Summary
Eosinophilic esophagitis (EoE) is an antigen‐driven T cell‐mediated chronic inflammatory disease where food and environmental antigens are thought to have a role. Human eosinophils express the immunoregulatory protein galectin‐10 and have T cell suppressive capacity similar to regulatory T cells (Tregs). We hypothesized that one function of eosinophils in EoE might be to regulate the T cell‐driven inflammation in the oesophagus. This was tested by evaluating the suppressive capacity of eosinophils isolated from the blood of adult EoE patients in a mixed lymphocyte reaction. In addition, eosinophilic expression of forkhead box protein 3 (FOXP3), the canonical transcription factor of Tregs, was determined by conventional and imaging flow cytometry, quantitative polymerase chain reaction (qPCR), confocal microscopy and immunoblotting. It was found that blood eosinophils from EoE patients had T cell suppressive capacity, and that a fraction of the eosinophils expressed FOXP3. A comparison of EoE eosinophils with healthy control eosinophils indicated that the patients' eosinophils had inferior suppressive capacity. Furthermore, a higher percentage of the EoE eosinophils expressed FOXP3 protein compared with the healthy eosinophils, and they also had higher FOXP3 protein and mRNA levels. FOXP3 was found in the cytosol and nucleus of the eosinophils from both the patients and healthy individuals, contrasting with the strict nuclear localization of FOXP3 in Tregs. To conclude, these findings suggest that the immunoregulatory function of eosinophils may be impaired in EoE.
Keywords: eosinophils, eosinophilic esophagitis, FOXP3, regulatory T cells, suppression
Introduction
Eosinophils have been regarded historically as potentially harmful cells due to an impressive capacity for producing reactive oxygen species and for secreting cytotoxic proteins. However, in the 1980s the eosinophil was proclaimed to be immunoregulatory, following experiments performed by Peterson and co‐workers, who observed that eosinophilic granule proteins could inhibit lymphocyte proliferation in vitro 1. Moreover, eosinophils have been shown to selectively inhibit T helper type 1 (Th1) cells and skew T cells towards a Th2‐like profile through indoleamine‐2,3‐dioxygenase‐mediated degradation of tryptophan, as tryptophan is more essential for the metabolism of Th1 than Th2 cells 2. Rofousse and co‐workers demonstrated that human eosinophils, when pre‐activated by immunoglobulin (Ig)A complexes, enhanced the proliferation of CD4+ Th cells but inhibited the activation of CD3–CD4+ T cells stimulated by dendritic cells 3. We have shown that eosinophils from haematopoietic stem cell recipients and healthy individuals suppress allogeneic T cell proliferation of both CD4+ and CD8+ T cells in a cell contact‐dependent manner 4. Recently, it was demonstrated that eosinophils can down‐regulate Th17 cells in the small intestine of mice 5. An intriguing finding is that regulatory T cells (Tregs) use galectin‐10, the prototypical protein of eosinophils, to suppress T cells 6. Collectively, these findings indicate that one function of eosinophils may be to preserve homeostasis by controlling T cells and their subsets, much like Tregs.
Eosinophilic oesophagitis (EoE) is one of the most common causes of swallowing and feeding difficulties in westernized countries 7. This disease was first recognized in the 1990s and its incidence appears to be escalating worldwide 8, 9. The barrier function of the oesophageal epithelium is defective in eosinophilic oesophagitis 10, which is likely to contribute either primarily or secondarily to the breach in tolerance that is at the centre of this chronic antigen‐driven inflammatory process. Most evidence points towards food allergens and/or airborne allergens as being the main driving antigens in EoE 11. The additional role of unknown antigens in the aetiology of this disease, as well as the interplay between gastroesophageal reflux and EoE, are still unresolved questions. Whereas the oesophagus of healthy individuals is devoid of eosinophils, pronounced oesophageal infiltration of eosinophils characterizes EoE and constitutes one diagnostic criterion 12, 13. EoE is presumed to be a T cell‐mediated inflammatory process, as experimental EoE cannot be triggered in T cell‐deficient mice 14. In line with this hypothesis, increased numbers of CD3+ T cells, with a preponderance of CD8+ T cells, as well as activated T cells have been demonstrated in the oesophageal epithelium of patients with active EoE 15, 16, 17, 18, 19. Moreover, elevated levels of IL5+CD4+ T cells in the circulation of children with EoE were found to correlate positively with oesophageal eosinophil counts, suggestive of co‐operation between these two cell types 20.
The aims of this study were threefold: (1) to assess if eosinophils derived from patients with EoE are capable of suppressing T cell activity, (2) to examine if eosinophils from EoE patients express the canonical transcription factor of Tregs, forkhead box protein 3 (FOXP3) and (3) to compare the eosinophils from EoE patients with healthy control eosinophils with regard to suppressive capacity and FOXP3 expression.
Materials and methods
Study participants
Thirty‐four adult patients with symptomatic EoE were recruited at the Head and Neck Surgery Departments at Sahlgrenska University Hospital, Gothenburg or at NÄL Hospital, Trollhättan, Sweden (Table 1). The 2011 EoE guidelines were followed to establish a correct diagnosis 21. An absolute requirement was the presence of ≥ 15 (peak count) eosinophils per high‐power field in one of the ≥ 4 biopsies collected from the oesophagus (whereby at least two samples were collected from the proximal and distal parts, respectively). Only patients who were refractory to treatment with proton pump inhibitors and/or had normal 24 h pH monitoring of the oesophagus were accepted, in order to exclude patients with gastroesophageal reflux disease. Biopsies from the duodenum and gastric antrum were also examined to ensure that patients with additional upper gastrointestinal diagnoses were not included. Thirty‐four healthy subjects without gastrointestinal symptoms or other inflammatory conditions (median age = 28 years; age range = 25–65 years; 59% men; 36% atopic) were also studied. Written informed consent was acquired from all the study participants. The study was approved by the Regional Ethical Review Board of Gothenburg, Sweden.
