Summary
Group 3 innate lymphoid cells (ILC3), defined by expression of the transcription factor retinoid‐related orphan receptor γt, play key roles in the regulation of inflammation and immunity in the gastrointestinal tract and associated lymphoid tissues. ILC3 consist largely of two major subsets, NCR + ILC3 and LTi‐like ILC3, but also demonstrate significant plasticity and heterogeneity. Recent advances have begun to dissect the relationship between ILC3 subsets and to define distinct functional states within the intestinal tissue microenvironment. In this review we discuss the ever‐expanding roles of ILC3 in the context of intestinal homeostasis, infection and inflammation – with a focus on comparing and contrasting the relative contributions of ILC3 subsets.
Keywords: group 3 innate lymphoid cells, innate lymphoid cell, lymphoid tissue‐inducer cells, mucosal immunology
Abbreviations
- CHILP
common helper ILC progenitor
- GM‐CSF
granulocyte–macrophage colony‐stimulating factor
- ID2
inhibitor of DNA‐binding protein
- IFN‐γ
interferon‐γ
- IL
interleukin
- ILC
innate lymphoid cell
- ILCP
ILC progenitor
- ILF
isolated lymphoid follicule
- LTi
lymphoid tissue‐inducer
- NCR+
natural‐cytotoxicity‐receptor‐positive
- NK cell
natural killer cell
- RORγt
retinoid‐related orphan receptor γt
- SFB
segmented filamentous bacteria
- Th
T helper
- TNF‐α
tumour necrosis factor‐α
The innate lymphoid cell family
The innate lymphoid cell (ILC) family has been the focus of intense investigation over recent years and ILC have emerged as key players in immune responses within multiple organs, particularly barrier surfaces such as the lung, skin and gastrointestinal tract.1, 2, 3, 4 The current nomenclature broadly divides ILC into three subgroups that are defined by their master transcription factor usage and cytokine‐producing capacity, and which closely mirror CD4+ T helper (Th) cell subsets.3 However, in contrast to T cells, ILC lack antigen‐specific receptors and do not require de novo proliferation and polarization for cytokine production, instead acting as ‘pre‐primed’ terminal effector cells that constitutively reside in barrier tissues and respond to alarmins and cytokine signals released following tissue damage.
Group 1 ILC (ILC1), which include classical natural killer (NK) cells as well as bona fide non‐NK ILC1, respond to interleukin‐12 (IL‐12), IL‐15 and IL‐18 and are defined by their expression of the transcription factor T‐bet, production of the cytokines interferon‐γ (IFN‐γ) and tumour necrosis factor‐α and mediate immune responses to intracellular pathogens and tumours.3, 5, 6, 7 Group 2 ILC (ILC2) respond to tissue‐derived alarmins including IL‐25 and IL‐33, are characterized by their expression of GATA‐3, produce the cytokines IL‐5, IL‐9 and IL‐13 and contribute to immune responses against multicellular pathogens (such as nematode parasites) as well as the pathogenesis of type 2 inflammatory diseases including asthma and atopic dermatitis.1, 2, 3
Retinoid‐related orphan receptor γt (RORγt) ‐expressing group 3 ILC (ILC3) respond to signals from tissue‐resident myeloid cells, such as IL‐1β and IL‐23, by producing cytokines including IL‐17A, IL‐17F and IL‐22.1, 2, 3, 4, 8 In the adult mouse, ILC3 can be further subdivided into at least two major subsets – the first is defined by the expression of the NK cell‐associated receptor NKp46 and has been termed natural‐cytotoxicity‐receptor‐positive ILC3 (NCR+ ILC3). The second major subset is defined by their expression of the chemokine receptor CCR6 and phenotypically mirrors fetal lymphoid tissue‐inducer cells (LTi), and so have been termed LTi‐like ILC3. Despite both subsets sharing significant functional overlap, increasing evidence suggests that these subsets also have distinct, non‐redundant functions.
