Abstract
Cancer cells exhibit dramatic alterations of chromatin organization at cis-regulatory elements, but the molecular basis, extent and impact of these alterations are still being unraveled. Here we identify extensive genome-wide modification of sites bearing the active histone mark H3K4me2 in primary human colorectal cancers (CRC), as compared to corresponding benign precursor adenomas. Modification of certain CRC sites highlighted the activity of the transcription factor CNOT3, which is known to control self-renewal of embryonic stem cells (ESC). In primary CRC cells, we observed a scattered pattern of CNOT3 expression, as might be expected for a tumor-initiating cell marker. CRC cells exhibited nuclear and cytoplasmic expression of CNOT3, suggesting possible roles in both transcription and mRNA stability. We found that CNOT3 was bound primarily to genes whose expression was affected by CNOT3 loss, and also at sites modulated in certain types of CRC. These target genes were implicated in ESC and cancer self-renewal and fell into two distinct groups: those dependent on CNOT3 and MYC for optimal transcription and those repressed by CNOT3 binding and promoter hypermethylation. Silencing CNOT3 in CRC cells resulted in replication arrest. In clinical specimens, early stage tumors that included >5% CNOT3+ cells exhibited a correlation to worse clinical outcomes compared to tumors with little to no CNOT3 expression. Together, our findings implicate CNOT3 in the coordination of colonic epithelial cell self-renewal, suggesting this factor as a new biomarker for molecular and prognostic classification of early-stage CRC.
Keywords: Colorectal cancer, cancer cell self-renewal, chromatin states in cancer, transcriptional control in cancer, cancer epigenome
INTRODUCTION
Cancer cells share with adult and embryonic stem cells (ESC) the property of vigorous self-renewal, and certain tumors – including those of the intestine – originate in native stem cells1,2. Moreover, ESC gene signatures are notably enriched in aggressive, poorly differentiated human cancers3–5, suggesting a shared transcriptional basis for normal and cancer cell self-renewal. MYC, in particular, activates many genes expressed normally in ESCs6,7 and aberrantly in cancers8, including colorectal cancer (CRC), where it is frequently overexpressed9 and essential for tumorigenesis10. ESC-like mRNA profiles, however, encompass both activated and repressed transcripts, and it is unclear how these necessary but opposite activities are coordinated. Aberrant gene activation and silencing in cancers in occur on a background of widespread chromatin alterations that are insufficiently characterized.
To investigate these alterations, we examined genome sites bearing the active histone mark H3K4me2 in human CRCs and in their precursor adenomas (Ad). Nucleosomes that carry specific covalent marks, such as H3K4me1/2/3 and H3K27ac, signify active cis-elements11–14 and the temporal dynamics of these marks (Fig. 1A) have revealed the underpinnings of differentiation in many cell types12,15. We identified thousands of cis-regulatory sites showing differential activity, and in some cases, these sites showed an abundance of the preferred binding motif for CNOT3, a transcription factor (TF) previously implicated in ESC self-renewal. CNOT3 is expressed in rare normal crypt cells and scattered CRC cells, suggesting a clonogenic subpopulation, and its overexpression is strongly associated with poor clinical outcomes. CNOT3 binding in primary CRCs and cell lines was highly enriched at genes that are affected by CNOT3 depletion. These genes correspond to both the active and silenced arms of the gene signature common to ESCs and aggressive carcinomas, and both arms responded to CNOT3 loss in coordinated, opposite fashion. Together, these findings implicate CNOT3 in control of cancer self-renewal and as a novel regulator of distinct transcriptional activities.
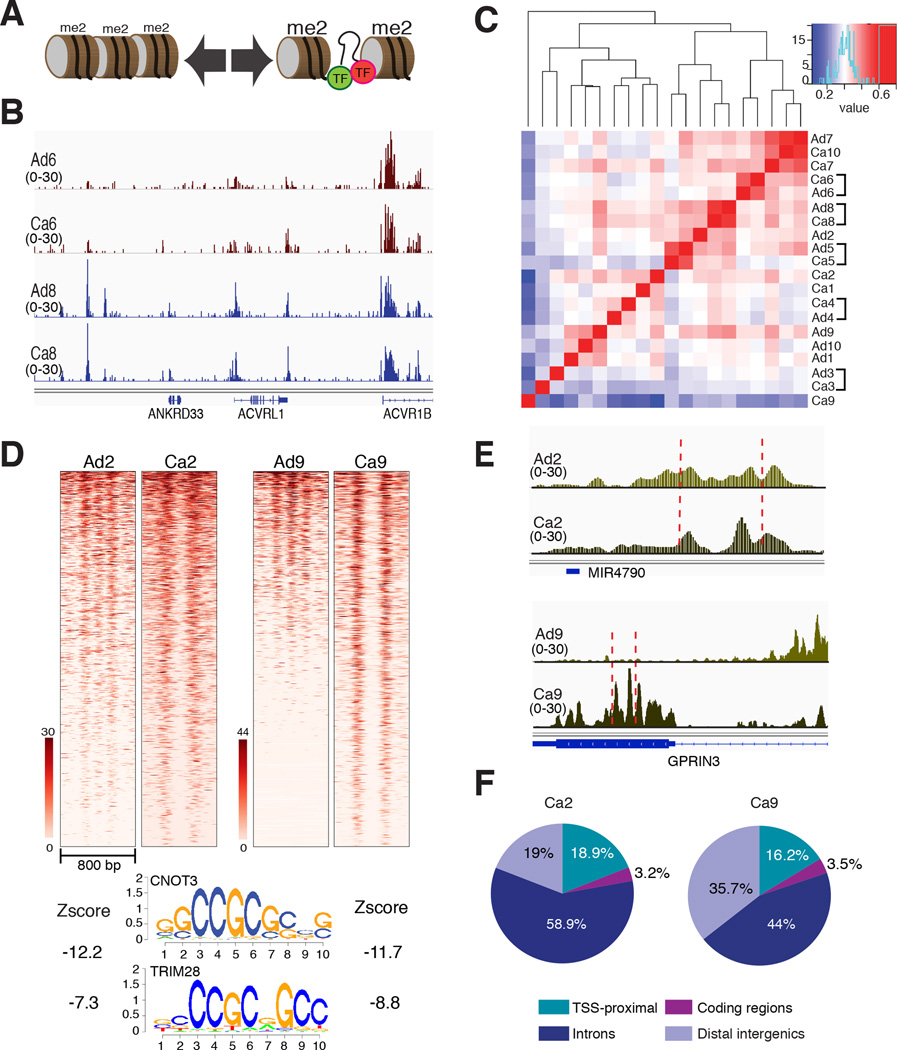
Figure 1. Analysis of H3K4me2-marked nucleosomes identifies CNOT3 as a candidate transcriptional regulator in colorectal cancer.
(A) Schema of the contrast in chromatin at inactive genomic regions (left) and marked nucleosome pairs flanking active, transcription factor (TF)-bound sites (right). (B) Integrative Genome Viewer (IGV) tracks showing H3K4me2+ nucleosomes shared across samples (e.g., at the ACVR1B locus) and others found specifically in certain Ad-Ca pairs (e.g., upstream of ANKRD33). (C) Heatmap representation of unsupervised clustering and Spearman correlations among genome-wide H3K4me2-marked nucleosomes in 10 pairs of human colon cancers (Ca) and the corresponding adenomas (Ad). Correlations are highest between individual Ad-Ca pairs. (D) Heatmap of nucleosomal H3K4me2 signals at the 1000 most differentially marked regions in Ca2 and Ca9, compared to their respective Ad. The sequence motifs most enriched in these Ca, shown below, are attributed to the TFs CNOT3 and TRIM28. (E) IGV traces of representative differential nucleosome pairs in Ad-Ca2 and Ad-Ca9. (F) Distribution of the 1000 top-scoring differential nucleosome pairs in Ca2 and Ca9, compared to their respective precursor Ad.
