Abstract
The Bromodomain and ExtraTerminal (BET) proteins are epigenetic ‘readers’ of acetylated histones in chromatin and have been identified as promising therapeutic targets in diverse cancers. However, it remains unclear how individual family members participate in cancer progression, and small molecule inhibitors such as JQ1 can target functionally independent BET proteins. Here we report a signaling pathway involving BRD4 and the ligand/receptor pair Jagged1/Notch1 that sustains triple-negative breast cancer migration and invasion. BRD4, but not BRD2 or BRD3, regulated Jagged1 expression and Notch1 signaling. BRD4-selective knockdown suppressed Notch1 activity and impeded breast cancer migration and invasion. BRD4 was required for interleukin-6-stimulated, Notch1-induced migration and invasion, coupling microenvironment inflammation with cancer propagation. Moreover, in patients, BRD4 and Jagged1 expression positively correlated with the presence of distant metastases. These results identify a BRD4/Jagged1/Notch1 signaling pathway that is critical for dissemination of triple-negative breast cancer.
Keywords: Bromodomain, breast cancer, triple-negative, metastasis, Notch1, Jagged1, breast cancer migration, breast cancer invasion, BRD4, BET protein
Introduction
The bromodomain and extra-terminal (BET) proteins form a family of chromatin-associated proteins that recognize epigenetic marks, such as N-ε-acetylated lysine residues in nucleosomal histones. BET proteins interact with chromatin and recruit to target promoters members of the transcription machinery to regulate gene expression (1,2). In mammals, the BET family is comprised of three ubiquitously expressed proteins: BRD2, BRD3, BRD4, and a testis-specific form, BRDT (3). We were the first to establish a link between expression and function of a BET protein and human cancer (3). A consistent literature has since reported essential roles of BRD2 and BRD4 in cell cycle control and cell proliferation, notably through their crucial role in regulating gene transcription (4-7). Like numerous other chromatin-regulatory proteins, the cancers in which BET proteins have been implicated are diverse. Small molecule BET-specific bromodomain inhibitors, e.g., (S)-JQ1 (8), highlight the therapeutic impact of targeting BET proteins in important malignancies, including acute myeloid leukemia (9,10), B cell lymphoma (11), lung (12), prostate (13), breast (14), pancreatic (15) and colorectal cancer (16). Several studies have reported that BET inhibition down-regulates the proto-oncogene MYC and its associated transcription (9,10,17,18). Moreover, BRD2 has been shown to associate with chromatin remodeling complexes to regulate transcription programs associated with cell proliferation (19). Interestingly, a recent study reported that BRD4 interacts with the transcription factor Twist to regulate Twist-dependent transcription programs and elicit breast cancer tumorigenesis (20). However, the molecular mechanisms by which BET proteins participate in cancer progression are not fully established. We build on these insights to develop a hypothesis that BET proteins play a role in the regulation of breast cancer aggressiveness.
Triple-negative breast cancer is an aggressive subtype that lacks estrogen receptor, progesterone receptor and epidermal growth factor receptor 2. These tumors are extremely challenging for therapy. Patients with triple-negative breast cancer cannot be treated by hormone therapy and present a worse outcome after conventional chemotherapy (21). Moreover, the lack of identified ‘druggable’ molecular targets severely limits the development of targeted therapeutic strategies. Triple-negative breast cancer cells activate multiple transcription programs, such as the epithelial-mesenchymal transition (EMT), which confers high plasticity, increased migration and invasion capabilities that can promote tumor dissemination (22). The breast tumor microenvironment is composed of various cell types including adipocytes; fibroblasts; infiltrating immune cells, such as macrophages and lymphocytes; and stem cells (23). Numerous factors secreted within this microenvironment have been reported to sustain tumor progression, including growth factors, hormones, cytokines and chemokines. Amid this complexity, inflammatory signaling molecules have focused investigators’ attention, because a direct association exists between chronic inflammation and cancer development (24). Notably, multiple pro-inflammatory mediators sustain the acquisition of an aggressive phenotype by tumor cells that can lead to resistance and dissemination. Significantly, BET protein inhibitors like (S)-JQ1 have anti-inflammatory properties (25,26), suggesting that BET protein targeting may disrupt inflammation-induced cancer aggressiveness. Here, we identify a role for BRD4 function in invasive properties of triple-negative breast cancer and reveal the underlying molecular mechanisms.
Materials and Methods
Cell Culture
Human breast cancer cell lines maintained at the NCI Office of Physical Sciences-Oncology Centers (PS-OC) Network Bioresource Core Facility (PBCF) were contractually obtained through the American Type Culture Collection, under a Material Transfer Agreement. The cell lines have been authenticated by the NIH Physical Sciences Oncology Consortium. MDA-MB-231 and MCF-7 were cultured in Dulbecco's modified Eagle's medium (DMEM). SUM149PT cells were cultured in DMEM/F12 + 5 μg/μl insulin and 0.5 μg/μl hydrocortisone (Sigma). All culture media were supplemented with 10% fetal bovine serum (FBS) or human serum (Millipore) where specified and 1% antibiotics (penicillin/streptomycin, Thermo Fischer Scientific). Cells were cultured at 37°C in a 5% CO2 atmosphere.
Antibodies and Reagents
The following antibodies were used: anti-BRD2, BRD3 and BRD4 (Bethyl Laboratories), anti-Jagged1 and anti-α-tubulin (Santa Cruz Biotechnology), anti-phosphoY705-STAT3, total STAT3 and anti-Notch1 Val1744 (Cell Signaling Technology). HRP-conjugated secondary antibodies were purchased from Bio-Rad. Fluorochrome-conjugated secondary antibodies were obtained from Jackson Laboratories. (R)- and (S)-JQ1 were purchased from Cayman Chemicals. Recombinant human IL-6 and DAPT were purchased from Sigma.
Plasmids, siRNAs and Transfection
Plasmids coding for His-tagged BET proteins, His-Jagged1 or control vector pReceiver-M01 were purchased from GeneCopoeia. ON-TARGETplus BET proteins siRNAs were obtained from Dharmacon and Jagged1 siRNA (sc-3702) from Santa Cruz Biotechnology. Cells were transfected with plasmids and siRNAs by Lipofectamine 2000 reagent (Thermo Fischer Scientific).