Table 1.
Clinical characteristics of the eosinophilic oesophagitis (EoE) patients
| Characteristic | Number (range) | % | |
|---|---|---|---|
| n | 34 | ||
| Age (years) | 44 (18–72)* | ||
| Male | 29 | 85 | |
| Symptoms | |||
| Dysphagia | 32/34 | 94 | |
| Food impaction | 26/34 | 76 | |
| Chest pain | 9/34 | 26 | |
| Nausea/vomiting | 6/34 | 18 | |
| Peak no. of eosinophils/high‐power field | 36 (15–100)* | ||
| Endoscopic findings | |||
| Trachealization | 23/34 | 68 | |
| Furrows | 14/34 | 41 | |
| White specks | 13/34 | 38 | |
| Strictures | 5/34 | 15 | |
| Mucosal shredding | 2/34 | 5.9 | |
| Sensitization † | |||
| Food and/or inhalant allergens | 23/34 | 68 | |
| No sensitization | 11/34 | 32 | |
*Median (range). †Specific serum immunoglobulin (Ig)E and/or prick test positive.
Blood samples
All participants donated 10 ml of ethylenediamine tetraacetic acid (EDTA)‐anticoagulated venous blood for flow cytometry analyses and 100 ml of heparin‐anticoagulated venous blood for quantitative polymerase chain reaction (qPCR), immunoblotting and mixed lymphocyte reactions (MLR). The maximum time that elapsed between the drawing of blood and flow cytometry was 24 h.
Eosinophil purification and handling
Eosinophils were isolated from lithium heparin‐treated blood by negative immunomagnetic depletion using magnetic activated cell sorting beads (MACS; Miltenyi Biotec, Bergisch Gladbach, Germany) coated with monoclonal antibodies (mAbs) against CD3, CD14, CD16 and CD19, as described previously 4, and (a) used directly for T cell suppression assays or (b) lysed using radioimmunoprecipitation assay (RIPA) lysis and extraction buffer (ThermoFisher Scientific, Waltham, MA, USA) supplemented with HaltTM protease and phosphatase inhibitor cocktail (ThermoFisher Scientific), followed by storage at −80°C until immunoblots were completed or (c) stored frozen at −20°C in RNAlater solution (Ambion, Paisley, UK) until qPCR was completed. Only eosinophil preparations of > 98% purity were used, as determined by Diff Quik staining (Polysciences Europe GmbH, Eppelheim, Germany) of cytospun cells.
RNA extraction and cDNA synthesis
Total RNA was prepared from eosinophils using the RNeasy Mini kit (Qiagen, Venlo, the Netherlands). RNA quality and concentration were evaluated using the Experion Automated Electrophoresis System (Bio‐Rad Laboratories, Berkeley, CA, USA). Total RNA (25 ng) was transcribed into cDNA (5 min at 20°C; 50 min at 42°C; 5 min at 70°C; and a final step at 4°C) using avian myeloblastosis virus (AMV) reverse transcriptase, PCR–deoxynucleotide (dNTP) mix, RNase inhibitor and the random primer pd(N)6 (all from Roche Applied Bioscience, Mannheim, Germany). cDNA was stored at −80°C until further use.
Real‐time (RT)–qPCR
TaqMan® gene expression assays (Applied Biosystems, Foster City, CA, USA) were used to analyse the expression of FOXP3 (Accession number Hs01085834_m1) and the hypoxanthine phosphoribosyltransferase 1 gene (HPRT1; Hs99999909_m1), the latter of which was used as an endogenous control, using the 7500 RT–PCR system and 7500 System SDS software (Applied Biosystems). The thermal cycling parameters were as follows: one cycle of 2 min at 50°C; one cycle of 10 min at 95°C; 40 cycles of 15 s at 95°C; and one cycle of 1 min at 60°C. The Pfaffl method was used to calculate the transcript levels of FOXP3 relative to those of HPRT1 22.
Flow cytometric analyses of eosinophils
EDTA‐treated blood samples were depleted of erythrocytes using distilled water. Unspecific binding of antibodies was blocked using 1 mg/ml Vivaglobin (CSL Behring GmbH, Marburg, Germany) and leucocytes were incubated with mAbs against CD16 [clone 3G8, fluorescein isothiocyanate (FITC)] and CCR3 [clone 5E8, Alexa Fluor (AF) 647], both from BD Biosciences (Franklin Lakes, NJ, USA). The cells were then permeabilized and fixed with the FOXP3 staining buffer set (Affymetrix eBioscience, San Diego, CA, USA) and labelled with α‐FOXP3 mAb [IgG1, clone 259D/C7, phycoerythrin (PE); BD Biosciences]. An isotype control antibody [mouse IgG1 kappa light chain isotype control, clone MOPC‐21, PE; BD Biosciences] and ‘fluorescence‐minus‐one’ controls were used to monitor background staining 23. Data were collected and analysed using the fluorescence activated cell sorter (FACS)Canto II Flow Cytometer (BD Biosciences) and FACSDiva software version 6.0 (BD Biosciences); 100 000 events were acquired and analysed with FlowJo version 7.6.4 software (TreeStar, Ashland, OR, USA). Eosinophils were defined by their high side‐scatter, low/no expression of CD16 and high expression of CCR3. The results are expressed as median fluorescence intensity (MFI) or %. Measurement of CCR3, which is expressed by > 98% of eosinophils, was used to calculate the proportion of eosinophils that expressed FOXP3.
