Abstract
Alveolar rhabdomyosarcoma (ARMS) is a devastating pediatric disease driven by expression of the oncogenic fusion gene PAX3-FOXO1A. In this study, we report overexpression of the nuclear receptor NR4A1 is rhabdomyosarcomas which is sufficient to drive high expression of PAX3-FOXO1A there. RNAi-mediated silencing of NR4A1 decreased expression of PAX3-FOXO1A and its downstream effector genes. Similarly, cell treatment with the NR4A1 small molecule antagonists 1,1-bis(3-indolyl)-1-(p-hydroxy or p-carbomethoxyphenyl)methane (C-DIM) decreased PAX3-FOXO1A. Mechanistic investigations revealed a requirement for the NR4A1/Sp4 complex to bind GC-rich promoter regions to elevate transcription of the PAX3-FOXO1A gene. In parallel, NR4A1 also regulated expression of β1-integrin which with PAX3-FOXO1A contributed to tumor cell migration that was blocked by C-DIM/NR4A1 antagonists. Taken together, our results provide a preclinical rationale for the use of NR4A1 small molecule antagonists to treat ARMS and other rhabdomyosarcomas driven by PAX3-FOXO1A.
Keywords: NR4A1, antagonists, PAX3-FOXO1A, rhabdomyosarcoma
INTRODUCTION
Rhabdomyosarcoma (RMS) is the most common soft tissue sarcoma that is primarily observed in children and adolescents, and accounts for 50% of all pediatric cancers and 50% of soft tissue sarcomas in children (1,2). The urgency for developing new and more effective drugs for treating RMS is demonstrated by a recent paper in JAMA (3) that reported at age 45, 95.5% of cancer survivors are afflicted with chronic health conditions due to prior treatment with cytotoxic drugs. This demonstrates the need for mechanism-based cancer therapeutics in order to decreased the use and/or dose of cytotoxic agents.
The orphan nuclear receptors NR4A1 (Nur77, TR3), NR4A2 (Nurr1) and NR4A3 (Nor1) play important roles in maintaining cellular homeostasis by their involvement in inflammation, immune and neuronal functions, metabolism, and differentiation (4,5). These receptors are early immediate genes induced by multiple stimuli and there is increasing evidence that NR4A receptors are potential drug targets for many diseases including cancer (4–7). Among the NR4A receptors, there has been extensive research on the expression and role of NR4A1 in cancer and one study found the loss of both NR4A1 and NR4A2 in mice results in hematological malignancies (8), suggesting tumor suppressor-like activity for NR4A1. In contrast, NR4A1 exhibits tumor promoter activity (6,7) in solid tumors. NR4A1 is also overexpressed in tumors from breast, lung, pancreatic, colon and ovarian cancer patients and is a negative prognostic factor for breast, lung and ovarian cancer patients (9–15).
Although endogenous ligands for NR4A1 and other NR4A receptors have not been identified, structurally-diverse compounds directly or indirectly target this receptor. Initial studies demonstrated that several apoptosis-inducing agents activated nuclear export of NR4A1 and formation of a pro-apoptotic complex with bcl-2 which subsequently disrupted mitochondria (16–18). Wu and coworkers identified cytosporone B and structural analogs as NR4A1 ligands and these compounds exhibited structure-dependent activation of nuclear NR4A1 and nuclear export (19–22). In contrast, studies in this laboratory have demonstrated that among a series of 1,1-bis(3'-indolyl)-1-(p-substituted phenyl)methanes (C-DIMs), several compounds, including the p-hydroxy (DIM-C-pPhOH) and p-carboxymethyl (DIM-C-pPhCO2Me) bound and activated nuclear NR4A1 and acted as NR4A1 antagonists (23).
In a series of studies, it was demonstrated that knockdown of NR4A1 (siRNA) by RNAi or treatment with C-DIM/NR4A1 ligands inhibits pancreatic, lung, kidney, colon, rhabdomyosarcoma (RMS), and breast cancer growth and induces apoptosis (9,13,14,23–28). Moreover, in breast cancer cells, these same treatments inhibit migration through downregulation of β1-integrin, an NR4A1-regulated gene (26). Pro-oncogenic NR4A1-regulated pathways/genes in RMS and other cancer cell lines are summarized in Figure 1A. NR4A1 activates TXNDC5 and IDH1 to decrease reactive oxygen species which facilitates activation of mTOR, and NR4A1 also regulates pro-survival and growth promoting genes through an NR4A1/Sp complex interacting with GC-rich gene promoters (9,13,27). The NR4A1/Sp gene regulation pathway does not require direct NR4A1 binding to promoter DNA and is commonly observed for several nuclear receptors including orphan receptors (29). Sp transcription factors (Sp1, Sp3 and Sp4) are highly overexpressed in RMS cell lines (28,30,31), and the anticancer agent tolfenamic acid decreased expression of Sp1, Sp3, Sp4 and pro-oncogenic Sp-regulated genes including PAX3-FOXO1A (30), a critical pro-oncogenic factor in alveolar RMS (ARMS), a deadly form of RMS. In this study, we hypothesize and subsequently confirm that PAX3-FOXO1A is an NR4A1/Sp-regulated gene and treatment with NR4A1 antagonists decreases expression of PAX3-FOXO1A and downstream genes in ARMS cell lines. Thus, NR4A1 antagonists represent a novel approach for treating ARMS patients that overexpress this receptor.
Figure 1.