Migration and Invasion Assays
For scratch assays, 100,000 cells were plated in 6-well plates. When confluence was reached, cell monolayers were scraped using a P200 pipet tip and washed to remove cell debris. Photos were taken during the subsequent 12 h to monitor scratch closure. For Transwell migration assay, cells were maintained in serum-free media for 3 h prior to the beginning of the assay to suppress any basal migratory/invasion signal. 150,000 cells were plated in Transwell inserts (pore size: 8 μm, Corning) and challenged for migration toward human serum for 6h in culture conditions. Cells that did not migrate were removed by scratching the upper side of the membrane with a cotton swab before fixation in absolute methanol for 5 min at −20°C. Cells were then stained with 1% crystal violet (Sigma) in 2% ethanol for 10 min. The percentage of migration was determined by calculating the sum of the area of total migrated cells on the entire membrane by using ImageJ software (National Institutes of Health, Bethesda, Maryland, USA). For invasion assay, Matrigel (Corning) was added onto the upper membranes prior to cell plating. Invasion was conducted for 16 h, then the protocol described above was used. Invaded cells were counted by using ImageJ software.
Immunoblotting
Cell pellets were lysed in RIPA buffer (50 mM Tris/HCl pH 7.5, 1 mM EDTA, 0.5 mM EGTA, 150 mM NaCl, 0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100). Samples containing 25 μg of protein were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. After saturation of non-specific binding sites in TBS-BSA 5%, membranes were probed with primary antibodies, then visualized with HRP-conjugated secondary antibodies. After incubation in ECL, membrane-associated light emission was quantified with a gel imager.
qRT-PCR
Total RNA was extracted using the RNEasy kit (Qiagen). Reverse transcription reactions were performed on 1 μg of RNA with the QuantiTect Reverse Transcription kit (Qiagen). The primers sequences used for this study are listed in Supplemental Table 1. PCR amplifications were performed with the MESA GREEN qPCR MasterMix (Eurogentec) on an ABI Prism 7500 thermal cycler.
The gene screening was conducted with the RT2 Profiler PCR EMT Array (Qiagen).
Immunocytochemistry Staining and Confocal Imaging
Cells were fixed in absolute methanol for 5 min at −20°C then permeabilized with 0.2% Triton X-100 in PBS buffer for 10 min. After saturation in blocking buffer (0.02% Triton X-100, 2% BSA in PBS) for 30 min, cells were incubated with primary antibodies then fluorochrome-conjugated secondary antibodies, both diluted in blocking buffer for 1 h. Finally, coverslips were mounted with ProLong Gold with DAPI (Thermo Fischer Scientific). Image acquisition was conducted using a Leica SP5 confocal microscope. For z-stack acquisition, a step of 0.3 μm was set. For calculation of NICD1 nuclear score, cells stained for NICD1 were entirely imaged in 3D then z-stacks were projected into a single image by sum projection using ImageJ software (NIH). A mask based on DAPI staining was created to isolate the nucleus of each cell. Based on that mask, nuclear background-corrected NICD1 intensity was measured. The cytoplasm area was determined by subtracting the nuclear mask from the entire cell area and cytoplasmic background-corrected NICD1 intensity was measured. The NICD1 nuclear score was then calculated for each cell as the ratio of nuclear and cytoplasmic NICD1 intensities.
Chromatin immunoprecipitation
MDA-MB-231 cells were treated for 24h with 400 nM (R)- or (S)-JQ1, then fixed in 1% formaldehyde at 37°C for 10 min, quenched 10 min with 125 mM glycine and lysed for chromatin immunoprecipitation (ChIP) as previously reported (26). Chromatin was precipitated with 2 μg anti-rabbit IgG (Cell Signaling Technology) or anti-BRD4 (Bethyl Laboratories) with Dynabead magnetic beads (Thermo Fischer Scientific). 5 ng of each sample was analyzed in triplicate by qPCR. The fold difference was calculated as 2^[Ct(input) - 2 Ct(ChIP)], and fold enrichment over anti-IgG was assessed. ChIP primers sequences used are listed in Supplemental Table 2.
MTT and Viability Assays
Cells were incubated with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for 3 h. The light absorbance was measured at 570 nm and corrected with background absorbance determined at 690 nm with a multiwell spectrophotometer. For viability assays, cells were co-stained for annexin V/propidium iodide (PI) using Dead Cell Apoptosis Kit for Flow Cytometry (Invitrogen) according to the manufacturer's instructions.
Meta-analysis of the association of BRD4 and JAG1 expression with distant metastasis-free survival
A meta-analysis of BRD4 and JAG1 expression among 664 breast cancer patients was performed using the Kaplan-Meier plotter online survival analysis software (www.kmplot.com) (27). Kaplan-Meier curves were generated using the optimal BRD4 (226054_at) and JAG1 (216268_s_at) probes. Two patient groups were obtained from the splitting of the entire dataset at the median of gene expression. The two groups were compared for distant metastasis-free survival and hazard ratio with 95% confidence interval (CI) and log-rank p value were calculated by the software.