Flow cytometric analyses of Tregs
Peripheral blood mononuclear cells (PBMC) were isolated from EDTA blood samples by Ficoll‐Hypaque (Amersham Bioscience, Little Chalfont, UK) density gradient centrifugation. PBMC were labelled with mAbs against CD25 (clone 2A3, allophycocyanin (APC)), CD4 (clone SK3, APC‐H7) and CD127 (clone HIL‐7R‐M21, FITC) (all from BD Biosciences), then permeabilized and fixed with the FOXP3 staining buffer set (Affymetrix), and incubated with α‐FOXP3 mAb (clone 259D/C7, PE; BD Biosciences). Tregs were defined as CD4+CD25+CD127neg.
Imaging flow cytometry
Eosinophils and Tregs were processed and stained as described above with the addition of 4′,6‐diamidino‐2‐phenylindole (DAPI) (140 nM) as a nuclear stain. Eosinophils and Tregs (1000 cells) were collected using an ImageStream®X Mark II (Amnis, Seattle, WA, USA) and analysed with IDEAS software (version 6.0). To determine what proportion of FOXP3 was present in the nucleus of eosinophils, a custom‐made analysis strategy was employed. First, a mask defining the nuclear region was created. The intensity of FOXP3 staining in this nuclear mask was divided by the total intensity of FOXP3 staining in the cell (as defined by the object mask in the FOXP3 channel), and the value was then multiplied by 100. The IDEAS feature used for the calculation was as follows: Intensity_Threshold (Object [M01, Ch01, Tight), DAPI, 50]_FOXP3/Intensity_Object (M03, FOXP3, Tight) FOXP3 × 100. Results are expressed as the median feature value from 1000 eosinophils for each donor.
FOXP3 immunoblot
Eosinophil lysates (17–40 µg protein) were separated on 4–15% Mini‐PROTEAN® TGX precast gels (Bio‐Rad Laboratories, Hercules, CA, USA) and transferred onto polyvinylidene fluoride (PVDF) membranes using the Trans‐Blot® TurboTM Transfer System (Bio‐Rad Laboratories). Membranes were blocked with 5% bovine serum albumin (BSA) in TBS‐Tween (0.1%), incubated with α‐FOXP3 antibodies (clone PCH101, diluted 1 : 200; eBioscience) overnight at 4°C, followed by incubation with horseradish peroxidase (HRP)‐conjugated goat anti‐rat IgG (1 : 6000; eBioscience) for 1 h. Blots were developed using the OPTI‐4CN substrate kit (Bio‐Rad Laboratories). Lysates of human FOXP3‐transfected 293T cells (Santa Cruz Biotechnology, Dallas, TX, USA) and purified Tregs (15–25 µg) were used as positive controls.
FOXP3 confocal microscopy
Purified eosinophils (106) and purified CD4+ T cells (106) were blocked using 1 mg/ml Vivaglobin and stained with mAbs against CD193 (clone 5E8; PE) and T cell receptor (TCR)‐α/β (clone IP26; AF488), respectively, for 30 min at 4°C. Next, cells were fixed and permeabilized using FOXP3 transcription factor fixation/permeabilization buffer (Affymetrix eBioscience). After a repeat blocking step, eosinophils and CD4+ T cells were stained with either mAb against FOXP3 (clone 259D/C7; AF647) or a mouse IgG1 isotype control antibody (clone MOPC21; AF647) for 30 min at 4°C. Lastly, cells were stained with Hoechst 34580 (10 µg/ml). Stained eosinophils and CD4+ T cells were cytospun onto microscope slides and slides mounted using ProLong Diamond Antifade Mountant (Molecular Probes, Eugene, OR, USA). Cells were visualized using a Plan‐Apochromat 63× oil‐immersion objective lens. Images were captured by laser scanning confocal microscopy (LSM700; Carl Zeiss, Oberkochen, Germany) using ZEN2010 software at the Center for Cellular Imaging at the Sahlgrenska Academy, University of Gothenburg.
PBMC suppression assay
A one‐way MLR composed of PBMC from a healthy blood donor (responder cells) mixed with a pool of γ‐irradiated PBMC from 11 donors (trigger cells) at a 1 : 1 ratio (105 : 105 cells) was started, as described previously 4. Three hours later, 105 eosinophils were added. Cell proliferation was measured after 6 days by pulsing with [3H]‐thymidine for 6 h and quantified as counts per minute (cpm).
T helper cell subset suppression assay
PBMC (105 cells) were added to α‐CD3‐coated (1 µg/ml, clone OKT‐3; Affymetrix), round‐bottomed 96‐well plates (TPP, Trasadingen, Switzerland), together with α‐CD28 mAb (2 µg/ml, clone CD28.2, Affymetrix). Three hours later, purified eosinophils (105 cells) were added to the CD3/CD28‐stimulated PBMC. PBMC:eosinophil co‐cultures were incubated for 2 days at 37°C. During the final 4 h of culture, cells were stimulated with phorbol myristate acetate (PMA) (50 ng/ml; Sigma‐Aldrich, St Louis, MO, USA) and ionomycin (1 µg/ml; Sigma‐Aldrich) in the presence of the Golgi transport inhibitor monensin (2 µM; BD Biosciences). After harvesting, cells were stained for viability (fixable viability dye eFluor780; Affymetrix eBioscience) and CD4 expression (clone RPA‐T4, FITC). Following fixation and permeabilization using the Cytofix/Cytoperm kit (BD Biosciences), cells were stained for the intracellular cytokines interferon [(IFN)‐γ, Brilliant Violet 421, clone B27], IL‐4 (APC, clone 8D4‐8) and IL‐17A (PE, clone N49‐653). All antibodies were from BD Biosciences. The fractions of Th1 (IFN‐γ+), Th2 (IL‐4+) and Th17 (IL‐17A+) cells among the CD4+ T cells present in cultures, with and without eosinophils, were determined.