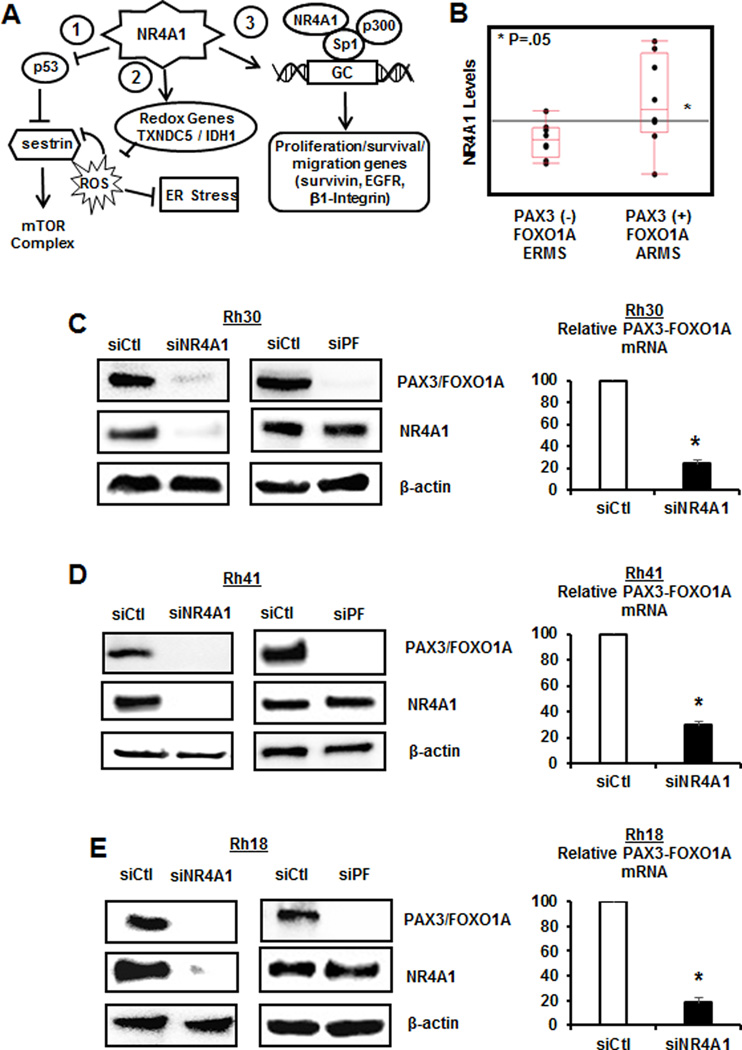
NR4A1 regulates PAX3-FOXO1A expression in ARMS. (A) Pro-oncogenic pathways regulated by NR4A1 in RMS and other cancer cell lines. (B) Analysis of PAX3-FOXO1A gene expression in ERMS and ARMS tumors expressing high and low levels of NR4A1 in patient-derived mRNA acquired from the NCBI GSE2851 dataset. Effects of knockdown of NR4A1 by RNAi (siNR4A1) in Rh30 (C), Rh41 (D) and Rh18 (E) on PAX3-FOXO1A protein and RNA was determined by western blot analysis of whole cell lysates and real time PCR of RNA extracts, respectively, as outlined in the Materials and Methods. Results were compared to cell lines transfected with a non-specific control oligonucleotide, and RNA results are means ± SE for three replicated determinations and significant (p<0.05) decreases are indicated (*).
MATERIALS AND METHODS
Cell lines, antibodies, chemicals, and other materials
RD, C2C12 and Rh30 cell lines were purchased from the American Type Culture Collection and were authenticated in 2014 (Promega Powerplex 18D) at the Duke University DNA Analysis Laboratory. Rh18 and Rh41 cells were received from Texas Tech University Health Sciences Center-Children’s Oncology Group in 2015. All cell lines were maintained at 37C in the presence of 5% CO2 with Rh30 maintained in RPMI-1640 Medium supplemented with 10% fetal bovine serum and 5% antibiotic. Rh41 and Rh18 cell lines were maintained in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 20% fetal bovine serum, 1× ITS (5 µg/mL insulin, 5 µg/mL transferrin, 5 ng/mL selenous acid), and 5% antibiotic. RPMI-1640 was purchased from Sigma-Aldrich (St. Louis, MO). IMDM was purchased from ThermoFisher Scientific (Waltham, MA) and ITS was purchased from Sigma Aldrich (St. Louis, MO). Lipofectamine 2000 was purchased from Invitrogen (Grand Island, NY). MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) was purchased from Sigma-Aldrich (St. Louis, MO). The C-DIM compounds were prepared as previously described. Summaries of the antibodies and oligonucleotides for RNAi, real time PCR and ChIP primers are summarized in Supplemental Table S1.
Total RNA expression analysis
Sample data of total RNA was acquired from NCBI GEO dataset GSE2787 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2787). In addition, transcription profiles of competitively hybridized microarray samples were quantified and lowess normalization for each spot was performed and subsequently converted in logarithmic scale, with submitted expression values corresponding to a log(2) ratio of normalized intensities. Expression values were listed into PAX3-FOXO1A positive (ARMS) and PAX3-FOXO1A negative (ERMS) groups in JMP® and a box plot was generated, from which a t-test was performed; significance was determined as a p-value less than 0.01, shown by an asterisk.
MTT assay
ARMS cells were plated in 24-well plates at 1 × 105 cells per well and allowed to adhere to the plate overnight. Cells underwent a 24-hr treatment with DIM-C-pPhCO2Me or siNR4A1 knockdown. Corresponding controls received DMSO. On the day of assay, treatment medium was replaced with fresh medium containing 0.83 mg of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) per ml and incubated for 4 hr. Medium was then aspirated off, and 200 µl of dimethyl sulfoxide was added to wells to solubilize crystals. The optical density of each sample was read on a microplate reader (SynergyMx, BioTek Instruments, Inc., Winooski, VT.) at 570 nm against a blank prepared from cell-free wells. Cell survival was expressed as a fraction of that of DMSO treated controls.
Tumor growth assay
Female athymic nude mice (6–8 weeks old) were obtained (Charles River Laboratory, Wilmington, MA) and maintained under specific pathogen-free conditions, housed in isolated vented cages, and allowed to acclimate for one week with standard chow diet. The animals were housed at Texas A&M University in accordance with the standards of the Guide for the Care and Use of Laboratory Animals and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). The protocol of the animal study was approved by the Institutional Animal Care and Use Committee (IACUC), Texas A&M University. Rh30 cell lines (4×106 cells) grown in RPMI media containing 10% FBS were detached, resuspended in 100 µl of phosphate-buffered saline with matrigel (BD Bioscience, Bedford, MA) (75:25), and implanted subcutaneously in the mice. When tumors reached about 40–50 mm3 size, the mice were randomized into control and treatment groups (6 animals per group) and treated with placebo or DIM-C-pPhCO2Me (40 mg/kg/d) in nano liquid carrier (administered in sodium carboxymethyl cellulose) by oral gavage every second day for 3 weeks. Tumor volumes and weights, and body weight were determined; the tumor size was measured using Vernier calipers, and the tumor volume was estimated by the formula: tumor volume (mm3) = (L × W2) × ½, where L is the length and W is the width of the tumor. All animals in the treatment group presented with an infiltrative, densely cellular neoplasm with similar histological features as observed in the control group. Multifocal areas of necrosis was also observed within the neoplasm in this group.
RESULTS
1. NR4A1 regulates PAX3-FOXO1A expression in ARMS cell lines
Previous analysis of publically available patient arrays shows that NR4A1 is upregulated in tumors from RMS patients and analysis of ARMS tumors showed that NR4A1 levels were also higher in PAX3-FOXO1A-positive (ARMS) vs. PAX3-FOXO1-negative (ERMS) tumors (Fig. 1B) (28,32). This analysis was limited by the few studies available; however, there was a trend between expression of NR4A1 and PAX3-FOXO1A. Previous studies show that NR4A1 antgonists or knockdown inhibits RD and Rh30 cell growth (28), and results illustrated in Supplemental Figure S1A show that these treatments have minimal effects on normal muscle C2C12 cells. We also observed that, in contrast to a recent study (33), TGFβ inhibits ARMS cell growth similar to that observed for NR4A1 knockdown and their combined effects are additive. Previous studies show that NR4A1 antagonists or knockdown inhibits RD and Rh30 cell growth (28) and results illustrated in Supplemental Figure S1A show that these treatments have minimal effects of normal muscle C2C12 cells. We also observed that in contrast to a recent study (33) TGFβ inhibits ARMS cell growth, similar to that observed for NR4A1 knockdown and their combined effects are additive. Transfection of Rh30 cell lines with siCtl (non-specific oligonucleotide) and siNR4A1 resulted in decreased expression of NR4A1 and PAX3-FOXO1A proteins and in a separate experiment, siNR4A1 also decreased PAX3-FOXO1A mRNA (Fig. 1C). Transfection of Rh30 cell lines with siPAX3-FOXO1A decreased expression of PAX3-FOXO1A but not NR4A1 protein confirming that NR4A1 regulated expression of the fusion gene in this cell line. We also carried out an identical set of experiments in Rh41 (Fig. 1D) and Rh18 (Fig. 1E) ARMS cell lines and the results were similar to that observed in Rh30 cell lines (Fig. 1C), confirming that NR4A1 regulates PAX3-FOXO1A expression.
