Abstract
Human cytomegalovirus (CMV), a ubiquitous herpesvirus, has been implicated in the etiology of breast cancer. It is clear that not all people exposed to CMV are equally likely to develop this malignancy, implying the presence of host genetic factors that might modulate the cancer-spurring properties of the virus. CMV has evolved sophisticated strategies for evading host immunosurveillance. One strategy involves encoding decoy Fcγ receptors (FcγR) that thwart the Fcγ-mediated effector functions, such as antibody-dependent cellular cytotoxicity. In this investigation, using an enzyme-linked immunosorbent assay (ELISA), we aimed to determine whether the decoy FcγR encoded by the CMV gene RL13 binds differentially to anti-CMV antibodies expressing different immunoglobulin GM (γ marker) allotypes, genetic markers of immunoglobulin G (IgG). Results of our ELISA binding studies showed that the absorbance values for the binding of the viral FcγR to the GM 17-expressing IgG antibodies were significantly higher than for the GM 3-expressing antibodies (0.60 vs. 0.36; p = 0.0019). These findings provide mechanistic insights into the modulating role played by the genetic variants of IgG in the generation of immunity to CMV in patients with breast cancer.
Keywords: GM allotypes, decoy Fcγ receptor, cytomegalovirus, immunoevasion
1. Introduction
Human cytomegalovirus (CMV), a common herpesvirus, has been implicated in the etiopathogenesis of many diseases, including breast cancer. One study found the evidence of viral expression in over 97% of neoplastic breast epithelium [1]. These findings were confirmed and extended in another investigation, which found 100% of primary breast cancer samples to be CMV positive, and also detected virus protein expression in neoplastic cells in sentinel lymph node metastases of breast cancer [2]. If CMV were involved in breast cancer pathogenesis, a strong immunological response to the virus by the host would be expected to be protective. To test this possibility, we characterized a large multiethnic cohort of patients with breast cancer and matched controls for antibodies to CMV glycoprotein B (gB), which is required for viral infectivity and plays an important role in viral attachment and entry. Results showed that cancer-free individuals had significantly higher levels of naturally occurring anti-gB IgG antibodies than patients with breast cancer. There was significant interindividual and interethnic variability in the magnitude of antibody responsiveness, and complex interactions between certain genes of the immune system contributed to this variability in both patients and controls [3].
The aim of the present investigation was to gain further mechanistic insights into the complex interplay between the virus and the immune system. In previous studies, we have shown that two CMV-encoded Fcγ receptors (FcγR), which the virus uses to evade Fcγ-mediated effector functions [4,5], bind differentially to non-immune IgG1 proteins expressing different immunoglobulin GM (γ marker) allotypes, genetic markers of IgG [6,7]. Here, we evaluated whether allotypically disparate, affinity purified anti-gB IgG antibodies from patients with breast cancer bind differentially to the decoy FcγR encoded by the CMV gene RL13.
2. Materials and methods
2.1. Study subjects
A total of 90 patients with invasive breast cancer (clinical or pathologic stage I-IV) were recruited at the Medical University of South Carolina (MUSC) Hollings Cancer Center (HCC) and MUSC outreach clinics. The study protocol was approved by the HCC Protocol Committee and the MUSC IRB. Peripheral blood samples were collected after informed consent. Race/ethnicity, age, and stage of disease of the patients were collected and are listed in Table 1.
Table 1.