Although investigation of CNOT3, a member of the CCR4-NOT protein complex, often focuses on its role in regulating mRNA stability, the protein was originally identified as a transcriptional regulator and considerable evidence supports this function from yeast to humans (reviewed in Ref.16). Some CNOT3 activities related to gene regulation, such as histone demethylation, seem to be indirect effects of transcript or protein stabilization17,18, but CNOT3-containing complexes also interact with RNA polymerase II (Pol2), the SAGA complex, and other factors to promote transcriptional initiation and elongation19,20. Other data suggest a repressive function in yeast and human cells, associated with Pol2 inhibition and histone deacetylation21,22, but specific mammalian CNOT3 activities and target genes are unknown. Cnot3−/− mouse embryos die before implantation owing to failure of inner cell mass expansion23, similar to impaired self-renewal of CNOT3-deficient ESCs7,24, and recurrent CNOT3 mutations in adult T-cell acute lymphoblastic leukemia (ALL)25 have unclear significance and tumorigenic mechanism. Our findings elucidate CNOT3 functions in CRC self-renewal, identify the likely underlying process, and demonstrate that CNOT3 expression in this disease carries significant prognostic value.
MATERIALS AND METHODS
Clinical materials and data analysis
For epigenome analysis, 10 pairs of frozen CRCs (stage III) and the precursor adenomatous polyp were obtained from a tumor bank at Brigham and Women's Hospital, Boston, MA. Tissue microarrays were prepared from 376 consecutive CRCs (Suppl. Table 5) banked at Hospital La Paz, Madrid, Spain. Human protection committees at each institution approved tissue use and informed consent was obtained from each subject. TF expression was associated with other variables using statistical package SPSS v19.0.0. Normalized RNA-seq data and tumor-specific deaths were compiled from 296 cases of MSI-negative CRCs from the TCGA collection (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix). ROC curve analyses defined 1,715 CNOT3 mRNA sequence tags as an optimal cut-off for sensitivity and specificity with respect to CRC-specific deaths. Kaplan-Meier plots were derived and groups compared using the log-rank test.
Chromatin immunoprecipitation (ChIP) and analysis
A pathologist selected tissue blocks of fresh-frozen adenomas and the corresponding CRCs containing >70% tumor cells. After washing in phosphate-buffered saline (PBS), 10 sections of 10 µm thickness were scraped into plastic tubes and treated with 0.2 U micrococcal nuclease (Sigma Aldrich) for 8 min at 37°C, followed by ChIP with 10 µg H3K4me2 antibody (Ab, Millipore) as described12. Twenty frozen tissue sections from 2 cases – Ca2 and Ca9 – were cross-linked with 1% formaldehyde for 20 min at 37°C, followed by ChIP with 10 µg CNOT3 Ab (Novus Biologicals, clone 4B8), and 10 paraffin-embedded tissue sections from one CNOT3hi CRC were deparaffinized, rehydrated and sonicated for 40 minutes, followed by ChIP with 10 µg H3K4me2 Ab. To detect CNOT3 and CNOT1 binding, respectively, HCT116 cells were cross-linked with 1% formaldehyde for 10 min or with 2 mM di(N-succinimidyl) glutarate for 45 min followed by 1% formaldehyde for 10 min. After quenching with 125 mM glycine for 5 min, cells were washed twice with ice-cold PBS and resuspended in sonication RIPA buffer (10 mM Tris pH7.4, 0.3 M NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 0.1% SDS, 10% Sarkosyl reagent, 1 mM DTT, and protease inhibitors). Chromatin (30 µg) was immunoprecipitated using 10 µg of CNOT3 (Novus Biologicals) or CNOT1 (Proteintech, 14276-1-AP) Ab.
DNA libraries were prepared using ThruPLEX-FD kits (Rubicon Genomics) and sequenced on a HiSeq2000 instrument (Illumina). H3K4me2+ nucleosomes were identified using Nucleosome Positioning from Sequencing (NPS) with default parameters26. For every nucleosome pair, relative depletion of a central nucleosome in each CRC against the matched Ad was estimated using the Nucleosome Stability-Destability (NSD) scoring scheme11. Clustering of called H3K4me2 peaks in Ad and Ca specimens was done by Spearman’s rank correlations of genome-wide ChIP-seq data. We used Cistrome tools (www.cistrome.org/) to call and annotate H3K4me2 and CNOT3 peaks, generate wiggle files and conservation plots, identify enriched sequence motifs, and compare data across ChIP-seq libraries. Wiggle tracks were visualized using the Integrative Genomics Viewer27. Heatmaps were prepared using Cistrome (http://www.cistrome.org/) or deepTools28 and SeqMiner29. ChIP-seq data for Pol2 (Gene Expression Omnibus, GSM803474), H3K4me3 (GSM955162), DNaseI hypersensitivity (GSM736493), DNA methylation (GSM955158), and for binding of YY1 (GSM803354), TCF4 (GSM782123), SP1 (GSM1010902), RAD21 (GSM1010848), JUND (GSM1010847), and MAX (GSM1010904) in HCT116 cells were analyzed in relation to CNOT3 binding. Functional annotation of CNOT3 binding sites was performed in GREAT (http://bejerano.stanford.edu/great/public/html/), using the association rule “Single Nearest Gene within 30 kb” and the whole human genome as a background30.
Immunodetection
For immunohistochemistry, we used Ab against CNOT3 (Novus Biologicals, clone 4B8, 1:50), TRIM28 (Abnova, clone 4E6, 1:250), KI67 (DAKO, clone MIB-1, 1:50) or CTNNB1 (BD Transduction Laboratories, clone 14/Beta-Catenin, 1:250) diluted in Tris-buffered saline (TBS, pH 8.0) containing 1% bovine serum albumin. Tumor sections were incubated with primary and secondary Ab for 30 min each at room temperature, washed in TBS, and incubated with the peroxidase-based EnVision kit (Dako). After accounting for technical failures, CNOT3 and TRIM28 expression could be evaluated in 374 and 307 of 376 consecutive cases, respectively. Expression was considered to be elevated when detected in the nuclei of ≥5% of tumor cells (CNOT3) or when global tumor levels were higher than those in adjacent normal epithelium (TRIM28). Mouse crypt and villus epithelial cells were separated using a modified EDTA-based chelation protocol31,32. Lgr5hi ISC were isolated from duodenal crypts of Lgr5GFP-IRES-CreER mice33. For immunocytochemistry, HCT116 and Caco-2 cell suspensions were fixed in 70% ethanol and centrifuged; pellets were dehydrated and embedded in paraffin. Sections were stained for CNOT3 as described above.
To prepare nuclear lysates, cells were suspended in lysis buffer (20 mM Tris pH 7.9, 1.5 mM MgCl2, 10 mM KCl), followed by nuclear lysis buffer (20 mM Tris pH 7.9, 1. 5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA), including protease and phosphatase inhibitors. Whole-cell extracts were prepared in parallel for immunoblotting controls. Subcellular protein fractions of HCT116 and Caco-2 cells were prepared using the FractionPREP system (BioVision). Proteins resolved by SDS–PAGE were transferred to nitrocellulose membranes, probed with CNOT3 (Novus Biologicals, clone 4B8), CNOT1 (Proteintech, 14276-1-AP), LAMIN B1 (Abcam, ab20396) or GAPDH (Bethyl laboratories, A300-641A) Ab, and detected by chemiluminescence after incubation with horseradish peroxidase-coupled secondary Ab (Santa Cruz).
Cell culture, shRNA treatment and rescue
HCT116 and Caco-2 cells were obtained from the American Type Culture Collection and authenticated in 2016 by high-resolution Small Tandem Repeat profiling (STR, GenePrint 10, Promega). Fragments were analyzed using GeneMapper v4.0 (Applied Biosystems) and alleles identified using GenePrint 10 custom bin files (Promega). Cells were infected with lentiviruses containing the pLKO.1 vector (Open Biosystems) and shRNAs against CNOT3 (#3: CGAGGAGAACGAGTTTCTCTA and #5: TGGAGCAGTTTGAAGATATTT) or a control non-toxic shRNA lacking complementarity with any human gene (NS: 5’-CCTAAGGTTAAGTCGCCCTCG-3’). Cells were selected in 2 µg/ml puromycin and CNOT3 depletion was assessed by immunoblotting. To rescue CNOT3 depletion, shRNA-treated HCT116 cells were selected for 5 days in puromycin, followed by infection with lentiviruses carrying the pULTRA-GFP vector (Addgene) with or without shRNA-resistant CNOT3 cDNA. Puromycin treatment was maintained during cell cycle analyses. For mouse xenografts, 5 × 106 HCT116 cells transduced with shRNA #3 or the control shRNA were suspended in a 1:1 mixture of PBS and growth factor-depleted Matrigel (BD Bioscience) and injected subcutaneously into the flanks of 6–8 week old NOD.CB17-Prkdcscid mice (The Jackson Laboratory). Mouse care followed protocols approved by the Animal Care and Use Committee at Ludwig-Maximilians University, Munich (Germany), and tumors were measured using calipers.