Group 3 innate lymphoid cell development
All ILC subsets (ILC1, ILC2 and ILC3), as well as adaptive lymphoid cells (B and T cells), are derived from a common lymphoid precursor (CLP) that is present in the fetal liver and adult bone marrow.9, 10 Recent studies have elegantly defined the presence of common ILC precursors that develop downstream of the common lymphoid precursor, and which have lost the potential to develop into T and B cells.5, 11, 12, 13 The initial differentiation of ILC‐restricted progenitors from multipotent common lymphoid precursors is dependent upon IL‐7 and characterized by expression of the inhibitor of DNA‐binding protein (ID2)14, 15 (Fig. 1). ID2+ progenitors give rise to all IL‐7Rα‐expressing ILC and have been termed the ‘common helper ILC progenitor’ (CHILP) to distinguish these cells from NK cell progenitors5 (Fig. 1). Progressive commitment to the ILC lineage is also dependent, to varying degrees, on a multitude of transcription factors including GATA‐3, TOX, TCF‐1 and NFIL316, 17, 18, 19, 20, 21 (and reviewed in detail in references 10 and 22).
Figure 1.
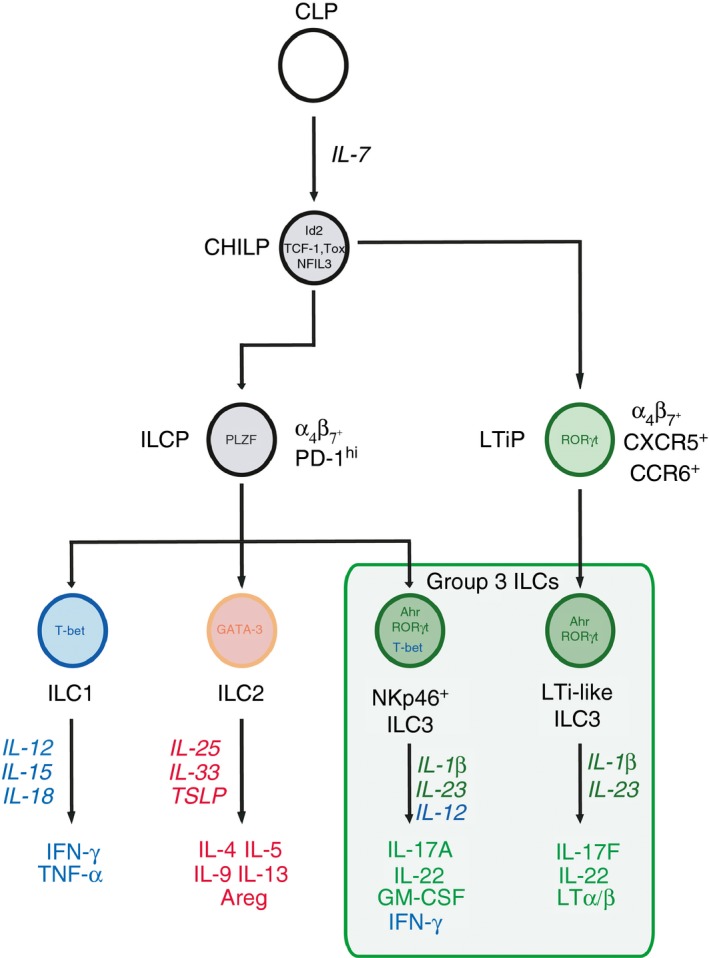
Ontogeny of group 3 innate lymphoid cells (ILC3). All ILC subsets are derived from a common lymphoid progenitor (CLP), Initial commitment towards an ILC fate is associated with up‐regulation of the transcription factor inhibitor of DNA‐binding protein (ID2) and is dependent upon interleukin‐7 (IL‐7). This progenitor population gives rise to all IL‐7receptor (IL‐7R) expressing ILC family members and has been termed the common ‘helper’ ILC progenitor (CHILP). Recent evidence suggests that bifurcation of progenitors occurs downstream of the CHILP. Acquisition of promyelocytic leukaemia zinc finger (PLZF) expression and expression of programmed cell death protein‐1 (PD‐1) mark further commitment to the ILC lineage (ILCP). However, while ILCP give rise to ILC1, ILC2 and NKp46+ ILC3s, these cells fail to generate lymphoid tissue‐inducer (LTi)‐like ILC3 progeny. In contrast, a distinct LTi‐precursor (LTiP) lacking PLZF‐expression is defined as CXCR5+ α 4 β 7 + and CCR6+ and gives rise to fetal LTi and adult LTi‐like ILC3. The bifurcation of group 3 ILC subsets during development may imprint differential transcriptional programmes that underlie the function of mature ILC3 subsets in the periphery.