Statistical Analyses
Statistical analyses were performed with Student's t test or ANOVA as indicated by using GraphPad Prism software. The following symbols were used to indicate significant differences: ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Results
BET protein inhibition reduces breast cancer cell migration and invasion
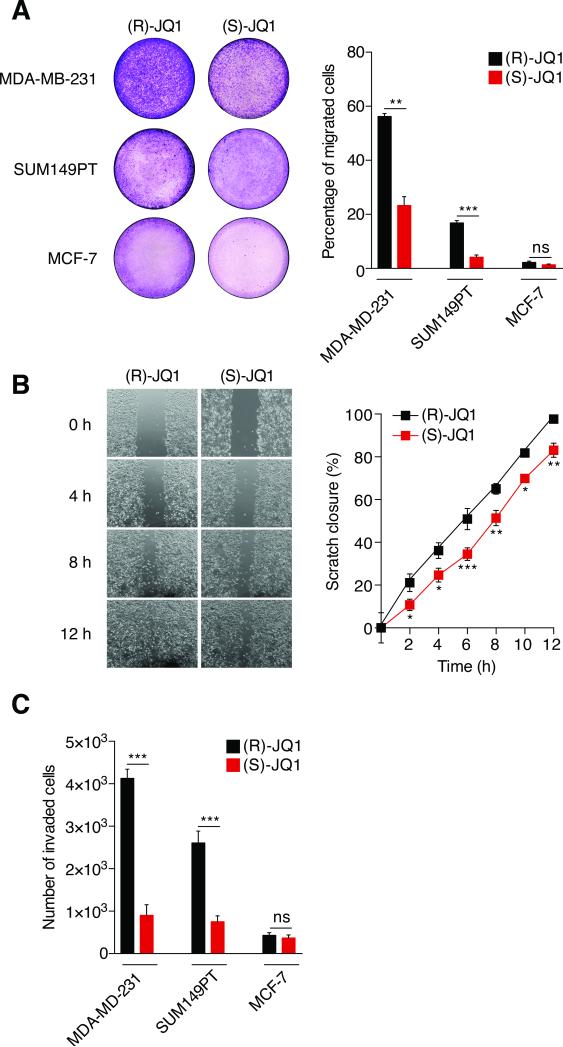
Triple-negative breast cancer is usually aggressive, with high histological grade. Based on published and preliminary data, we hypothesized that BET proteins regulate breast cancer cell migration and invasion. Our strategy was first to use (S)-JQ1, a pan-BET-bromodomain inhibitor that targets BRD2, BRD3 and BRD4 (8) and investigate its effects on cell migration and invasion, with the inactive enantiomer (R)-JQ1 as the negative control. We pre-treated human breast cancer cell lines for 3 h with either (S) or (R)-JQ1, then assayed migration with a Transwell system. Pan-BET inhibition significantly reduced migration of aggressive cancer cells (Figure 1A). (S)-JQ1 reduced migration of MDA-MB-231 and SUM149PT cells by 60% and 75%, respectively. We also tested MCF-7 cells, which are not highly migratory, consistent with their luminal-A phenotype. As expected, neither MCF-7 cells treated with (S)-JQ1 nor negative controls migrated. We assessed whether differences in migration were due to cell death or cell cycle perturbation. Viability of control and (S)-JQ1-treated cells determined by either MTT assay or annexin V/propidium iodide co-staining was comparable in all cases (Figures S1A-B). (S)-JQ1 increased the fraction of cells in G1 phase by about 10% and reduced the proportion in G2/M (Figure S1C) as previously reported (13). We also conducted scratch assays to assess cell motility and 2D migration (Figure 1B). Control MDA-MB-231 cells fully recovered the scratched area within 12 h. However, (S)-JQ1-treated cells were 20-25% slower and unable to close the wound by the endpoint. We then measured cell invasion through Matrigel upon BET protein inhibition (Figure 1C). (S)-JQ1 reduced the number of invaded MDA-MB-231 and SUM14PT cells by 80% and 70%, respectively. No difference was observed for the poorly invasive MCF-7 cells. Taken together, our results show that pan-BET protein inhibition reduces breast cancer cell migration and invasion.
Figure 1. BET protein inhibition reduces breast cancer cell migration and invasion.
(A) MDA-MB-231, SUM149PT and MCF-7 cells were pre-treated with either 400 nM (R)-JQ1 or (S)-JQ1 for 3 h then challenged for migration for 6 h with 10% FBS, using a Transwell system. Results are presented as percentage of migrated area in comparison to total membrane area. Left panel: representative photographs of the total membrane area showing migrated cells stained with crystal violet. Right panel: bars show means ± SEM of four independent experiments. Statistical analyses were performed by using Student's t test.
(B) MDA-MB-231 confluent monolayer cell cultures were pre-treated with either 400 nM (R)-JQ1 or (S)-JQ1 for 3 h, then scratched with a pipet tip. Photographs were taken every two hours and the percentage of the scratch closure was determined. Left panel: representative photographs of scratched monolayer cultures of MDA-MB-231 cells treated with either (R)-JQ1 or (S)-JQ1 at different times. Right panel: means ± SEM of two independent experiments were plotted. Statistical analyses were performed by using two-way ANOVA.
(C) MDA-MB-231, SUM149PT and MCF-7 cells were pre-treated with either 400 nM (R)-JQ1 or (S)-JQ1 for 3 h under conditions of serum deprivation, then plated onto Matrigel in Transwell inserts and challenged for invasion for 16 h with 10% FBS. Results are presented as the total number of invading cells. Bars show means ± SEM of four independent experiments. Statistical analyses were performed by using Student's t test.
The following symbols were used to indicate significant differences: ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
BRD4 regulates breast cancer cell migration and invasion
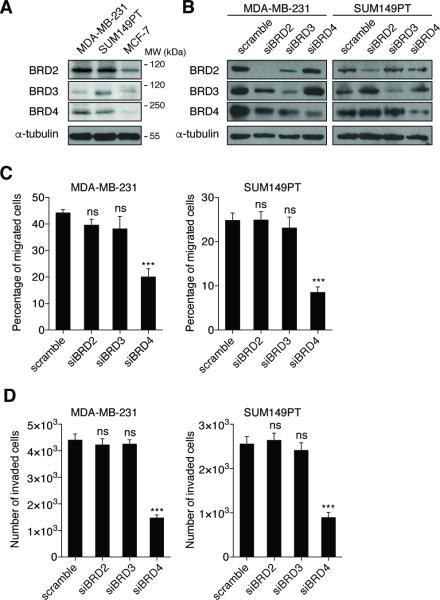
In order to explore the pathway and BET proteins more deeply, we undertook a specific siRNA-knockdown strategy to investigate the requirement for each BET protein separately, because (S)-JQ1 is not selective and inhibits all BET family members (8). All of the breast cancer cell lines used in this study express BRD2, BRD3 and BRD4 (Figure 2A). We measured mRNA expression of each BET gene to validate knockdown specificity and efficiency for each siRNA. We confirmed that each siRNA only ablates its targeted sequence and does not reduce mRNA of other BET family members (Figures S2A-C). Transfection for 72h with 50 nM siRNA achieved selective depletion of each BET protein (Figure 2B). We next challenged BET protein-depleted MDA-MB-231 cells with migration and invasion stimuli in the Transwell system (Figures 2C-D). Neither BRD2 nor BRD3 depletion impacted migration and invasion. Significantly, however, BRD4 depletion reduced migration by 50% and invasion by 40%. Similar results were obtained with SUM149PT cells (Figures 2C-D). These differences were not indirect consequences of cell cycle perturbation, proliferation or apoptosis, because no knockdown significantly altered viability (Figures S2D and S2F). Interestingly, BRD2 or BRD4 depletion led to a small increase (5-11%) of the G1 fraction (Figure S2E). Taken together, these results newly identify BRD4 as a regulator of breast cancer cell migration and invasion.
Figure 2. BRD4 regulates breast cancer cell migration and invasion.
(A) Endogenous expression of BET proteins in several breast cancer cell lines was detected by immunoblotting. Molecular weights (MW) in kDa corresponding to the immunoblotted proteins are indicated.
(B) siRNA-mediated BET protein depletion was validated by immunoblotting. Cells were transfected for 72h with the indicated siRNAs (50 nM). Blots shown are representative of three independent experiments.
