Abstract
PURPOSE
To determine if the cif gene is present in pathogenic Pseudomonas aeruginosa isolates from bacterial keratitis patients at the Aravind Eye Hospital, a referral eye care center in southern India, and from corresponding environmental isolates.
METHODS
Polymerase chain reaction amplification was performed on strains of P. aeruginosa isolated from ocular infections and environmental soil samples collected from the area surrounding Aravind Eye Hospital. DNA sequencing of 16S ribosomal DNA amplicons was carried out to verify strain identity.
RESULTS
We determined that 45 of 48 patient isolates carry a genomic copy of cif. Analysis of a catalogue of environmental strains previously isolated from the surrounding area revealed that only 4 of 10 P. aeruginosa strains and 1 of 14 strains of related species carry the cif gene.
CONCLUSIONS
This is the first study to show P. aeruginosa strains with ocular pathogenicity carry the cif gene, and that the presence of this gene may be enriched over its prevalence in the environment. Taken together, these results suggest a potential role for Cif in acute bacterial keratitis.
Keywords: Cif, epoxide hydrolase, virulence factor, Pseudomonas aeruginosa, bacterial keratitis
Introduction
Infectious keratitis remains a leading cause of worldwide visual impairment. In the developing world, the incidence of corneal ulcers is tenfold higher than in the United States.1 Pseudomonas aeruginosa is the most common Gram-negative isolate in keratitis studies and is often the first or second most common bacterial pathogen overall.2–4 Moreover, it is responsible for a disproportionate amount of morbidity and mortality from bacterial keratitis (BK).5 Ocular Pseudomonas infections generally develop acutely within 24 hours of exposure.6 Although many agree that disruption of the corneal epithelium due to trauma or contact lens wear contributes a mechanical component to the pathogenesis of BK,7 the mechanisms that result in rapid onset infection and severe ulceration remain an area of active research interest.
In addition to ocular infections, P. aeruginosa also commonly colonizes the urinary tract, burn wounds, and the lung.8 To establish chronic infections of the lung, P. aeruginosa secretes a variety of virulence factors that affect surrounding microflora and host innate immune barriers.9 CFTR inhibitory factor (Cif) is an epoxide hydrolase virulence factor that is delivered into the cytoplasm of host airway epithelial cells and decreases levels of CFTR (cystic fibrosis transmembrane conductance regulator), an ATP binding cassette transporter responsible for maintaining properly hydrated airway-surface liquid in the lung.10 By removing CFTR, Cif may facilitate biofilm formation and infection. This hypothesis is supported by the observation that P. aeruginosa cif gene expression persists longitudinally in cystic fibrosis (CF) patients with chronic lung infection.10,11 Further study of the role of this virulence factor in BK could elucidate previously undiscovered epoxide signaling mechanisms on the ocular surface. In this retrospective cross-sectional study, we examined clinical isolates of P. aeruginosa BK to determine if the presence of the cif gene is correlated with ocular infectivity.
Materials and Methods
Clinical Isolates
Strains of P. aeruginosa were isolated from ocular infections presenting to Aravind Eye Hospital, Madurai, Tamil Nadu, India. Per institutional protocol, all corneal ulcers were referred to the Cornea Department, where corneal scrapings and bacterial culture were performed under anesthesia with published methods.12 All samples were obtained in accordance with procedures approved by the Aravind Institutional Review Board. This study has been approved as exempt human subjects research by the Dartmouth Committee on the Protection of Human Subjects.
Environmental isolates
Environmental soil samples were collected from the area surrounding Aravind Eye Hospital, Madurai, Tamil Nadu, India. For each soil sample collected, sterile phosphate buffered saline was added; samples were inverted and incubated for 2 hours at 37°C. Samples were centrifuged and 100 μL of each supernatant was plated on PA isolation agar.13 Samples were incubated overnight at 37°C. Two colonies from each isolate plate were selected and sub-cultured into 5 mL of Miller’s lysogeny broth (LB).14 In addition to the newly cultured environmental isolates, 10 isolates of P. aeruginosa were obtained from Madurai Kamaraj University. All environmental isolates were sub-cultured into 5 mL of LB. All samples were 16S rRNA gene sequenced, and sequencing results were entered into the NIH BLAST (Basic Local Alignment Sequence Tool) database. Results were used to determine species identity.