Characteristics of breast cancer patients
| Race | Stage | No. of subjects | Age, median (range), years |
|---|---|---|---|
| AA | I | 12 | 59.0 (37–70) |
| II | 11 | 55.0 (40–70) | |
| III | 5 | 53.0 (32–72) | |
| IV | 7 | 60.0 (48–64) | |
| All patients | 35 | 58.0 (32–72) | |
| CA | I | 24 | 59.5 (31–82) |
| II | 13 | 52.0 (44–73) | |
| III | 1 | 56.0 (56.0) | |
| IV | 16 | 58.5 (38–81) | |
| All patients | 54 | 57.5(31–82) | |
| Hispanic | |||
| I | 0 | 0 (0) | |
| II | 1 | 61.0 (61.0) | |
| III | 0 | 0 (0) | |
| IV | 0 | 0 (0) | |
| All patients | 1 | 0 (0) | |
| Total | 90 | ||
AA: African American; CA: Caucasian American
2.2. GM genotyping
GM allotypes are encoded by immunoglobulin heavy chain G1 (IGHG1), IGHG2, and IGHG3 genes on chromosome 14. They are localized on the constant region of γ1, γ2, and γ3 chains.
IgG1 markers GM 3 and 17 (arginine to lysine), were genotyped by a pre-designed TaqMan® genotyping assay from Applied Biosystems Inc. (Foster City, CA), employing the following primers and probes:
Forward primer: 5′ CCCAGACCTACATCTGCAACGTGA-3′
Reverse primer: 5′ CTGCCCTGGACTGGGACTGCAT-3′
Reporter 1 (GM 17-specific): VIC-CTCTCACCAACTTTCTTGT-NFQ
Reporter 2 (GM 3-specific): FAM-CTCTCACCAACTCTCTTGT-NFQ
IgG2 markers GM 23− and 23+ (valine to methionine), were genotyped by a TaqMan® genotyping assay from Applied Biosystems Inc., employing the following primers and probes:
Forward primer: 5′ CCCGAGGTCCAGTTCAACT-3′
Reverse primer: 5′ CGTGGCTTTGTCTTGGCATTATG-3′
Reporter 1 (GM 23-specific): VIC-CACCTCCACGCCGTC-NFQ
Reporter 2 (GM 23+specific): FAM- CACCTCCATGCCGTC -NFQ
For the determination of IgG3 markers GM 5 and 21, a previously described PCR-RFLP method was used [8].
2.3. Determination of anti-CMV gB antibodies
IgG antibodies to CMV gB in the sera of patients were determined by a previously described ELISA [9].
2.4. Affinity purification of anti-gB IgG1 antibodies expressing GM 3 or GM 17 allotypes
Sera from 17 subjects who were homozygous for either GM 3 or GM 17 allele, and who had high titers (> 1:400) of antibodies to CMV gB, were used for affinity purifications. Briefly, 1 ml of serum from each individual was mixed with 4 ml of PBS and subjected to 40% ammonium sulfate fractionation. The contents were centrifuged and dialyzed against acetate buffer (pH 4.5), and the precipitated protein (predominantly serum albumin) was removed, with the contents then being further dialyzed against PBS. CMV gB (Sino Biological) was coupled to Pierce™ NHS-activated magnetic beads according to the manufacturer’s protocol. The total IgG was passed through this column and washed. The bound proteins were then eluted with glycine HCl buffer (pH 2.5), neutralized with 1M Tris HCl (pH 8.00), and concentrated. The preparation was passed repeatedly through beads coupled with a mixture of anti-human IgG2, IgG3 and IgG4, to remove the antibodies of these subclasses from the preparation. (For IgG Fc-viral FcγR binding studies, it is necessary to use affinity purified IgG antibodies, which would ensure that any interaction between the CMV FcγR proteins and IgG antibodies involves the portion of the IgG molecule responsible for the effector functions, and not the antigen-binding region of the molecule.)