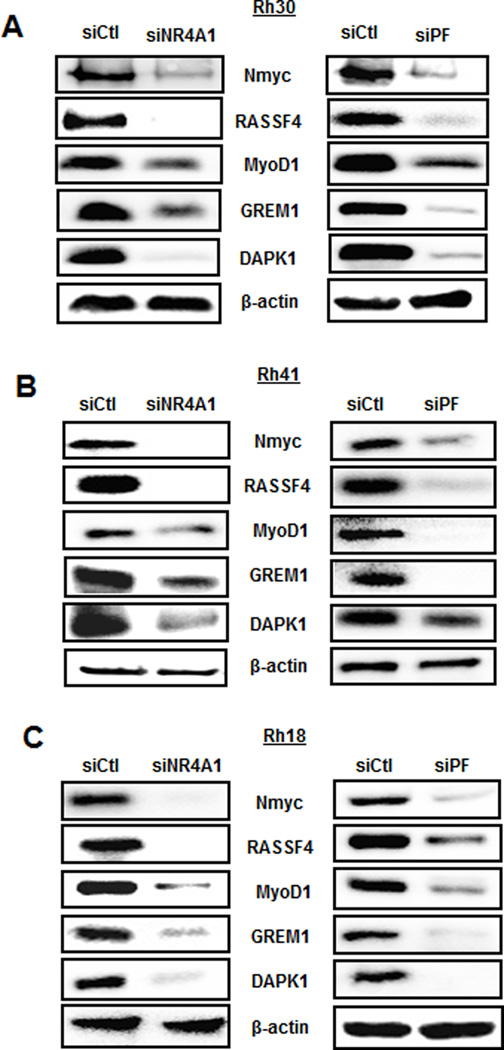
The highly pro-oncogenic activity of PAX3-FOXO1A is primarily due to regulation of downstream genes which include the oncogene NMyc, ras-association domain family 4 (RASSF4), myogenic differentiation-1 (MYOD1), gremlin 1 (GREM1) and death-associated protein kinase-1 (DAPK1) (33–36). Knockdown of NR4A1 in an Rh30 cell line resulted in decreased expression of NMyc, Rassf4, MyoD1, Grem1 and DAPK1 and similar results were observed in cell lines transfected with siPAX3-FOXO1A (Fig. 2A). Moreover, comparable results were observed in Rh41 (Fig. 2B) and Rh18 (Fig. 2C) cell lines confirming that NR4A1 downregulation results in decreased expression not only of PAX3-FOXO1A but also PAX3-FOXO1A-regulated genes in ARMS cell lines.
Figure 2.
NR4A1 regulates expression of PAX3-FOXO1A-dependent genes. Rh30 (A), Rh41 (B) and Rh18 (C) cell lines were transfected with siCtl, siNR4A1 or siPAX3-FOXOA1 and after 72 hr, whole cell lysates were analyzed by western blots. Common lysates were used for western blots illustrated in Figures 1 and 2.
2. NR4A1 antagonists decrease expression of PAX3-FOXO1A in ARMS cell lines
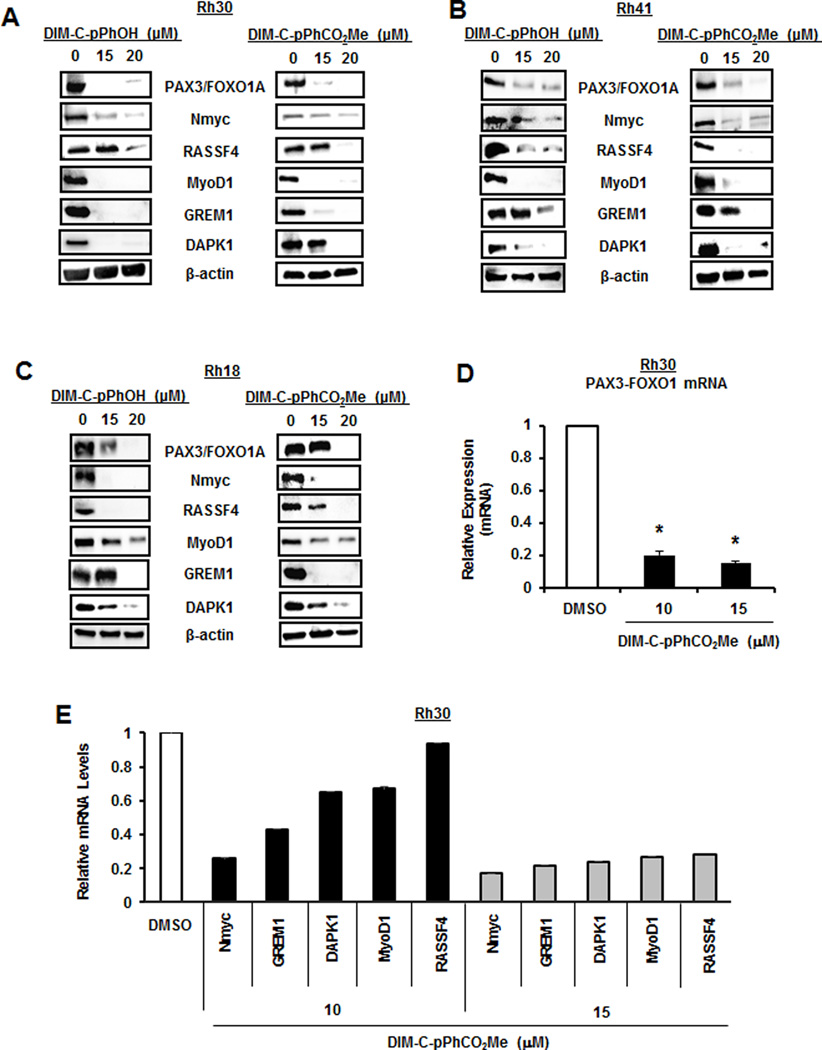
Previous studies show that DIM-C-pPhOH and DIM-C-pPhCO2Me bind the ligand binding domain of NR4A1 (23) and act as NR4A1 antagonists in pancreatic, colon, breast, kidney and RMS cells (13,23–28). Treatment of Rh30 cell lines with DIM-C-pPhOH and DIM-C-pPhCO2Me decreased expression of PAX3-FOXO1A protein and this was accompanied by downregulation of NMyc, Rassf4, MyoD1, Grem1 and DAPK1 proteins (Fig. 3A). Similar results were observed in Rh41 (Fig. 3B) and Rh18 (Fig. 3C) cell lines, confirming that knockdown of NR4A1 by RNAi (Figs. 1 and 2) or inactivation of NR4A1 by treatment with NR4A1 antagonists resulted in the abrogation of the PAX3-FOXO1A transcriptional program in ARMS cell lines. Using Rh30 cell lines as a model, we also observed that DIM-C-pPhCO2Me decreased expression of PAX3-FOXO1A (Fig. 3D) and NMyc, Grem1, DAPK1, MyoD1 and Rassf4 (Fig. 3E) mRNA levels in Rh30 cell lines. This is consistent with the potential role of NR4A1 as a regulator of PAX3-FOXO1A gene expression and PAX3-FOXO1A-mediated regulation of the downstream genes.