Statistical methods
The Wilcoxon matched‐pairs signed‐rank test or the Mann‐Whitney U‐test was used to determine statistical significance when comparing paired and non‐paired groups, respectively. GraphPad Prism version 5.0 software (GraphPad, San Diego, CA, USA) was used. P < 0·05 was considered statistically significant.
Results
Eosinophils from EoE patients are suppressive, but less so than eosinophils from healthy individuals
First, we evaluated the capacity of blood eosinophils from EoE patients to suppress T cell proliferation in an MLR, and found that they could suppress T cells (Fig. 1a). However, their suppressive capacity was inferior to that of blood eosinophils from healthy individuals: eosinophils from the EoE patients inhibited T cell proliferation by a median of 51% (25–75 percentile = 28–63%), whereas eosinophils from healthy donors inhibited T cell proliferation by a median of 90% (25–75 percentile = 56–98%), P = 0·0001 (Fig. 1b). We also evaluated the capacity of eosinophils from EoE patients and healthy controls to suppress different Th cell subsets. It appears that eosinophils from EoE patients (n = 2) and healthy controls (n = 2) alike could suppress all Th cell subsets analysed, i.e. IFN‐γ+ Th1 cells, IL‐4+ Th2 cells and IL‐17A+ Th17 cells (Fig. 1c).
Figure 1.
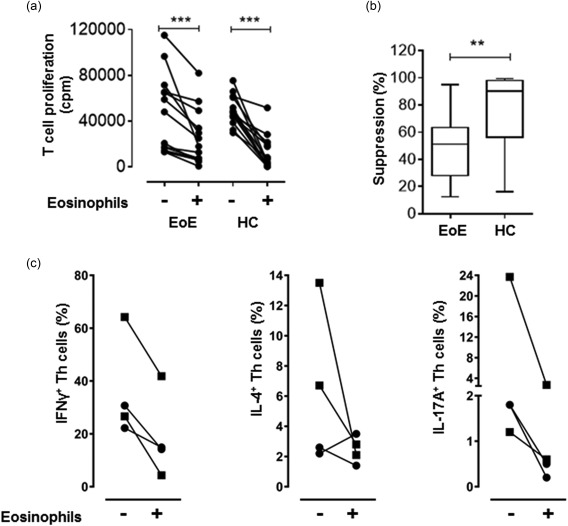
(a) Eosinophils from eosinophilic oesophagitis (EoE) patients and healthy controls (HC) suppress polyclonal T cell proliferation. Mixed lymphocyte reactions (MLRs) were performed for 6 days without or with eosinophils from EoE patients (n = 14) or from healthy individuals (n = 14). An eosinophil to peripheral blood mononuclear cell (PBMC) ratio of 1 : 1 was used. ***P = 0·0001 (Wilcoxon matched‐pairs signed‐rank test). (b) The suppressive capacity of eosinophils from EoE patients is inferior to that of healthy controls. Boxes with whiskers indicate medians, quartiles and min/max range. **P <0·01 (Mann‐Whitney test). (c) Eosinophils from EoE patients demonstrate the same T helper (Th) cell suppressive pattern as eosinophils from healthy controls. The fraction of interferon (IFN)‐γ‐, interleukin (IL)‐4 and IL‐17A‐producing Th cells in CD3/CD28‐stimulated PBMC cultures, with or without eosinophils from EoE patients (n = 2, squares) or from healthy individuals (n = 2, circles), was determined after 2 days of culture.
Eosinophils from EoE patients and healthy individuals express FOXP3, but a higher fraction of the EoE eosinophils are FOXP3+
Next, we investigated if the eosinophils from EoE patients expressed the regulatory transcription factor FOXP3. To begin with, Tregs from a healthy subject were stained for FOXP3 (Fig. 2a). Next, eosinophils from a healthy individual were stained using the same protocol and were also found to express FOXP3 (Fig. 2b). Representative zebra and histogram plots indicating the percentage of cells that expressed FOXP3 and their intensities of staining (MFI) among CD4+ T cells and eosinophils from two healthy individuals are shown in Fig. 2a,b, respectively. It may be seen that whereas the FOXP3‐expressing T cells (Tregs) seem to form a distinct subset (Fig. 2a), this was not evident among the eosinophils (Fig. 2b). Compiled data from the EoE patients revealed that a higher percentage of their eosinophils expressed FOXP3 (median = 5·4%; 25–75 percentile = 2·7–21·7%) compared with the healthy eosinophils (median = 2·7%; 25–75 percentile = 1·0–10·4%), P = 0·047 (Fig. 3a). Moreover, half the EoE patients (eight of 17) had > 10% of FOXP3+ eosinophils. Approximately twice as high levels of FOXP3 protein were detected in the eosinophils from the EoE patients compared with the healthy individuals' eosinophils, measured as MFI (Fig. 3b). As a further verification of these findings, the levels of FOXP3 mRNA in highly purified eosinophils were determined by qPCR. Eosinophils derived from the blood of EoE patients and healthy donors alike expressed FOXP3 mRNA, but the FOXP3 mRNA levels were 3·6‐fold higher in the EoE patients (Fig. 3c).
Figure 2.
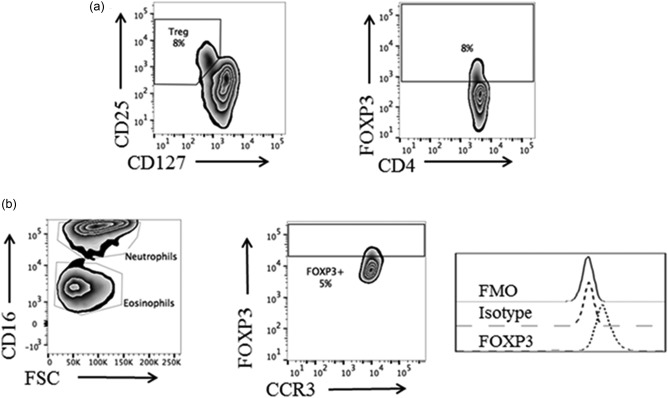
Identification of forkhead box protein 3 (FOXP3)‐expressing eosinophils in whole blood by flow cytometry. (a) Gating and staining strategy to identify FOXP3‐expressing regulatory T cells (Tregs) in whole blood. (b) Gating strategy and representative flow cytometry plots derived from a healthy donor showing the percentage of FOXP3+ eosinophils and FOXP3 staining intensity, measured as median fluorescence intensity (MFI). Isotype and fluorescence‐minus‐one (FMO) controls are included.