Cell cycle analyses
Triplicate samples of 5,000 cells were seeded in 96-well plates in medium containing puromycin for 48 h before proliferation was assessed using CellTiter 96 (Promega) at 570 nm absorbance using a Synergy-2 multi-detection plate reader (BioTek). Alternatively, cells were incubated with 10 µM bromodeoxyuridine (BrdU) for 2 h, washed in PBS, and fixed in 70% ethanol. DNA was denatured in 2 N HCl at room temperature for 15 min and resuspended in 0.1 M Na2B4O7, followed by staining with FITC-conjugated BrdU Ab (BD Pharmingen), washing in PBS, additional staining with 10 mg/ml propidium iodide for 30 min, and analysis on a FACScan instrument (Becton-Dickinson). For analysis of apoptosis, we used a FITC Annexin V Apoptosis detection kit I (BD Pharmingen) and analyzed cells on a FACScan instrument (Becton-Dickinson). Phospho-histone H3 was analyzed as described previously34, using Histone H3 (Cell Signaling, cat. no. 9715) and Phospho-histone H3 (Cell Signaling, 3377) Ab.
RNA expression and analysis
Total RNA was isolated using Trizol (Invitrogen) and treated sequentially with the RNeasy Mini Kit (Qiagen) and Turbo DNA Free (Ambion). RNA-seq libraries were prepared from 300 ng RNA using the TruSeq RNA Sample Preparation kit (Illumina) and 75-bp single-end sequences were obtained on a NextSeq 500 Instrument (Illumina). Sequence tags were mapped to reference genome Hg19 using TopHat v2.0.6 and transcript levels were calculated on data from triplicate samples as fragments per kb per 106 mapped reads (FPKM) using Cufflinks v2.0.2. Differential expression was determined with CuffDiff, using Chi-square tests with 1 degree of freedom and tow-tailed P values to assess statistical significance35. Log2 (FPKM+1) values for control and CNOT3-depleted samples were plotted for display. Functional cluster analysis of differentially expressed genes was performed using Ingenuity Pathways Analysis (Ingenuity Systems) to identify the most relevant molecular interactions, functions and pathways linking them. Gene Set Enrichment Analysis was performed with GSEA software (http://www.broadinstitute.org/gsea/index.jsp), by comparing triplicate control and CNOT3-depleted HCT116 cell samples. Gene sets (Suppl. Table 2) derived from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp) were considered enriched at FDR <5%, using Signal2Noise as the metric, with the meandiv normalization mode and 1,000 permutations of gene sets.
Quantitative RT-PCR and immunoblotting
Total RNA was isolated using Trizol (Invitrogen, Grand Island, NY), treated with the RNeasy Mini Kit (Qiagen), and DNA was eliminated using Turbo DNA Free (Ambion). For quantitative RT–PCR, 1 µg total RNA was reverse transcribed with Superscript III First Strand Synthesis System (Invitrogen); cDNA was amplified using SYBRGreen PCR Master Mix (Applied Biosystems) and the following oligonucleotide primers:
CCND1 (fw: GGTGGTGCTGGGGAAGTTGAAGTG, rev: TCGACGGTGGGTACATGGCAAACT); CNOT3 (fw: GAGGGAGCAGCAGCAGTAGT, rev: GGGAGGATTCACAGGCAGT); DAPK1 (fw: TTTTTATTTATTTTTTAGTTGTGTTTT, rev: CCTTAACCTTCCCAATTACTC); FANC (fw: GCTAGTCCACTGGCTTCTGG, rev: GGACTCAGTTCCAACCCAAA)
GAPDH (fw: GAGTCAACGGATTTGGTCGT, rev: AATGAAGGGGTCATTGATGG); GATA4 (fw: CTCTACCACAAGATGAACGGC, rev: TTCCGTTTTCTGGTTTGGATCC); GATA5 (fw: CTCTACCACAAGATGAATGGCG, rev: TGCTTTCCTTCTTCATAGCCAG); HIC1 (fw: ATCTGCGGGAAGAAGTTCAC, rev: GCATCTTCATGTGGCTGATC); HNF1 (fw: GTGGCGAAGATGGTCAAGTCC, rev: CCCTTGTTGAGGTGTTGGG); PYCARD (fw: CTGCACTTTATAGACCAGCACC, rev: CTGGTGTGAAACTGAAGAGCTT); SFRP1 (fw: CAAGAAGAAGAAGCCCCTGAAG, rev: CAAGTACTGGCTCTTCACCTTG); SFRP2 (fw: AAGCCTGCAAAAATAAAAATGATG; TGTAAATGGTCTTGCTCTTGGTCT); SFRP4 (fw: GCCCCTCATGAAGATGTACAAC, rev: GCCTTTCCTGTACCATCATGTC); TIMP3 (fw: GCAACTCCGACATCGGTAAG, rev: GCCCAAGGACATCGAGTTT); WNT3A (fw: TCCGCTTCTGCAGGAACTAC, rev: AACTGGTGCTGGCACTCCT).
Accession codes
All gene expression and ChIP-seq data from this study are deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with accession code GSE66216.
RESULTS
Dynamic nucleosomes in human CRCs suggest activation of an ESC self-renewal pathway
We used chromatin immunoprecipitation (ChIP-seq) of mononucleosome fractions from 10 primary human CRCs and their respective precursor Ad to identify an average of 219,400 well-positioned nucleosomes carrying H3K4me2, a hallmark of active promoters and enhancers12,13 (Suppl. Table 1). CRC epigenomes tended to resemble those of their Ad precursors more than those of other CRCs (Fig. 1B–C), implying that tumors acquire hundreds of distinctive alterations at presumptive cis-elements. Using nucleosome stability-destability (NSD) analysis11,12, we further identified sites where individual Ad showed weak or no H3K4me2 marking, while the paired Ca specimen showed strong marks and apparent displacement of a central nucleosome (Fig. 1D–E, Suppl. Fig 1A). In these regions, most cases showed enrichment of the motifs for TFs such as FOS and JUN (Suppl. Fig. 1A), which mediate growth factor signaling in CRC36, indicating common pathway activation across tumors. However, two sample pairs, Ad-Ca2 and Ad-Ca9, lacked AP1 motif enrichment; instead, differentially active sites in these CRCs were highly enriched for other motifs (Fig. 1D), which ChIP studies in mouse ES cells attribute to the TFs CNOT3 and TRIM28 (Ref. 7). Dynamic nucleosomes in these tumors mapped near transcription start sites (TSSs) as well as in introns and intergenic regions that likely represent distant enhancers (Fig. 1F). The same sites largely lacked H3K4 methylation in normal colonic epithelium (Suppl. Fig. 1B), indicating selective activation in some CRCs.
Based in part on TF binding data, a module containing CNOT3, TRIM28, ZFX and MYC is implicated in ESC self-renewal7. ZFX also controls hematopoietic stem cell renewal37 and MYC has an established role in ESCs6,8, CRC9,10, and other cancers. In Ca2 and Ca9 – and only in these cases – regions of dynamic nucleosome activity were also enriched for MYC and ZFX motifs (Suppl. Fig. 1C), but the tumors did not share salient CRC mutations (Suppl. Table 1). Thus, some CRCs may activate regions marked with H3K4me2 that bind ESC self-renewal factors. Moreover, CNOT3 mutations occur in 8% of T-cell ALL25 and at least 2.4% of CRCs (http://cancer.sanger.ac.uk/cosmic).