Figure 3.
NR4A1 antagonists downregulate PAX3-FOXO1A. Rh30 (A), Rh41 (B) and Rh18 (C) cell lines were treated with the NR4A1 antagonists DIM-C-pPhOH and DIM-C-pPhCO2Me for 24 hr and whole cell lysates were analyzed by western blots as outlined in the Materials and Methods. Rh30 cells were treated with 10 or 15 µM DIM-C-pPhCO2Me for 24 hr and RNA extracts were examined by real time PCR for expression of PAX3-FOXO1A (D) and PAX3-FOXO1A-regulated genes (E). Results (D and E) are expressed as means ± SE for 3 replicate determinations and significantly (p<0.05) decreased expression compared to DMSO (control) is indicated (*).
3. Mechanism of NR4A1 regulation of PAX3-FOXO1A
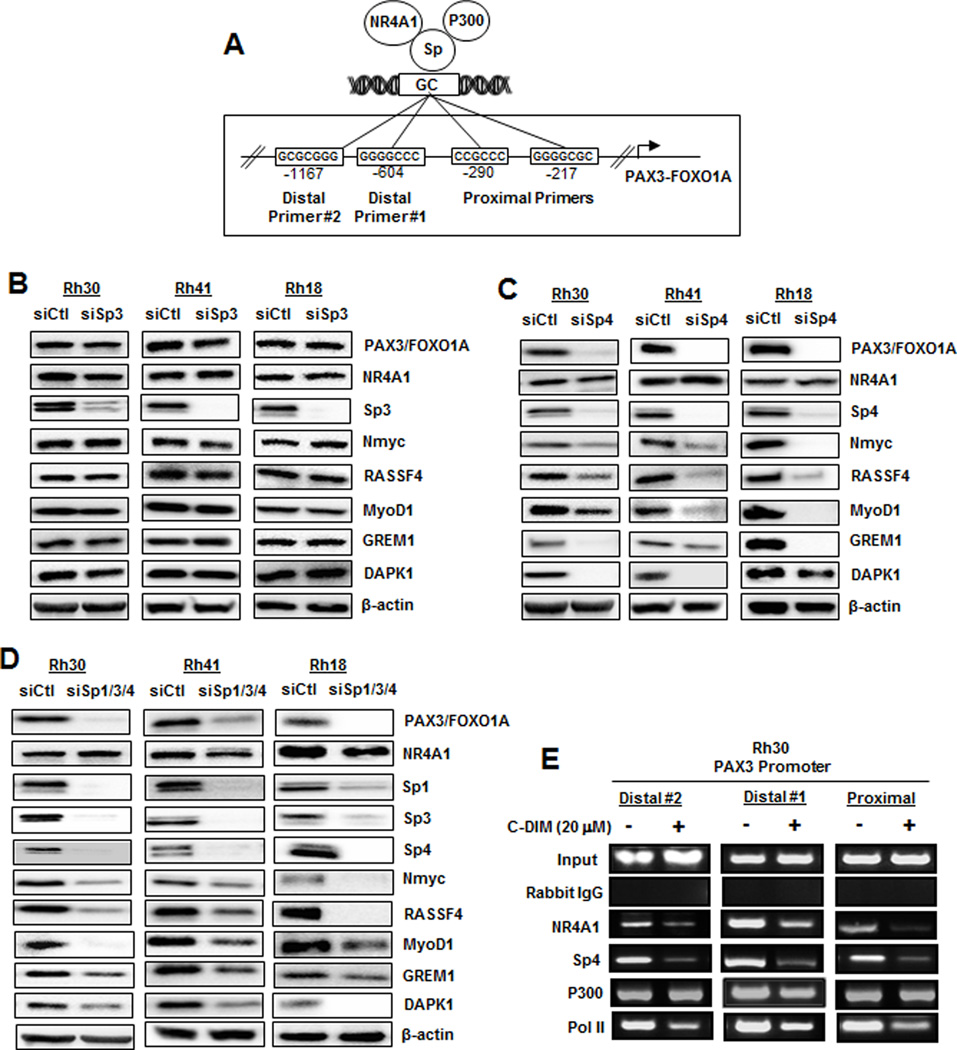
Previous studies show that NR4A1, in combination with p300, act as nuclear cofactors for expression of some Sp1-regulated genes including survivin, TXNDC5, IDH1, α5-integrin and β1-integrin (9,26,28). The PAX3-FOXO1A promoter has several GC-rich binding sites (Fig. 4A), and we therefore investigated the role of Sp1 in regulating expression of PAX3-FOXO1A and downstream genes by RNAi. Knockdown of Sp1 decreased Sp1 and p300 proteins but did not affect expression of PAX3-FOXO1A or downstream genes in Rh30, Rh41 or Rh18 cell lines (Suppl. Fig. S1A), suggesting that in contrast to previous studies on NR4A1/Sp1-regulated genes (9,26,28), neither Sp1 nor p300 were required. This was confirmed by knockdown of p300 in ARMS cell lines which did not affect expression of PAX3-FOXO1A and downstream genes (Suppl. Fig. S1B). Since Sp3 and Sp4 also bind GC-rich promoter sites and are overexpressed in RMS cell lines (30,31), we investigated the effects of Sp3 and Sp4 knockdown and downregulation of Sp1/3/4 (combined) (Figs. 4B–4D, respectively). Knockdown of Sp3 had minimal effects on expression of PAX3-FOXO1A and downstream genes; however, knockdown of either Sp4 or Sp1/3/4 resulted in decreased expression of PAX3-FOXO1A, NMyc, Rassf4, Grem1, MyoD1 and DAPK1. Results of these RNAi experiments indicated that Sp4 interactions with NR4A1 regulated PAX3-FOXO1A expression and therefore we carried out ChIP assays on the three different GC-rich regions of the PAX3-FOXO1A gene promoter (Fig. 4A) to determine NR4A1/Sp4 promoter interactions. In untreated Rh30 cell lines, NR4A1, Sp4, p300 and pol II were associated with the promoter and treatment with 20 µM DIM-C-pPhOH for 6 hr decreased interactions of pol II, NR4A1 and Sp4 with the two distal and proximal regions of the PAX3-FOXO1A gene promoter (Fig. 4E). P300 and other Sp proteins also interacted with the PAX3-FOXO1A promoter (data not shown); however, these proteins did not play a functional role in regulation of PAX3-FOXO1A. We also showed by RNAi that CBP knockdown did not alter expression of PAX3-FOXO1A (Suppl. Fig. S1C) and current studies are investigating other cofactors which may coregulate NR4A1/Sp4-dependent expression of PAX3-FOXO1A.