Further commitment towards individual ILC subgroups is associated with transition through a phase characterized by promyelocytic leukaemia zinc finger (PLZF) expression (encoded by Zbtb16) and high surface expression of programmed cell death protein‐1 (PD‐1) (Fig. 1).11, 23 Whereas ID2+ CHILP give rise to all major ILC subsets, PLZF‐expressing progenitors (ILCP) gave rise to ILC1, ILC2 and NKp46+ ILC3 – but not LTi‐like ILC3 – suggesting that PLZF+ ILCP have lost the capacity to generate LTi‐like ILC3 progeny.11 In line with this finding, a bifurcation of ILC progenitor programming occurs before acquisition of PLZF expression that generates a distinct LTi precursor that is Id2+ PLZF− and characterized by the expression of CXCR5, α 4 β 7 and CCR6 (Fig. 1).12, 13 These studies suggest that adult LTi‐like ILC3 may be developmentally divergent from other ILC, including NKp46+ ILC3, and indicate that these two ILC3 subsets may differ to a greater degree than currently appreciated.
Expression of RORγt is essential for the development and maturation of all members of the ILC3 family, including fetal LTi, adult LTi‐like ILC3 and NKp46+ ILC3.24, 25, 26, 27 RORγt‐expressing progenitors are present in the fetal liver of mice.14 Although much of ILC3 development is thought to take place in this compartment before birth, ILC3 may also develop from adult bone marrow ILCP and immature committed ILC3 precursors have the capacity to migrate from the bone marrow to the periphery where they undergo final maturation.14, 28, 29, 30 Similarly, human adult CD34+ haematopoietic precursor cells have been described that exhibit RORγt expression and an ILC3 transcriptional signature,29 while human CD34+ RORγt+ precursors have been found in adult tonsils and the gut.28, 29, 31 Hence, in addition to the generation of fully committed mature ILC3 subsets during fetal development, further ILC3 maturation of precursors may occur locally in the tissue following birth.
Tissue‐resident ILC3: from cradle to grave
Group 3 ILC are among the first immune cells to seed intestinal tissue. Before birth, fetal LTi‐like ILC3 are crucial initiators of peripheral secondary lymphoid tissue organogenesis and, in concert with endothelial lymphoid tissue organizer populations,32 are required for the generation of lymphoid tissues, including the mesenteric lymph node and Peyer's patches in the small intestine24, 33 (reviewed in detail in ref. 34). LTi act to orchestrate the generation of nascent lymphoid tissue through a process that requires the interaction between LTi cells and stromal organizer cells and that is dependent upon LTi‐expressed lymphotoxin.34, 35
Similarly, interactions between adult LTi‐like ILC3 and stromal cells are also important for lymphoid tissue formation later in life, as evident from the formation of discrete LTi‐like ILC3 clusters termed cryptopatches in the intestinal lamina propria that form 1–2 weeks after birth.34, 36 Cryptopatches predominantly contain LTi‐like ILC3 and dendritic cells organized around stromal cell populations, and are situated close to the bottom of intestinal crypt structures. Cryptopatch‐resident ILC3 maintain the local crypt stem cell pool and regulate tissue repair following inflammation, chemotherapy or transplantation.37, 38, 39 In addition, cryptopatches act as a site for B‐cell recruitment to form mature tertiary lymphoid structures known as isolated lymphoid follicules (ILFs).36, 40, 41 ILFs are important sites for production of IgA against intestinal bacteria and ILF hyperplasia occurs in the context of inflammation or bacterial outgrowth,41 suggesting that ILFs are ILC3‐regulated inducible lymphoid tissues required for orchestration of local responses to commensal bacteria.