2.5. Expression of recombinant RL13-encoded ectodomain of FcγR in mammalian cells and purification from culture supernatant
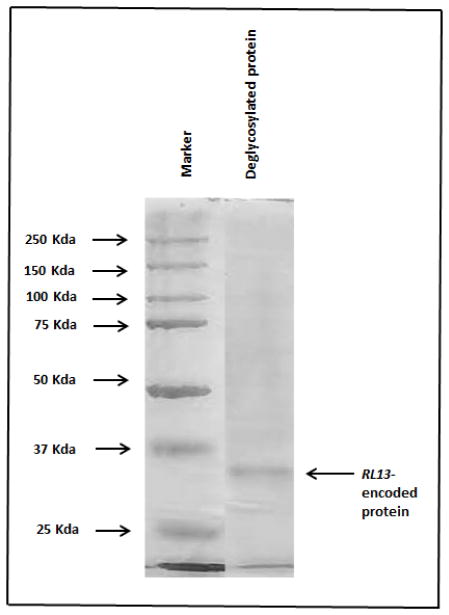
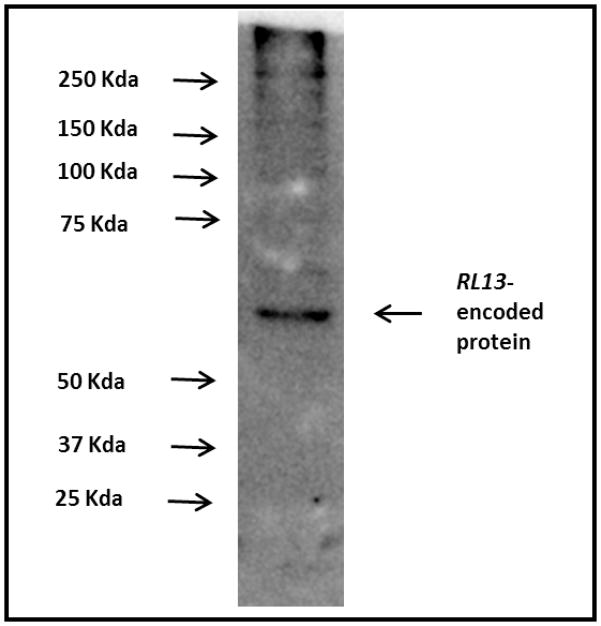
The gene encoding the 248-amino acid sequence of the extracellular domain of RL13-encoded protein of CMV [10] was synthesized by GenScript (Piscataway, NJ) and inserted in a pcDNA3.1/V-His between HindIII and BamHI restriction sites. The recombinant protein was expressed in HEK-293 cells (human embryonic kidney cells; ATCC # CRL1573), using standard methods. It was affinity purified by passing through Ni NTA agarose affinity matrix (Qiagen, Germantown, MD) and eluted with phosphate buffer containing 250 mM imidazole. This preparation was deglycosylated with PNGase F (NEB, Ispwich, MA) and tested for purity by staining with 0.25% Coomassie Brilliant Blue (R-250) after SDS–polyacrylamide gel electrophoresis (SDS–PAGE) on 12% polyacrylamide gel. Figure 1 shows a single band corresponding to RL13 migrating between 25 KDa and 37 KDa molecular weight markers. Figure 2 shows the immunoblot analysis with anti-4x-histag antibody.
Figure 1.
Coomassie Brilliant Blue staining of affinity purified, deglycosylated RL13 protein separated on 12% SDS-PAGE.
Figure 2.
Immunoblot analysis of RL13-encoded protein, using a mouse monoclonal antibody to 4x his tag.
2.6. ELISA for the binding of RL13-encoded FcγR to IgG1 proteins
The binding of allotypically different (GM 3/GM 17) IgG1 proteins to the RL13-encoded protein was normalized by determining their binding to a polyclonal sheep anti-human IgG (Fc) (Sigma Chemical Co., St. Louis, MO) by ELISA. Half of the microtiter plate wells were coated with the recombinant RL13-encoded protein (10μg/ml) and the other half with the sheep anti-human IgG (Fc) (100ng/ml), which had no specificity for any GM allotypes or IgG subclasses. Bound IgG1 proteins expressing different allotypes were detected by HRP conjugate of an anti-human IgG (Fab) (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) antibody. To determine the dilution suitable for the binding studies, a full titration curve was generated for each affinity purified IgG1 preparation on coated ELISA plates, and the dilution required to give the absorbance at the midpoint of the titration curve (mid-OD) was determined in a manner similar to that described by Shields et al. [11]. The absorbance value for the binding of each IgG1 protein to the sheep anti-human IgG (Fc) under the same dilution was brought to 1 absorbance unit, and the binding of the RL13 -encoded protein was then expressed relative to its binding to the sheep anti-human IgG (Fc). Each experiment was replicated 6 times.