Figure 4.
Role of p300/NR4A1/Sp in regulation of PAX3-FOXO1A in ARMS cells. (A) GC-rich Sp binding sites in the proximal and two distal regions of the PAX3-FOXO1A gene promoter. ARMS cell lines were transfected with siSp3 (B), siSp4 (C), and siSp1/3/4 (D). Whole cell lysates were analyzed by western blots as outlined in the Materials and Methods. (E) Rh30 cells were treated with 20 µM DIM-C-pPhCO2Me for 6 hr and association of various factors with the proximal and two distal regions of the PAX3-FOXO1A promoter were determined in a ChIP assay as outlined in the Materials and Methods. Sp1 and Sp3 also bound the PAX3-FOXO1A gene promoter (data not shown).
4. Functional and in vivo studies
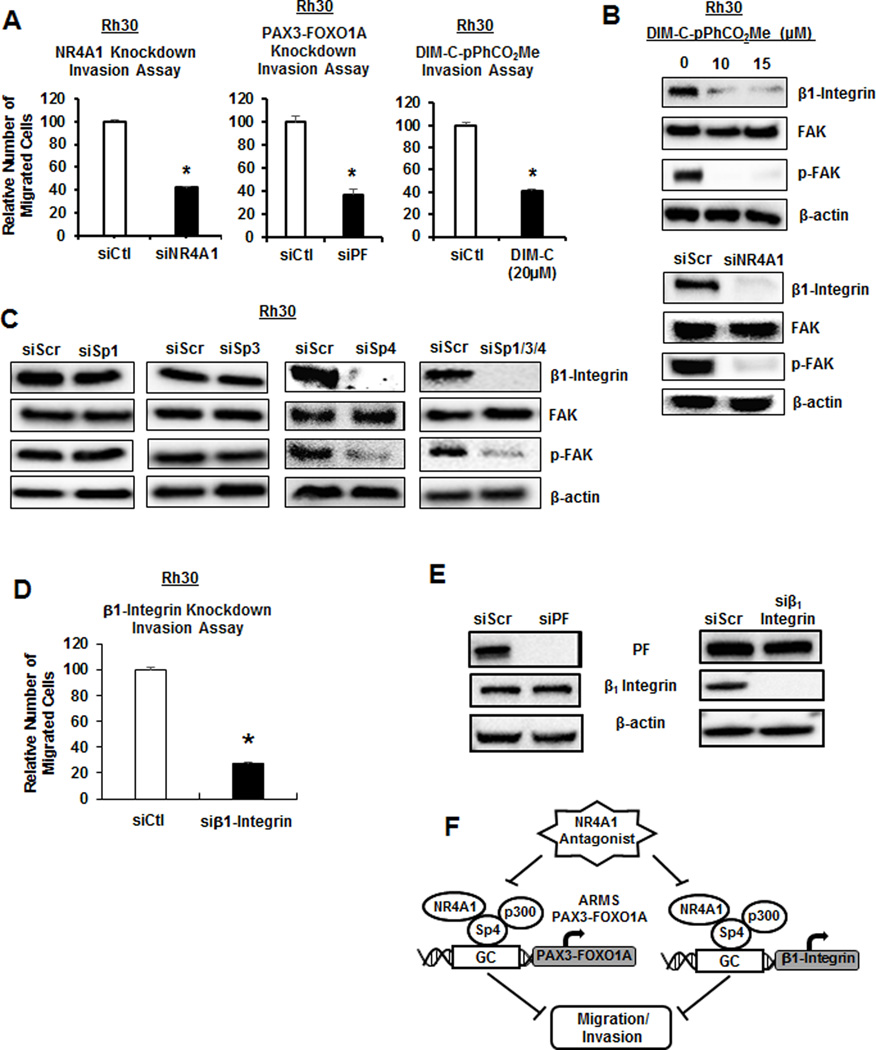
Previous studies showed that PAX3-FOXO1A plays an important role in ARMS cell migration/invasion (37,38) and this is confirmed in Rh30 cell lines where transfection of siPAX3-FOXO1A decreased migration in a Boyden chamber assay (Fig. 5A). Moreover, transfection of Rh30 cell lines with siNR4A1 or treatment with the NR4A1 antagonist DIM-C-pPhCO2Me also inhibited Rh30 cell line migration, confirming that either direct (siPAX3-FOXO1A) or indirect downregulation of the fusion gene by inactivation of NR4A1 decreased Rh30 cell line migration. We recently observed that knockdown of β1-integrin by siNR4A1 or treatment with DIM-C-pPhCO2Me decreased migration of breast cancer cells and this involved an NR4A1/Sp1 complex binding to GC-rich elements in the β1-integrin promoter (26). DIM-C-pPhCO2Me also decreased expression of β1-integrin and phosphorylation of FAK (pFAK downstream from β1-integrin) in Rh30 cell lines and knockdown of NR4A1 by RNAi also gave similar results (Fig. 5B). Moreover, using lysates from the Sp knockdown studies (Fig. 4 and Suppl. Fig. S2), we also observed that only siSp4 and siSp1/3/4 decreased β1-integrin and pFAK (Fig. 5C), suggesting that NR4A1/Sp4 regulated both β1-integrin and PAX3-FOXO1A, and knockdown of β1-integrin also decreased Rh30 cell line migration (Fig. 5D). Furthermore, knockdown of PAX3-FOXO1A has no effect on Beta-1 Integrin and vice versa (Fig. 5E). We conclude that NR4A1-mediated migration of Rh30 cell lines is dependent on both PAX3-FOXO1A and β1-integrin (Fig. 5F) which can be targeted simultaneously by C-DIM/NR4A1 antagonists.
Figure 5.
