Abstract
MYC is a pleiotropic transcription factor that activates and represses a wide range of target genes and is frequently deregulated in human tumors. While much is known about the role of MYC in transcriptional activation and repression, MYC can also regulate mRNA cap methylation through a mechanism that has remained poorly understood. Here it is reported that MYC enhances mRNA cap methylation of transcripts globally, specifically increasing mRNA cap methylation of genes involved in Wnt/β-catenin signaling. Elevated mRNA cap methylation of Wnt signaling transcripts in response to MYC leads to augmented translational capacity, elevated protein levels, and enhanced Wnt signaling activity. Mechanistic evidence indicates that MYC promotes recruitment of RNA methyltransferase (RNMT) to Wnt signaling gene promoters by enhancing phosphorylation of serine 5 on the RNA Polymerase II Carboxy-Terminal Domain, mediated in part through an interaction between the TIP60 acetyltransferase complex and TFIIH.
Implications
MYC enhances mRNA cap methylation above and beyond transcriptional induction.
Keywords: mRNA cap methylation, MYC, canonical Wnt signaling
Introduction
MYC is essential for normal cell growth, development, and differentiation and plays an important role in cell metabolism, protein synthesis, and cell cycle regulation (1, 2). When MYC is deregulated or constitutively activated via overexpression, amplification, or chromosomal translocation, cells can be transformed and proliferate in the absence of extracellular mitogenic input (3). MYC overexpression is a hallmark of approximately 70% of cancers (4), and inhibition of MYC has been shown to result in tumor regression in a host- or cell-type dependent manner (5). MYC is a pleiotropic transcription factor that induces genome-wide transcriptional amplification (6, 7) by interacting with its cofactor transactivation/transformation-associated protein (TRRAP) (8) though there have been multiple interpretations of the data regarding MYC’s role in transcriptional gene activation and repression (9, 10). TRRAP is found in complex with either TIP60 or GCN5, histone acetyltransferases (HATs) that induce an open chromatin conformation and transcription (11). Despite many advances in the MYC field, functions of MYC independent of transactivation and repression of genes have not been completely evaluated.
Canonical Wnt signaling can promote transcription and elevated expression of MYC (12, 13). Conversely, MYC has been shown to activate Wnt signaling by repressing the Wnt signaling inhibitors, DKK1 and SFRP1, indicating interplay between MYC and Wnt signaling (14). Canonical Wnt/β-catenin signaling has been implicated in tissue homeostasis and development and regulates numerous cell processes via mitogenic stimulation, including cell differentiation, cell polarity and motility (15, 16). Wnt signaling is associated with a number of diseases, including coronary artery disease, osteoporosis, diabetes, obesity, and cancer (17).
Recent studies have revealed that MYC possesses the ability to post-transcriptionally promote protein accumulation without altering mRNA levels by stimulating mRNA cap methylation (18-20). This effect is independent of MYC induced transcription as mRNA cap methylation occurs in the absence of DNA and/or MAX binding. mRNA capping and methylation occur during transcriptional initiation (21, 22). Phosphorylation of serine-5 (phospho-S5) on the Carboxy-Terminal Domain (CTD) of RNA Polymerase II (RNA Pol II) leads to the production of nascent mRNA and recruitment of RNA guanylyltransferase (RNGTT) and RNA (guanine-7-)-methyltransferase (RNMT) (23-25). RNGTT adds an inverted guanosine cap and RNMT, in combination with RNMT-Activating Mini protein (RAM), methylates the cap on mRNA (20, 26-28). mRNA cap methylation further stabilizes the nascent mRNA, protects from exonuclease attack, and is required for efficient cap-dependent mRNA translation and protein production (29, 30). MYC was shown to promote mRNA cap methylation of select MYC transcriptional target genes and cooperates with RNMT to promote cell transformation (19, 20). Thus far, the extent to which MYC mediates mRNA cap methylation is unknown. The mechanism behind MYC-mediated mRNA cap methylation and its impact in the context of normal and transformed cells are not well characterized.
Herein, we report that MYC promotes mRNA cap methylation and protein production of Wnt/β-catenin signaling transcripts through recruitment of cyclin-dependent kinase 7 (CDK7) and consequently RNMT to gene promoters. Interactions between MRG15, a component of the NuA4 TRRAP complex, and XPB, a component of TFIIH, are necessary for MYC-mediated CDK7 recruitment. These findings demonstrate that MYC employs multiple mechanisms, including mRNA cap methylation, to promote protein production, proliferation, and transformation.
Materials and Methods
Cell lines, drug treatment, and Western blotting
For c-MYC induction experiments, the conditional Tet-MYC vector in P493-6 cells was repressed with 0.1 μg/ml tetracycline (Sigma) for 72 h (31). P493-6 cells were kindly provided by Chi Van Dang, University of Pennsylvania. MYC expression was induced for 1 h or 24 h by washing the cells three times with RPMI-1640 medium containing 10% tetracycline system-approved FBS (Clontech). Immortalized mammary epithelial cells (IMECs) and MCF10A cells were cultured in DMEM:F12 50:50 medium supplemented with 10 ng/mL EGF, 5 ng/mL insulin, 0.5 μg/mL hydrocortisone, and 5% FBS as previously described (32). MCF7, HeLa, HEK293T, and MDA-MB-231 cells were cultured in DMEM/10% FBS. MM1.S cells were propagated in RPMI-1640 medium containing 15% FBS. For SNS-032 and THZ1 (APExBIO) treatments, MYC expression was repressed in P493-6 cells for 72 h and restored through tetracycline withdrawal for 0 or 23 h prior to treatment. Cells were treated with 100 nM SNS-032 and 50 nM or 250 nM THZ1 for 24 h and 1 h, respectively, followed by F buffer (10 mM Tris pH 7.05, 50 mM NaCl, 30 mM Na4P2O7, 5 μM ZnCl2, 50 mM NaF, 10% Glycerol, 0.5% Triton X-100) lysis for immunoblotting or TRIzol RNA extraction for mRNA analysis. Western blots were imaged using the BioRad Molecular Imager ChemiDoc XRS+ System and band intensity was quantified using ImageJ.
Plasmids and RNA interference
Plasmid transfections (vector, HA-RNMT, FLAG-RAM, CBP-MYC) were performed using LipoD293™ In Vitro DNA Transfection Reagent per protocol (SignaGen). Cells were harvested for protein or RNA 24 h post-transfection. Silencer Select Pre-designed siRNAs were obtained from Life Technologies and transfected using RNAiMax (Invitrogen; 20nM). Cells were harvested for protein or RNA 48 h post-transfection. Silencer Select negative control #2 (Life Technologies) was used as a transfection control.