Colony PCR
For each PCR reaction, a final concentration of 0.2 μM forward and reverse primer, 25 μL Quick-Load Taq 2x Master mix (NEB M0271S) to a final volume of 50 μL with ddH2O. Colonies measuring 1 mm in diameter were picked up with a sterilized pipette tip and directly transferred to the PCR tube as DNA template. The thermal cycle program consisted of one cycle of 95 °C for 10 minutes, 95 °C for 30 seconds, 62 °C for 30 seconds, 68 °C for 2 minutes, return to 95 °C and repeat from step 2 for 32 additional cycles, 68 °C for 10 minutes, and a final incubation at 4 °C. Primers used were as follows: cif_Forward 5′ CTG AGG AAG TCG ATC ACC AG 3′, cif_Reverse 5′ GAT CCT CGA TAG ACT CTG CC 3′, rplU_Forward 5′ TTA CCG GTG GCA AGC AGC ACA AAG 3′, rplU_Reverse 5′ TTC ACG TCT TCG CCA TTG GCA ACC 3′. For all experiments, strain PA14 was used as a positive control and the presence of the Pseudomonas aeruginosa rplU gene sequence was used to confirm bacterial species identity. PCR-amplified DNA fragments were observed by 1% agarose gel electrophoresis (w/v, Invitrogen 16500) and SYBR Safe DNA stain (Invitrogen S33102). The amplified DNA fragments were visualized by UV illumination.
Strain Sequencing
DNA from isolates was purified using PUREGENE kit (QIAGEN 158522). PCR targeting 16S rDNA was carried out on both environmental and clinical P. aeruginosa isolates. For a 50 μl reaction, 1 μl of 200 mM dNTPs, 5 μl of Reaction buffer (Tris with MgCl2) 10 pM forward primer (U2F : 5′ GGC GTG CTT AAC ACA TGC AAG TCG 3′) and Reverse Primer (Ru3R : 5′ GCG GCT GGC ACG TAG TTA G 3′). 1.2 U/μl of Taq polymerase were used. Amplification was carried out in a thermal cycler (PTC 200) yielding a 470 base pair product. DNA sequencing of 16S rDNA amplicons was carried out commercially after purification of amplified products (Bio basic inc, Bangalore Genei, India). Cyclic PCR amplification was carried out using Big Dye Terminator Ver 3.1 followed by sequencing using Genetic Analyzer 3130. The sequences obtained were then compared with the BLASTn database at the National Centre for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/BLAST/). BLAST outputs were sorted based on maximum identity, and identifications were made when BLAST searches yielded at least 85% and for closely related species with more than 90% of query coverage.
Results
The cif gene is present in the majority of clinical isolates of P. aeruginosa
In total, we collected 48 clinical BK isolates from the Aravind Eye Hospital in Madurai, India from 2006 to 2010.5 Clinical records were available for 15 of these isolates. These records show an average patient age of 46 with a male predominance (66%) comparable to 2011 Madurai census data.15 A small subset of patients were immunosuppressed secondary to corticosteroid use (n=5) or poorly controlled diabetes mellitus (n=5) at time of infection. Additionally, three patients among the cohort self-reported a history of corneal trauma.
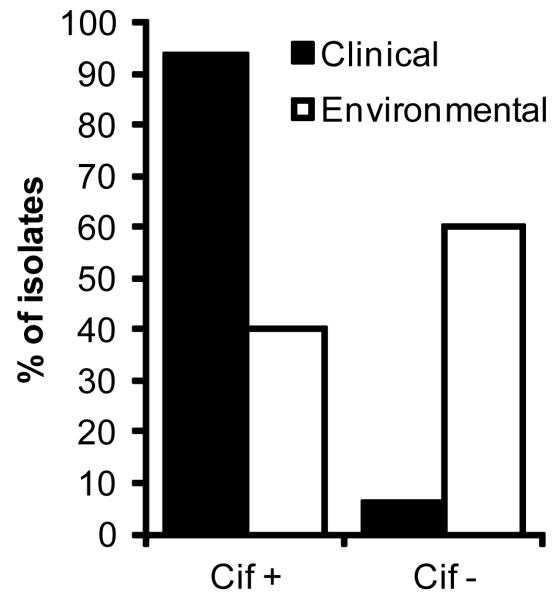
To test the hypothesis that the cif gene may also be present in acutely pathogenic strains, such as those responsible for BK, we performed colony PCR on subcultures of the BK isolates. We used previously validated primers targeting the cif gene as well as P. aeruginosa 50S ribosomal subunit L21 (rplU). P. aeruginosa strain PA14 was included as a positive control in all experiments. Analysis of 48 unique samples revealed 94% of isolates were cif positive, with only 3 pathogenic samples failing to amplify the cif gene (Figure 1). For all isolates, PCR was run in experimental triplicates. All strains were rplU positive, consistent with the species identification as P. aeruginosa.