PAX-FOXO1A and β1-integrin are regulated by NR4A1/Sp4 and are both pro-migration genes. (A) Knockdown of PAX3-FOXO1A (siPF) or NR4A1 by RNAi and treatment with DIM-C-pPhCO2Me decreased Rh30 migration in a Boyden chamber assay as outlined in the Materials and Methods. (B) Rh30 cells were treated with DIM-C-pPhCO2Me or transfected with siNR4A1, and whole cell lysates were analyzed for β1-integrin/FAK expression by western blots. (C) Rh30 cells were transfected with oligonucleotides targeted to Sp1, Sp3, Sp4 and Sp1/3/4 (combined), and whole cell lysates were analyzed by western blots. Lysates were obtained from studies outlined in Supplemental Figure 1 and Figures 2B–2D. (D) Rh30 cells were transfected with β1-integrin and cell migration was determined in a Boyden chamber assay. (E) Rh30 cells were transfected with siRNA targeting either PAX3-FOXO1A or β1-integrin and whole cell lysates were analyzed by western blot. (F) Model for NR4A1/Sp4 regulation of PAX3-FOXO1A and β1-integrin. Results (A and D) are expressed as means ± SE for 3 replicate determinations and significant (p<0.05) decreases are indicated (*).
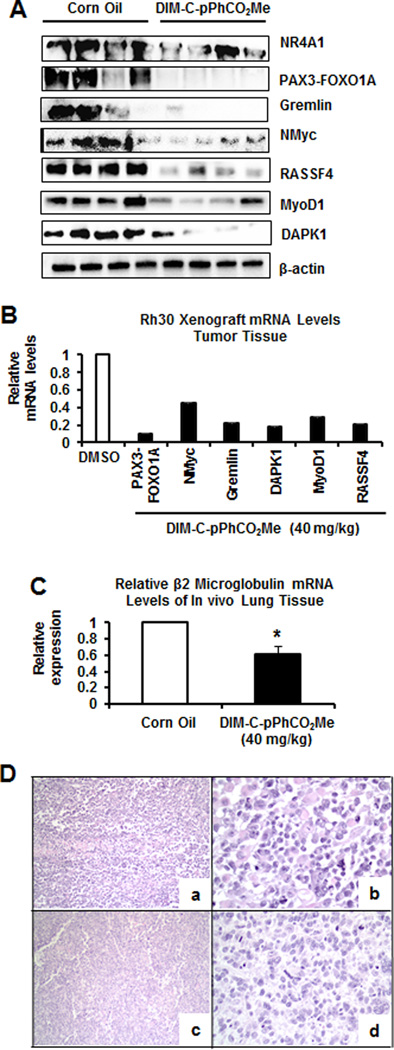
Previous studies showed that the NR4A1 antagonist DIM-C-pPhCO2Me (40 mg/kg/day) inhibited tumor growth in athymic nude mice bearing Rh30 cells as xenografts (28). We examined lysates from tumors treated with corn oil (control) or DIM-C-pPhCO2Me and the treatment significantly decreased expression of PAX3-FOXO1A mRNA and protein and downstream genes NMyc, Rassf4, MyoD1 and DAPK1 (Fig. 6A and 6B). This is consistent with the results of in vitro studies (Fig. 3). Moreover, using human B2-microglobulin mRNA as a unique marker, we observed decreased expression in lungs of mice bearing Rh30 cells as xenografts; this was consistent with decreased lung metastasis (Fig. 6C).
Figure 6.
In vivo studies. Tumors from mice treated with corn oil or DIM-C-pPhCO2Me (40 mg/kg/d) (25) were extracted for protein (A) and mRNA (B) analysis by western blots and real-time PCR, respectively. (C) Human β2-microglobulin mRNA expression in lungs from control and DIM-C-pPhCO2Me-treated mice were determined by real time PCR. (D) Representative images of rhabdomyosarcoma observed on control (a, b) and treatment groups (c, d). In both groups, the neoplasm was characterized by a population of round and pleomorphic cells arranged in sheets, with moderate to abundant cytoplasm and round nuclei. Scattered multinucleated neoplastic cells were observed. The neoplasm had multifocal necrotic areas in both the control and treatment groups. Hematoxylin and eosin; 100× (a, c), 400× (b, d).
DISCUSSION
RMS is primarily observed in children and adolescents and accounts for 5% of all pediatric cancers and 50% of soft tissue sarcomas in children (39). Although ERMS patients respond well to current therapies which include surgery and radiotherapy combined with treatment with cytotoxic drug combinations, patients with ARMS have a poor diagnosis. Cytogenetic analysis has demonstrated that 2;13 and 1;13 chromosomal translocations generating PAX3-FOXO1A and PAX7-FOXO1A fusion genes, respectively, are highly prevalent (55% and 22%, respectively) in tumors from ARMS patients. The PAX3-FOXO1A fusion gene is the critical prognostic marker for ARMS patients with metastatic disease, with an estimated overall 4 year survival of 8% compared to 75% survival rate of patients with PAX7-FOXO1A-expressing tumors (39–41). Unfortunately, RMS patients that survive current cytotoxic drug therapies have an increased risk for several diseases later in life (3), emphasizing the critical need for development of new mechanism-based therapies which have fewer long term adverse effects. Results of PAX3-FOXO1A knockdown or overexpression studies in RMS and other cell lines demonstrate the functional importance of this fusion gene in maintaining the aggressive cancer cell phenotype and this is due, in part, to the pro-oncogenic PAX3-FOX01-regulated genes (39,42). Development of agents that target PAX3-FOXO1A is ongoing and includes thapsigargin, fenretinide, HDAC inhibitors, and polo-like kinase inhibitors; however, the efficacy of these compounds for clinical applications in ARMS chemotherapy is not known (43–47).
There is evidence that ROS-inducing anticancer agents such as HDAC inhibitors are effective anticancer agents against RMS in both laboratory rodent and cell models and that ROS decreases expression of Sp1, Sp3, Sp4 and pro-oncogenic Sp-regulated genes (31,43,46). In addition, ROS-independent downregulation of Sp transcription factors in RMS cells treated with tolfenamic acid demonstrates that several pro-oncogenic genes in RMS including c-Met, CXCR4, insulin-like growth factor receptor (IGFR), and platelet-derived growth factor receptor α (PDGFRα) are Sp-regulated. Tolfenamic acid also decreases PAX3-FOXO1A in Rh30 cell lines (30). Knockdown of Sp transcription factors or NR4A1 in RMS cell lines resulted in decreased cell proliferation, induction of apoptosis, and inhibition of cell migration (30,31). The comparable functions of Sp transcription factors and NR4A1 are due, in part, to coregulation of genes by NR4A1/Sp complexes that bind GC-rich gene promoters such as survivin, TXNDC5, IDH1 and β1-integrin (9,26,28). Ongoing RNAseq and array studies also show that there is a considerable overlap between genes coregulated by NR4A1 and Sp transcription factors, and we hypothesized that PAX3-FOXO1A is also an NR4A1/Sp-regulated gene.