GST-eIF4E protein production and purification
E. coli optimized for efficient protein production, (BL21 (De3); Protein Express), were transformed with a pGEX-GST-eIF4E plasmid. Bacteria were spun down at 8,300 × g for 15 min and lysed in 20 mL of LCB buffer (0.1 M KCl, 20 mM HEPES, 0.2 mM EDTA) plus 1% NP40, 1 mg/mL lysozyme, and protease inhibitors. The lysate was kept on ice for 30 min, sonicated at 70% amplitude for 3 min (1 sec ON, 3 sec OFF), and spun at 27,000 × g for 30 min. The supernatant was run over a column containing 0.3 mL immobilized 2'/3'-EDA-m7GTP agarose beads (Jena Bioscience) two times, followed by three washes with Buffer A (20 mM HEPES pH 7.5, 10% glycerol, 5 mM 2-mercaptoethanol). GST-eIF4E was eluted three times with 1 μM m7GTP (Jena Bioscience) in Buffer A (600 μL each). The protein was dialyzed overnight in PBS plus 0.5 μM EDTA and 5% glycerol at 4°C.
GST-eIF4E immobilization and m7G-mRNA isolation
GST-eIF4E (~7 μg) was added to a 1.5 mL microcentrifuge tube containing 0.5 mL F buffer. Magnetic glutathione (GSH) beads (~20 μL) pre-blocked in binding buffer (7 mM HEPES pH 7.9, 300 mM NaCl, 0.08 mM EDTA, 0.8 mM DTT, 17% glycerol, 0.1% NP40, 1.1% polyvinyl alcohol, 400 μg/mL polyuridylic acid, 17 μg/mL poly(deoxyguanylic-deoxycytidylic) acid) overnight were subsequently added to each tube and tubes were rotated for 2 h at 4°C followed by two washes with F buffer (modified from McCracken, et al.) (23). pppN-mRNA, GpppN-mRNA, and m7GpppN-mRNA were in vitro transcribed from pKS-EZH2 using the MaxiScript kit (Ambion). MaxiScript mRNA (1 ng) or total RNA (1 μg) was added to each tube containing 0.3 mL binding buffer plus 1 μL RNasin Plus RNase Inhibitor (Promega) and rotated for 1.5 h at 4°C in order to isolate m7G-mRNA. This rotation was followed by two washes with wash buffer (20 mM HEPES pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 1 mM DTT, 20% glycerol, 0.5% NP40) where the beads were transferred to a new siliconized microcentrifuge tube in between the first and second wash. Following the second wash, mRNA bound to the beads was removed using TRIzol. mRNA was precipitated overnight in isopropanol (20 μg glycogen) at −20°C and was resuspended in 30 μL RNase-free water. m7G-mRNA isolated from the pulldown was used as a substrate for RT-qPCR and signals were normalized to total input mRNA for each condition.
RNA sequencing and analysis
RNA isolated from P493-6 cells at different time points following MYC induction (0 and 24 h) was subjected to a m7G-mRNA pulldown using GST-eIF4E (1 μg RNA). Input RNA and mRNA isolated from the pulldown (four samples in total; two samples from each time point, one input and one pulldown) were subjected to 100 bp paired-end sequencing using the Illumina-HiSeq platform. Reads were aligned to the human genome (hg38) using STAR aligner version 2.3.0.1 (default setting), and reads were quantified using the human genome in Partek Flow. Total mapped reads ranged from 13 to 17 million per sample for inputs (total mRNA) and from 3 to 19 million per sample for m7G-mRNA pulldowns. Genes with <5 total mapped reads were discarded to eliminate false positives. For input samples, gene reads were normalized to total mapped reads and cell number. For m7G-mRNA pulldown samples, reads were normalized to total mapped reads and cell number followed by normalization to input to eliminate MYC induced effects at the transcriptional level. Reads were normalized to cell number since P493-6 cells have increased cell size and RNA content per cell upon MYC induction. Unbiased clustering was performed to generate heat maps. The clusters were created by mean centering the log2 values, and the clustering was performed using a single linkage method (for input) or an average linkage method (for m7G-mRNA pulldown) with a Pearson un-centered correlation similarity metric.
RT-qPCR
Total RNA from log phase cultures or m7G-mRNA from GST-eIF4E pulldowns was isolated with TRIzol (Life Technologies), and cDNA was synthesized using the iScript cDNA Synthesis Kit (BioRad). Two-step real-time PCR was performed using SYBR Green Mix on a C1000 Thermal cycler (BioRad). Gene expression was normalized to cell number or total input mRNA and expressed as fold change relative to control.
Chromatin Immunoprecipitation (ChIP)
ChIPs were performed according to the Upstate ChIP Kit protocol (Millipore). Briefly, log phase cells were crosslinked with 1% formaldehyde at 37°C and sonicated twice after lysis using a water bath sonicator (BioRuptor) on high intensity for 10 minutes at 30-second intervals. qPCR was used to measure enrichment and data are presented as a proportion of bound DNA normalized to input for each sample (i.e. % input) relative to an IgG control.
Polysome Profiling
Log phase cells were treated with 100 μg/mL cycloheximide for 1 min, washed in PBS (100 μg/mL cycloheximide) and lysed in 200 μL lysis buffer (20 mM Tris pH 7.5, 2.5 mM MgCl2, 120 mM KCl, 0.3% NP40, 100 μg/mL cycloheximide). Extracts were centrifuged at 19,400 × g for 15 min. Supernatants (100 μg total RNA) were layered over a 20-mL gradient of 10-50% sucrose, 20 mM Tris pH 7.5, 2.5 mM MgCl2, 60mM KCl, 100 μg/mL cycloheximide. Gradients were centrifuged at 166,880 × g for 4 h. A Biocomp Gradient Master was used to collect 300-μL fractions and absorbance was measured at 260 nM. RNA was then purified from pooled fractions by Acid:Phenol:Chloroform (Ambion) extraction, and used as a substrate for GST-eIF4E m7G-mRNA pulldown. RNA was analyzed by RT-qPCR.
Protein synthesis assay
P493-6 cells were labeled with 50μM Click-iT AHA (L-azidohomoalanine) (Invitrogen) for 2 h at 37°C in methionine-free RPMI/1640 medium at two time points for MYC induction (0 h – MYC off, 24 h – MYC on). Manufacturer’s Click-iT labeling and protein detection protocols were followed with minor modification. Cells were lysed in 100 μL lysis buffer (1% SDS in 50 mM Tris-HCl, pH 8.0) for 15 min on ice, sonicated with a probe sonicator, and centrifuged at 19,400 × g at 4°C for 5 min. Supernatant containing protein was saved, and 60 μg of protein from each sample was added to the biotin alkyne detection reaction as specified in the protocol. A total of 30 μL (stock 200 μL) was run on an SDS-PAGE gel, and a Western blot was performed with probing using HRP-conjugated streptavidin (BioLegend) in 5% BSA for 1 h. Intensity was quantified using ImageJ.