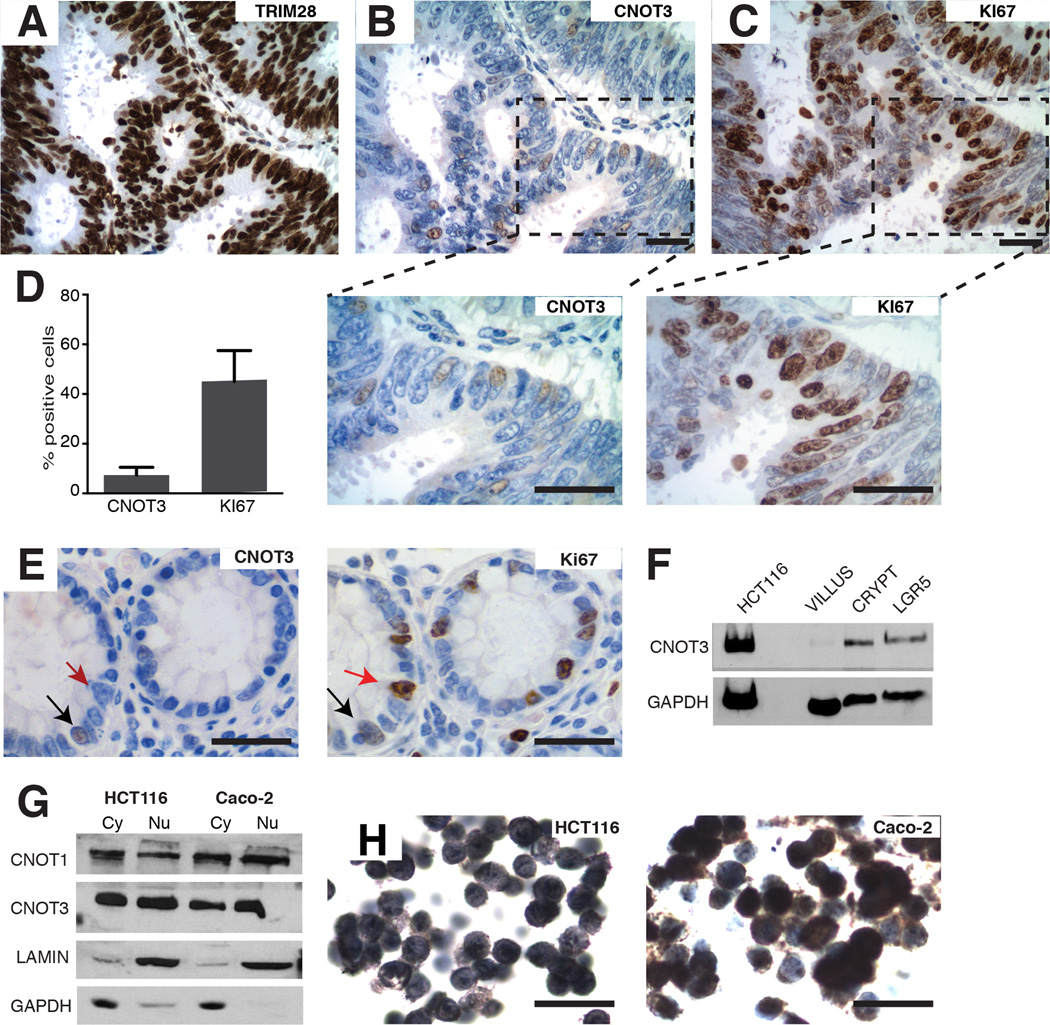
A separate, large series of primary CRCs, tested with highly active and specific CNOT3 and TRIM28 antibodies (Ab, Suppl. Fig. 2A), revealed markedly different expression patterns. TRIM28 was present in every epithelial cell nucleus (Fig. 2A). In contrast, CNOT3 appeared in a distinctive, scattered distribution (Fig. 2B) and in many fewer cells than the proliferation marker KI67, though the two proteins overlapped modestly (Fig. 2B–D). Unlike other heterogeneously distributed proteins, such as nuclear β-catenin – which is often highest at the invasive front38 – CNOT3 expression was scattered throughout the tumor mass (Suppl. Fig. 2B). Thus, CNOT3 marks a minor population of tumor cells, and considering CNOT3’s role in ESC self-renewal, this may be a clonogenic pool. Indeed, expression in normal human colon was weak and confined to rare crypt epithelial cells (Fig. 2E). Moreover, although IHC failed to detect CNOT3 in normal mouse intestines, immunoblots showed absence in post-mitotic villi and restricted expression in self-renewing crypts, including LGR5+ intestinal stem cells (Fig. 2F).
Figure 2. CNOT3 is expressed in a scattered subpopulation of CRC cells.
(A–C) Immunohistochemistry (IHC) for TRIM28 (A), CNOT3 (B) and KI67 (right) in serial sections of a representative CRC specimen. While TRIM28 is expressed in all tumor cells, CNOT3 is expressed in a minority of scattered nuclei, compared to a much larger fraction of KI67+ cells, with modest overlap of CNOT3 and KI67. The areas boxed in b and c are magnified below. (D) Mean counts (± SD) of CNOT3+ and KI67+ nuclei from 500 cells in each of 3 independent CNOT3+ CRCs. (E) IHC for CNOT3 (left) and KI67 (right) in serial sections of normal human colon mucosa, showing CNOT3 in some (black arrows) but not most (red arrows, e.g.) KI67+ crypt epithelial cells. (F) Immunoblot analysis of CNOT3 in murine intestinal Lgr5+ cells sorted by flow cytometry and in isolated intestinal crypts and villi. HCT116 human CRC cells serve as a positive control. (G) Immunoblot analysis of cytoplasmic (Cy) and nuclear (Nu) fractions of HCT116 and Caco-2 cells, showing presence of CNOT1 and CNOT3 in both fractions. LaminB1 and GAPDH serve as positive controls for the Nu and Cy fractions, respectively. (H) CNOT3 immunocytochemistry in HCT116 and Caco-2 cells, showing both Nu and Cy staining. All scale bars, 50 µm.
Sub-cellular localization of CNOT3 and requirements in CRC growth and cell cycle progression
Although CNOT3 was originally recognized as a transcription factor, recent attention has focused on its role in regulating mRNA stability. Our identification of CNOT3 as a protein that may bind genomic sites that are dysregulated in CRC prompted us to examine its sub-cellular location. Immunoblotting of CRC cell extracts revealed presence of CNOT3 and CNOT1 in both the nuclear and cytoplasmic fractions (Fig. 2G), and immunocytochemistry confirmed this distribution for CNOT3 (Fig. 2H). The CNOT3 Ab reacts in immunoblots with a single protein of the expected molecular mass (Suppl. Fig. 2A), and in further confirmation of sub-cellular localization and Ab specificity, both cytoplasmic and nuclear signals were abrogated in HCT116 cells treated with CNOT3-specific shRNA (Fig. 3A–B).
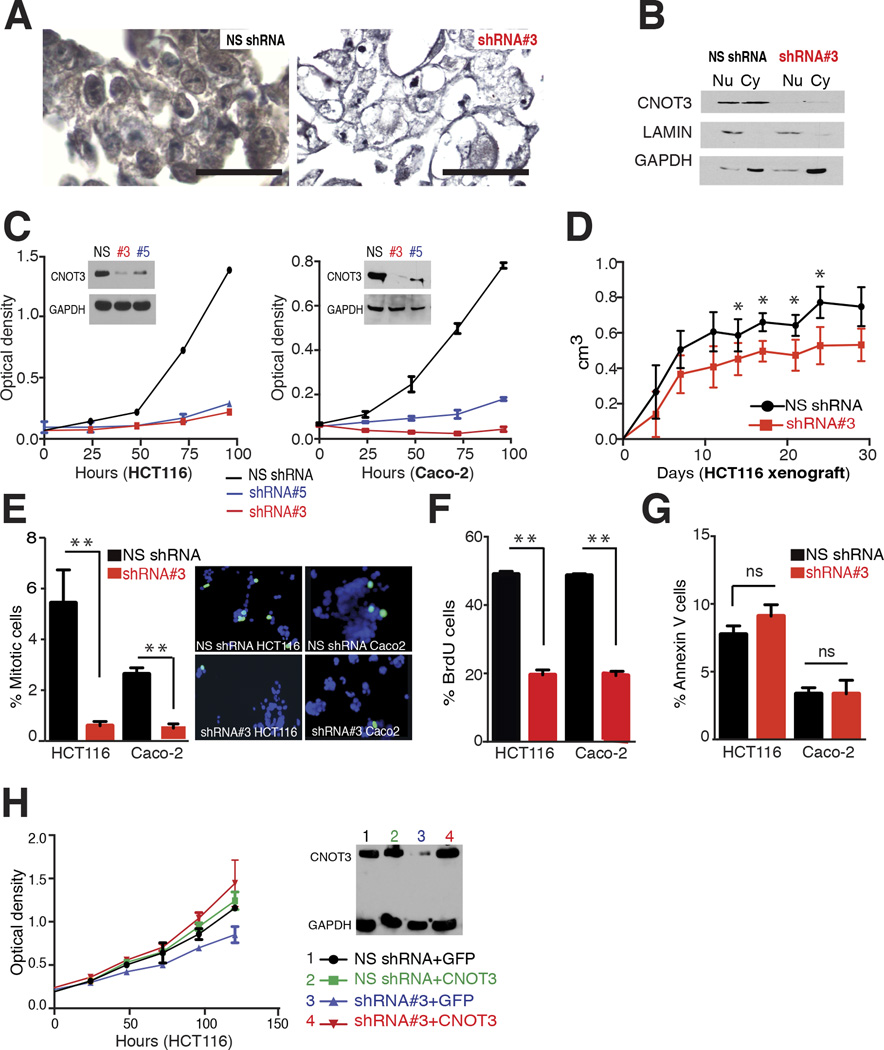
Figure 3. Effects of CNOT3 depletion on CRC cells.
