehicle - control (p=0.03184). The volume reduction was even more pronounced in mice treated with BYL719 in combination with aspirin (BYL719 + aspirin vs. control: FDR p-value= 0.00224) (Figure 6B). No statistically significant changes were observed in tumor volume between mice treated with BYL719 alone vs. the combination of BYL719 with aspirin (FDR p-value=n.s.) at this single time-point (day 33). However, based on our mixed effects model of tumor growth and, independently of the time point selected, we found a significant decrease in the tumor growth rate of approximately 0.21% (p=0.02) with the addition of aspirin to BYL719. The combination treatment group also showed a decrease of 44% (p<0.0001) of tumor growth rate relative to the vehicle control group. Notably, aspirin alone did not result in a reduction in tumor growth as previously observed in Figure 1E. This is likely due to the lower frequency of aspirin administration, which was initially employed to minimize any toxicity or negative side effects from daily treatment of the above drug combination. This observation reinforces epidemiological findings that highlight the importance of duration and dosage of aspirin used, in order to obtain a chemotherapeutic benefit (11).
Figure 6. Aspirin augments tumor growth suppression induced by BYL719.
(A) Schematic showing treatment regime for administration of aspirin and BYL719 in SUM159-PT xenografted mice. (B) Growth kinetics indicating average tumor volume at each time point in cohorts of mice treated with vehicle (0.5% methylcellulose, DMSO/PBS), Aspirin (100mg/kg), BLY719 (10mg/kg), or Aspirin + BYL (n=10 tumors/group; 5 mice each containing bilateral tumors). Aspirin or DMSO vehicle was administered via intraperitoneal injection while BYL719 was administered via oral gavage. Statistical analysis was performed using a pairwise student t-test (with FDR correction), for comparison of tumor volume at day 33; * p<0.05, ** p<0.01.
Discussion
Although aspirin is primarily administered as an antipyretic and analgesic, numerous observational and randomized clinical trials have suggested a potential chemotherapeutic use in cancer patients (9,10,45). Two independent studies have demonstrated that aspirin use is associated with increased survival in colorectal cancer patients with mutant-PIK3CA but not among those with the wild-type gene (12,13). Despite these observations, the molecular basis for this phenomenon remains poorly understood. By contrast, there are few well-characterized studies on a potential therapeutic benefit from aspirin intake in breast cancer. However, somatic PIK3CA mutations are highly prevalent in breast cancer, and aspirin use is associated with a decrease in breast cancer mortality and distant recurrence (11).
In this study, we investigated whether adjuvant aspirin treatment could improve the efficacy of specific PIK3CA and PI3K inhibitors currently under clinical trial evaluation for the treatment of breast cancer. To date, many of these inhibitors display limited clinical efficacy. Our data indicate that aspirin decreases the growth and viability of mutant PIK3CA breast cancer cells both in vitro and in vivo. Cells expressing mutant PIK3CA are more sensitive to aspirin compared to cells expressing wild-type PIK3CA in vitro. Notably, relatively lower concentrations of aspirin robustly blocked the growth of PIK3CA mutant cells grown in soft agar, suggesting that under conditions of stress, PIK3CA mutant cells may be more sensitized to aspirin treatment.
Our findings demonstrate that the cell growth inhibitory effects of aspirin on mutant PIK3CA cells are unlikely to be mediated exclusively by induction of NF-κB or COX-2. Instead, the aspirin-induced growth suppression phenotype can be ascribed to activation of AMPK and inhibition of mTORC1 signaling. While COX-2 expression strongly associates with the benefit of aspirin intake in colorectal cancer, epidemiological studies have shown that improved breast cancer survival associated with aspirin use is independent of COX-2 status (46). This is consistent with mechanistic studies, which demonstrated that the antitumor effects of aspirin are COX-2 independent and are mediated by other pathways, including Wnt/β-catenin signaling (47,48).
Although our data point to COX-2 and NF-κB-independent mechanism(s) in the cell autonomous effects of aspirin in PIK3CA mutant breast cancer, we cannot completely rule out the contribution of these two aspirin targets in PI3K-driven breast cancer. In the context of the tumor microenvironment, attenuation of COX-2 activity and NF-κB signaling blocks cancer growth and metastasis via several mechanisms that target the immune system, platelet activation/angiogenesis and other components of the tumor milieu (49–51). This notion is consistent with the finding that mutant PIK3CA activates IKKβ/NF-κB which in turn plays a critical role in the regulation of inflammatory genes, including the PTGS2 gene which encodes for COX-2. Therefore, the multiple direct targets of aspirin, including COX-2, NF-κB as well as AMPK likely explain the sensitivity of mutant PIK3CA cancers to this drug.