Knockdown of NR4A1 or treatment of Rh30, Rh41 and Rh18 ARMS cell lines with NR4A1 antagonists decreased expression of PAX3-FOX01A (protein and mRNA) and downstream genes (Figs. 1–3) and similar results were observed after PAX3-FOXO1A knockdown by RNAi. In vivo studies also showed that DIM-C-pPhCO2Me decreased PAX3-FOXO1A and downstream genes in Rh30-derived tumors (Fig 6). The role of Sp transcription factors in mediating this response was investigated by RNAi (Fig. 4) and the results indicated that Sp4 and not Sp1 or Sp3 was involved in expression of PAX3-FOXO1A. A recent study showed that Sp1, Sp3 and Sp4 regulate expression of several genes in common; however, all three transcription factors also regulate unique sets of genes (48), and in ARMS cell lines PAX3-FOXO1A expression is dependent on NR4A1 and Sp4; this was confirmed in ChIP assays (Fig. 4). We did not observe that p300 (or CBP) was required for NR4A1/Sp4-mediated regulation of PAX3-FOXO1A. This differed from NR4A1/Sp1-dependent genes, and current studies are investigating the identity of factors that may be involved.
In summary, this study shows that the critical pro-oncogenic PAX3-FOXO1A fusion gene is regulated by NR4A1/Sp4 interactions with GC-rich gene promoter elements. PAX3-FOXO1A gene expression can be inhibited by targeting either the receptor or Sp4 since knockdown of NR4A1 or Sp4 by RNAi blocks PAX3-FOXO1A gene expression. This study shows for the first time that C-DIM/NR4A1 antagonists represent a new class of mechanism-based agents that target PAX3-FOXO1A and other pro-oncogenic genes/pathways (Fig. 1A). These antagonists also simultaneously decrease expression of PAX3-FOXO1A and β1-integrin genes (Fig. 5F) which play a role in ARMS cell line migration/invasion. Current studies are focused on development of more efficacious C-DIM/NR4A1 antagonists for clinical applications in ARMS chemotherapy and for combination therapies that will reduce requirements for cytotoxic drugs and thereby decrease the incidence of health effects later in life (3).
Supplementary Material
Acknowledgments
Funding: Funding was provided by the National Institutes of Health (P30-ES023512), the Sid Kyle Endowment, and Texas AgriLife Research.
Footnotes
Conflict of Interest: There are no conflicts of interest to declare.
LITERATURE CITED
- 1.Paulino AC, Okcu MF. Rhabdomyosarcoma. Current Problems in Cancer. 2008;32:7–34. doi: 10.1016/j.currproblcancer.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Parham DM, Ellison DA. Rhabdomyosarcomas in adults and children: an update. Archives of Pathology and Laboratory Medicine. 2006;130:1454–1465. doi: 10.5858/2006-130-1454-RIAACA. [DOI] [PubMed] [Google Scholar]
- 3.Hudson MM, Ness KK, Gurney JG, Mulrooney DA, Chemaitilly W, Krull KR, et al. Clinical ascertainment of health outcomes among adults treated for childhood cancer. JAMA. 2013;309:2371–2381. doi: 10.1001/jama.2013.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Molecular Endocrinology. 2010;24:1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safe S, Jin UH, Morpurgo B, Abudayyeh A, Singh M, Tjalkens RB. Nuclear receptor 4A (NR4A) family - orphans no more. Journal of Steroid Biochemistry and Molecular Biology. 2016;157:48–60. doi: 10.1016/j.jsbmb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safe S, Jin UH, Hedrick E, Reeder A, Lee SO. Minireview: role of orphan nuclear receptors in cancer and potential as drug targets. Molecular Endocrinology. 2014;28:157–172. doi: 10.1210/me.2013-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nature Medicine. 2007;13:730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 8.Delgado E, Boisen MM, Laskey R, Chen R, Song C, Sallit J, et al. High expression of orphan nuclear receptor NR4A1 in a subset of ovarian tumors with worse outcome. Gynecologic Oncology. 2016 doi: 10.1016/j.ygyno.2016.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SO, Abdelrahim M, Yoon K, Chintharlapalli S, Papineni S, Kim K, et al. Inactivation of the orphan nuclear receptor TR3/Nur77 inhibits pancreatic cancer cell and tumor growth. Cancer Research. 2010;70:6824–6836. doi: 10.1158/0008-5472.CAN-10-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muscat GE, Eriksson NA, Byth K, Loi S, Graham D, Jindal S, et al. Research resource: nuclear receptors as transcriptome: discriminant and prognostic value in breast cancer. Molecular Endocrinology. 2013;27:350–365. doi: 10.1210/me.2012-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JR, Gan WJ, Li XM, Zhao YY, Li Y, Lu XX, et al. Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP-9 and E-cadherin. Carcinogenesis. 2014;35:2474–2484. doi: 10.1093/carcin/bgu157. [DOI] [PubMed] [Google Scholar]
- 12.Zhou F, Drabsch Y, Dekker TJ, de Vinuesa AG, Li Y, Hawinkels LJ, et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-beta signalling. Nat Commun. 2014;5:3388. doi: 10.1038/ncomms4388. [DOI] [PubMed] [Google Scholar]
- 13.Lee SO, Andey T, Jin UH, Kim K, Singh M, Safe S. The nuclear receptor TR3 regulates mTORC1 signaling in lung cancer cells expressing wild-type p53. Oncogene. 2012;31:3265–3276. doi: 10.1038/onc.2011.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho SD, Yoon K, Chintharlapalli S, Abdelrahim M, Lei P, Hamilton S, et al. Nur77 agonists induce proapoptotic genes and responses in colon cancer cells through nuclear receptor-dependent and nuclear receptor-independent pathways. Cancer Research. 2007;67:674–683. doi: 10.1158/0008-5472.CAN-06-2907. [DOI] [PubMed] [Google Scholar]
- 15.Wu H, Lin Y, Li W, Sun Z, Gao W, Zhang H, et al. Regulation of Nur77 expression by beta-catenin and its mitogenic effect in colon cancer cells. FASEB Journal. 2011;25:192–205. doi: 10.1096/fj.10-166462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Kolluri SK, Gu J, Dawson MI, Cao X, Hobbs PD, et al. Cytochrome c release and apoptosis induced by mitochondrial targeting of nuclear orphan receptor TR3. Science. 2000;289:1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 17.Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 18.Kolluri SK, Zhu X, Zhou X, Lin B, Chen Y, Sun K, et al. A short Nur77-derived peptide converts Bcl-2 from a protector to a killer. Cancer Cell. 2008;14:285–298. doi: 10.1016/j.ccr.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu JJ, Zeng HN, Zhang LR, Zhan YY, Chen Y, Wang Y, et al. A unique pharmacophore for activation of the nuclear orphan receptor Nur77 in vivo and in vitro. Cancer Research. 2010;70:3628–3637. doi: 10.1158/0008-5472.CAN-09-3160. [DOI] [PubMed] [Google Scholar]
- 20.Zhan YY, Chen Y, Zhang Q, Zhuang JJ, Tian M, Chen HZ, et al. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nature Chemical Biology. 2012;8:897–904. doi: 10.1038/nchembio.1069. [DOI] [PubMed] [Google Scholar]