TCF Reporter Assay
For T-cell factor (TCF) reporter assays, low-passage cells (105) were plated onto six-well plates. One day following plating, each well was transfected with 0.9 μg 14X Super TOP or 8X FOP FLASH plasmid (33) and 0.09 μg pRL-SV40 using LipoD293™ In Vitro DNA Transfection Reagent per protocol (SignaGen). Two days following transfection, cells were lysed, and luciferase and Renilla activity were measured with Dual-Glo luciferase reagents (Promega). Luciferase readings were normalized against Renilla readings, and TOP FLASH/FOP FLASH ratios were calculated.
Statistics
All experiments were repeated 3 times. An unpaired student’s t-test was performed with Welch’s correction to determine standard deviation and statistical significance. p ≤ 0.05 was considered statistically significant. Error bars represent SEM.
Further methodologic details are included in Supplementary Information.
Results
MYC promotes mRNA cap methylation of essential Wnt pathway transcripts
MYC was previously shown to promote mRNA cap methylation of a number of transcriptionally activated target genes in c-myc −/− (HO 15.19) rat fibroblast cells expressing exogenous MYC (18, 19). To explore this activity further, we optimized and validated a capturing tool to isolate m7G-mRNA for use in a transcriptome-wide study (Supplementary Fig. S1). In our analysis, we used P493-6 cells, a model of Burkitt’s lymphoma expressing a doxycycline-repressible MYC transgene. RNA was isolated under MYC-repressed and MYC-induced conditions for subsequent analysis of both total mRNA and mRNA cap methylation. We captured m7G-mRNA using recombinant GST-eIF4E and performed RNA-Seq on both input and eIF4E-bound mRNAs to identify the MYC transcriptional targets as well as mRNAs with enhanced cap methylation over and above any transcriptional induction (Fig. 1A; see Methods). As previously described, induction of MYC induced > 1.2 fold transactivation of approximately 80% of the 13,500 genes analyzed (Supplementary Fig. S2A), which is consistent with recent publications (4, 6, 7, 14). Transcriptional induction was validated using NanoString analysis (Supplementary Table S1). By comparing the relative induction of mRNA in the m7G fraction to the mRNA inputs, we determined the net effect on enhanced cap methylation following 24 h of MYC activation. Our analyses revealed that roughly 30% of analyzed transcripts (~1,200) are induced > 1.2 fold at the mRNA cap methylation level, over and above the transcriptional induction.
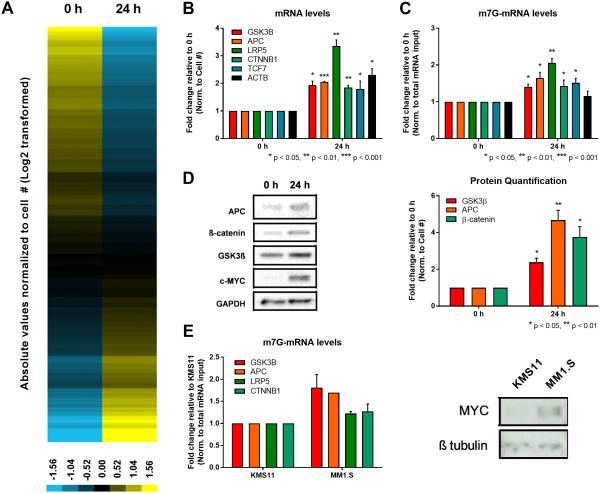
Figure 1.
MYC-mediated global induction of mRNA cap methylation including canonical Wnt/β-catenin signaling pathway transcripts. (A) Unbiased clustering of m7G-mRNA pulldowns from P493-6 cells normalized to cell number and changes at the level of total mRNA in response to MYC. Total of 3,294 genes analyzed. Values transformed by log2 and mean centered (yellow – up, blue – down). (B) Total mRNA levels of Wnt signaling transcripts in P493-6 cells normalized to cell number determined using RT-qPCR (n=3, biological). (C) m7G-mRNA levels of Wnt signaling transcripts in P493-6 cells normalized to total mRNA input (n=3, biological). (D) Western blot to detect protein levels of Wnt signaling factors in P493-6 cells. Quantification in right panel (n=3, biological). (E) m7G-mRNA levels of Wnt signaling transcripts in KMS11 and MM1.S multiple myeloma cells normalized to total mRNA input. Western blot in right panel. Error bars indicate SEM and asterisks indicate significance (*p < 0.05, **p < 0.01, ***p < 0.001).
To validate the RNA-Seq data and explore mechanism, we focused on transcripts that displayed a robust increase in mRNA cap methylation since these transcripts appeared most responsive to MYC for this modification. We found that MAX, a protein that heterodimerizes with MYC (34), was upregulated by MYC at the mRNA cap methylation and protein levels, above and beyond the small induction at the mRNA level (Supplementary Fig. S3). Furthermore, we noted that many key components in the canonical Wnt signaling pathway (14 transcripts) were among the fraction of genes with MYC-induced cap methylation, including GSK3B, APC, LRP5, CTNNB1, and TCF7. We analyzed expression of these five genes/proteins since they are direct mediators downstream of Wnt-receptor signaling. MYC induction resulted in increased total mRNA levels of all five Wnt signaling transcripts, as well as ACTB (a control transcript independent of Wnt signaling) (Fig. 1B), indicating that these genes are MYC transcriptional targets. More importantly, all five Wnt transcripts displayed a MYC-mediated increase in mRNA cap methylation, significantly above and beyond the effect on transcription, whereas there was no detectable increase in mRNA cap methylation of ACTB (Fig. 1C). This observation was validated for the same genes in two additional MYC-transduced systems, myc-null rat fibroblasts and immortalized human mammary epithelial cells (IMECs), +/− exogenous MYC. In both systems, MYC overexpression led to enhanced total mRNA and Wnt-transcript specific mRNA cap methylation in MYC overexpressing cells (Supplementary Fig. S2B-E). We also analyzed the protein levels of three of these genes (GSK3β, APC, and β-catenin) and found that the increase in mRNA cap methylation corresponded to an increase in protein levels above the expected effect due to enhanced transcription alone (Fig. 1D). Moreover, siRNA depletion of RNMT in IMEC:MYC cells resulted in diminished protein levels of GSK3β and APC relative to control siRNA (Supplementary Fig. S2F). Collectively, these data suggest that MYC post-transcriptionally induces an increase in protein levels from genes involved in Wnt signaling. To investigate whether or not MYC-mediated mRNA cap methylation occurs in cancer, we tested multiple myeloma cancer cell lines with varying MYC levels. mRNA cap methylation of Wnt pathway transcripts was most elevated in the cell line that had the highest MYC level, MM1.S cells, relative to KMS11 cells (Fig. 1E). We also found that mRNA cap methylation of Wnt signaling transcripts was enhanced in MM1.S cells compared to additional cancer cell lines with lower MYC expression and control, MCF-10A cells (Supplementary Fig. S2G).