We also demonstrate that aspirin in combination with PI3K inhibitors results in enhanced growth suppression of PIK3CA/PTEN mutant breast cancer cells. This growth suppression phenotype is due to enhanced AMPK activation and inhibition of mTORC1 signaling. We further show that mutant PIK3CA inhibits AMPK activity but activates mTORC1 signaling, most likely due to mutant PIK3CA-induced activation of AKT. AKT phosphorylation of AMPKα1 at pS487 has been shown to inhibit AMPK-α1 activation and phosphorylation at pT172 (52). In the case of mTORC1, AKT activation promotes mTORC1 signaling through several mechanisms including phosphorylation of TSC2 and PRAS40 (53,54). Although activation of AMPK has been shown to inhibit mTORC1 signaling, future studies aimed at deciphering whether aspirin-induced inhibition of mTORC1 signaling is AMPK dependent, are warranted.
Aspirin is a readily available and inexpensive drug already approved by regulatory agencies worldwide and with many potential chemotherapeutic properties. In contrast to other drugs being tested for cancer treatment, it has fewer severe side effects and due to its pleiotropic targets, may be effective at targeting both the primary tumor and distant metastases. Taken together, our findings provide a mechanistic rationale for the use of aspirin in combination with PI3K inhibitors for the treatment of breast cancers that show PI3K-pathway dependency. It also highlights the importance of assessing whether PIK3CA mutation status would be a reliable biomarker for identifying breast cancer patients who are most likely to benefit from adjuvant aspirin therapy. Our study reinforces the need in performing clinical trials to evaluate the potential chemotherapeutic effects of aspirin in breast cancer patients, and whether any beneficial outcome outweighs side effects due to chronic aspirin intake, and ultimately how this ancient drug compares to other chemotherapeutic agents currently under clinical evaluation.
Supplementary Material
Acknowledgments
This study was supported in part by grants from the National Institutes of Health (A.T., CA177910), DOD/CDMRP/BCRP Era of Hope Scholar Award (S.S.M.), and a Howard Hughes Medical Institute International Student pre-doctoral fellowship (W. S. H.).
We thank Drs. John Rinn, Carl Novina, Lewis Cantley, Michele Holmes and members of the Toker laboratory for productive discussions; Joan Brugge, Kornelia Polyak, Albert Baldwin and Brendan Manning for providing reagents; Amey Barakat for technical assistance.
Footnotes
The authors disclose no potential conflict of interest
References
- 1.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 2.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 3.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer research. 2005;65:10992–1000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 4.Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–20. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandolfi PP. Breast cancer--loss of PTEN predicts resistance to treatment. N Engl J Med. 2004;351:2337–8. doi: 10.1056/NEJMcibr043143. [DOI] [PubMed] [Google Scholar]
- 6.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levesque H, Lafont O. Aspirin throughout the ages: a historical review. La Revue de medecine interne/fondee par la Societe nationale francaise de medecine interne. 2000;21(Suppl 1):8s–17s. doi: 10.1016/s0248-8663(00)88720-2. [DOI] [PubMed] [Google Scholar]
- 8.Hennekens CH, Dyken ML, Fuster V. Aspirin as a therapeutic agent in cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation. 1997;96:2751–3. doi: 10.1161/01.cir.96.8.2751. [DOI] [PubMed] [Google Scholar]
- 9.Baron JA, Cole BF, Sandler RS, Haile RW, Ahnen D, Bresalier R, et al. A randomized trial of aspirin to prevent colorectal adenomas. N Engl J Med. 2003;348:891–9. doi: 10.1056/NEJMoa021735. [DOI] [PubMed] [Google Scholar]
- 10.Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet. 2012;379:1591–601. doi: 10.1016/S0140-6736(12)60209-8. [DOI] [PubMed] [Google Scholar]
- 11.Holmes MD, Chen WY, Li L, Hertzmark E, Spiegelman D, Hankinson SE. Aspirin intake and survival after breast cancer. J Clin Oncol. 2010;28:1467–72. doi: 10.1200/JCO.2009.22.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo E, Church DN, Sieber O, Ramamoorthy R, Yanagisawa Y, Johnstone E, et al. Evaluation of PIK3CA Mutation As a Predictor of Benefit From Nonsteroidal Anti-Inflammatory Drug Therapy in Colorectal Cancer. J Clin Oncol. 2013;31:4297–305. doi: 10.1200/JCO.2013.50.0322. [DOI] [PubMed] [Google Scholar]