(A) CNOT3 immunocytochemistry in HCT116 cells treated with control, non-specific (NS) shRNA (left) or with CNOT3-specific shRNA #3 (right), confirming CNOT3 loss and Ab specificity and showing typical changes in morphology of CNOT3-depleted cells. (B) Immunoblot analysis of nuclear (Nu) and cytoplasmic (Cy) fractions of HCT116 cells treated with non-specific (NS) or CNOT3-specific shRNA #3. LaminB1 and GAPDH serve to mark the Nu and Cy fractions, respectively. (C) Reduced CNOT3 levels, achieved with two independent shRNAs (#3 and #5, immunoblot in the inset), impaired proliferation of CRC cell lines HCT116 (left) and Caco-2 (right), compared to treatment with a non-specific (NS) control shRNA. Each value represents the mean ± SD optical density from triplicate samples; other cell lines are shown in Suppl. Fig. 3C. (D) Reduced growth of CNOT3-depleted HCT116 cells in murine xenografts. Each plotted value represents the mean ± SD from 5 replicates. (E–G) Significantly reduced phospho-Histone H3 staining (E, determined manually in blind counts) and BrdU uptake (F, determined by flow cytometry) in CNOT3-depleted HCT116 and Caco-2 cells, without concomitant change in apoptotic AnnexinV+ cells (G, flow cytometry). Graphs represent the mean ± SD from 3 replicates. (F) An shRNA-resistant CNOT3 cDNA construct, but not a GFP cDNA control, restored cell replication in HCT116 cells depleted of endogenous CNOT3 using shRNA #3. The immunoblot shows relative CNOT3 and GAPDH levels in total cell lysates.
We detected CNOT3 in all CRC cell lines (Suppl. Fig 3A), and the Cancer Cell Line Encyclopedia indicated that CNOT3 mRNA levels are similar in lines derived from most cancers (data not shown). Using lentiviruses to deliver specific shRNAs, we depleted >70% of CNOT3 mRNA in each of four CRC cell lines (Suppl. Fig 3B) and confirmed protein loss in HCT116 and Caco-2 cells (Fig. 3C). Compared to cells treated with non-specific shRNA, two independent CNOT3 shRNAs markedly impaired growth of each cell line (Fig. 3C, Suppl. Fig. 3C), similar to the effect of CNOT3 loss in ES cells7,24. CNOT3 deficiency also impaired growth of HCT116 cell xenografts in mice (Fig. 3D) and specifically reduced the fraction of cells in M or S phases in cultured CRC cells (Fig. 3E–F), without inducing apoptosis (Fig. 3G). Re-expression of CNOT3 in depleted cells restored protein levels and growth kinetics (Fig. 3H), indicating that the shRNA effects were on-target. Thus, CRC cells express CNOT3 in the cytoplasm as well as the nucleus and require this factor to sustain growth in culture and in xenografts in vivo.
CNOT3 binding in CRC cells and its relation to chromatin states
Having identified CNOT3 because its preferred motif was the most enriched within dynamic genomic regions in Ca2 and Ca9, we undertook ChIP-seq analysis in these primary tumors and in HCT116 cells. Binding sites in each case outnumbered the few sites reported previously in ESCs (Ref. 7) and were substantially enriched for the CNOT3 consensus motif (Fig. 4A, Suppl. Fig. 4A). About 40% of all binding occurred at promoters (−2 kb to +1 kb from TSSs, Fig. 4B), but most genes that bound CNOT3 at the promoter showed additional distant binding. CNOT3 bound many of the same sites in primary tumors as in HCT116 cells, with significant overlap at distant sites and nearly total overlap at promoters (Fig. 4C). Between 52% (HCT116 cells) and 76% (Ca9) of binding occurred within CpG islands, both at promoters and elsewhere (Suppl. Fig. 4B). Importantly, we detected CNOT3 occupancy between 33% (Ca2) and 40% (Ca9) of the 1,000 most dynamic nucleosome pairs (Suppl. Fig. 4C), coinciding significantly with well-positioned, H3K4me2-marked nucleosomes in Ca2, Ca9, and selected other CRCs (Fig. 4D). Archival tissue from another CRC with a high CNOT3+ cell fraction also showed a profile of dynamic regions resembling that of Ca9 (Fig. 4E). Notably, up to 1/3 of binding occurred at sites that lack H3K4me2 (Fig. 4D), indicating association with both active and inactive genes.
Figure 4. Features of CNOT3 binding in primary CRCs and cell lines and the relation to chromatin.
(A) Representative IGV traces of CNOT3 binding sites identified by ChIP-seq at promoters and intergenic regions in Ca9, Ca2 and HCT116 and corresponding motif analysis and z-scores. (B) Distribution of CNOT3 binding sites in Ca9, Ca2, and HCT116. (C) Overlap of CNOT3 binding sites in the 3 sources of data at TSSs (top) and intergenic regions (bottom). (D) Heatmap of H3K4me2-marked nucleosomes in three CRCs at CNOT3 binding sites identified in Ca9. K-means clustering (k=3) reveals groups of sites with or without marked flanking nucleosomes; IGV traces representing the first cluster and the two other largely similar clusters are shown to the right. (E) Average distribution of H3K4me2 signals in an archival CNOT3hi CRC (inset: CNOT3 IHC; scale bar, 50 µm) flanking the 1,000 most accessible regions in Ca9. (F) GREAT analysis of CNOT3 binding sites ≤30 kb from TSSs, showing the top over-represented categories from the MSigDB ontology of genetic and chemical perturbations. Binomial corrected (FDR) p-values are shown at log10 scale; all GREAT-derived ontologies are listed in Suppl. Table 2.
Because CNOT3 binds tightly to CNOT1 and CNOT2 (Ref.39), presence of these factors might further increase confidence in the CNOT3 cistrome. CNOT2 Ab gave no ChIP signals in HCT116 cells, but CNOT1 binding coincided significantly with that of CNOT3 (Suppl. Fig. 4D). GREAT analysis30 revealed that genes with nearby CNOT3 binding serve in transcriptional control and embryogenesis (Suppl. Table 2); they are enriched for (a) ESC transcripts activated by MYC in aggressive, poorly differentiated cancers3,5 and (b) targets of Polycomb/EZH2 regulation (Fig. 4F). These findings evoke CNOT3 requirements in ESC self-renewal7, and Polycomb targets are especially interesting because many genes silenced by this pathway in ESCs tend to have silent hypermethylated promoters in CRC, through unknown mechanisms40–42.