2.7. Statistical analysis
For comparison of the absorbance values between the two groups (GM 3/3 vs. GM 17/17), general linear mixed regression models were used. These types of models are ideal for handling within-subject repeated observations [12]. The model included a random subject effect with a compound symmetry covariance structure, in order to account for the intra-class correlation among individual subjects’ six repeated measurements. The model also included a fixed effect for experiment number (1–6), to account for any potential systematic differences from one experiment to another. All tests were two-tailed, and the statistical significance was defined as p < 0.05. Analyses were conducted using SAS v9.4 Proc Mixed (Cary, NC).
3. Results and discussion
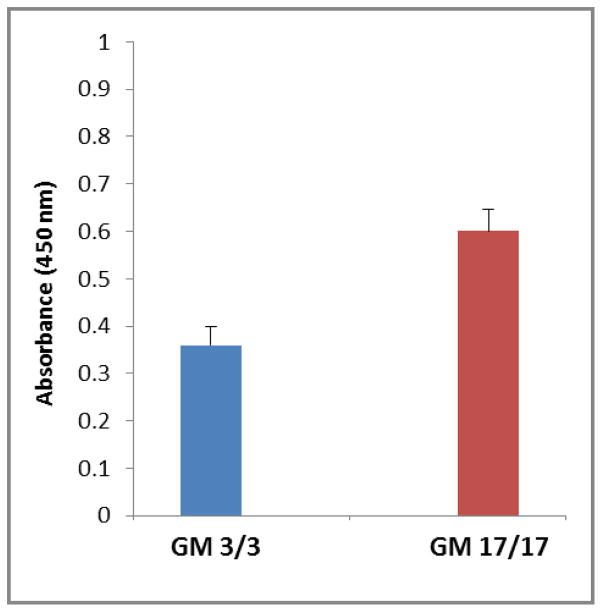
As presented in Figure 3, the ELISA absorbance values for the viral FcγR and anti-CMV gB IgG antibody binding in the GM 3/3 group were significantly lower than the absorbance values in the GM 17/17 group (GM 3/3: mean = 0.36, 95% CI = 0.26 to 0.47; GM 17/17: mean = 0.60, 95% CI = 0.51 to 0.69; p = 0.0019). These results show that RL13-encoded FcγR had higher affinity for IgG1 proteins expressing the GM 17 allotype than those expressing the allelic GM 3 allotype. The GM 3 and 17 allotypes are characterized by an arginine to lysine substitution in the CH1 region of the γ1 chain. Although viral FcγRs have been shown to bind the CH2-CH3 interface of immunoglobulin γ chains, it is possible that amino acid substitutions distant from the binding site itself could influence the protein conformation and thus indirectly affect its binding affinity. The importance of GM allotypes expressed in the CH1 region of γ1 chain for the viral FcγR binding has been conclusively shown for the herpes simplex virus type 1[13].
Figure 3.
Absorbance values (450 nm) for the binding of IgG1 proteins to the RL13-encoded FcγR protein in subjects of GM 3/3 (n = 7) and GM 17/17 (n = 10) genotypes. The bars and whiskers represent the means and standard errors, respectively, which were obtained using a general linear mixed model with a random subject effect to control for within-subject correlation.