Figure 1. The majority of P. aeruginosa keratitis clinical isolates carry a genomic copy of the cif gene.
PCR amplification of cif gene loci in 48 clinical isolates revealed 45 strains were cif positive. In contrast, only 4 of 10 environmental P. aeruginosa isolates were cif positive. All tested strains tested positive for 16S ribosomal subunit (rplU) control.
Cif is less prevalent in environmental isolates
Since the cif gene was present in the vast majority of clinical Pseudomonas isolates tested from Aravind, we next sought to determine the prevalence of this gene in strains isolated from the surrounding environment. We analyzed 10 strains of P. aeruginosa previously isolated from local soil samples. By colony PCR, all 10 isolates tested positive for rplU, corroborating species identity. In contrast to the patient samples, only 4 of the 10 environmental strains were positive for the cif gene. We also isolated and analyzed additional soil strains, and identified 14 non-aeruginosa Pseudomonads, of which only one tested cif positive (Table 1). These results suggest that P. aeruginosa isolated from keratitis patients may be enriched for the presence of the cif gene in comparison to environmental Pseudomonas strains.
Table 1. Environmental isolates cultured on PA agar.
PCR amplification was performed on 14 species from soil. None corresponded to P. aeruginosa, although many were Pseudomonads. Only strain 2 was found to be cif positive.
| Strain Species | |
|---|---|
| 1 | Pseudomonas putida |
| 2 | Pseudomonas spp. |
| 3 | Pseudomonas spp. (fluorescens) |
| 4 | Pseudomonas putida |
| 5 | Aeromonas sanarelli |
| 6 | Pseudomonas nitroreducens |
| 7 | Pseudomonas spp (putida) |
| 8 | Uncultured bacterium. |
| 9 | Pseudomonas nitroreducens |
| 10 | Pseudomonas spp. |
| 11 | Uncultured Achromobacter |
| 12 | Pseudomonas spp. |
| 13 | Pseudomonas spp. |
| 14 | Uncultured bacterium |
Discussion
In this study, we provide the first evidence that the cif gene is present and its frequency is most likely enriched in the genomes of pathogenic P. aeruginosa strains isolated from BK patients, suggesting a potential role for cif in acute infection. The cif gene was less prevalent in a collection of environmental P. aeruginosa when compared to the near ubiquity of cif in clinical strains. Additionally, we identified 14 non-aeruginosa Pseudomonas species, and interestingly, one of these strains harbored the cif gene. The lower prevalence of cif in the genomes of the environmental isolates compared to patient isolates may indicate that Cif facilitates colonization or maintenance of infection.
There is strong evidence to suggest that Cif serves as a virulence factor in the colonization and infection of the lung by P. aeruginosa.16–21 While lung infection has been the predominant focus of Cif research, there is limited evidence to suggest this virulence factor may play a role during infection at other sites and in patients without CF. Inoculation of P. aeruginosa strain PA14 in rat peritoneum induces a 16-fold increase in cif mRNA expression.22 Our observations extend the range of opportunistic niches in which Cif may contribute to the infectious potential of P. aeruginosa.
Cif is an epoxide hydrolase with enzymatic activity that is required for its cellular effect on CFTR.23–26 In addition, Cif has been shown to exert effects on other ATP binding cassette (ABC) transporters: the transporter associated with antigen processing 1 (TAP1)27 and the drug efflux pump p-glycoprotein in kidney and intestinal epithelium.28 Thus, there may be additional ABC transporters affected by Cif that remain to be identified, which play a role in ocular infection by P. aeruginosa. Furthermore, endogenous epoxides, including epoxyeicosatrienoic acids, are lipid mediators that serve as regulators of inflammation and hypertension.29 Thus, based on its enzyme activity, an alternative role for Cif in ocular pathogenesis could be to dysregulate immune signaling mechanisms.