(C-D) MDA-MB-231 (left panel) and SUM149PT (right panel) cells were transfected with either control siRNA (scramble) or BET-targeted siRNAs for 72h prior to migration (C) or invasion (D) assays. Bars show means ± SEM of three independent experiments. Statistical analyses were performed by using two-way ANOVA.
The following symbols were used to indicate significant differences: ns, p > 0.05; ***, p < 0.001.
Identification of JAG1 as a BRD4 target gene
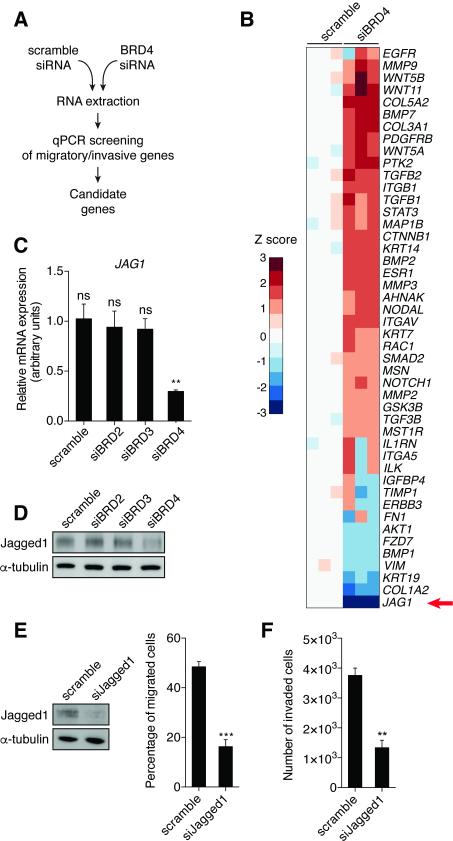
In order to unravel cellular pathways involved in BRD4-mediated migration and invasion, we performed gene expression analysis in control and BRD4-depleted breast cancer cell lines (Figure 3A). We selected a panel of 46 genes involved in regulation of migration and invasion. Knockdown of BRD4 in MDA-MB-231 cells resulted in significantly altered expression of 7 genes (4 up-regulated, 3 down-regulated, Z score ≥ 2 or ≤ −2, p-value < 0.05) (Figure 3B). The most down-regulated gene was JAG1, which encodes Jagged1 protein, one of the canonical ligands for the Notch receptor family (28,29). Similar results were found in SUM149PT cells (Figure S3A). As a transmembrane protein, Jagged1 binds Notch1 and Notch3 receptors and triggers their activation, inducing receptor proteolysis by γ-secretase and release of Notch intracellular domain (NICD), which translocates to the nucleus and associates with a transcription complex to regulate downstream target genes. Jagged1 regulates multiple cancer-associated processes including proliferation, survival, EMT, metastasis and angiogenesis (29). Interestingly, triple-negative cancers generally have higher levels of Jagged1 expression, which is associated with reduced disease-free survival (30).
Figure 3. Identification of JAG1 as a BRD4-dependent gene.
(A) Screening strategy.
(B) Heatmap representing relative expression of 46 genes involved in regulation of migration and invasion in response to BRD4 depletion in MDA-MB-231 cells. Z scores are represented using a color code. Results for separate knockdown experiments are shown (n = 3).
(C-D) Jagged1 mRNA (C) and protein (D) expression were measured in MDA-MB-231 cells 72h after transfection with the indicated siRNAs (50 nM).
(E) Immunoblot validates Jagged1 depletion in MDA-MB-231 48h after transfection with the indicated siRNAs (20 nM) (left panel). Migration assay was conducted with scramble or siJagged1-depleted MDA-MB-231 cells (right panel). Bars show means ± SEM of three independent experiments. Statistical analyses were performed by using Student's t test.
(F) Invasion assay was conducted with scrambled siRNA- or siJagged1-depleted MDA-MB-231 cells. Bars show means ± SEM of three independent experiments. Statistical analyses were performed by using Student's t test.
The following symbols were used to indicate significant differences: ns, p > 0.05; **, p < 0.01; ***, p < 0.001
We confirmed that BRD4 knockdown down-regulated both Jagged1 mRNA and protein (Figures 3C-D; Figures S3B-C). Notably, Jagged1 expression was stable in BRD2- or BRD3-depleted cells, suggesting that JAG1 is a BRD4-specific target gene. Jagged1/Notch signaling modulates cancer cell migration and invasion (31,32). We therefore evaluated migration and invasion of MDA-MB-231 and SUM149PT cells upon Jagged1 depletion (Figures 3E-F; Figures S3D). Jagged1 knockdown reduced both migration and invasion by 65%. These results identify JAG1 as a BRD4 target gene that regulates breast cancer cell migration and invasion.
BRD4 regulates migration and invasion through Jagged1/Notch1 signaling
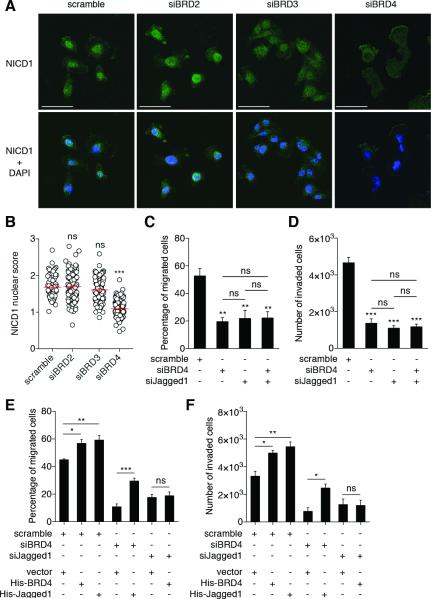
BRD4 regulation of Jagged1 expression led us to hypothesize that BRD4 regulates the Jagged1/Notch pathway. To test this hypothesis, we measured Notch1 activation by immunostaining. NICD1 translocates to the nucleus after the Jagged1/Notch1 interaction to induce target gene transcription (28). Thus, we expect to detect increased nuclear NICD1 upon Notch1 activation; calculation of nuclear:cytoplasmic ratio is described in Methods. We measured NICD1 nuclear enrichment in MDA-MB-231 cells upon BET protein depletion (Figures 4A-B). In control scramble cells, NICD1 was preferentially localized within nuclei, reflecting basal Notch1 activity. NICD1 distribution was unchanged in BRD2- or BRD3-depleted cells; calculated nuclear scores were similar for control (1.69 ± 0.04), BRD2- (1.64 ± 0.03) and BRD3-depleted cells (1.58 ± 0.02). However, BRD4-depleted cells exhibited more homogenous NICD1 distribution within cytoplasm and nuclei. Some cells even showed nuclear exclusion of NICD1. Thus, the NICD1 nuclear score of BRD4-depleted cells was significantly reduced compared to control (1.2 ± 0.02). These results indicate that BRD4 depletion diminished Notch1 activation, thus BRD4 regulates the Jagged1/Notch1 pathway.