The affinity of the RL13-encoded FcγR to the allotypically disparate IgG1 proteins is similar to that of UL119-UL118-encoded FcγR [7], but different from that of the TRL11/IRL11-encoded FcγR, which has a higher affinity for the GM 3-expressing IgG1 [6]. It is important to note that the studies involving the three viral FcγRs are not comparable. In addition to the differences in sample sizes, the studies involving the TRL11/IRL11- and UL119-UL118-encoded FcγRs used allotypically disparate non-immune IgG1 proteins, while the current study measured the binding affinity of the RL13-encoded FcγR to the affinity purified anti-CMV gB IgG1 antibodies isolated from patients with invasive breast cancer.
Higher affinity of GM 17-expressing IgG1 to the viral FcγR would imply that subjects with the GM 17 allotype would be more likely to have the Fcγ domains of their anti-CMV IgG antibodies scavenged, thereby reducing their immunological competence to eliminate the virus through Fcγ-mediated effector mechanisms, such as antibody-dependent cellular cytotoxicity, antibody-dependent cellular phagocytosis, and antibody-dependent complement-dependent cytotoxicity. Consequently, subjects possessing the GM 17 allotype would be expected to be at an increased risk of developing CMV-associated diseases, such as breast cancer, while those carrying the GM 3 allotype (because of the lower affinity to the viral FcγR) would be expected to have a reduced risk.
The results presented here, together with the racial restriction in the expression of GM alleles, might shed some light on the mechanisms underlying the racial disparities in breast cancer prognosis and survival. It is well established that breast cancer mortality rates are higher among African American (AA) women than among Caucasian American (CA) women. The frequency of the GM 17 allele is significantly higher in people of African descent than in other populations [14]. In fact, almost all AA women, without genetic admixture, are positive for this allele. Additionally, CMV seropositivity is significantly associated with female sex and seroprevalence is higher in AA than in CA people [15]. Collectively, these observations suggest that the higher affinity of the GM 17-expressing anti-CMV IgG antibodies in AA patients might, at least partially, contribute to the worse disease outcome in this group.
As noted above, the TRL11/IRL11-encoded FcγR has higher affinity to the IgG1 proteins expressing the GM 3 allele than to those expressing the GM 17 allele [6]. The GM 3 allele is very common in the populations of European descent and absent in those of African descent. We have reported a significant association between GM 3 and susceptibility to breast cancer in a Brazilian population of European descent [16]. It is possible that during the CMV-human co-evolution, the virus has employed different decoy FcγRs to counteract the effects of immune effector mechanisms in different population groups. It is, however, important to emphasize that breast cancer is a multifactorial, polygenic disease, and the contribution of immunoglobulin GM genes to the immunoevasion strategies of CMV is unlikely to account for the total interindividual and interracial variability in breast cancer outcomes.
4. Conclusions
This is the first report presenting evidence that CMV RL13-encoded FcγR discriminates between immunoglobulin GM alleles. It should be replicated by independent studies. Additionally, large-scale multiethnic studies need to be conducted to gain further mechanistic insights into the interplay between CMV and immunoglobulin genes and breast cancer outcomes.
Acknowledgments
We thank Ms. Leah Davis, Amanda McCain and Stacy Stewart for their invaluable assistance in obtaining the blood samples from patients. This work was supported in part by the Avon Foundation. Dr. Nietert’s effort on this project was supported by grants from the National Institutes of Health (NCATS grant # UL1TR001450, NIAMS grant # 5P60AR062755, and NIGMS Grant # U54-GM104941).