P. aeruginosa interacts indirectly with the epithelium during chronic infection.10 Cif is transported into airway epithelial cells via outer membrane vesicles (OMVs) secreted by P. aeruginosa. These OMVs diffuse through overlying mucus and fuse with membrane lipid rafts, delivering virulence factors into the host cytoplasm.30 OMVs are proinflammatory and cytotoxic to both the pulmonary and corneal epithelia.31 The immune response to P. aeruginosa strains in the eye is dominated by dense neutrophil infiltrates forming neutrophil extracellular traps.30,31 Cytotoxic strains that are associated with higher rates of morbidity, including vision impairment, are able to evade this neutrophil capture mechanism by secreting high amounts of OMVs.31 This mechanism suggests OMVs provide necessary factors for P. aeruginosa to establish corneal infection.
In principle, another explanation for the observed enrichment of cif+ strains could be the routine transfer of P. aeruginosa BK strains among patients. Indeed, in the context of CF, recent large-scale genomic sequencing of patient isolates of P. aeruginosa identified ten bacterial clone types present at initial colonization in multiple subjects, several of whom may have experienced direct patient-to-patient transmission during overlapping hospital visits, while others may have been independently infected by closely related environmental strains.32 However, unlike CF airway infections, P. aeruginosa keratitis is most often a community-acquired infection and not transmitted in patient-care settings. In a number of studies, distinct subpopulations of P. aeruginosa have been isolated from keratitis patients and are characterized by expression of specific virulence factors, suggesting that one or more subset(s) of environmental bacteria are also responsible for the majority of BK cases.33–35 Thus, the relative enrichment of strains bearing the cif gene most likely reflects a selective advantage in the context of keratitis.
There are a few caveats to our conclusions. Sequencing of clinical and environmental isolates was performed with primers specific to the wild-type cif gene. Thus, a silent mutation at the primer-annealing site could produce a false-negative result; although this variability would have to preferentially affect the environmental strains in order to skew these data. Additionally, our data only address the presence of the cif gene, not gene expression. Marvig et al. sequenced serial CF patient isolates and identified convergent evolution of a large number of genes within distinct P. aeruginosa clones, suggesting regulatory network remodeling over time within each individual patient.32 Their conclusion suggests the presence of an evolutionary selective pressure to maintain genes that confer an advantage, regardless of gene expression throughout the course of infection. Many virulence factors vital to P. aeruginosa infection are maintained in highly conserved pathogenicity islands.36 However, the cif gene does not reside within any established island, arguing for a specific functional role of the gene in acute ocular infection.
Taken together, these data suggest a potential role for Cif in the ocular infectivity of P. aeruginosa and thus lay the groundwork for follow-up studies. Ideally, future experiments will yield primary clinical isolates that can be analyzed not only for cif gene presence, but also gene expression and Cif protein levels. Given the very high prevalence of cif genes among BK isolates (94% of patients), a larger sample cohort, as well as detailed medical records, will be required to determine the extent of Cif's contribution to the course of infection and ultimate outcomes. Additional insights may be provided by analyzing the prevalence of Cif in cases where ocular infections with P. aeruginosa do not lead to BK. In parallel, we have developed a variety of matched strains of P. aeruginosa with Cif deletions or knock-ins of inactive mutations.37,38 Together with animal models of BK,39 these strains will provide an excellent basis for validating the impact of Cif on colonization, persistence, antibiotic resistance, and severity of infection. In addition, the availability of recently-developed Cif inhibitor molecules offers the prospect of developing therapeutic interventions or preventing infection for high-risk populations.25,32
Acknowledgements
We would like to thank Madurai Kamaraj University for providing P. aeruginosa isolates, the Aravind Microbiology and Ophthalmology laboratories for providing clinical P. aeruginosa isolates and Dr. Kyle Cady for helpful discussion.
Grant Information
Funding support was provided by a Dartmouth-Fogarty International Fellowship and by NIH grants R01-AI091699, P20-GM113132, T32-AI007519, and T32-DK007301
Footnotes
Competing Interests
CB and DM are co-inventors on patent-pending inhibitors of Cif
Consent
Ethical approval for the use of patient isolates was obtained through the Aravind Institutional Review Board. The Dartmouth CPHS reviewed the study and determined that it constitutes exempt research.
Author Contributions
CB, RSGK, and LP collected the data. LP provided the clinical isolates and patient information. CB designed the project with contribution from DM, MZ and LP. CB and JSL analyzed the data. CB, JSL, DM, and MZ wrote the manuscript.