MYC enhances translational capacity of Wnt pathway genes by inducing mRNA cap methylation
We next investigated the effect of MYC-mediated mRNA cap methylation on translation and protein synthesis. mRNA capping and methylation are required for efficient binding of translation initiation factors, and then m7G-mRNA is recruited to the 40S ribosomal subunit for translation (35, 36). Therefore, mRNA that is m7G-capped in response to MYC is predicted to have increased loading of ribosomes (i.e. polysomes) and an increased rate of translation. To assess polysome loading, mRNA bound to ribosomes was extracted from P493-6 cells at 0, 1, and 24 h post-MYC induction, and fractions were collected for analysis. Polysome profiling revealed that P493-6 cells with maximal MYC levels 24 h post-induction had a dramatic increase in the amount of mRNA in late polysome fractions (Fig. 2A), indicative of a higher translational capacity. The gradual shift of mRNA from the monosomal 80S peak to late polysomal peaks corresponded with the increase in MYC levels. Isolation and analysis of m7G-mRNA from polysome fractions showed that m7G-Wnt transcripts were present in late polysome fractions when MYC levels were highest at 24 h (Fig. 2B). Our results parallel previously published data from rat fibroblast cells in which MYC overexpressing cells had RNA profiles shifted to the late polysome fractions (20). Our data suggest that the increase in translation of Wnt pathway transcripts in P493-6 cells is in part due to increased mRNA cap methylation and more efficient loading of Wnt signaling mRNA on ribosomes.
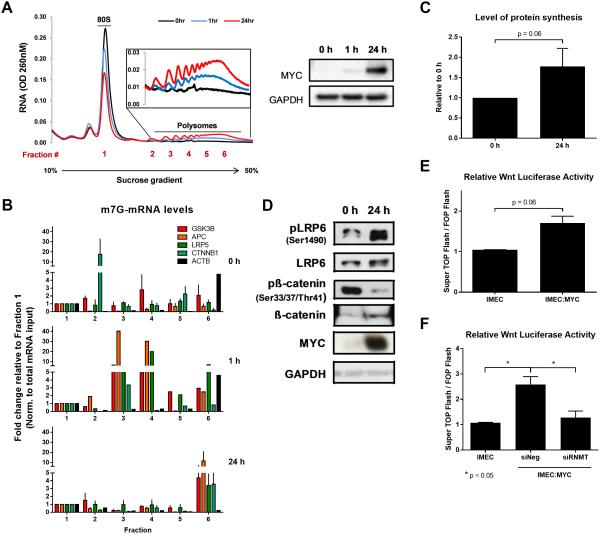
Figure 2.
MYC promotes translation and total protein synthesis by augmenting mRNA cap methylation of Wnt/β-catenin signaling pathway transcripts. (A) Polysome profiling of ribosome-bound mRNA from P493-6 cells at three time points of MYC induction (0, 1, and 24 h). Data shown are from one representative sample. (B) m7G-mRNA levels of Wnt signaling transcripts in each collected fraction (1 through 6) normalized to total mRNA input and fraction 1. (0 h – top panel, 1 h – middle panel, 24 h – bottom panel) (n=3, biological). (C) Quantification of total protein synthesis detected with HRP-streptavidin (n=3, biological). (D) Western blot detecting phosphorylation status and total LRP6 and β-catenin in P493-6 cells at 0 h and 24 h. (E) Relative Wnt signaling activity measured in IMEC and IMEC:MYC cells using a TCF/LEF luciferase reporter assay (n=3, biological). Expressed as ratio (TOP Flash/FOP Flash). (F) Relative Wnt signaling activity measured in IMEC and IMEC:MYC cells following RNMT depletion with siRNA using a TCF/LEF luciferase reporter assay (n=3, biological). Expressed as ratio (TOP Flash/FOP Flash). Error bars indicate SEM and asterisks indicate significance (*p < 0.05).
To confirm that increased mRNA cap methylation and polysome loading leads to a general increase in protein synthesis, a protein synthesis assay was performed. Increased incorporation of an azide-tagged amino acid analog was detected in P493-6 cells 24 h after MYC was induced compared to baseline (Fig. 2C). This increased amino acid incorporation is indicative of enhanced total protein synthesis, partly as a result of increased mRNA cap methylation, polysome loading on mRNA and augmented translational capacity.
To explore the net effect of MYC on Wnt signaling, we assayed phosphorylation of LRP6 and β-catenin in P493-6 cells. MYC induction led to increased levels of both LRP6 and β-catenin and an even greater induction of p-LRP6 and reduction of p-β-catenin, consistent with increased Wnt signaling (Fig. 2D, Supplementary Fig. S4A-B). Similarly, transient transfection of TOP-Flash/FOP-Flash TCF/LEF reporter plasmids into IMECs showed increased Wnt signaling with constitutive MYC overexpression (Fig. 2E). Conversely, depletion of RNA methyltransferase (RNMT) in MYC overexpressing cells resulted in reduced Wnt activity (Fig. 2F), suggesting that MYC-mediated induction of Wnt signaling activity is in part dependent on RNMT and mRNA cap methylation.
Depletion of RNMT or TRRAP complexes reduces mRNA cap methylation of Wnt signaling transcripts
The addition of a cap prevents degradation of a nascent mRNA (37, 38), while the addition of a methyl group enables effective binding of eIF4E and recruitment of ribosomes for translation (30). While RNA methyltransferase (RNMT) has been shown to promote cap methylation of Cyclin D1 mRNA in combination with MYC (20), the capping enzyme, RNA guanylyltransferase (RNGTT), and its effect on mRNA cap methylation has not been studied. To assess a role for RNGTT and/or RNMT/RNMT-Activating Mini protein (RAM) complex in MYC-mediated mRNA cap methylation of Wnt transcripts, we depleted each protein in IMECs using siRNA. Depletion of RNMT or RAM resulted in a statistically significant reduction in mRNA cap methylation of Wnt signaling transcripts (Fig. 3A), while depletion of RNGTT had no effect (discussed in more detail below). The reduction in mRNA cap methylation of Wnt transcripts following depletion of RNMT or RAM mimicked the results observed with MYC depletion, suggesting that MYC mediates mRNA cap methylation through modification or recruitment of RNMT and/or RAM. Conversely, increased mRNA cap methylation was observed when RNMT, RAM, or MYC was overexpressed in IMECs (Fig. 3B).