- 14.Goel A, Chang DK, Ricciardiello L, Gasche C, Boland CR. A novel mechanism for aspirin-mediated growth inhibition of human colon cancer cells. Clin Cancer Res. 2003;9:383–90. [PubMed] [Google Scholar]
- 15.Hu M, Peluffo G, Chen H, Gelman R, Schnitt S, Polyak K. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3372–7. doi: 10.1073/pnas.0813306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 17.Hutti JE, Pfefferle AD, Russell SC, Sircar M, Perou CM, Baldwin AS. Oncogenic PI3K mutations lead to NF-kappaB-dependent cytokine expression following growth factor deprivation. Cancer research. 2012;72:3260–9. doi: 10.1158/0008-5472.CAN-11-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown KK, Montaser-Kouhsari L, Beck AH, Toker A. MERIT40 Is an Akt Substrate that Promotes Resolution of DNA Damage Induced by Chemotherapy. Cell reports. 2015;11:1358–66. doi: 10.1016/j.celrep.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuznetsov HS, Marsh T, Markens BA, Castano Z, Greene-Colozzi A, Hay SA, et al. Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer discovery. 2012;2:1150–65. doi: 10.1158/2159-8290.CD-12-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laajala TD, Corander J, Saarinen NM, Makela K, Savolainen S, Suominen MI, et al. Improved statistical modeling of tumor growth and treatment effect in preclinical animal studies with highly heterogeneous responses in vivo. Clin Cancer Res. 2012;18:4385–96. doi: 10.1158/1078-0432.CCR-11-3215. [DOI] [PubMed] [Google Scholar]
- 21.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. 2014 arXiv preprint arXiv:14065823. [Google Scholar]
- 22.Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nature cell biology. 2001;3:785–92. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman LS, Hardman JG, Limbird LE, Gilman AG. Goodman & Gilman’s the pharmacological basis of therapeutics. New York: McGraw-Hill; 2001. p. xxvii.p. 2148. [Google Scholar]
- 24.Juarez Olguin H, Flores Perez J, Lares Asseff I, Loredo Abdala A, Carbajal Rodriguez L. Comparative pharmacokinetics of acetyl salicylic acid and its metabolites in children suffering from autoimmune diseases. Biopharmaceutics & drug disposition. 2004;25:1–7. doi: 10.1002/bdd.379. [DOI] [PubMed] [Google Scholar]
- 25.Yin MJ, Yamamoto Y, Gaynor RB. The anti-inflammatory agents aspirin and salicylate inhibit the activity of I(kappa)B kinase-beta. Nature. 1998;396:77–80. doi: 10.1038/23948. [DOI] [PubMed] [Google Scholar]
- 26.Vane JR. Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nature: New biology. 1971;231:232–5. doi: 10.1038/newbio231232a0. [DOI] [PubMed] [Google Scholar]
- 27.Roth GJ, Majerus PW. The mechanism of the effect of aspirin on human platelets. I. Acetylation of a particulate fraction protein. The Journal of clinical investigation. 1975;56:624–32. doi: 10.1172/JCI108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecomte M, Laneuville O, Ji C, DeWitt DL, Smith WL. Acetylation of human prostaglandin endoperoxide synthase-2 (cyclooxygenase-2) by aspirin. The Journal of biological chemistry. 1994;269:13207–15. [PubMed] [Google Scholar]
- 29.Kopp E, Ghosh S. Inhibition of NF-kappa B by sodium salicylate and aspirin. Science. 1994;265:956–9. doi: 10.1126/science.8052854. [DOI] [PubMed] [Google Scholar]
- 30.Kaltschmidt B, Linker RA, Deng J, Kaltschmidt C. Cyclooxygenase-2 is a neuronal target gene of NF-kappaB. BMC molecular biology. 2002;3:16. doi: 10.1186/1471-2199-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto K, Arakawa T, Ueda N, Yamamoto S. Transcriptional roles of nuclear factor kappa B and nuclear factor-interleukin-6 in the tumor necrosis factor alpha-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. The Journal of biological chemistry. 1995;270:31315–20. doi: 10.1074/jbc.270.52.31315. [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Oh JM, No JH, Bang YJ, Juhnn YS, Song YS. Involvement of NF-kappaB and AP-1 in COX-2 upregulation by human papillomavirus 16 E5 oncoprotein. Carcinogenesis. 2009;30:753–7. doi: 10.1093/carcin/bgp066. [DOI] [PubMed] [Google Scholar]