Commensal microbiota‐derived signals are critical in driving ILC3 recruitment and expansion in the intestine.27, 30 Before birth, ILC3 seeding of the fetal intestine may be modulated by passively transmitted maternal signals, including those derived from the microbiota.42 Aryl hydrocarbon receptor (Ahr) ligands bound by maternal antibodies cross the placenta and increase the number of NKp46+ ILC3 in the uncolonized fetal intestine.42 Additional environmental signals experienced in utero can further modulate ILC3. Similarly, maternal retinoids were found to be critical for the development of LTi cells in the developing fetus by regulating transcription of RORγt.43 Dietary metabolites also play key roles in supporting ILC3 function and stability. Vitamin A deprivation results in a loss of intestinal ILC3 in adult mice, which can be reversed upon supplementation of diet with the vitamin A‐derived metabolite retinoic acid.44, 45 Similarly, Ahr acts to sense soluble aromatic hydrocarbons – present in the diet and produced by commensal bacteria – and is essential for the development of both LTi‐like and NKp46+ ILC3, as well as IL‐22 production.14, 27, 28, 46, 47, 48, 49, 50, 51 Together these signals tune ILC3 numbers and responses and establish bidirectional interactions between ILC3 and the commensal microbiota during the initial colonization of barrier tissue sites.
In contrast, relatively little is known about the fate or lifespan of ILC3 subsets. ILC3 have a long half‐life in tissues and lymph nodes and are largely replenished locally within the tissue.30, 52 Surprisingly, ILC3 can maintain their phenotype and function even in the absence of RORγt expression and persist for a significant length of time.53 Nonetheless, changes in dietary metabolites, chronic inflammation or infectious insult result in disruption of ILC3 effector functions or even irreversible depletion of ILC from the intestinal tissue altogether.54, 55 Further studies are required to understand how ILC3 numbers and functions are maintained and how they can be restored following damage or infection‐induced perturbation.
ILC3 plasticity and heterogeneity
Although the developmental networks that drive ILC3 ontogeny have been extensively defined in recent years, the degree to which mature ILC3 subsets differ transcriptionally and functionally in peripheral tissues is only now beginning to emerge. Recent high‐resolution epigenetic and transcriptomic analyses of human and murine ILC3 have begun to clarify subset‐specific functional and transcriptional phenotypes, while revealing that ILC3 exhibit a higher degree of plasticity and heterogeneity than previously appreciated.56, 57, 58, 59, 60 Despite sharing many overlapping transcriptional pathways LTi‐like and NKp46+ ILC3 subsets exhibit considerable differences in chromatin accessibility and predicted transcription factor occupancy among accessible gene regulatory elements.58, 59, 60 Differences between these two canonical ILC3 subsets are further highlighted on the transcriptional level.56, 60 Despite significant overlap in expression of core ILC3 member genes, including Rorc and Il23r, NKp46+ ILC3 and LTi‐like ILC3 also displayed subset‐restricted gene expression56 (Fig. 2). NKp46+ ILC3 are characterized by subset‐restricted expression of multiple genes required for their development and maturation, such as Il12rb2, Tbx21 and Notch1,51, 61 and are defined by expression of eponymous NK cell‐associated receptors. Recent evidence suggests that NKp46 expression may act as a novel pattern recognition molecule to facilitate responses to fungal pathogens,62 whereas engagement of NKp44 on human ILC3 results in pro‐inflammatory cytokine production.63 Although NCR+ ILC3 are largely thought to be non‐cytotoxic cells, NKp46+ ILC3 express Gzmc – which encodes a murine granzyme molecule known to induce cell death.60, 64 This observation is in line with a considerable transcriptional overlap between NKp46+ ILC3 and ILC1, which also exhibit a degree of cytotoxic capacity.65 Indeed NKp46+ ILC3 were found to share more transcripts with ILC1s than with LTi‐like ILC3.56
Figure 2.