Figure 3.
MYC promotes mRNA cap methylation of canonical Wnt signaling pathway transcripts through TRRAP-containing cofactor complexes by utilizing RNMT. (A) m7G-mRNA levels of Wnt signaling transcripts in IMEC cells following siRNA depletion of RNGTT, RNMT, RAM, or MYC (normalized to total mRNA input). mRNA levels of each factor following siRNA knockdown in right panel (normalized to ACTB) (n=3, biological). (B) m7G-mRNA levels of Wnt signaling transcripts in IMECs following transient transfection with HA-RNMT, FLAG-RAM, HA-RNMT and FLAG-RAM, or CBP-MYC (normalized to total mRNA input). Protein levels of each factor following transfection in right panel (n=2, biological). (C) m7G-mRNA levels of Wnt signaling transcripts in IMEC cells following siRNA depletion of TIP60 or TRRAP (normalized to total mRNA input). Protein levels of each factor following siRNA knockdown in right panel (n=3, biological). Error bars indicate SEM and asterisks indicate significance (*p < 0.05, **p < 0.01, ****p < 0.0001).
Previous data and results presented herein with respect to Wnt pathway transcripts (Supplementary Fig. S2D) have shown that MYC partially mediates mRNA cap methylation of specific transcripts via MYC Box II (MBII), a conserved domain within all MYC proteins (19). Since transactivation is mediated by the MBII-dependent interaction of MYC with TRRAP and either TIP60 or GCN5 (8, 11), we tested whether MYC mediates mRNA cap methylation via TRRAP. Depletion of TRRAP and TIP60 resulted in reduced mRNA cap methylation of Wnt signaling transcripts (Fig. 3C), recapitulating the results detected with depletion of MYC itself. Together, the data indicate that MYC induces mRNA cap methylation of Wnt signaling transcripts in part through its cofactor TRRAP, complexed with TIP60.
MYC promotes recruitment of RNMT to Wnt signaling gene promoters
To determine whether MYC plays a role in RNMT recruitment to Wnt signaling promoters, chromatin immunoprecipitation (ChIP) was performed in HEK293FT cells expressing FLAG-tagged RNMT and RNGTT. MYC was subsequently depleted using siRNA in the same cell lines. MYC depletion resulted in a statistically significant reduction in recruitment of RNMT to Wnt signaling promoters (GSK3B and APC, Fig. 4A). MYC and TIP60 depletion in HEK293FT cells overexpressing RNMT, but not RNGTT, brought about a statistically significant reduction in mRNA cap methylation of all Wnt signaling transcripts analyzed (Fig. 4B). This provides additional evidence that RNMT, but not RNGTT, is the limiting factor in MYC-mediated mRNA cap methylation.
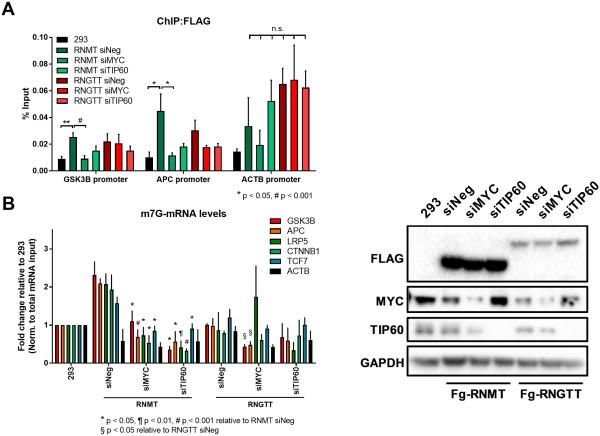
Figure 4.
MYC and TIP60 promote RNMT recruitment to canonical Wnt signaling pathway gene promoters and mRNA cap methylation. (A) FLAG chromatin immunoprecipitations (ChIPs) performed in HEK293FT cells overexpressing FLAG-tagged RNMT or RNGTT where MYC or TIP60 were depleted using siRNA. qPCR performed to detect levels of each factor at GSK3B and APC promoters (normalized to input, ACTB control) (n=3, biological). (B) m7G-mRNA levels of Wnt signaling transcripts in RNMT or RNGTT overexpressing cells following MYC or TIP60 depletion (normalized to total mRNA input) (n=3, biological). siRNA depletion detected using Western blot in right panel. Error bars indicate SEM and asterisks indicate significance (*/§p < 0.05, ¶p < 0.01, #p < 0.001).
To further validate a role for TIP60 in MYC-mediated mRNA cap methylation, we depleted TIP60 using siRNA in cells expressing FLAG-tagged RNMT or RNGTT. Consistent with MYC depletion, reducing TIP60 diminished recruitment of RNMT to Wnt signaling promoters, supporting a model that MYC facilitates mRNA cap methylation via MBII and its cofactors, TRRAP and TIP60.
CDK7 is required for MYC-dependent mRNA cap methylation of Wnt signaling transcripts
CDK7 phosphorylates S5 on the RNA Pol II CTD, which recruits both RNGTT and RNMT to promoters (23-25). Hence, this modification is a strong candidate to mediate MYC activity. To explore this, CDK7 and CDK9 (a kinase responsible for phosphorylation of S2 on the CTD) were depleted in IMECs using siRNA, and m7G-mRNA was isolated for analysis. Depleting CDK7, but not CDK9, led to a statistically significant reduction in mRNA cap methylation of Wnt signaling transcripts (Fig. 5A). To confirm that this observation was due to CDK7 activity as part of the TFIIH basal transcription factor, another component specific to TFIIH, XPD, was depleted using siRNA. XPD depletion mimicked CDK7 depletion, resulting in reduced mRNA cap methylation (Supplementary Fig. S5). Importantly, siRNA depletion of key factors involved in transcription did not result in significant cell cycle arrest, senescence or apoptosis (data not shown).
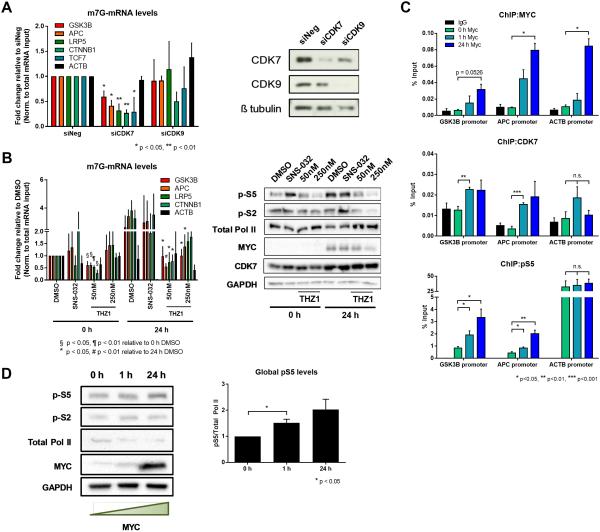
Figure 5.