Figure 3.
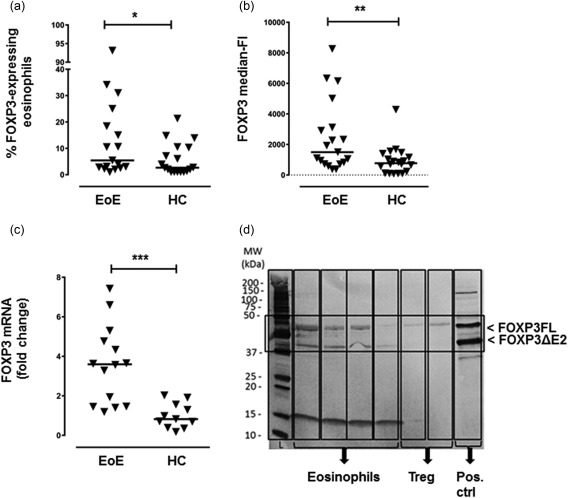
Forkhead box protein 3 (FOXP3) is expressed by blood eosinophils derived from patients with eosinophilic esophagitis (EoE) and healthy controls (HC). (a) Fraction of blood eosinophils that express the FOXP3 protein among EoE patients (n = 17) and healthy controls (n = 18). Each symbol denotes one individual. (*P = 0·047, Mann‐Whitney U‐test). (b) Levels of intracellular FOXP3 protein in eosinophils analysed by flow cytometry measured as median fluorescence intensity. Each symbol denotes one individual (**P = 0·01, Mann‐Whitney U‐test). (c) FOXP3 mRNA levels in blood eosinophils derived from EoE patients (n = 14) and healthy controls (n = 11) were analysed by real‐time quantitative polymerase chain reaction (RT‐qPCR). Each symbol denotes one individual. Data are presented as mean fold change in mRNA levels relative to those of the healthy subjects (***P = 0·0002, Mann‐Whitney U‐test). (a–c) Horizontal bars represent the median. (d) Human eosinophils express two isomers of FOXP3. Immunoblots of eosinophilic lysates derived from four individual eosinophilic oesophagitis (EoE) patients probed with α‐FOXP3 antibodies. Purified regulatory T cells (Tregs) and lysates of a FOXP3‐transfected cell line served as positive controls. The two bands corresponding to the FOXP3FL and FOXP3ΔE2 isomers are indicated by the box.
Eosinophils from EoE patients express two isoforms of human FOXP3
Immunoblot analyses were also performed using eosinophil lysates from four EoE patients to study FOXP3 expression further. The two major bands that correspond to two isoforms of human FOXP3, the full‐length isoform (FOXP3FL) and the isoform lacking exon 2 (FOXP3ΔE2), were both detected in all samples (Fig. 3d).
FOXP3 is found in the cytosol of eosinophils
Next, we wanted to determine the subcellular localization of FOXP3 protein in the eosinophils of EoE patients, i.e. if FOXP3 was localized in the nucleus or in the cytosol. To this end, 1000 eosinophils from EoE patients (n = 4) and from healthy controls (n = 6) were analysed using an imaging flow cytometer. Gating was based on in‐focus single cells and eosinophils were identified as side‐scatterhigh, CCR3high, CD16low cells with a lobulated nucleus (determined visually). For comparison, 1000 Tregs from healthy donors (n = 4) were also analysed, and the images showed that all the FOXP3 was found in the nucleus of the Tregs (Fig. 4a), as expected 24. In contrast, images and quantitative analysis of FOXP3 distribution revealed that more than half of the eosinophilic FOXP3 was localized to the cytosol (median = 65%; min–max = 58–66%), with the remainder detected in the nucleus (median = 35%; min–max = 34–42%) among the EoE patients (Fig. 4b). A similar distribution was noted for the eosinophils from the healthy donors, i.e. a median of 62% (min–max = 56–67%) of FOXP3 staining was found in the cytosol and the remainder (median = 38%; min–max = 33–44%) in the nucleus (Fig. 4c). To verify further the cellular distribution of eosinophilic FOXP3, we performed confocal microscopy on stained, cytospun eosinophils. We indeed observed FOXP3 in the eosinophilic cytosol (Fig. 5a).
Figure 4.