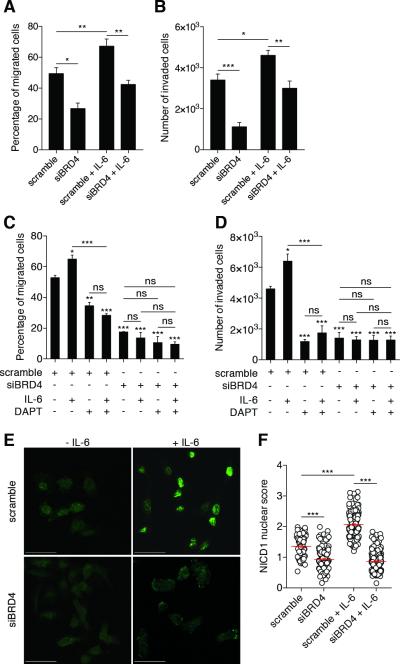
Figure 4. Jagged1/Notch1 signaling pathway acts downstream of BRD4 to regulate breast cancer cell migration and invasion.
(A) Representative images from immunofluorescence experiments showing the cellular NICD1 distribution. NICD1 is stained in green and nuclei in blue, with DAPI in merged images (lower panels). Scale bar: 50 μm.
(B) NICD1 nuclear score (for calculation details, see Experimental Procedures section). One dot represents a measurement of one cell. Bars show means ± SEM of three independent experiments. Statistical analyses were performed by using one-way ANOVA.
(C-D) MDA-MB-231 were transfected with the indicated siRNAs (scramble, siBRD4: 50 nM; siJagged1: 20 nM) for 72h prior to conduct migration (C) and invasion (D) assays. Bars show means ± SEM of three independent experiments. Statistical analyses were performed by using one-way ANOVA.
(E-F) MDA-MB-231 were transfected with the indicated siRNAs (scramble, siBRD4: 50 nM; siJagged1: 20 nM) for 72h and plasmids for 24h prior to conduct migration (E) and invasion (F) assays. Bars show means ± SEM of three independent experiments. Statistical analyses were performed by using one-way ANOVA.
The following symbols were used to indicate significant differences: ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We then hypothesized that BRD4 regulates breast cancer cell migration and invasion through Jagged1/Notch1 signaling. BRD4 and Jagged1/Notch1 may act in a single signaling pathway. If BRD4 acts upstream of Jagged1, we would expect that ablation of BRD4 or Jagged1 individually impairs migration and invasion to a similar degree, compared to co-depletion. We conducted migration and invasion assays under these conditions (Figures 4C-D). As shown above, individual ablation of BRD4 or Jagged1 significantly reduced migration and invasion. In support of the hypothesis, co-depletion of BRD4 and Jagged1 did not cause additive inhibition. Furthermore, if BRD4 acts upstream of Jagged1, Jagged1 overexpression in BRD4-depleted cells should restore cell migration and invasion. Similarly, BRD4 overexpression in Jagged1-depleted cells should fail to rescue. We found that overexpression of either BRD4 or Jagged1 increased migration and invasion by about 20% in siRNA control cells (Figures 4E-F). Critically, Jagged1 overexpression rescued migration (Figure 4E) and invasion (Figure 4F) in BRD4-depleted cells. BRD4 overexpression in Jagged1-depleted cells failed to restore migration and invasion, confirming that Jagged1 acts downstream of BRD4. Taken together, our results indicate that BRD4 regulates migration and invasion through Jagged1/Notch1 signaling.
BRD4 and JAG1 expression correlate with disseminated human breast cancer in vivo
To test the hypothesis that BRD4/Jagged1/Notch1 signaling is crucial for cancer propagation, we determined whether BRD4 and JAG1 expression correlate in vivo with breast cancer dissemination. We performed a meta-analysis among 664 breast cancer patients using the Kaplan-Meier plotter database (27). We analyzed distant metastasis-free survival, defined by the interval between initial diagnosis and time when metastases are detected: a good indicator of tumor dissemination. The dataset was equally divided, based on median expression of both genes, to obtain ‘high expression’ (BRD4high, JAG1high (n=332)) and ‘low expression’ groups (BRD4low, JAG1low (n=332)). Higher expression of BRD4 and JAG1 was significantly associated with shorter distant metastasis-free survival in breast cancer patients (Figure S4; hazard ratio = 1.505, 95% CI = 1.084 – 2.069, log-rank p value = 0.0147). Thus, we conclude that differential expression of BRD4/Jagged1/Notch1 signaling significantly influences metastasis-free survival in breast cancer patients.
Interleukin-6 induces BRD4-dependent Notch1 signaling to regulate breast cancer aggressiveness
Numerous pro-inflammatory cytokines, such as interleukin (IL)-6 or tumor necrosis factor (TNF)-α and their signaling pathways, have been implicated in the acquisition of migratory and invasive properties and cancer dissemination (23,24). BET proteins regulate pro-inflammatory cytokine signaling and BET protein inhibitors exhibit anti-inflammatory properties (25,26). Jagged1 expression is regulated by pro-inflammatory cytokines, including IL-6, TNF-α and NF-κB signaling (33,34). Thus, we developed a hypothesis that IL-6 induces BRD4-dependent Notch1 activation to regulate migration and invasion.
We depleted BET proteins in MDA-MB-231 cells with siRNA as above, treated them with recombinant human IL-6 for 24h, then conducted migration assays (Figure 5A). IL-6 treatment increased MDA-MB-231 cell migration by 35% in scramble control cells compared to untreated controls. Crucially, BRD4 depletion ablates IL-6-dependent migration. Similar results were obtained for invasion assays (Figure 5B). We observed comparable results in SUM149PT cells (data not shown). No differences in MCF-7 cells migration or invasion were observed upon BRD4 overexpression or IL-6 treatment (Figures S5A-B). These results suggest that BRD4 is required as an effector of IL-6-dependent migration and invasion in triple-negative breast cancer cells. As shown in Figures 5C-D, we observed that DAPT, a γ-secretase inhibitor, reverted the pro-migratory and pro-invasive effects of IL-6, confirming that Notch signaling is required to mediate these IL-6 functions. We did not observe cumulative effects on migration and invasion using DAPT in BRD4-depleted cells. Finally, DAPT treatment reverted the increase of migration and invasion induced by BRD4 overexpression (Figure S5C), reinforcing the idea that BRD4 and Notch1 act through the same signaling pathway.