- 33.Ziegelbauer K, Gantner F, Lukacs NW, Berlin A, Fuchikami K, Niki T, et al. A selective novel low-molecular-weight inhibitor of IkappaB kinase-beta (IKK-beta) prevents pulmonary inflammation and shows broad anti-inflammatory activity. British journal of pharmacology. 2005;145:178–92. doi: 10.1038/sj.bjp.0706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Din FV, Valanciute A, Houde VP, Zibrova D, Green KA, Sakamoto K, et al. Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology. 2012;142:1504–15. e3. doi: 10.1053/j.gastro.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawley SA, Fullerton MD, Ross FA, Schertzer JD, Chevtzoff C, Walker KJ, et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–22. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes & development. 2011;25:1895–908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, Yan H, Frost P, Gera J, Lichtenstein A. Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade. Mol Cancer Ther. 2005;4:1533–40. doi: 10.1158/1535-7163.MCT-05-0068. [DOI] [PubMed] [Google Scholar]
- 39.Wan X, Harkavy B, Shen N, Grohar P, Helman LJ. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene. 2007;26:1932–40. doi: 10.1038/sj.onc.1209990. [DOI] [PubMed] [Google Scholar]
- 40.Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, et al. PTEN-deficient cancers depend on PIK3CB. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13057–62. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10:1533–41. doi: 10.1158/1535-7163.MCT-11-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. The Journal of clinical investigation. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 44.Nyfeler B, Bergman P, Wilson CJ, Murphy LO. Quantitative visualization of autophagy induction by mTOR inhibitors. Methods Mol Biol. 2012;821:239–50. doi: 10.1007/978-1-61779-430-8_14. [DOI] [PubMed] [Google Scholar]
- 45.Rothwell PM, Fowkes FG, Belch JF, Ogawa H, Warlow CP, Meade TW. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377:31–41. doi: 10.1016/S0140-6736(10)62110-1. [DOI] [PubMed] [Google Scholar]
- 46.Holmes MD, Chen WY, Schnitt SJ, Collins L, Colditz GA, Hankinson SE, et al. COX-2 expression predicts worse breast cancer prognosis and does not modify the association with aspirin. Breast Cancer Res Treat. 2011;130:657–62. doi: 10.1007/s10549-011-1651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bos CL, Kodach LL, van den Brink GR, Diks SH, van Santen MM, Richel DJ, et al. Effect of aspirin on the Wnt/beta-catenin pathway is mediated via protein phosphatase 2A. Oncogene. 2006;25:6447–56. doi: 10.1038/sj.onc.1209658. [DOI] [PubMed] [Google Scholar]
- 48.Hanif R, Pittas A, Feng Y, Koutsos MI, Qiao L, Staiano-Coico L, et al. Effects of nonsteroidal anti-inflammatory drugs on proliferation and on induction of apoptosis in colon cancer cells by a prostaglandin-independent pathway. Biochemical pharmacology. 1996;52:237–45. doi: 10.1016/0006-2952(96)00181-5. [DOI] [PubMed] [Google Scholar]
- 49.Xu L, Stevens J, Hilton MB, Seaman S, Conrads TP, Veenstra TD, et al. COX-2 inhibition potentiates antiangiogenic cancer therapy and prevents metastasis in preclinical models. Sci Transl Med. 2014;6:242ra84. doi: 10.1126/scitranslmed.3008455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markosyan N, Chen EP, Evans RA, Ndong V, Vonderheide RH, Smyth EM. Mammary carcinoma cell derived cyclooxygenase 2 suppresses tumor immune surveillance by enhancing intratumoral immune checkpoint activity. Breast Cancer Res. 2013;15:R75. doi: 10.1186/bcr3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang DJ, Ratnam NM, Byrd JC, Guttridge DC. NF-kappaB functions in tumor initiation by suppressing the surveillance of both innate and adaptive immune cells. Cell reports. 2014;9:90–103. doi: 10.1016/j.celrep.2014.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hawley SA, Ross FA, Gowans GJ, Tibarewal P, Leslie NR, Hardie DG. Phosphorylation by Akt within the ST loop of AMPK-alpha1 down-regulates its activation in tumour cells. The Biochemical journal. 2014;459:275–87. doi: 10.1042/BJ20131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nature cell biology. 2007;9:316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 54.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature cell biology. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.