Footnotes
Conflict of interest
The authors declare no financial or commercial conflict of interest
This is the first report presenting evidence that cytomegalovirus RL13-encoded Fcγ receptor discriminates between immunoglobulin GM (γ marker) allotypes, genetic markers of IgG.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harkins LE, Matlaf LA, Soroceanu L, Klemm K, Britt WJ, Wang W, Bland KI, Cobbs CS. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taher C, de Boniface J, Mohammad AA, Religa P, Hartman J, Yaiw KC, Frisell J, Rahbar A, Söderberg-Naucler C. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One. 2013;8:e56795. doi: 10.1371/journal.pone.0056795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey JP, Gao G, Namboodiri AM, Iwasaki M, Kasuga Y, Hamada GS, Tsugane S. Humoral immunity to cytomegalovirus glycoprotein B in breast cancer patients and matched controls: contribution of immunoglobulin γ, κ, and Fcγ receptor gene. J Infect Dis. 2016;213:611–617. doi: 10.1093/infdis/jiv472. [DOI] [PubMed] [Google Scholar]
- 4.Atalay R, Zimmermann A, Wagner M, Borst E, Benz C, Messerle M, Hengel H. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs. J Virol. 2002;76:8596–8608. doi: 10.1128/JVI.76.17.8596-8608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corrales-Aguilar E, Trilling M, Hunold K, Fiedler M, Le VT, Reinhard H, Ehrhardt K, Mercé-Maldonado E, Aliyev E, Zimmermann A, Johnson DC, Hengel H. Human cytomegalovirus Fcγ binding proteins gp34 and gp68 antagonize Fcγ receptors I, II and III. PLoS Pathog. 2014;10:e1004131. doi: 10.1371/journal.ppat.1004131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Namboodiri A, Pandey JP. The human cytomegalovirus TRL11/IRL11-encoded FcγR binds differentially to allelic variants of immunoglobulin G1. Arch Virol. 2011;156:907–910. doi: 10.1007/s00705-011-0937-8. [DOI] [PubMed] [Google Scholar]
- 7.Pandey JP, Namboodiri AM, Radwan FF, Nietert PJ. The decoy Fcγ receptor encoded by the cytomegalovirus UL119-UL118 gene has differential affinity to IgG proteins expressing different GM allotypes. Hum Immunol. 2015;76:591–594. doi: 10.1016/j.humimm.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balbín M, Grubb A, de Lange GG, Grubb R. DNA sequences specific for Caucasian G3m(b) and (g) allotypes: allotyping at the genomic level. Immunogenetics. 1994;39:187–193. doi: 10.1007/BF00241259. [DOI] [PubMed] [Google Scholar]
- 9.Hackett DJ, Zhang C, Stefanescu C, Pass RF. Enzyme-linked immunosorbent assay for measurement of cytomegalovirus glycoprotein B antibody in serum. Clin Vaccine Immunol. 2010;17:836–839. doi: 10.1128/CVI.00422-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortese M, Calò S, D’Aurizio R, Lilja A, Pacchiani N, Merola M. Recombinant human cytomegalovirus (HCMV) RL13 binds human immunoglobulin G Fc. PLoS One. 2012;7:e50166. doi: 10.1371/journal.pone.0050166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, Fox JA, Presta LG. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem. 2001;276:6591–6604. doi: 10.1074/jbc.M009483200. [DOI] [PubMed] [Google Scholar]
- 12.McCulloch C, Searle SR. Generalized, Linear, and Mixed Models. John Wiley & Sons Inc; New York: 2001. [Google Scholar]
- 13.Atherton A, Armour KL, Bell S, Minson AC, Clark MR. The herpes simplex virus type 1 Fc receptor discriminates between IgG1 allotypes. Eur J Immunol. 2000;30:2540–2547. doi: 10.1002/1521-4141(200009)30:9<2540::AID-IMMU2540>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 14.Steinberg AG, Cook CE. The Distribution of Human Immunoglobulin Allotypes. Oxford University Press; New York: 1981. [Google Scholar]
- 15.Bate SL, Dollard SC, Cannon MJ. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin Infect Dis. 2004;50:1439–1447. doi: 10.1086/652438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey JP, Kistner-Griffin E, Iwasaki M, Bu S, Deepe R, Black L, Kasuga Y, Hamada GS, Tsugane S. Genetic markers of immunoglobulin G and susceptibility to breast cancer. Hum Immunol. 2012;73:1155–1158. doi: 10.1016/j.humimm.2012.07.340. [DOI] [PubMed] [Google Scholar]