MYC mediates mRNA cap methylation of Wnt/β-catenin signaling pathway transcripts by recruiting CDK7 to gene promoters. (A) m7G-mRNA levels of Wnt pathway transcripts in IMECs following CDK7 or CDK9 siRNA depletion normalized to total mRNA input (n=3, biological). Depletion was confirmed using Western blot in right panel. Error bars indicate SEM and asterisks indicate significance (*p < 0.05, **p < 0.01). (B) m7G-mRNA levels of Wnt pathway transcripts in P493-6 cells at the 0 h (MYC off) or 24 h time point (MYC on) following treatment with either 100nM SNS-032 or 50nm/250nM THZ1. Normalized to total mRNA input (n=3, biological). Inhibition was confirmed using Western blot in right panel. Error bars indicate SEM and asterisks indicate significance (§p < 0.05, ¶p < 0.01 relative to 0 h DMSO and *p < 0.05, #p < 0.01 relative to 24 h DMSO). (C) Chromatin immunoprecipitations (ChIPs) for MYC, CDK7, and phospho-S5 RNA Pol II were performed in P493-6 cells at three time points of MYC activation (0, 1, and 24 h). qPCR performed to detect levels of each factor at GSK3B and APC promoters (normalized to input, ACTB control) (n=3, biological). (D) Western blot detecting global phospho-S5 levels in P493-6 cells. Phospho-S5 levels quantified in right panel (n=3, biological). Error bars indicate SEM and asterisks indicate significance (*p < 0.05, **p < 0.01, ***p < 0.001)
As an independent approach to study the role of CDK7 in MYC activities, P493-6 cells were treated with a pan-CDK inhibitor (SNS-032) or a CDK7-specific inhibitor (THZ1) with or without MYC induction. THZ1 treatment led to inhibition of CDK7 (confirmed by RNA Pol II phospho-S5 reduction) and resulted in significantly reduced mRNA cap methylation of Wnt signaling transcripts even in the presence of maximal MYC levels 24 h post-MYC induction (Fig. 5B). These data support the concept that MYC mediates mRNA cap methylation through CDK7.
To investigate whether MYC recruits CDK7 to Wnt signaling promoters and thereby enhances RNA Pol II phospho-S5 levels, ChIP for CDK7 and phospho-S5 was performed. Occupancy of CDK7 and phospho-S5 on GSK3B and APC promoters was measured by qPCR using P493-6 cells. Interestingly, binding of CDK7 to the GSK3B and APC promoters paralleled MYC binding from the 0 h to 24 h time point, with a modest but statistically significant increase at 1 h of MYC induction (Fig. 5C). Consequently, phospho-S5 also accumulated significantly at both the GSK3B and APC promoters corresponding to increased MYC levels. No statistical change was observed between time points at the ACTB (β-actin) promoter, a negative control. A global increase in phospho-S5 levels has been observed in cell types overexpressing MYC (18). Accordingly, phospho-S5 was elevated on a global scale in P493-6 cells, analogous to a global increase in MYC levels (Fig. 5D).
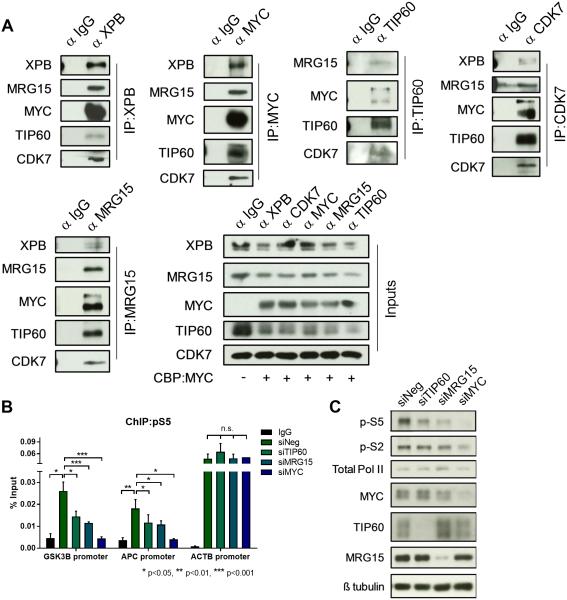
MYC recruits CDK7 to Wnt signaling gene promoters through interaction between MRG15 and XPB
Previous observations suggested that MYC may interact with CDK7 (18), but we were unable to confirm this in immunoprecipitations from native cell extracts. We, therefore, considered other mechanisms by which MYC could recruit CDK7. One candidate, MRG15, is a component of the NuA4 complex containing TRRAP/TIP60 and has recently been shown to interact with factors containing a small conserved amino acid sequence, FxLP (39). Analysis of various components in the TFIIH complex revealed that XPB, a factor in the core complex, contains a FxLP amino acid sequence. We asked whether there was an interaction between MRG15 and XPB which could bring MYC and CDK7 in proximity. Immunoprecipitation of XPB revealed an interaction between XPB, MRG15, MYC, TIP60, and CDK7 in HEK293T and IMEC cells (Fig. 6A, Supplementary Fig. S6). In support of an interaction, independent immunoprecipitation of MRG15, MYC, TIP60, or CDK7 resulted in co-immunoprecipitation of the above mentioned factors within both the NuA4 and TFIIH complexes. Furthermore, depleting MRG15 using siRNA knockdown in IMECs resulted in diminished phospho-S5 levels globally and at Wnt signaling gene promoters, consistent with a reduction in phospho-S5 following TIP60 and MYC depletion (Fig. 6B and 6C).
Figure 6.
MYC mediates mRNA cap methylation of Wnt/β-catenin signaling pathway transcripts through an interaction between NuA4 and TFIIH. (A) Immunoprecipitation of XPB, MYC, TIP60, CDK7, or MRG15 from HEK293T cells overexpressing MYC. Co-immunoprecipitated proteins detected using Western blot. (B) ChIP for phospho-S5 RNA Pol II was performed in IMECs where TIP60, MRG15, or MYC were depleted using siRNA. qPCR performed to detect levels of each factor at GSK3B and APC promoters (normalized to input, ACTB control) (n=3, biological). (C) Western blot to detect global phospho-S5 levels following depletion of TIP60, MRG15, or MYC. Error bars indicate SEM and asterisks indicate significance (*p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
Our previous study found that MYC promotes an increase in the protein levels of some targets in the absence of a change at the mRNA level (18). Additional analysis revealed that MYC stimulates an increase in the protein levels of target genes above and beyond the change in mRNA levels by enhancing mRNA cap methylation (20). Our study uses transcriptome-wide expression analysis to identify an important signaling pathway, the Wnt/β-catenin signaling pathway, affected by MYC at the post-transcriptional level of mRNA cap methylation. Our findings provide novel evidence for a role of TRRAP complexes, specifically TIP60 and MRG15, in MYC-mediated mRNA cap methylation through their interaction with XPB and CDK7 in TFIIH.