Effects of CNOT3 binding on gene expression
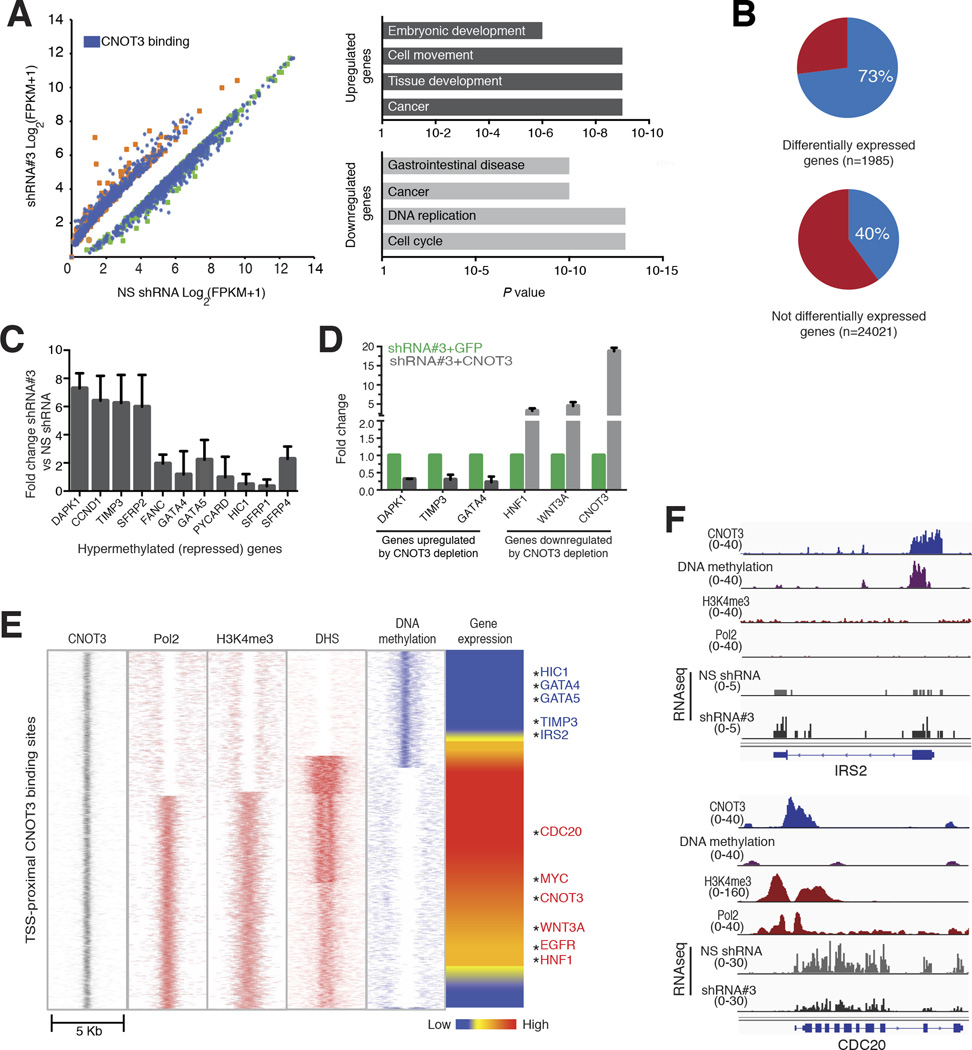
CNOT3 depletion by specific shRNAs affected more than 5,000 transcripts (false discovery rate, FDR <0.05). Among the most significant changes (FDR <0.01), many more transcripts (~2,000) were reduced than were increased (Fig. 5A), which is notable because CNOT3’s role in mRNA deadenylation17 should be unrelated to DNA binding and should stabilize, not lower, transcript levels when CNOT3 is absent. Cell replication genes predominated among down-regulated transcripts, whereas up-regulated genes were enriched for functions in embryonic and tissue development (Fig. 5A), indicating CNOT3 activity beyond cell replication per se. At least 73% of affected genes had nearby CNOT3 binding, whether expression was reduced or increased in CNOT3-depleted cells, compared to 40% of unaffected genes (Fig. 5B, P <0.0001). Moreover, RT-PCR analysis verified that representative silent transcripts were indeed reactivated in CNOT3-deficient HCT116 cells (Fig. 5C). shRNA-resistant CNOT3 reduced expression of up-regulated genes and increased the levels of down-regulated genes (Fig. 5D). These data confirm that our findings are specific and reveal opposing and seemingly direct effects of CNOT3 on both active and silent genes.
Figure 5. Relation of CNOT3 binding to gene regulation.
(A) Scatter plot of RNA-seq analysis showing genes differentially expressed (q <0.001) in triplicate samples of HCT116 cells treated with a non-specific shRNA (NS, x-axis) or CNOT3 shRNA #3 (y-axis). Genes with (blue dots) and without (green or orange dots) nearby CNOT3 binding are colored and the top Gene Ontology terms (derived from Ingenuity analysis) enriched among genes affected by CNOT3 deficiency are shown. (B) Proportions of CNOT3 binding near genes that are differentially expressed (top) or unaffected (bottom) in CNOT3-depleted cells. (C) qRT-PCR verification of increased transcript levels of selected genes derepressed in CNOT3-deficient HCT116 cells. (D) Representative gene expression changes occurring in CNOT3-depleted HCT116 cells are rescued by shRNA-resistant cDNA, indicating shRNA specificity. (E) Heatmaps showing presence or absence of RNA polymerase II (Pol2), H3K4me3, DHS, DNA methylation and mRNA expression in parental HCT116 cells at CNOT3-bound promoters. Selected genes affected by CNOT3 depletion are listed. (F) Representative IGV traces for each class of CNOT3-bound promoters, showing various chromatin features and RNA-seq data from control and CNOT3-deficient cells.
In agreement with this possibly dual role, we observed two classes of CNOT3-bound promoters (Fig. 5E). About 65% of the promoters showed strong H3K4me3 marking, DNaseI hypersensitivity (DHS), and presence of Pol2 in HCT116 cells; the corresponding genes lacked DNA methylation and were generally expressed at high levels. The remaining promoters showed no H3K4me3, Pol2 or DHS, but were heavily methylated, and most associated transcripts were barely detectable (Fig. 5E–F). In general, when affected by CNOT3 loss, bound genes that are expressed in the parental cells showed lower levels; only silent genes were increased in transcript levels (Fig. 5E–F), again implying direct CNOT3 effects on both gene activation and silencing.
Key transcriptional targets and pathways of CNOT3 regulation
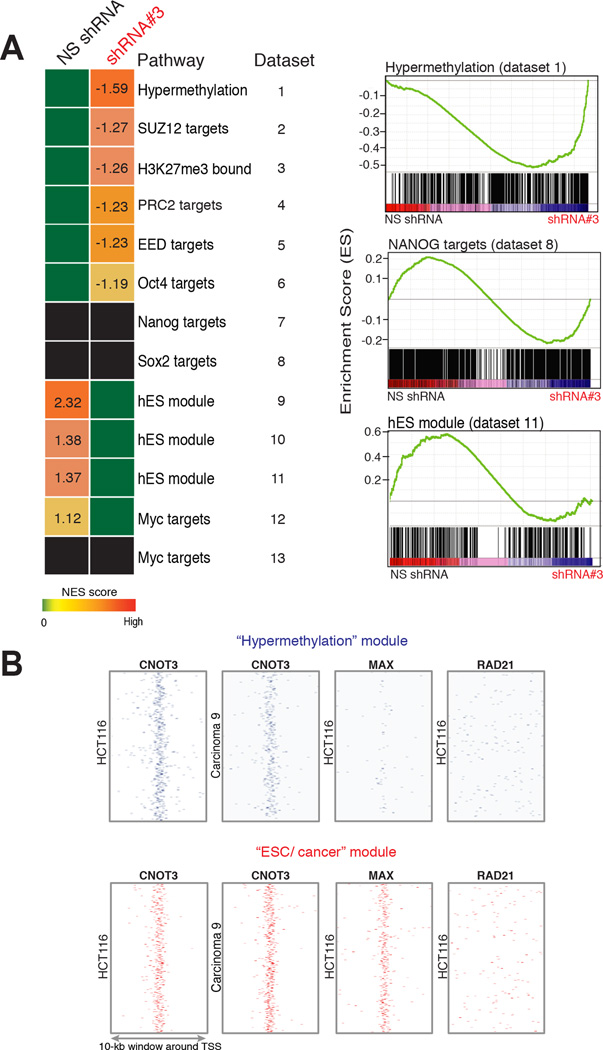
ESC genes that are recapitulated in cancer and Polycomb-regulated genes comprised the functions most enriched among CNOT3-bound genes (Fig. 4F, Suppl. Table 3). Furthermore, the increased or reduced expression of these gene groups in CNOT3-deficient cells suggested that they might belong to the modules repressed or activated, respectively, by CNOT3. Indeed, directed examination of CNOT3-dependent genes using Gene Set Enrichment Analysis (GSEA)43 gave a striking pattern: H3K27me3/Polycomb target genes were notably derepressed, whereas genes active in ESC and aggressive carcinomas were coordinately down-regulated in CNOT3-deficient cells (Fig. 6A). Multiple independent GSEA datasets gave concordant results, and the modest overlap with MYC-regulated genes was statistically significant. Because cell replication genes were excluded from the ESC/cancer signature3, these overlaps are likely not a trivial consequence of reduced cell division. Moreover, CNOT3-dependent genes reduced in HCT116 cells belong in the ESC/cancer module, but not the ESC pluripotency module associated with SOX2 or NANOG (Fig. 6A). Genes derepressed upon CNOT3 loss belong to a separate “hypermethylation module” that is silenced by virtue of H3K27me3 marks in ESC and by promoter hypermethylation in HCT116 cells and primary CRCs41,42.