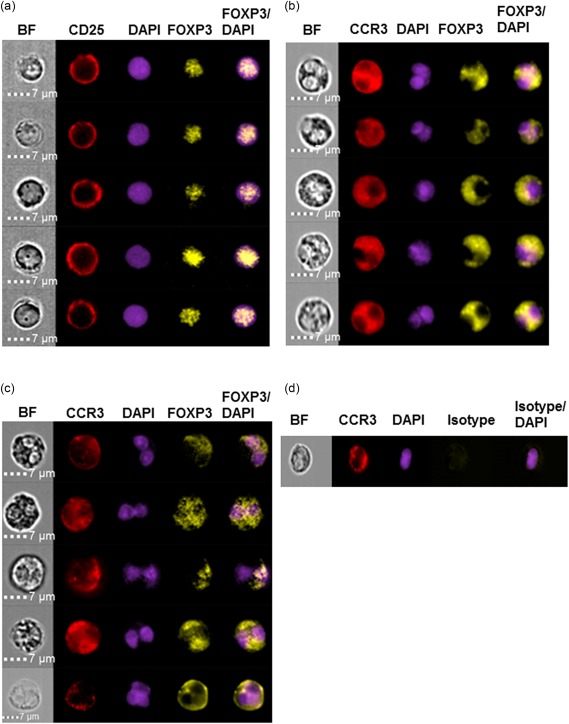
Intracellular distribution of forkhead box protein 3 (FOXP3) differs in regulatory T cells (Tregs) and eosinophils. ImageStream images of individual leukocytes stained for CD25, CCR3 and FOXP3, as well as overlay images, are shown. Bright field (BF) images and images of 4′,6‐diamidino‐2‐phenylindole (DAPI)‐stained cells are provided to identify the cytosol and nucleus in individual cells. (a) Representative ImageStream images depicting Tregs (n = 5) from a healthy donor showing the nuclear localization of FOXP3. (b,c) Representative ImageStream images of eosinophils (n = 5) from (b) an eosinophilic oesophagitis patient and (c) a healthy control. Most of the eosinophilic FOXP3 stain is found in the cytosol. (d) FOXP3 isotype control stain of an eosinophil from a healthy control.
Figure 5.
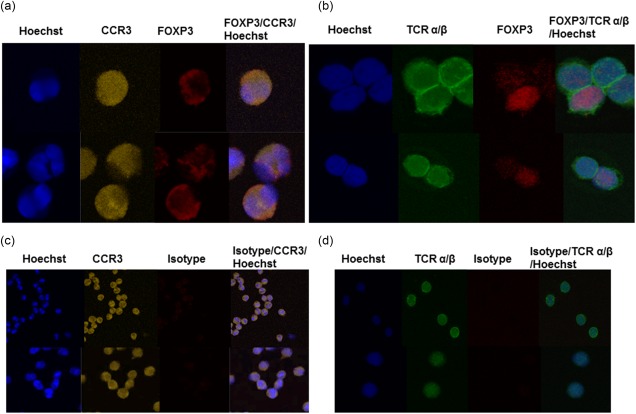
Confocal microscopy reveals cytosolic forkhead box protein 3 (FOXP3) in eosinophils. Eosinophils and CD4+ T cells from healthy individuals, stained for FOXP3, and CCR3 (eosinophils) or T cell receptor (TCR)‐α/β (CD4+ T cells), were cytopsun onto glass slides and FOXP3 expression detected by laser scanning confocal microscopy. Shown are representative images of (a) FOXP3+ eosinophils and (b) FOXP3+CD4+ T cells. Isotype control‐stained eosinophils (c) and CD4+ T cells (d) are shown for comparative purposes.
Discussion
In this study, we have shown that blood‐derived eosinophils from EoE patients have T cell suppressive capacity. We have shown previously that the eosinophils in the circulation of EoE patients have an altered phenotype compared with eosinophils from both healthy subjects and people with airborne allergy, which indicates that the cells in the bloodstream have received signals from the oesophagus and modified their molecular patterns accordingly 25. Moreover, the eosinophils in the blood of EoE patients essentially maintain their distinctive phenotype even after successful treatment with corticosteroids 26. Bullock et al. demonstrated a strong relationship between the levels of activated CD4+ T cells in the periphery and peak numbers of eosinophils in the oesophagus of EoE patients 20. Together, these findings indicate that EoE may, to a certain extent, be viewed as a systemic disease, and that it is relevant to study the function of eosinophils not only in the oesophageal mucosa but also in the circulation of EoE patients.
EoE is viewed currently as an emerging allergic disease, a condition characterized by dysregulated immune responses to food and/or airborne allergens 11; in addition, as‐yet unidentified antigens may be contributory. Against this background, our finding that the eosinophils from the EoE patients had reduced T cell suppressive capacity compared with the healthy eosinophils is intriguing. However, there are caveats to this observation: the difference in suppressive capacity between EoE patients and healthy individuals (51 versus 90%) might not be physiologically relevant, and the most suppressive eosinophils may have migrated from the blood into the oesophagus in the EoE patients. Furthermore, it is possible that there are T cell suppressive subsets among eosinophils, similar to the Treg subsets that exist among T cells.
Continuing our exploration of the immunoregulatory functions of eosinophils in EoE, we investigated whether eosinophils could express the canonical Treg transcription factor FOXP3. Realizing the controversial nature of this hypothesis, we used different methods to exclude the possibility of experimental artefacts. First, by using conventional flow cytometry, we demonstrated FOXP3‐expressing cells within the granulocyte gate. This was verified by imaging flow cytometry, where we could ascertain that each of the FOXP3+ cells were eosinophils, based on the lobular shape of the nucleus, together with a high side‐scatter and expression of the eotaxin receptor CCR3. We could also demonstrate eosinophilic FOXP3 mRNA expression by using qPCR. Our data indicate that, similar to human Tregs and conventional T cells, it is only a fraction of the eosinophil population that expresses FOXP3. Approximately 5% of the eosinophils expressed FOXP3 among the EoE patients, although the variation was considerable; half the patients had more than 10% FOXP3‐expressing eosinophils. This is comparable to the frequency of FOXP3+ Tregs in peripheral blood, which is normally 5–10% of all CD4+ T cells 27. FOXP3‐positive eosinophils were less frequent among the healthy controls, constituting approximately 3% of all eosinophils. We also report that eosinophils akin to Tregs express two isoforms of human FOXP3 protein, both of which appear to mediate suppressive activity in human Tregs, and to act in concert 28.