References
- 1.Sommer A, Taylor HR, Ravilla TD, et al. Challenges of ophthalmic care in the developing world. JAMA Ophthalmol. 2014;132:640–644. doi: 10.1001/jamaophthalmol.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourcier T, Thomas F, Borderie V, et al. Bacterial keratitis: predisposing factors, clinical and microbiological review of 300 cases. Br J Ophthalmol. 2003;87:834–838. doi: 10.1136/bjo.87.7.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrakis G, Alfonso EC, Miller D. Shifting trends in bacterial keratitis in south Florida and emerging resistance to fluoroquinolones. Ophthalmology. 2000;107:1497–1502. doi: 10.1016/s0161-6420(00)00179-2. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan M, Muthiah S, Jeena M, et al. Subgroup Analysis in the Steroids for Corneal Ulcers Trial—Reply. Arch Ophthal. 2012;130 doi: 10.1001/archophthalmol.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green M, Matthew G, Andrew A, et al. Risk Factors and Causative Organisms in Microbial Keratitis. Cornea. 2008;27:22–27. doi: 10.1097/ICO.0b013e318156caf2. [DOI] [PubMed] [Google Scholar]
- 6.Burns RP. Pseudomonas aeruginosa Keratitis: Mixed Infections of the Eye. Am J Ophthalmol. 1969;67:257–262. doi: 10.1016/0002-9394(69)93156-0. [DOI] [PubMed] [Google Scholar]
- 7.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 8.Costerton J. Introduction to biofilm. Int J Antimicrob Agents. 1999;11:217–221. doi: 10.1016/s0924-8579(99)00018-7. [DOI] [PubMed] [Google Scholar]
- 9.Bleves S, Viarre V, Salacha R, et al. Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons. Int J Med Microbiol. 2010;300:534–543. doi: 10.1016/j.ijmm.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 10.MacEachran DP, Ye S, Bomberger JM, et al. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect Immun. 2007;75:3902–3912. doi: 10.1128/IAI.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballok AE, Bahl CD, Dolben EL, et al. Epoxide-Mediated CifR Repression of cif Gene Expression Utilizes Two Binding Sites in Pseudomonas aeruginosa. J Bacteriol. 2012;194:5315–5324. doi: 10.1128/JB.00984-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wihelmus KR, L.T., Ostao MS, et al. Laboratory Diagnosis of Ocular Infections. American Society for Microbiology; 1994. [Google Scholar]
- 13.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 14.Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Data CP . Madurai District : Census 2011 data. Census 2011; [Accessed December 6, 2016]. 2015. http://www.census2011.co.in/census/district/45-madurai.html. Published 2011. [Google Scholar]
- 16.Bomberger JM, Ely KH, Bangia N, et al. Pseudomonas aeruginosa Cif protein enhances the ubiquitination and proteasomal degradation of the transporter associated with antigen processing (TAP) and reduces major histocompatibility complex (MHC) class I antigen presentation. J Biol Chem. 2014;289:152–162. doi: 10.1074/jbc.M113.459271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballok AE, O’Toole GA. Pouring salt on a wound: Pseudomonas aeruginosa virulence factors alter Na+ and Cl− flux in the lung. J Bacteriol. 2013;195:4013–4019. doi: 10.1128/JB.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bomberger JM, Siying Y, MacEachran DP, et al. A Pseudomonas aeruginosa Toxin that Hijacks the Host Ubiquitin Proteolytic System. PLoS Pathog. 2011;7:e1001325. doi: 10.1371/journal.ppat.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bomberger JM, Maceachran DP, Coutermarsh BA, et al. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 2009;5:e1000382. doi: 10.1371/journal.ppat.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacEachran DP, Ye S, Bomberger JM, et al. The Pseudomonas aeruginosa secreted protein PA2934 decreases apical membrane expression of the cystic fibrosis transmembrane conductance regulator. Infect Immun. 2007;75:3902–3912. doi: 10.1128/IAI.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swiatecka-Urban A, Moreau-Marquis S, Maceachran DP, et al. Pseudomonas aeruginosa inhibits endocytic recycling of CFTR in polarized human airway epithelial cells. Am J Physiol Cell Physiol. 2006;290:C862–C872. doi: 10.1152/ajpcell.00108.2005. [DOI] [PubMed] [Google Scholar]