To determine which transcripts are targeted by MYC for mRNA cap methylation transcriptome-wide, we performed RNA-Seq on m7G-mRNA pulldowns compared to RNA inputs from P493-6 cells at two time points following MYC induction (0 h and 24 h). Roughly 30% of the transcripts analyzed were elevated at the mRNA cap methylation level in response to MYC, over and above any transcriptional induction, while the remaining fraction of the transcriptome exhibited only transcriptional induction with no net change in relative cap methylation. A smaller fraction of mRNAs exhibit transcriptional induction with an apparent reduction in the extent of cap methylation. We have focused this study on increased cap methylation, and exploration of any reduction in mRNA cap methylation will require further experimentation. It is notable that MAX was among the fraction of transcripts targeted by MYC for increased mRNA cap methylation, presumably providing more MAX protein to heterodimerize with increased levels of MYC protein.
The Wnt/β-catenin signaling pathway is important in embryonic development, normal cell proliferation, and differentiation; however, dysregulation of factors within the pathway due to mutations leads to aberrant activation of the pathway and carcinogenesis (15, 16, 40). The Wnt/β-catenin signaling pathway is stimulated in the presence of mitogens and results in transcription of MYC itself (13). Conversely, MYC has been shown to promote activation of canonical Wnt signaling transcripts by repressing Wnt inhibitors DKK1 and SFRP1 (14). In our dataset, we found that MYC mediates mRNA cap methylation of canonical Wnt signaling transcripts and that these transcripts represent an important pathway upregulated by MYC using this post-transcriptional modification. At one level, MYC upregulates general translation as demonstrated by an increase in mRNA ribosome loading and total protein synthesis. While an increase in translation and total protein production in response to MYC can be attributed to a number of mechanisms (41-45), a greater amount of m7G-Wnt signaling mRNAs were found in late polysome fractions following MYC induction. This suggests that MYC increases translation and protein synthesis of Wnt pathway transcripts in part by increasing mRNA cap methylation, which enhances the efficiency of ribosome loading. Furthermore, MYC induction increases Wnt signaling activity. Regulation of Wnt/β-catenin signaling by MYC using multiple mechanisms demonstrates how crucial MYC-induced Wnt signaling is in normal cell development and tumorigenesis.
RNMT and RAM are both required for methylation of the mRNA cap (27, 46). We have demonstrated that RNMT and RAM, but not the capping enzyme, RNGTT, promote mRNA cap methylation of Wnt/β-catenin signaling pathway transcripts. Since methylation is not possible without prior guanylylation by RNGTT, we presume that RNGTT is not rate limiting. Moreover, we find that MYC promotes RNMT recruitment to Wnt signaling promoters and mRNA cap methylation of these transcripts. We presume this recruitment is indirect since we do not observe a direct interaction between RNMT and MYC. This implies that RNMT is the limiting factor in mRNA cap methylation and suggests that MYC mediates mRNA cap methylation of Wnt transcripts by inducing recruitment of RNMT to their promoters. Furthermore, RNMT appears to be required for the increase in MYC-induced Wnt signaling activity suggesting that MYC-mediated mRNA cap methylation plays a critical role in promoting Wnt signaling. RNMT is sufficient to transform mammary epithelial cells (20) and therefore appears to be a better potential target of MYC regulation than RNGTT. Independently, MYC has been shown to promote mRNA cap methylation of select transcripts, and this is partially MBII dependent (19). We show that MYC cofactor complexes, including TRRAP and TIP60, are required to stimulate mRNA cap methylation of Wnt signaling transcripts suggesting that MYC indirectly mediates mRNA cap methylation via its cofactor complexes. Our results demonstrate that TRRAP cofactor complexes have dual roles in transcriptional and post-transcriptional activation of genes in response to MYC.
CDK7 enhances RNGTT and RNMT recruitment to promoters by phosphorylating S5 on the RNA Pol II CTD (23, 47). Our data show that CDK7, not CDK9, is required for mRNA cap methylation of canonical Wnt pathway transcripts, and the active form of CDK7 is necessary for MYC-mediated mRNA cap methylation. Recent studies have demonstrated that CDK7 is critical for normal cell growth and, when deregulated, for carcinogenesis. CDK7 inhibition using THZ1 has revealed integral roles for CDK7 in both transcriptional pausing and mRNA cap methylation (25). These data have established that therapeutically targeting CDK7 with THZ1, a CDK7 specific inhibitor, leads to tumor regression and reduces levels of MYC in MYC overexpressing tumors (48, 49). In our study, CDK7 recruitment and accumulation of phospho-S5 at both the GSK3B and APC promoters correlate with MYC levels and MYC recruitment to the same promoters. We suggest that MYC recruits CDK7 to promoters due to an interaction between the MYC/TRRAP/TIP60 complex and the TFIIH/CDK7/XPB module. We postulate that MRG15, a component of the NuA4/TRRAP complex, interacts with XPB, a core component of the TFIIH complex, bringing MYC in close proximity to CDK7 (Fig. 7). Consequently, MYC mediates phosphorylation of S5 on the CTD of RNA Pol II through CDK7, which leads to the recruitment of RNGTT and RNMT resulting in augmented mRNA capping and methylation.
Figure 7.
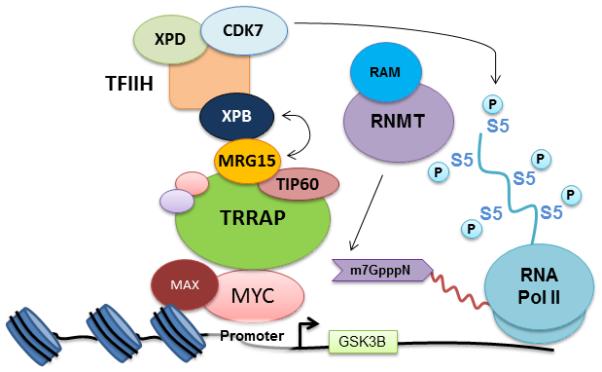
Model for MYC-mediated mRNA cap methylation of Wnt/β-catenin signaling pathway transcripts. MYC interacts with NuA4, composed of TRRAP, TIP60, and MRG15. MRG15 interacts with XPB, a component of TFIIH, bringing CDK7 in proximity to promoters and RNA Pol II. CDK7 phosphorylates S5 on the RNA Pol II CTD, which leads to the recruitment of RNGTT and RNMT/RAM. RNGTT caps nascent mRNA and RNMT mediates mRNA cap methylation.