Figure 5. IL-6 induces BRD4-dependent Notch1 signaling to regulate breast cancer aggressive properties.
(A-B) MDA-MB-231 cells were treated with recombinant human IL-6 (50 ng/ml) for 24h under either siBRD4 or siRNA scramble control conditions, to conduct migration (A) and invasion (B) assays. Bars show means ± SEM of three independent experiments. Statistical analyses were performed with one-way ANOVA.
(C-D) MDA-MB-231 cells were treated with recombinant human IL-6 (50 ng/ml, 24h) and/or with DAPT (10 μM, 16h) under either siBRD4 or siRNA scramble control conditions, to conduct migration (C) and invasion (D) assays. Bars show means ± SEM of three independent experiments. Statistical analyses were performed with one-way ANOVA.
(E) Representative images from immunofluorescence experiments showing cellular NICD1 distribution. NICD1 is stained in green and nuclei in blue with DAPI in merged images (lower panels). Scale bar: 50 μm.
(F) NICD1 nuclear score. One dot represents a measurement of one cell. Statistical analyses of three independent experiments were performed with one-way ANOVA.
The following symbols were used to indicate significant differences: ns, p > 0.05; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We measured Notch1 activation as above after treatment with human recombinant IL-6 in scramble- or BRD4-depleted MDA-MB-231 cells (Figures 5E-F). As shown above, untreated scramble cells exhibited high nuclear NICD1 staining (nuclear score = 1.47 ± 0.03), indicating basal activation of Notch1 signaling. Consistent with previous results, untreated BRD4-depleted cells showed homogeneous NICD1 staining through cytoplasm and nuclei (nuclear score = 0.99 ± 0.04), reflecting reduced Notch1 activation. After IL-6 treatment, nuclear translocated NICD1 was increased in scramble cells (nuclear score = 2.07 ± 0.04). This Notch1 activation was not observed in BRD4-depleted cells, where the NICD1 distribution remained homogeneous after IL-6 treatment (nuclear score = 0.95 ± 0.04). Taken together, these results indicate that IL-6 induces a BRD4-dependent Notch1 activation to regulate breast cancer migration and invasion.
BRD4 associates with the Jagged1 promoter
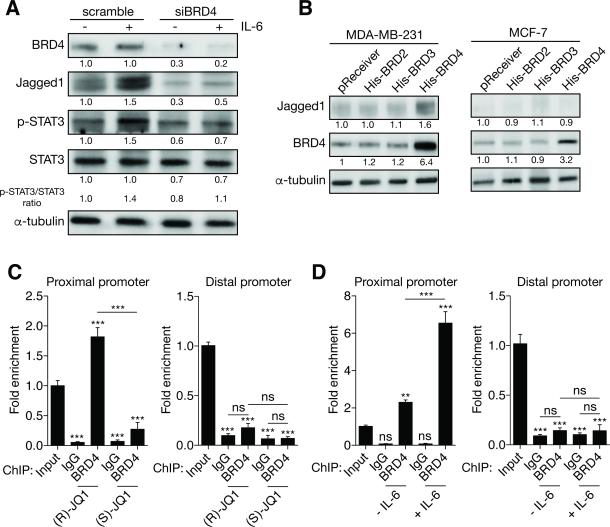
To precise the molecular relationship between IL-6, BRD4 and Jagged1, we examined Jagged1 expression in control or BRD4-depleted MDA-MB-231 upon IL-6 stimulation (Figure 6A). As shown above, BRD4 depletion leads to Jagged1 down-regulation. We found that IL-6 treatment increased Jagged1 expression in control cells but not in BRD4-depleted cells, where IL-6 signaling is impaired, as we detected no phosphorylation of STAT3 compared to the control condition. We confirmed that BRD4 plays a central role in the regulation of Jagged1 expression by overexpressing BRD4 in our cells lines. We showed that BRD4 overexpression leads to an up-regulation of Jagged1 mRNA (Figure S5D) and protein (Figure 6B) in MDA-MB-231 and in SUM149PT (data not shown) but not in MCF-7 cells. To precise the molecular mechanisms of this regulation, we assessed if BRD4 associates with the Jagged1 promoter by ChIP. We investigated the binding of BRD4 at two different sites described as proximal and distal regions of the Jagged1 promoter (35). We found that BRD4 modestly interacts with the proximal region of the Jagged1 promoter where no interaction was detected at the distal site (Figure 6C). (S)-JQ1 disrupts BET protein/chromatin interactions by competing with the acetylated histones binding sites (8). As expected, (S)-JQ1 treatment prevents association of BRD4 with the Jagged1 promoter. Interestingly, we detected a significant enrichment of BRD4 at the proximal site upon IL-6 treatment (Figure 6D). These results provide a mechanistic explanation for the control of Jagged1 expression by BRD4. Altogether, our results show that BRD4 plays a pivotal role to couple IL-6 signaling and Jagged1 expression to regulate breast cancer cell migration and invasion.
Figure 6. BRD4 interacts with the JAG1 promoter.
(A) Immunoblots of BRD4, Jagged1, phospho-STAT3 (p-STAT3) and STAT3 in MDA-MB-231 cells treated or not with IL-6 (50 μg/ml, 24h) under either siBRD4 or siRNA scramble control conditions. Quantifications relative to control with normalization using α-tubulin levels as a loading control are indicated. The ration p-STAT3/STAT3 is also indicated to illustrate STAT3 activation.
(B) Immunoblots of BRD4 and Jagged1 in MDA-MB-231 or MCF-7 cells under overexpression of the BET proteins for 48h. Quantifications relative to control with normalization using α-tubulin levels as a loading control are indicated.
(C-D) MDA-MB-231 cells were treated with either 400 nM (R)- or (S)-JQ1 for 24h as indicated then harvested for ChIP. (C) BRD4 interacts with the proximal JAG1 promoter but is absent from its distal promoter. (D) BRD4 is enriched at the proximal JAG1 promoter under IL-6 treatment (50 ng/ml for 24h).
The following symbols were used to indicate significant differences: ns, p > 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
The BET family is comprised in mammals of three ubiquitously expressed members: BRD2, BRD3 and BRD4. These proteins share two tandem bromodomains in their N terminus, which contain anchoring motifs for binding to nucleosomal histones. In their carboxyl-terminus, they also possess different domains involved in interactions with chromatin-modifying enzymes and transcription factors (2). Despite their close homology, BET proteins do not share identical biological functions. Several processes are differentially regulated by BRD2, BRD3 and BRD4, including insulin production and sensitivity (26,36,37). BET bromodomain-specific inhibitors are available, but it is important to note that these compounds, including (S)-JQ1, do not discriminate between the different BET proteins (8). Thus, new small molecules that are selective for each BET family member are urgently needed.