Another interesting finding was that the subcellular distribution of FOXP3 differed between eosinophils and Tregs such that a significant proportion of eosinophilic FOXP3 was localized to the cytosol, whereas all the FOXP3 in the Tregs was confined to the nucleus; the latter is already well known 24. The same pattern of FOXP3 distribution as from the healthy controls was seen among the eosinophils from the EoE patients. The predominantly cytosolic distribution of FOXP3 in eosinophils is reminiscent of that seen in human effector T cells (CD4+CD25–), which can express FOXP3 transiently upon activation 24. Importantly, it seems that it is only nuclear FOXP3 that endows T cells with suppressive capacity 24. Thus, our findings of a predominantly cytoplasmic location of eosinophilic FOXP3 is in line with what has been shown for human effector T cells 24, 29, and could signify that FOXP3 may also be an activation marker in eosinophils.
The presence of FOXP3 mRNA and protein in eosinophils has not been reported previously. However, FOXP3 mRNA has been identified in leucocytes besides T cells, namely in cell lines derived from B, NK and non‐specified myeloid cells, although FOXP3 protein was not detected in the non‐T cells 30. In addition, FOXP3 expression has been demonstrated in various epithelial cell types in mice 31; if this holds true for humans is yet to be determined. A recent genomewide analysis identified more than 5000 potential target genes of human FOXP3 solely in human natural Tregs 32, illustrating the extreme complexity of elucidating the putative functions of this transcription factor in human health and disease.
FOXP3 expression has been investigated in the context of EoE prior to this study, but with a focus upon T cells. Two studies reported that the numbers of FOXP3‐expressing T cells were increased in the esophagi of children with EoE relative to control children 33, 34, whereas a study of adult patients with EoE reported a relative lack of FOXP3‐expressing T cells in the patients compared with controls 35. Nevertheless, it is not possible to draw conclusions regarding the suppressive capacity of cells based on phenotype, and hence it is still unclear if FOXP3‐expressing Tregs are quantitatively and/or qualitatively deficient in EoE.
The strength of this study is our well‐characterized cohort, which may be said to be representative for EoE patients, as the majority of the study participants were male and had a high incidence of sensitization to food and inhalant allergens. Further, the recruitment of our cohort from ear, nose and throat (ENT) departments is reflected in the predominance of patients suffering from dysphagia. Moreover, we present new and original data suggesting that the immunoregulatory capacity of eosinophils may be impaired in EoE, adding to the knowledge regarding possible immunopathogenetic mechanisms behind this new disease. A limitation of the study is that we have focused exclusively upon blood eosinophils. However, it is somewhat challenging to conduct functional studies using eosinophils isolated from oesophageal biopsies in view of the size of the biopsies that may be collected from patients and the paucity of cells therein. Moreover, it is impossible to conduct functional studies of oesophageal eosinophils from healthy individuals for comparative purposes, as the oesophagus is the only part of the GI tract that is naturally devoid of eosinophils 21.
To conclude, eosinophils in the circulation of EoE patients have T cell suppressive capacity and express FOXP3. However, these eosinophils appear to have impaired T cell suppressive capacity compared with healthy eosinophils. Whether or not oesophageal eosinophils in EoE patients also have reduced suppressive capacity remains to be determined. Finally, we report that human eosinophils can express FOXP3, but that it might be primarily an activation marker, rather than a suppressive molecule in patients with EoE, based on its cytosolic distribution.
Disclosure
The authors declare no financial or commercial disclosures.
Author contributions
C. L. performed most experiments, compiled data and contributed to the paper. J. W. performed some of the experiments, compiled data and wrote parts of the paper. H. B. and M. B. recruited the patients and provided the clinical data. M. I. designed the PCR and contributed to the paper. A. W. was instrumental in the imaging flow cytometry experiments. R. S. participated in the planning of the paper. C. W. planned and supervised the entire study and wrote the paper.
Acknowledgements
This work was funded by grants from the LUA‐ALF (71580), Strategic ALF/Transplantation (74080), Cancer and Allergy Foundation research grant (149781), Health and Medical Care Committee of the Regional Executive Board of Region Västra Götaland (96490), Inga Britt and Arne Lundberg Research Foundation and the University of Gothenburg.
References
- 1. Peterson CG, Skoog V, Venge P. Human eosinophil cationic proteins (ECP and EPX) and their suppressive effects on lymphocyte proliferation. Immunobiology 1986; 171:1–13. [DOI] [PubMed] [Google Scholar]
- 2. Odemuyiwa SO, Ghahary A, Li Y et al Cutting edge: human eosinophils regulate T cell subset selection through indoleamine 2,3‐dioxygenase. J Immunol 2004; 173:5909–13. [DOI] [PubMed] [Google Scholar]
- 3. Harfi I, Schandene L, Dremier S, Roufosse F. Eosinophils affect functions of in vitro‐activated human CD3‐CD4+ T cells. J Transl Med 2013; 11:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersson J, Cromvik J, Ingelsten M et al Eosinophils from hematopoietic stem cell recipients suppress allogeneic T cell proliferation. Biol Blood Marrow Transplant 2014; 20:1891–8. [DOI] [PubMed] [Google Scholar]
- 5. Sugawara R, Lee EJ, Jang MS et al Small intestinal eosinophils regulate Th17 cells by producing IL‐1 receptor antagonist. J Exp Med 2016; 213:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kubach J, Lutter P, Bopp T et al Human CD4+CD25+ regulatory T cells: proteome analysis identifies galectin‐10 as a novel marker essential for their anergy and suppressive function. Blood 2007; 110:1550–8. [DOI] [PubMed] [Google Scholar]