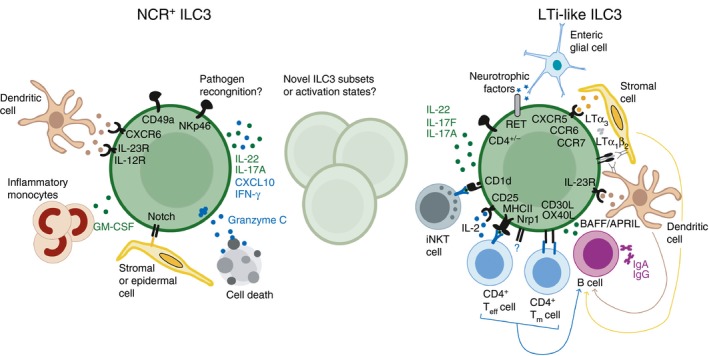
Group 3 innate lymphoid cell (ILC3) subset‐defining phenotypes and functions. Fully mature ILC3 subsets in peripheral tissues are broadly categorized into two major subsets; NKp46+ ILC3 (left panel) and lymphoid tissue‐inducer (LTi) ‐like ILC3 (right panel). Although both subsets express retinoid‐related orphan receptor γt (ROR γt) and share some core functions, emerging evidence suggests subset‐restricted functions. Phenotypic surface markers that typically used to define ILC3 subsets (bold) include natural killer (NK) ‐cell‐associated receptors, such as NKp46 and CD49a, for natural‐cytotoxicity‐receptor‐positive (NCR +) ILC3s, whereas LTi‐like ILC3s are primarily characterized by their expression of CCR6 and variable expression of CD4. Differential localization of NKp46+ ILC3 and LTi‐like ILC3 to the lamina propria and lymphoid tissues, respectively, is determined by differential chemokine receptor profiles. The difference in ILC3 subset localization is indicative of functional specialization. LTi‐like ILC3 express multiple molecules that endow this subset with the ability to indirectly and directly modulate innate‐like and adaptive immune responses in lymphoid tissues. In contrast, NCR + ILC3 appear to largely lack the ability to modulate adaptive immune responses but exhibit unique interactions with populations of myeloid and stromal cells in the tissue that drive the relatively pro‐inflammatory phenotype of this subset. Finally, although the majority of mature ILC3s exhibit NCR + or LTi‐like phenotypes recent transcriptomic studies indicate that multiple intermediate activation states may exist, which are likely determined by transition through metabolic states and differing environmental niches.
The LTi‐like ILC3 demonstrate subset‐specific expression of characteristic genes such as Cd4 and Ccr6, but also express additional genes indicative of novel functionality in LTi‐like ILC3 (Fig. 2). Among these is the gene encoding RET – a neuroregulatory receptor – which is important for LTi‐mediated formation of Peyer's patches.66 LTi‐like ILC3 expression of RET endows them with the ability to respond to neurotrophic factors released from enteric glial cells in response to microbial signals. Neurotrophic factor signalling reinforced production of IL‐22 and modulation of epithelial barrier integrity,67 suggesting that LTi‐like ILC3 may have the unique potential to respond to environmental cues from the enteric nervous system. Indeed, inflammation‐induced tertiary lymphoid tissue formation – which is regulated by ILC3 – requires vagal nerve innervation in the intestine.68 In addition, LTi‐like ILC3 were also found to specifically express Neuropilin‐1, an immunoregulatory factor that is expressed by subsets of regulatory T cells,56, 69, 70 which may indicate additional novel and subset‐restricted roles for LTi‐like ILC3.
Despite the identification of ILC3 subset‐specific functions, recent evidence has also highlighted a significant degree of heterogeneity among tissue‐resident ILC3, which blurs the boundaries of current subset classification. Single cell‐sequencing analyses of all ILC present in the small intestinal lamina propria of mice distinguished five discrete transcriptional states among ILC3.60 Several transcripts identified have not previously been associated with a function in ILC3 and encode genes involved in metabolism, as well as chemokines and growth factors (Fig. 2 and60). Nonetheless, comparison mapping of bulk sequencing data of sorted NKp46+ ILC3 and LTi‐like ILC3 subsets demonstrated that the two major ILC3 subsets span several transcriptional states, suggesting a dynamic interchange between transcriptional or functional states in response to environmental cues and/or cellular interactions.60 This is in line with the finding that intestinal RORγt‐expressing ILC demonstrate significant plasticity (Fig. 3). In addition to CCR6+ LTi‐like ILC3 and NKp46+ ILC3, the small intestine contains a population of ‘double‐negative’ RORγt+ CCR6− NKp46− ILC3.61, 71 ‘Double‐negative’ ILC3 represent intestinal precursors of the NKp46+ ILC3 subset, and acquire NKp46 and IFN‐γ expression in a process driven by up‐regulation of T‐bet and dependent upon IL‐23, Notch signalling and the microbiota.51, 61, 71 In addition, fate‐mapping studies revealed the presence of NKp46+ T‐bet+ ‘ex‐ILC3’ that had lost RORγt‐expression in response to pro‐inflammatory cytokine stimulation and that functionally and phenotypically resemble ILC1 5, 61, 71 (Fig. 3). Complimentary findings in human tissue samples demonstrated reversible conversion between NCR+ ILC3 subsets and ILC1‐like ‘ex‐ILC3’ dependent on the local cytokine milieu, which was determined by tissue‐resident myeloid populations.72, 73 These findings demonstrate significant plasticity among CCR6− ILC3, which is both tuneable and reversible. However, the relationship of this plastic ILC3 lineage to LTi‐like ILC3 is unclear.