Here we report a new BRD4-Jagged1/Notch1 signaling axis in breast cancer cells (Figure 7). We show that BRD4 regulates Jagged1 expression and Notch1 signaling to regulate triple-negative breast cancer migration and invasion. Interestingly, JAG1 is a BRD4-specific gene, because BRD2 or BRD3 knockdown does not affect Jagged1 expression (Figures 3C-D). No association between BRD4 and Jagged1 expression was observed in luminal-A cells. Thus, it is possible that the BRD4-Jagged1 relationship we unraveled here is limited to triple-negative breast cancers which undergo deep chromatin remodeling, notably through EMT, to acquire invasive properties. The acquisition of new epigenetics marks, including acetylated histones, provides new docking sites for chromatin ‘readers’ like BET proteins at the newly activated promoters. Recently, it has been reported that BRD4-Twist interaction is crucial for the regulation of Twist-dependent transcription programs (20). We showed that BRD4 associates with the proximal region of Jagged1 promoter (Figures 6C-D). Twist is recruited to the JAG1 promoter and is a direct activator of Jagged1 transcription (35). Thus, it is likely that BRD4 and Twist are part of a transcription complex that positively regulates Jagged1 expression. Disruption of Twist-BRD4 interactions impairs WNT5A expression and tumor progression of basal-like breast cancer (20). Twist is a master transcription factor involved in EMT and its association with BRD4 strongly suggests a role for BET proteins in regulation of this essential process. A growing body of evidence supports this hypothesis, especially arising from developmental research. For example, BRD4 controls self-renewal and pluripotent functions of embryonic stem cells, and BRD4 inhibition promotes expression of EMT markers and differentiation (38).
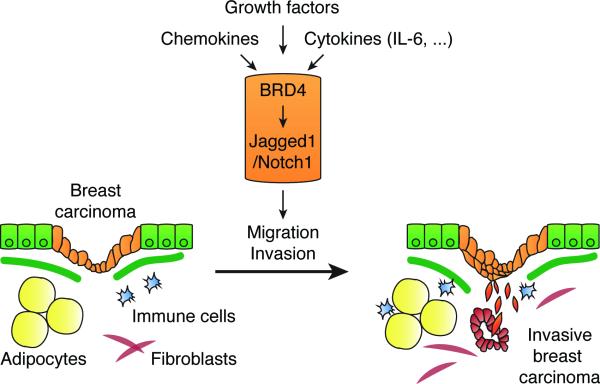
Figure 7. Proposed model: a BRD4/Jagged1/Notch1 signaling pathway to regulate breast cancer dissemination.
Breast cancer microenvironment is composed of diverse cell types including notably adipocytes, fibroblasts or immune cells, secreting numerous chemoattractant factors such as growth factors, chemokines or cytokines, including IL-6, that sustain cell migration or invasion. We identified BRD4 as a regulator of Jagged1, a Notch1 ligand, which regulates Notch1 signaling activity in breast cancer cells to modulate their migration and invasion. The pro-inflammatory cytokine IL-6 enriches BRD4 at the Jagged 1 promoter to stimulate its expression and promote breast cancer cell migration and invasion through Notch1 signaling. A combined elevated expression of BRD4 and Jagged1 in breast cancer associates in vivo with the presence of distant metastases.
Interestingly, we also found that BRD4 depletion alters expression of multiple genes involved in extracellular matrix (ECM) regulation such as COL1A2, COL3A1, COL5A2 or KRT19 (Figures 3 and S3). These findings are consistent with previous results showing that BRD4 ectopic expression in a mouse mammary tumor cell line also alters ECM regulatory genes (39). As an important component of the breast cancer microenvironment, the ECM participates in several steps of cancer progression, but also is involved in trafficking of stromal and immune cells in the vicinity of the tumor (23). Deciphering the roles of BET proteins in ECM modulation is crucial to better understand their exact roles within the tumor microenvironment.
The Notch signaling pathway is commonly activated and altered in cancer. Jagged1 has been implicated in regulation of multiple processes in cancer, including proliferation, survival, EMT, angiogenesis, metastasis, cancer cell ‘stemness’ and therapy resistance in several cancer types (29). More importantly, Jagged1 is not solely expressed by cancer cells; its expression and roles have been reported in diverse cell types present in the tumor microenvironment, including endothelial cells and pericytes, where it plays crucial roles in angiogenesis (40,41); dendritic cells (42,43), and regulatory T (Treg) cells (42,44,45). In many cancer types, regulatory T cell expansion inhibits tumor-specific immune responses and helps tumor cells evade immunosurveillance (46,47). Altogether, Jagged1 represents an attractive target in cancer therapy because of its important roles in cancer cells, but also in vasculature and immune cells. A classical therapeutic strategy to block Jagged1/Notch signaling is to use γ-secretase inhibitors (GSIs). GSIs are extensively investigated in Phase I/II clinical trials for solid tumors including breast cancer and show promising results (48). Whereas single-drug strategies are ineffective for long-term treatment, combination therapies are often necessary. Here, we offer evidence that BRD4 targeting may be useful to down-regulate the Jagged1/Notch1 pathway and block triple-negative breast cancer dissemination.
Triple-negative breast tumors are characterized by aggressive behavior and reduced sensitivity to classical chemotherapy. Within the breast tumor microenvironment, multiple cell types promote tumor progression through the production of messengers, including growth factors, hormones, cytokines and chemokines. These messengers activate diverse signaling cascades and transcription programs in cancer cells to increase migration and invasion, facilitating tumor progression and dissemination. Inflammatory mediators are well-known contributors to these processes. A recent study published by Boelens et al. presented the concept that, within the inflammatory breast tumor microenvironment, stromal cells can up-regulate Notch signaling in cancer cells (49). Our results are an important contribution to this study as we report strong interconnections between IL-6 signaling and BRD4 to control Jagged1 expression, Notch1 activation and breast cancer cell migration and invasion. We provide evidence that BRD4 targeting in cancer cells uncouples pro-migratory/invasive signals, including IL-6, from their biological effects. Significantly, we have also previously reported that either BRD2 deletion or BET protein inhibition reduces IL-6 and TNF-α production in lipopolysaccharide-challenged macrophages (26). Other studies have reported that BET targeting leads to diminished production of pro-inflammatory cytokines in models of various pathologies (25,50). Therefore, BET proteins constitute a promising therapeutic target within the breast tumor microenvironment to resolve inflammation and disrupt de facto its effects on tumor progression and dissemination.