- 21.Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nature Chemical Biology. 2008;4:548–556. doi: 10.1038/nchembio.106. [DOI] [PubMed] [Google Scholar]
- 22.Wang WJ, Wang Y, Chen HZ, Xing YZ, Li FW, Zhang Q, et al. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nature Chemical Biology. 2014;10:133–140. doi: 10.1038/nchembio.1406. [DOI] [PubMed] [Google Scholar]
- 23.Lee SO, Li X, Hedrick E, Jin UH, Tjalkens RB, Backos DS, et al. Diindolylmethane analogs bind NR4A1 and are NR4A1 antagonists in colon cancer cells. Molecular Endocrinology. 2014;28:1729–1739. doi: 10.1210/me.2014-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedrick E, Lee SO, Doddapaneni R, Singh M, Safe S. Nuclear receptor 4A1 as a drug target for breast cancer chemotherapy. Endocrine-Related Cancer. 2015;22:831–840. doi: 10.1530/ERC-15-0063. [DOI] [PubMed] [Google Scholar]
- 25.Hedrick E, Lee SO, Kim G, Abdelrahim M, Jin UH, Safe S, et al. Nuclear receptor 4A1 (NR4A1) as a drug target for renal cell adenocarcinoma. PloS One. 2015;10:e0128308. doi: 10.1371/journal.pone.0128308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hedrick E, Lee SO, Doddapaneni R, Singh M, Safe S. NR4A1 antagonists inhibit β1-integrin-dependent breast cancer cell migration. Molecular and Cellular Biology. 2016;36:1383–1394. doi: 10.1128/MCB.00912-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SO, Jin UH, Kang JH, Kim SB, Guthrie AS, Sreevalsan S, et al. The orphan nuclear receptor NR4A1 (Nur77) regulates oxidative and endoplasmic reticulum stress in pancreatic cancer cells. Molecular Cancer Research. 2014;12:527–538. doi: 10.1158/1541-7786.MCR-13-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacey A, Hedrick E, Li X, Patel K, Doddapaneni R, Singh M, et al. Nuclear receptor 4A1 (NR4A1) as a drug target for treating rhabdomyosarcoma (RMS) Oncotarget. 2016;7:31257–31269. doi: 10.18632/oncotarget.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safe S, Kim K. Nuclear receptor-mediated transactivation through interaction with Sp proteins. Progress in Nucleic Acid Research and Molecular Biology. 2004;77:1–36. doi: 10.1016/S0079-6603(04)77001-4. [DOI] [PubMed] [Google Scholar]
- 30.Chadalapaka G, Jutooru I, Sreevalsan S, Pathi S, Kim K, Chen C, et al. Inhibition of rhabdomyosarcoma cell and tumor growth by targeting specificity protein (Sp) transcription factors. International Journal of Cancer. 2013;132:795–806. doi: 10.1002/ijc.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedrick E, Crose L, Linardic CM, Safe S. Histone deacetylase inhibitors inhibit rhabdomyosarcoma by reactive oxygen species-dependent targeting of specificity protein transcription factors. Molecular Cancer Therapeutics. 2015;14:2143–2153. doi: 10.1158/1535-7163.MCT-15-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarver A. Endothelial and alveolar rhabdomyosarcoma mRNA expression. NCBI GEO database. 2011 [Google Scholar]
- 33.Ahn EH, Mercado GE, Lae M, Ladanyi M. Identification of target genes of PAX3-FOXO1 in alveolar rhabdomyosarcoma. Oncology Reports. 2013;30:968–978. doi: 10.3892/or.2013.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crose LE, Galindo KA, Kephart JG, Chen C, Fitamant J, Bardeesy N, et al. Alveolar rhabdomyosarcoma-associated PAX3-FOXO1 promotes tumorigenesis via Hippo pathway suppression. Journal of Clinical Investigation. 2014;124:285–296. doi: 10.1172/JCI67087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercado GE, Xia SJ, Zhang C, Ahn EH, Gustafson DM, Lae M, et al. Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes, Chromosomes and Cancer. 2008;47:510–520. doi: 10.1002/gcc.20554. [DOI] [PubMed] [Google Scholar]
- 36.Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, et al. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:13264–13269. doi: 10.1073/pnas.96.23.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall AD, Lagutina I, Grosveld GC. PAX3-FOXO1 induces cannabinoid receptor 1 to enhance cell invasion and metastasis. Cancer Research. 2011;71:7471–7480. doi: 10.1158/0008-5472.CAN-11-0924. [DOI] [PubMed] [Google Scholar]
- 38.Thuault S, Hayashi S, Lagirand-Cantaloube J, Plutoni C, Comunale F, Delattre O, et al. P-cadherin is a direct PAX3-FOXO1A target involved in alveolar rhabdomyosarcoma aggressiveness. Oncogene. 2013;32:1876–1887. doi: 10.1038/onc.2012.217. [DOI] [PubMed] [Google Scholar]
- 39.Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nature Reviews: Cancer. 2015;15:426–439. doi: 10.1038/nrc3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fredericks WJ, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr FG, et al. The PAX3-FKHR fusion protein created by the t(2;13) translocation in alveolar rhabdomyosarcomas is a more potent transcriptional activator than PAX3. Molecular and Cellular Biology. 1995;15:1522–1535. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. Journal of Clinical Oncology. 2012;30:1670–1677. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 42.Zhu B, Davie JK. New insights into signalling-pathway alterations in rhabdomyosarcoma. British Journal of Cancer. 2015;112:227–231. doi: 10.1038/bjc.2014.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abraham J, Nunez-Alvarez Y, Hettmer S, Carrio E, Chen HI, Nishijo K, et al. Lineage of origin in rhabdomyosarcoma informs pharmacological response. Genes and Development. 2014;28:1578–1591. doi: 10.1101/gad.238733.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jothi M, Mal M, Keller C, Mal AK. Small molecule inhibition of PAX3-FOXO1 through AKT activation suppresses malignant phenotypes of alveolar rhabdomyosarcoma. Molecular Cancer Therapeutics. 2013;12:2663–2674. doi: 10.1158/1535-7163.MCT-13-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrero Martin D, Boro A, Schafer BW. Cell-based small-molecule compound screen identifies fenretinide as potential therapeutic for translocation-positive rhabdomyosarcoma. PloS One. 2013;8:e55072. doi: 10.1371/journal.pone.0055072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24:710–724. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thalhammer V, Lopez-Garcia LA, Herrero-Martin D, Hecker R, Laubscher D, Gierisch ME, et al. PLK1 phosphorylates PAX3-FOXO1, the inhibition of which triggers regression of alveolar rhabdomyosarcoma. Cancer Research. 2015;75:98–110. doi: 10.1158/0008-5472.CAN-14-1246. [DOI] [PubMed] [Google Scholar]
- 48.Hedrick E, Cheng Y, Jin UH, Kim K, Safe S. Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget. 2016;7:22245–22256. doi: 10.18632/oncotarget.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.