Figure 3.
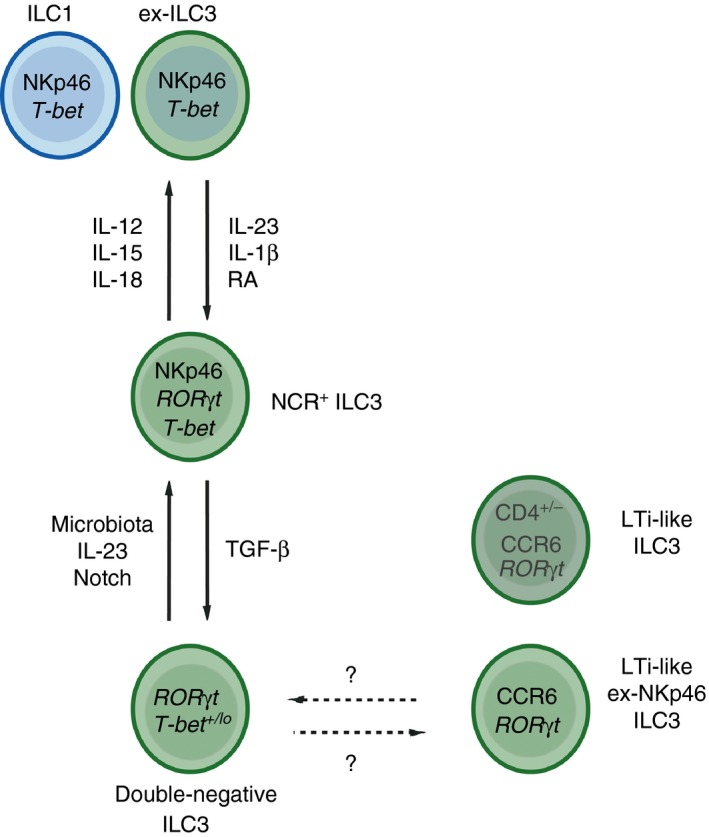
Plasticity and heterogeneity of intestinal group 3 innate lymphoid cell (ILC3). In addition to NKp46+ ILC3 and CCR6+ lymphoid tissue‐inducer (LTi) ‐like ILC3 a ‘double‐negative’ NKp46− CCR6− ILC3 population is present in the intestine. These cells act as a plastic precursor pool that gives rise to NKp46+ ILC3s in a reversible manner in response to local signals, which is regulated by a gradient of T‐bet expression. In addition, NKp46+ ILC3s may further lose retinoid‐related orphan receptor γt (ROR γt) expression in response to inflammatory cytokines and take on a phenotype that mirrors ILC1. These ‘ex‐ILC3’ retain their plasticity and are able to revert to an NKp46+ ILC3 or ‘double‐negative’ precursor phenotype upon alterations in the local cytokine milieu. LTi‐like ILC3 are thought to develop from a distinct progenitor. However, a small proportion of CCR6+ ILC3 display a history of NKp46 expression (ex‐NKp46 ILC3), suggesting that they may also be derived from tissue‐resident plastic ILC3s, although these cells largely lack expression of other LTi‐associated markers such as CD4 and MHCII. These findings blur the boundaries of ILC3 subset ontogeny and necessitate further studies to determine the relationship between bona fide LTi‐like ILC3 and those derived from plastic precursors. The high degree of reversible plasticity among ILC3s suggests a highly dynamic regulation of phenotype and function in response to environmental and infectious cues. This may be beneficial and allow transient increases in ILC3s with a pro‐inflammatory capacity when required, while allowing reversion to a more tolerogenic balance of ILC3 subsets following resolution of the infection or inflammatory insult.