Supplementary Material
Acknowledgements
We thank Boston University-Boston Medical Center Cancer Center faculty R. Flynn, N. Ganem and H. Feng, and B. Nikolajczyk for helpful comments and suggestions. We thank Anna C. Belkina, the BUMC Flow Cytometry and Cellular Imaging Core Facilities for technical assistance.
Grant Support. This work was supported by a grant from NIH, U01CA182898 to G.V. Denis.
Footnotes
Conflict of interest. The authors affirm that they have no conflicts to disclose.
References
- 1.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 2.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nature reviews Cancer. 2012;12(7):465–77. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denis GV, Green MR. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10(3):261–71. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 4.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000;11(8):417–24. [PMC free article] [PubMed] [Google Scholar]
- 5.Guo N, Faller DV, Denis GV. Activation-induced nuclear translocation of RING3. J Cell Sci. 2000;113(Pt 17):3085–91. doi: 10.1242/jcs.113.17.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20(23):4899–909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, et al. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22(18):6509–20. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y, et al. Blockade of oncogenic IkappaB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc Natl Acad Sci U S A. 2014;111(31):11365–70. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimamura T, Chen Z, Soucheray M, Carretero J, Kikuchi E, Tchaicha JH, et al. Efficacy of BET bromodomain inhibition in Kras-mutant non-small cell lung cancer. Clin Cancer Res. 2013;19(22):6183–92. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510(7504):278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529(7586):413–7. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene. 2015 doi: 10.1038/onc.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Zhou J, Ye F, Xiong H, Peng L, Zheng Z, et al. BRD4 inhibitor inhibits colorectal cancer growth and metastasis. Int J Mol Sci. 2015;16(1):1928–48. doi: 10.3390/ijms16011928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5(3):502–11. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25(2):210–25. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 22.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468(7327):1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. Journal of immunology. 2013;190(7):3670–8. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123(3):725–31. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 28.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Masiero M, Banham AH, Harris AL. The notch ligand JAGGED1 as a target for anti-tumor therapy. Front Oncol. 2014;4:254. doi: 10.3389/fonc.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9(1):R6. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MH, Kim HB, Yoon SP, Lim SC, Cha MJ, Jeon YJ, et al. Colon cancer progression is driven by APEX1-mediated upregulation of Jagged. The Journal of clinical investigation. 2013 doi: 10.1172/JCI65521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai Y, Wilson G, Huang B, Peng M, Teng G, Zhang D, et al. Silencing of Jagged1 inhibits cell growth and invasion in colorectal cancer. Cell Death Dis. 2014;5:e1170. doi: 10.1038/cddis.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. The Journal of clinical investigation. 2007;117(12):3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto M, Taguchi Y, Ito-Kureha T, Semba K, Yamaguchi N, Inoue J. NF-kappaB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun. 2013;4:2299. doi: 10.1038/ncomms3299. [DOI] [PubMed] [Google Scholar]
- 35.Chen HF, Huang CH, Liu CJ, Hung JJ, Hsu CC, Teng SC, et al. Twist1 induces endothelial differentiation of tumour cells through the Jagged1-KLF4 axis. Nat Commun. 2014;5:4697. doi: 10.1038/ncomms5697. [DOI] [PubMed] [Google Scholar]
- 36.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. The Biochemical journal. 2010;425(1):71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F, Deeney JT, Denis GV. Brd2 gene disruption causes “metabolically healthy” obesity: epigenetic and chromatin-based mechanisms that uncouple obesity from type 2 diabetes. Vitamins and hormones. 2013;91:49–75. doi: 10.1016/B978-0-12-407766-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Micco R, Fontanals-Cirera B, Low V, Ntziachristos P, Yuen SK, Lovell CD, et al. Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep. 2014;9(1):234–47. doi: 10.1016/j.celrep.2014.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, et al. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008;105(17):6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.High FA, Lu MM, Pear WS, Loomes KM, Kaestner KH, Epstein JA. Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2008;105(6):1955–9. doi: 10.1073/pnas.0709663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Kennard S, Lilly B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed JAGGED1. Circ Res. 2009;104(4):466–75. doi: 10.1161/CIRCRESAHA.108.184846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gopisetty A, Bhattacharya P, Haddad C, Bruno JC, Jr., Vasu C, Miele L, et al. OX40L/Jagged1 cosignaling by GM-CSF-induced bone marrow-derived dendritic cells is required for the expansion of functional regulatory T cells. Journal of immunology. 2013;190(11):5516–25. doi: 10.4049/jimmunol.1202298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bugeon L, Gardner LM, Rose A, Gentle M, Dallman MJ. Cutting edge: Notch signaling induces a distinct cytokine profile in dendritic cells that supports T cell-mediated regulation and IL-2-dependent IL-17 production. Journal of immunology. 2008;181(12):8189–93. doi: 10.4049/jimmunol.181.12.8189. [DOI] [PubMed] [Google Scholar]
- 44.Vigouroux S, Yvon E, Wagner HJ, Biagi E, Dotti G, Sili U, et al. Induction of antigen-specific regulatory T cells following overexpression of a Notch ligand by human B lymphocytes. J Virol. 2003;77(20):10872–80. doi: 10.1128/JVI.77.20.10872-10880.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yvon ES, Vigouroux S, Rousseau RF, Biagi E, Amrolia P, Dotti G, et al. Overexpression of the Notch ligand, Jagged-1, induces alloantigen-specific human regulatory T cells. Blood. 2003;102(10):3815–21. doi: 10.1182/blood-2002-12-3826. [DOI] [PubMed] [Google Scholar]
- 46.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews Cancer. 2012;12(4):252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motz GT, Coukos G. Deciphering and reversing tumor immune suppression. Immunity. 2013;39(1):61–73. doi: 10.1016/j.immuni.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan X, Wu H, Xu H, Xiong H, Chu Q, Yu S, et al. Notch signaling: an emerging therapeutic target for cancer treatment. Cancer letters. 2015;369(1):20–7. doi: 10.1016/j.canlet.2015.07.048. [DOI] [PubMed] [Google Scholar]
- 49.Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159(3):499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nadeem A, Al-Harbi NO, Al-Harbi MM, El-Sherbeeny AM, Ahmad SF, Siddiqui N, et al. Imiquimod-induced psoriasis-like skin inflammation is suppressed by BET bromodomain inhibitor in mice through RORC/IL-17A pathway modulation. Pharmacol Res. 2015;99:248–57. doi: 10.1016/j.phrs.2015.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.