Figure 6. Genes bound and regulated by CNOT3 correspond to ESC and hypermethylated gene modules.
(A) Gene Set Enrichment Analysis of CNOT3-regulated genes with respect to distinct components of the ESC program. The table expresses color-coded normalized enrichment scores (NES) and representative GSEA plots for modules increased (top), unaffected (center) or decreased (bottom) in expression in CNOT3-depleted HCT116 cells are shown. All listed NES values reflect significant (P <0.0001) enrichments. (B) Heatmaps of CNOT3-bound genes in the hypermethylated (top) and ESC (bottom) modules in HCT116 and Ca9, in relation to RAD21 (as a control) and MAX binding in HCT116 cells.
To determine if these salient effects on transcription occur at CNOT3-occupied genes, we examined CNOT3-bound promoters identified in Ca9 and HCT116 cells. These promoters were highly represented among the ESC/cancer and hypermethylation modules, segregating clearly into each group (Fig. 6B); both modules were significantly enriched in CNOT3 binding relative to the genome background (Suppl. Table 4). To establish the specificity of CNOT3 in this effect, we considered other TFs that occupy about as many sites in HCT116 cells. Binding of TCF4 and YY1 was slightly enriched at ESC/cancer module genes, but not at genes in the hypermethylation module; three other factors – SP1, RAD21, JUND1 – lacked enrichment at either module (Fig. 6B, Suppl. Table 4). Although HCT116 cells have minimal MYC, genes in the ESC/cancer – but not those in the hypermethylation – module commonly bind the MYC partner factor MAX. These findings agree with joint roles for CNOT3 and MYC in ESC self-renewal7 and with co-occurrence of CNOT3 and MYC motifs at cis-regulatory sites active in Ca2 and Ca9 (Suppl. Fig. 1B). These data suggest that CNOT3 collaborates with MYC-family TFs to activate genes and has a separate, MYC-independent role in gene silencing. The two groups of CNOT3-regulated genes together constitute the full transcriptional program associated with cellular self-renewal.
CNOT3 has a distinctive distribution in human CRCs and signifies poor prognosis
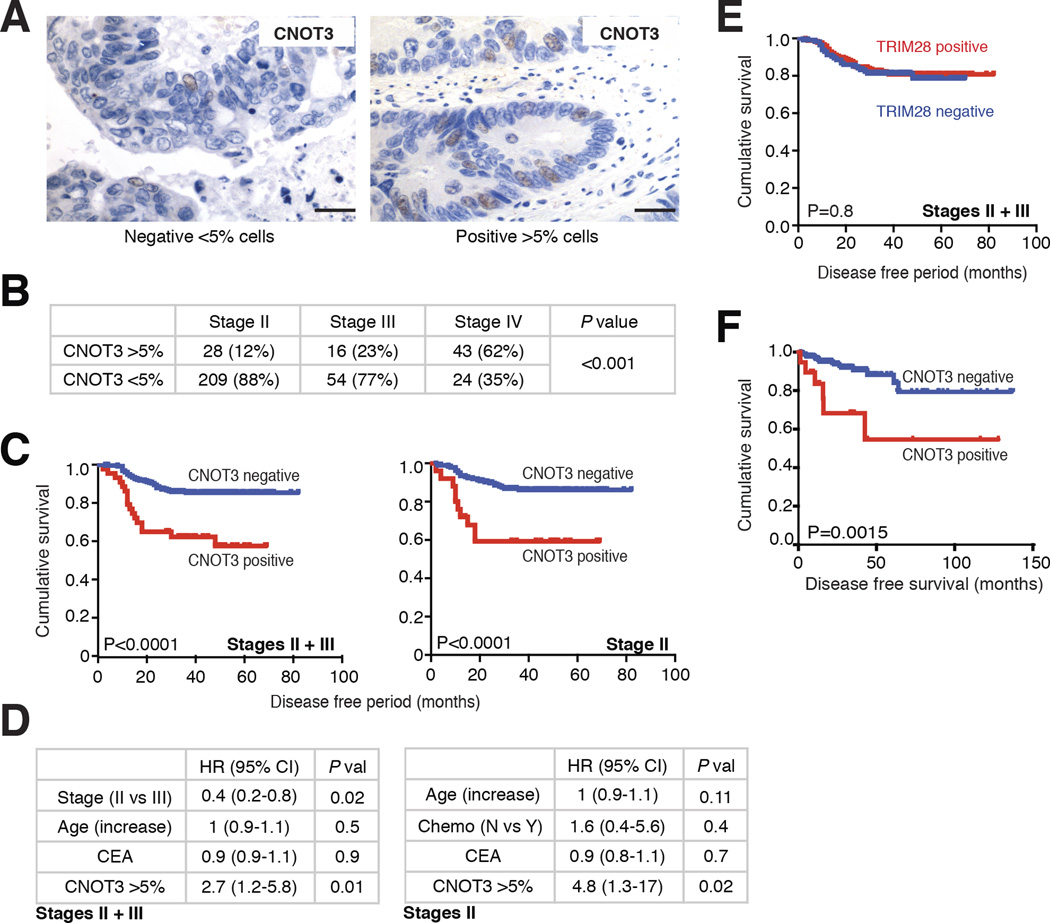
Based on the importance and role of CNOT3 revealed in these studies, we examined the significance of CNOT3 expression in a collection of 376 primary human CRC specimens (Suppl. Table 5). Most samples expressed CNOT3 in <1% of tumor cells, as noted above (Fig. 2). However, 87 of the cases (23%) showed CNOT3 in 5% to 20% of cancer cells (Fig. 7A, Suppl. Table 5). In line with these findings, CNOT3 mRNA levels in tumor cells exceeded those in normal colonic mucosa when immunohistochemistry (IHC) showed >5% CNOT3+ cells, but not in cases with low CNOT3+ cell fractions (Suppl. Fig. 5A); we therefore considered 5% of stained cells as the criterion for CNOT3 overexpression. In this large collection of primary CRCs, high CNOT3+ cell fractions were progressively higher in disease stages II, III and IV (Fig. 7B).
Figure 7. CNOT3 overexpression is associated with advanced CRC and high risk of relapse.
(A) IHC for CNOT3 in representative CRC specimens with typical (<1% to 5% of stained cells) or high (>5% to 20% of stained cells) CNOT3+ cell fractions. Scale bar, 50 µm. (B) Association of high CNOT3+ cell fractions with increasing CRC stage. (C) Kaplan-Meier survival analysis of patients with CRC stages II and III (combined, left, N=307) or stage II only (right, N=237), showing reduced long-term disease-free survival in cases with excess CNOT3. (D) Multivariate analysis in stages II and III (combined) or in stage II CRC of the influence of various factors, including high CNOT3+ cell fraction, on patient survival. (E) Kaplan-Meier survival analysis of patients whose CRCs did (red) or did not (blue) overexpress TRIM28. (F) Kaplan-Meier survival analysis of 296 patients with microsatellite-stable (MSI-negative) tumors with low or high CNOT3 mRNA levels in CRC stages II and III from The Cancer Genome Atlas collection.
About 1/2 to 4/5 of patients with CRC stages III and II, respectively, are cured by surgery, while the remaining patients eventually succumb to metastatic disease44. A high CNOT3+ cell fraction was strongly associated with disease relapse and survival (Fig. 7C–D), predicting relapse even better than disease stage (Fig. 7D). This association was especially pronounced in Stage II disease (hazard ratio 3.7, P <0.0001) and independent of other factors, such as patient age, tumor size, or exposure to adjuvant chemotherapy (Fig. 7D, Suppl. Fig. 5C–D). By contrast, TRIM28 overexpression, which occurred in about half of the same case series (Suppl. Table 5B, Suppl. Fig. 5B), showed no association with any pathological or clinical variable (data not shown) and no relation to patient survival (Fig. 7E).
KRAS (40%) and BRAF (7%) mutations were as frequent in cancers with high CNOT3+ cell fractions as in sporadic CRCs. Although CNOT3 is overexpressed in cases with microsatellite instability (MSI; e.g., Ca2), which account for 15% of CRCs in most series, only 3.2% of CRCs with a high CNOT3+ cell fraction were MSI+. Data from 296 CIN+ cases in The Cancer Genome Atlas45 replicated the strong association of high CNOT3 mRNA levels with poor clinical outcomes (Fig. 7F). Thus, CNOT3 is necessary for cellular self-renewal and also a novel, independent marker of especially aggressive CRCs in two large patient cohorts.