One aspect of our proposed mechanism that remains unclear is why particular promoters are responsive to MYC-mediated mRNA cap methylation since we were unable to find a common motif. In addition, further experiments are necessary to confirm a direct interaction between MRG15 and XPB, to verify that members within NuA4 and TFIIH exist in one large complex, and to determine the specific motifs required for this interaction. Nevertheless, our study presents a novel set of transcripts involved in the Wnt/β-catenin signaling pathway that are affected by MYC at the level of mRNA cap methylation, above and beyond changes at the transcriptional level of mRNA.
Supplementary Material
Acknowledgements
The authors thank Joanna Hamilton, Xiangjun Xiao, and Carol Ringelberg for help with RNA-Seq data analysis, and members of the Cole lab for helpful discussions and feedback. This work was supported by a grant from the National Cancer Institute (CA080320; MDC).
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Posternak et al., 2016) and are accessible through GEO Series accession number GSE76614 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76614).
Funding Support: This study was supported by a grant from the National Cancer Institute (CA055248; MDC). CC is supported by the NIH Centers of Biomedical Research Excellence (COBRE; GM103534), the Dartmouth Clinical and Translational Science Institute (UL1TR001086) and the National Center for Advancing Translational Sciences (KL2TR001088).
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414(6865):768–73. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008 Dec;8(12):976–90. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Seminars in cancer biology. 2006;16(4):253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Felsher DW. MYC Inactivation Elicits Oncogene Addiction through Both Tumor Cell-Intrinsic and Host-Dependent Mechanisms. Genes & cancer. 2010 Jun;1(6):597–604. doi: 10.1177/1947601910377798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012 Sep 28;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012 Sep 28;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94(3):363–74. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 9.Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014 Jul 24;511(7510):483–7. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014 Jul 24;511(7510):488–92. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Molecular and cellular biology. 2000 doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998 Sep 4;281(5382):1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 13.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111(2):241–50. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 14.Cowling VH, D'Cruz CM, Chodosh LA, Cole MD. c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1. Molecular and cellular biology. 2007;27(14):5135–46. doi: 10.1128/MCB.02282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nusse R. Wnt signaling in disease and in development. Cell research. 2005;15(1):28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- 16.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012 Jun 8;149(6):1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Molecular and cellular biology. 2007;27(6):2059–73. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole MD, Cowling VH. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene. 2009;28(9):1169–75. doi: 10.1038/onc.2008.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowling VH. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene. 2010;29(6):930–6. doi: 10.1038/onc.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sims RJ, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Current Opinion in Cell Biology. 2004;16(3):263–71. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Current Opinion in Cell Biology. 2005;17(3):251–6. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 23.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, et al. 5'-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & development. 1997;11(24):3306–18. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure (London, England : 1993) 2004;12(11):2067–79. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Nilson KA, Guo J, Turek ME, Brogie JE, Delaney E, Luse DS, et al. THZ1 Reveals Roles for Cdk7 in Co-transcriptional Capping and Pausing. Molecular Cell. 2015;59(4):576–87. doi: 10.1016/j.molcel.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ensinger MJ, Moss B. Modification of the 5' terminus of mRNA by an RNA (guanine-7-)-methyltransferase from HeLa cells. The Journal of biological chemistry. 1976;251(17):5283–91. [PubMed] [Google Scholar]
- 27.Gonatopoulos-Pournatzis T, Dunn S, Bounds R, Cowling VH. RAM/Fam103a1 is required for mRNA cap methylation. Mol Cell. 2011 Nov 18;44(4):585–96. doi: 10.1016/j.molcel.2011.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aregger M, Cowling VH. Human cap methyltransferase (RNMT) N-terminal non-catalytic domain mediates recruitment to transcription initiation sites. The Biochemical journal. 2013;455(1):67–73. doi: 10.1042/BJ20130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shatkin AJ. Capping of eucaryotic mRNAs. Cell. 1976;9(4):645–53. doi: 10.1016/0092-8674(76)90128-8. PT 2. [DOI] [PubMed] [Google Scholar]
- 30.Schwer B, Saha N, Mao X, Chen HW, Shuman S. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics. 2000;155(4):1561–76. doi: 10.1093/genetics/155.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, et al. Control of cell growth by c-Myc in the absence of cell division. Current biology : CB. 1999;9(21):1255–8. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 32.DiRenzo J, Signoretti S, Nakamura N, Rivera-Gonzalez R, Sellers W, Loda M, et al. Growth factor requirements and basal phenotype of an immortalized mammary epithelial cell line. Cancer research. 2001;62(1):89–98. [PubMed] [Google Scholar]
- 33.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997 Mar 21;275(5307):1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 34.Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–51. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 35.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annual review of biochemistry. 1999;68:913–63. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 36.von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nature structural & molecular biology. 2004;11(6):503–11. doi: 10.1038/nsmb779. [DOI] [PubMed] [Google Scholar]
- 37.Schwer B, Mao X, Shuman S. Accelerated mRNA decay in conditional mutants of yeast mRNA capping enzyme. Nucleic acids research. 1998;26(9):2050–7. doi: 10.1093/nar/26.9.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yasuhiro F, Alba L, Aaron JS. 5′-Terminal structure and mRNA stability. Nature. 1977;266(5599):235–9. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 39.Xie T, Graveline R, Kumar GS, Zhang Y, Krishnan A, David G, et al. Structural basis for molecular interactions involving MRG domains: implications in chromatin biology. Structure (London, England : 1993) 2012;20(1):151–60. doi: 10.1016/j.str.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. Journal of mammary gland biology and neoplasia. 2004;9(2):119–31. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- 41.Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(21):11198–202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, et al. Identification of c-myc responsive genes using rat cDNA microarray. Cancer research. 2000;60(21):5922–8. [PubMed] [Google Scholar]
- 43.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. The EMBO journal. 2001;20(6):1383–93. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuhmacher M, Kohlhuber F, Hölzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic acids research. 2001;29(2):397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23(18):3217–21. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 46.Dunn S, Cowling VH. Myc and mRNA capping. Biochimica et biophysica acta. 2015;1849(5):501–5. doi: 10.1016/j.bbagrm.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larochelle S, Amat R, Glover-Cutter K, Sansó M, Zhang C, Allen JJ, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nature structural & molecular biology. 2012;19(11):1108–15. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014 Nov 20;159(5):1126–39. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, et al. CDK7-Dependent Transcriptional Addiction in Triple-Negative Breast Cancer. Cell. 2015;163(1):174–86. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.