- 22.Mashburn LM, Jett AM, Akins DR, et al. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahl CD, Morisseau C, Bomberger JM, et al. Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor. J Bacteriol. 2010;192:1785–1795. doi: 10.1128/JB.01348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahl CD, Madden DR. Pseudomonas aeruginosa Cif defines a distinct class of α/β epoxide hydrolases utilizing a His/Tyr ring-opening pair. Protein Pept Lett. 2012;19:186–193. doi: 10.2174/092986612799080392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahl CD, Hvorecny KL, Bomberger JM, et al. Inhibiting an Epoxide Hydrolase Virulence Factor from Pseudomonas aeruginosa Protects CFTR. Angew Chem Int Ed Engl. 2015;54:9881–9885. doi: 10.1002/anie.201503983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bahl CD, Hvorecny KL, Morisseau C, et al. Visualizing the Mechanism of Epoxide Hydrolysis by the Bacterial Virulence Enzyme Cif. Biochemistry. 2016;55:788–797. doi: 10.1021/acs.biochem.5b01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bomberger JM, Ely KH, Bangia N, et al. Pseudomonas aeruginosa Cif Protein Enhances the Ubiquitination and Proteasomal Degradation of the Transporter Associated with Antigen Processing (TAP) and Reduces Major Histocompatibility Complex (MHC) Class I Antigen Presentation. J Biol Chem. 2013;289:152–162. doi: 10.1074/jbc.M113.459271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye S, MacEachran DP, Hamilton JW, et al. Chemotoxicity of doxorubicin and surface expression of P-glycoprotein (MDR1) is regulated by the Pseudomonas aeruginosa toxin Cif. Am J Physiol Cell Physiol. 2008;295:C807–C818. doi: 10.1152/ajpcell.00234.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inceoglu B, Bora I, Schmelzer KR, et al. Soluble epoxide hydrolase inhibition reveals novel biological functions of epoxyeicosatrienoic acids (EETs) Prostaglandins Other Lipid Mediat. 2007;82:42–49. doi: 10.1016/j.prostaglandins.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballok AE, Filkins LM, Bomberger JM, et al. Epoxide-mediated differential packaging of Cif and other virulence factors into outer membrane vesicles. J Bacteriol. 2014;196:3633–3642. doi: 10.1128/JB.01760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan Q, Dwyer M, Rahman S, et al. Distinct susceptibilities of corneal Pseudomonas aeruginosa clinical isolates to neutrophil extracellular trap-mediated immunity. Infect Immun. 2014;82:4135–4143. doi: 10.1128/IAI.02169-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marvig RL, Sommer LM, Molin S, et al. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 33.Lomholt JA, Poulsen K, Kilian M. Epidemic population structure of Pseudomonas aeruginosa: evidence for a clone that is pathogenic to the eye and that has a distinct combination of virulence factors. Infect Immun. 2001;69:6284–6295. doi: 10.1128/IAI.69.10.6284-6295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart RMK, Wiehlmann L, Ashelford KE, et al. Genetic Characterization Indicates that a Specific Subpopulation of Pseudomonas aeruginosa Is Associated with Keratitis Infections. J Clin Microbiol. 2011;49:993–1003. doi: 10.1128/JCM.02036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stapleton F, Dart JKG, Seal DV, et al. Epidemiology of Pseudomonas aeruginosa keratitis in contact lens wearers. Epidemiol Infect. 1995;114:395. doi: 10.1017/s0950268800052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kung VL, Ozer EA, Hauser AR. The Accessory Genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2010;74:621–641. doi: 10.1128/MMBR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacEachran DP, Stanton BA, O’Toole GA. Cif is negatively regulated by the TetR family repressor CifR. Infect Immun. 2008;76:3197–3206. doi: 10.1128/IAI.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bahl CD, Hvorecny KL, Bomberger JM, et al. Inhibiting an Epoxide Hydrolase Virulence Factor from Pseudomonas aeruginosa Protects CFTR. Angew Chem Int Ed Engl. 2015;54:9881–9885. doi: 10.1002/anie.201503983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marquart ME. Animal Models of Bacterial Keratitis. J Biomed Biotechnol. 2011;2011:1–12. doi: 10.1155/2011/680642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamura S, Hvorecny KL, Niu J, et al. Rational Design of Potent and Selective Inhibitors of an Epoxide Hydrolase Virulence Factor from Pseudomonas aeruginosa. J Med Chem. 2016;59:4790–4799. doi: 10.1021/acs.jmedchem.6b00173. [DOI] [PMC free article] [PubMed] [Google Scholar]