Although CCR6+ LTi‐like ILC3 are thought to be relatively stable and ontologically distinct, recent evidence suggests that a proportion of ILC3 expressing a CCR6+ LTi‐like phenotype may also be derived from the plastic population of ILC3 in the intestine. Fate‐mapping of the NKp46 locus revealed populations of ‘ex‐NKp46 ILC3’ that no longer expressed NKp46, many of which had reverted to a ‘double‐negative’ ILC3 phenotype.74, 75 Transition between phenotypes was regulated locally via Notch and suppressed by transforming growth factor‐β, suggesting that the NKp46+ ILC3 phenotype is unstable and dynamically regulated.74, 75 Surprisingly, a small proportion of CD4− CCR6+ ILC3 also exhibited a history of NKp46 expression, suggesting that at least some ILC3 with an LTi‐like phenotype may be derived from the same precursor as NKp46+ ILC3, rather than from a pure LTi‐precursor.74 Taken together, these findings highlight the heterogeneity and plasticity of ILC3 in the intestine, challenge the distinction between mature ILC3 subsets and highlight the difficulty of categorizing ILC3 subsets based on a limited numbers of surface markers.
In humans it has been challenging to address the full extent of ILC3 heterogeneity and to identify human ILC3 subsets that are directly analogous to murine LTi‐like ILC3 and NKp46+ ILC3 subsets. Recent single‐cell sequencing studies using non‐biased clustering of transcriptome data among human ILC identified three distinct RORγt‐expressing ILC3 subsets.57 Although all human ILC3 subsets shared expression of Rorc, Tox, Ahr, Tcf7 and Notch‐induced genes, distinct subsets could be distinguished by the expression of either NKp44, HLA‐associated genes or SELL (which encodes CD62L).57 The latter subset also expressed CD45RA and responded poorly to re‐stimulation, suggesting the presence of a ‘naive’ or undifferentiated ILC3 phenotype in humans. Interestingly, murine ‘ex‐NKp46 ILC3’ that have reverted to a ‘double‐negative’ phenotype also demonstrate relatively suppressed cytokine‐producing capacity.74 Hence, further investigation is required to determine the relationship of this ‘naive’ human ILC3 subset to other established human ILC3 phenotypes, particularly whether this population is analogous to the plastic ‘double‐negative’ ILC3 precursor found in mice.
Both mouse and human single‐cell sequencing approaches highlighted expression of MHCII as a defining marker of a distinct ILC3 subset.56, 60 MHCII has previously been identified on adult LTi‐like ILC3 in mice and humans,76 suggesting that bona fide LTi‐like ILC3 may be distinguished functionally from the plastic pool of CCR6− ILC3 (including NCR+ ILC3) by their ability to interact with adaptive immune cells. Hence, it is tempting to speculate that human NKp44+ and HLA+ ILC3 subsets may be analogous to murine NKp46+ and MHCII+ LTi‐like ILC3 populations, respectively – although further studies are required to define these populations further in multiple human tissues.
ILC3 functions under homeostatic conditions
ILC3 play critical roles in maintaining intestinal tissue health and homeostasis, predominantly through the regulation of steady‐state interactions with commensal bacteria. This is achieved by constitutive production of cytokines that act to regulate epithelial barrier function and commensal bacterial diversity, as well as through interactions with cells of the adaptive immune system that further enforce a tolerogenic environment to prevent commensal bacteria‐driven inflammation. Emerging evidence suggests that ILC3 subsets may differentially contribute to homeostasis in a subset‐specific manner.