DISCUSSION
This study reveals that individual CRC epigenomes differ substantially from those of their Ad precursors, reflecting extensive reprogramming of regions marked with H3K4me2. Thus, progression from Ad to CRC is accompanied by aberrant activation and inactivation of thousands of presumptive cis-elements, disclosing one basis for widespread gene dysregulation. Although dynamic regions in several CRCs were enriched for the AP1 consensus motif, in keeping with a dominant role for Epidermal Growth Factor signaling, tumors generally differed from each other in the complement of activated regulatory sites, and 2 of 10 cases showed new sites highly enriched for the CNOT3 motif. Underscoring this observation, CNOT3 is overexpressed in about 23% of CRCs and binds many of the same regions of dynamic chromatin in tumors and cell lines. CNOT3 is usually expressed in rare crypt cells and <1% of scattered CRC cells, and when this domain expands, it remains patchy among largely non-replicating cells, as expected of a clonogenic population that might fuel tumor growth1,2,46,47. Although early studies proposed that such clonogenic cells express particular surface proteins, marker profiles and stem-cell activity seem fluid48,49; the hallmark feature may instead be a transcriptional or chromatin state that favors self-renewal, with elements that persist in daughter cells. Based on our combined data from CNOT3 ChIP and mRNA expression in CNOT3-silenced cells, we nominate CNOT3 as a regulator of such a gene signature in a sizeable fraction of CRCs.
We considered this possibility because the CNOT3 motif defined by ChIP in mouse ESCs (Ref. 7) is enriched at sites that aberrantly acquire H3K4me2 in some CRCs. Subsequently, we observed enrichment of the same motif in independent ChIP experiments, using a specific Ab that revealed both nuclear CNOT3 and robust binding at the same CRC cis-regulatory regions where we had detected CNOT3 motif enrichment. CNOT1, another member of the CCR4-NOT complex that also appears in the nuclear fraction, bound many of the same sites. Thus, although some transcriptional functions attributed to CNOT3 may be better explained as indirect effects of RNA or protein stabilization17,18, our data cross-validate exceptionally well and are most compatible with CNOT3’s well-studied, bona fide role in transcriptional control. We cannot, however, conclude that CNOT3 contacts DNA directly, and neither its primary sequence nor the crystal structure of its C-terminal 150 residues (representing 1/5 of the protein) complexed with portions of CNOT1 and CNOT2 reveals a conventional DNA-binding domain39. It is indeed possible that the transcriptional functions we ascribe to CNOT3 occur through interacting proteins that impart DNA sequence specificity.
CRCs show global DNA hypomethylation and CpG island hypermethylation50. Many of the hypermethylated genes are also targets of Polycomb regulation in ESCs40–42, reinforcing the idea that certain transcriptional modules underlie both ESC and cancer cell behaviors; these modules include activated and silenced genes. Whereas MYC and other sequence-specific TFs activate cancer and ESC genes8, repression is commonly attributed to methylation of DNA or Histone 3 Lysine 27, activities that lack intrinsic sequence specificity. Some other factor must therefore direct these modifications to selected genes, and the signals that coordinate opposing actions at different gene sets associated with self-renewal are not well understood. Therefore, a crucial finding is that CNOT3 binds both active (H3K4me3+ Pol2+ DHS+) and inactive (methylated, H3K4me3− Pol2− DHS−) promoters, with CNOT3 loss reducing expression of bound active genes and increasing levels of bound silent genes. Even after excluding genes associated with cell replication3, both groups of seemingly direct CNOT3 targets belong in the ESC signature that is replicated in poorly differentiated cancers. Our findings hence suggest that CNOT3 directly influences transcription of particular gene sets that are activated or inactivated, as part of a self-renewal program, in CRC.
Importantly, CNOT3 overexpression in CRC is associated with worse clinical outcomes, independent of traditional factors such as disease stage or adjuvant chemotherapy. This is especially the case in Stage II CRC, where there is a particular paucity of prognostic markers51,52, and we validated the association in the independent TCGA cohort. Taken together, our data hence implicate CNOT3 in conferring aggressive CRC behaviors through a transcriptional program for self-renewal and as a factor with prognostic value in CRCs.
Supplementary Material
Acknowledgments
Financial support:
Supported by the DFCI-Novartis Drug Discovery Program (R.A.S.); a gift from the Lind family (R.A.S.); NIH grants P50CA127003 (Dana-Farber/Harvard), R01CA72851 and CA181572 (A.G.); Instituto de Salud Carlos III, Spanish Economy and Competitiveness Ministry (PI13-01818, P.C.); and fellowships from Asociación Española Contra el Cáncer (Programa de Formación Avanzada en Oncología 2010) and Fundación Caja Madrid (P.C.) and the American-Italian Cancer Foundation (A.C.).
We thank Jingxuan Liu for constructing plasmids; Anne Küchler, Esther Díaz and María de Miguel for technical assistance; and Clifford Meyer, Lewyn Li, Henry Long and Myles Brown for valuable discussions.
Footnotes
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
P.C. and R.A.S. conceived and designed the study; P.C., J.M., D.H., M.M. and A.G. obtained data; P.C., A.C., C.Y., V.M. and L.T. performed computational and statistical analyses; E.B. reviewed CRC pathology; J.F. supervised clinical collections and correlations; R.A.S. provided overall supervision; P.C. and R.A.S. drafted the manuscript, with input from all authors.
REFERENCES
- 1.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 2.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 5.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 7.Hu G, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 10.Sansom OJ, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 11.He HH, et al. Positioned nucleosomes flanking a labile nucleosome characterize transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verzi MP, et al. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martynoga B, et al. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collart MA, Panasenko OO, Nikolaev SI. The Not3/5 subunit of the Ccr4-Not complex: a central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell Signal. 2013;25:743–751. doi: 10.1016/j.cellsig.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 17.Doidge R, Mittal S, Aslam A, Winkler GS. Deadenylation of cytoplasmic mRNA by the mammalian Ccr4-Not complex. Biochem Soc Trans. 2012;40:896–901. doi: 10.1042/BST20120074. [DOI] [PubMed] [Google Scholar]
- 18.Mersman DP, Du HN, Fingerman IM, South PF, Briggs SD. Polyubiquitination of the demethylase Jhd2 controls histone methylation and gene expression. Genes Dev. 2009;23:951–962. doi: 10.1101/gad.1769209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue T, et al. CNOT3 contributes to early B cell development by controlling Igh rearrangement and p53 mRNA stability. J Exp Med. 2015;212:1465–1479. doi: 10.1084/jem.20150384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. doi: 10.1101/gad.2020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collart MA, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 22.Winkler GS, Mulder KW, Bardwell VJ, Kalkhoven E, Timmers HT. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–3099. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neely GG, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng X, et al. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem Cells. 2012;30:910–922. doi: 10.1002/stem.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye T, Ravens S, Krebs AR, Tora L. Interpreting and visualizing ChIP-seq data with the seqMINER software. Methods Mol Biol. 2014;1150:141–152. doi: 10.1007/978-1-4939-0512-6_8. [DOI] [PubMed] [Google Scholar]
- 30.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim TH, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiser MM. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J Biol Chem. 1973;248:2536–2541. [PubMed] [Google Scholar]
- 33.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 34.Sulahian R, et al. An integrative analysis reveals functional targets of GATA6 transcriptional regulation in gastric cancer. Oncogene. 2014;33:5637–5648. doi: 10.1038/onc.2013.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 37.Galan-Caridad JM, et al. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brabletz T, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci USA. 2001;98:10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boland A, et al. Structure and assembly of the NOT module of the human CCR4-NOT complex. Nature structural & molecular biology. 2013;20:1289–1297. doi: 10.1038/nsmb.2681. [DOI] [PubMed] [Google Scholar]
- 40.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 41.McGarvey KM, et al. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 45.Network, T.C.G.A. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 48.Kreso A, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:54354–54358. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaffer CL, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Sousa EMF, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 52.Dalerba P, et al. CDX2 as a Prognostic Biomarker in Stage II and Stage III Colon Cancer. N Engl J Med. 2016;374:211–222. doi: 10.1056/NEJMoa1506597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.