- 7. Furuta GT, Katzka DA. Eosinophilic esophagitis. N Engl J Med 2015; 373:1640–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Attwood SE, Smyrk TC, Demeester TR, Jones JB. Esophageal eosinophilia with dysphagia. A distinct clinicopathologic syndrome. Dig Dis Sci 1993; 38:109–16. [DOI] [PubMed] [Google Scholar]
- 9. Giriens B, Yan P, Safroneeva E et al Escalating incidence of eosinophilic esophagitis in Canton of Vaud, Switzerland, 1993–2013: a population‐based study. Allergy 2015; 70:1633–9. [DOI] [PubMed] [Google Scholar]
- 10. Simon D, Radonjic‐Hosli S, Straumann A, Yousefi S, Simon HU. Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy 2015; 70:443–52. [DOI] [PubMed] [Google Scholar]
- 11. Cianferoni A, Spergel J. Eosinophilic esophagitis: a comprehensive review. Clin Rev Allergy Immunol 2016; 50:159–74. [DOI] [PubMed] [Google Scholar]
- 12. Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras CA, Katzka DA. ACG clinical guideline: evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE). Am J Gastroenterol 2013; 108:679–92. [DOI] [PubMed] [Google Scholar]
- 13. Papadopoulou A, Koletzko S, Heuschkel R et al Management guidelines of eosinophilic esophagitis in childhood. J Pediatr Gastroenterol Nutr 2014; 58:107–18. [DOI] [PubMed] [Google Scholar]
- 14. Mishra A, Schlotman J, Wang M, Rothenberg ME. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J Leukoc Biol 2007; 81:916–24. [DOI] [PubMed] [Google Scholar]
- 15. Desai TK, Stecevic V, Chang CH, Goldstein NS, Badizadegan K, Furuta GT. Association of eosinophilic inflammation with esophageal food impaction in adults. Gastrointest Endosc 2005; 61:795–801. [DOI] [PubMed] [Google Scholar]
- 16. Le‐Carlson M, Seki S, Abarbanel D, Quiros A, Cox K, Nadeau KC. Markers of antigen presentation and activation on eosinophils and T cells in the esophageal tissue of patients with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2013; 56:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lucendo AJ, Navarro M, Comas C et al Immunophenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am J Surg Pathol 2007; 31:598–606. [DOI] [PubMed] [Google Scholar]
- 18. Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2‐type allergic inflammatory response. J Allergy Clin Immunol 2001; 108:954–61. [DOI] [PubMed] [Google Scholar]
- 19. Teitelbaum JE, Fox VL, Twarog FJ et al Eosinophilic esophagitis in children: immunopathological analysis and response to fluticasone propionate. Gastroenterology 2002; 122:1216–25. [DOI] [PubMed] [Google Scholar]
- 20. Bullock JZ, Villanueva JM, Blanchard C et al Interplay of adaptive th2 immunity with eotaxin‐3/c‐C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2007; 45:22–31. [DOI] [PubMed] [Google Scholar]
- 21. Liacouras CA, Furuta GT, Hirano I et al Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol 2011; 128:3–20. [DOI] [PubMed] [Google Scholar]
- 22. Pfaffl MW. A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roederer M. Compensation in flow cytometry. Curr Protoc Cytom 2002; Chapter 1:1.14.1‐1.14.20. [DOI] [PubMed] [Google Scholar]
- 24. Magg T, Mannert J, Ellwart JW, Schmid I, Albert MH. Subcellular localization of FOXP3 in human regulatory and nonregulatory T cells. Eur J Immunol 2012; 42:1627–38. [DOI] [PubMed] [Google Scholar]
- 25. Johnsson M, Bove M, Bergquist H et al Distinctive blood eosinophilic phenotypes and cytokine patterns in eosinophilic esophagitis, inflammatory bowel disease and airway allergy. J Innate Immun 2011; 3:594–604. [DOI] [PubMed] [Google Scholar]
- 26. Lingblom C, Bergquist H, Johnsson M et al Topical corticosteroids do not revert the activated phenotype of eosinophils in eosinophilic esophagitis but decrease surface levels of CD18 resulting in diminished adherence to ICAM‐1, ICAM‐2, and endothelial cells. Inflammation 2014; 37:1932–44. [DOI] [PubMed] [Google Scholar]
- 27. Ito T, Hanabuchi S, Wang YH et al Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity 2008; 28:870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allan SE, Passerini L, Bacchetta R et al The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest 2005; 115:3276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morgan ME, van Bilsen JH, Bakker AM et al Expression of FOXP3 mRNA is not confined to CD4+CD25+ T regulatory cells in humans. Hum Immunol 2005; 66:13–20. [DOI] [PubMed] [Google Scholar]
- 30. Yamamoto M, Tsuji‐Takayama K, Suzuki M et al Comprehensive analysis of FOXP3 mRNA expression in leukemia and transformed cell lines. Leuk Res 2008; 32:651–8. [DOI] [PubMed] [Google Scholar]
- 31. Chen GY, Chen C, Wang L, Chang X, Zheng P, Liu Y. Cutting edge: broad expression of the FoxP3 locus in epithelial cells: a caution against early interpretation of fatal inflammatory diseases following in vivo depletion of FoxP3‐expressing cells. J Immunol 2008; 180:5163–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sadlon TJ, Wilkinson BG, Pederson S et al Genome‐wide identification of human FOXP3 target genes in natural regulatory T cells. J Immunol 2010; 185:1071–81. [DOI] [PubMed] [Google Scholar]
- 33. Fuentebella J, Patel A, Nguyen T et al Increased number of regulatory T cells in children with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2010; 51:283–9. [DOI] [PubMed] [Google Scholar]
- 34. Tantibhaedhyangkul U, Tatevian N, Gilger MA, Major AM, Davis CM. Increased esophageal regulatory T cells and eosinophil characteristics in children with eosinophilic esophagitis and gastroesophageal reflux disease. Ann Clin Lab Sci 2009; 39:99–107. [PubMed] [Google Scholar]
- 35. Stuck MC, Straumann A, Simon HU. Relative lack of T regulatory cells in adult eosinophilic esophagitis – no normalization after corticosteroid therapy. Allergy 2011; 66:705–7. [DOI] [PubMed] [Google Scholar]