Steady‐state ILC3 are the dominant source of IL‐22, a cytokine that acts on non‐haematopoietic cells including intestinal epithelial cells.8 IL‐22 signalling induces a cascade of gene expression in epithelial cells typified by the secretion of antimicrobial peptides such as RegIII proteins, S100a8 and S100a9.8, 77 These antimicrobial peptides act to regulate spatial segregation of commensal bacteria from the epithelial barrier by establishing a gradient of bactericidal activity, which has been termed the ‘demilitarized zone’.77 Production of IL‐22 further reinforces spatial segregation of commensal bacteria by driving transcription of membrane‐bound mucins and goblet cell hyperplasia.78, 79
Mice with disrupted ILC3‐derived IL‐22 responses exhibit altered gut microbiota and increased susceptibility to experimentally induced colitis.49, 80, 81, 82 Most notably, expansion of segmented filamentous bacteria (SFB) – a canonical Th17‐inducing commensal species – occurs in the absence of ILC3 responses.49 ILC3‐derived IL‐22 induced by SFB colonization stimulates serum amyloid protein production from epithelial cells that drive the recruitment of Th17 cells to the ileum, which subsequently act to control SFB burdens.83 Both intestinal NKp46+ ILC3 and LTi‐like ILC3 produce IL‐22 but inhabit distinct niches within the lamina propria and intestinal lymphoid structures. Interestingly, mice specifically lacking NKp46+ ILC3 do not demonstrate dysbiosis, outgrowth of SFB or changes in Th17 cell numbers in the small intestine at steady state.84 Further studies specifically depleting LTi‐like ILC3 are needed to determine whether these findings merely reflect redundancy among IL‐22‐producing ILC3 or subset‐specific roles in the regulation of commensal bacterial species. ILC3‐derived IL‐22 also results in fucosylation of intestinal epithelial cell‐associated carbohydrate chains following colonization of the intestinal tract by commensal bacteria.85, 86, 87 Whereas, certain species of mutualistic bacteria possess the necessary machinery to use carbohydrate fucose as an energy source, many pathogenic bacteria lack this ability and are outcompeted for energy sources. IL‐22‐dependent fucosylation favours the occupancy of mutualistic species in intestinal niches close to the epithelial barrier and was shown to protect against infections by Salmonella spp., Citrobacter rodentium and the opportunistic pathogen Enterococcus faecalis.85, 86, 87
In addition, ILC3‐derived IL‐22 is essential to prevent bacterial translocation and infections with opportunistic pathogens present in the commensal flora. Mice lacking homoeostatic IL‐22 production or depleted for ILC3 demonstrate bacterial translocation to the peripheral organs as a result of disrupted barrier integrity and a failure to contain lymphoid tissue‐resident commensal bacteria species.88, 89, 90 Thus, ILC3 play critical roles during homeostasis by regulating epithelial barrier responses and the balance of commensal bacterial species, largely through the production of IL‐22.
ILC3 interactions with adaptive immunity
NKp46+ ILC3 are largely dispersed in the intestinal lamina propria, whereas LTi‐like ILC3 are found clustered within lymphoid tissues where they have the potential to influence the adaptive immune system. This localization is determined in part through subset‐specific chemokine receptor expression. Localization of NKp46+ ILC3 is dictated by expression of CXCR6 resulting in retention in the lamina propria in response to CXCL16‐producing dendritic cell subsets.91, 92 LTi‐like ILC3 localize to secondary and tertiary lymphoid tissues driven by expression of CXCR5, CCR6 and CCR7 and cluster within interfollicular regions where they are ideally poised to interact with both T and B cells. Indeed, LTi‐like ILC3 express multiple proteins that enable interactions with adaptive immune cells93, 94 (Fig. 2).
MHCII is highly expressed on CCR6+ LTi‐like ILC3 resident in lymph nodes, but absent on NKp46+ ILC3.55, 76, 94, 95 Under homeostatic conditions MHCII+ ILC3 act to limit intestinal inflammation by deletion of commensal bacteria‐specific CD4+ T cells from the intestine and associated lymphoid tissues.55, 76, 96 This pathway may have implications for human disease as HLA‐DR expression on human intestinal ILC3 is reduced in paediatric Crohn's disease patients.55 MHCII+ ILC3 migrate from the intestine to the mesenteric lymph node in a CCR7‐dependent manner, suggesting that antigens may be acquired locally in the tissue before migration to lymph nodes.94 Surprisingly, a dichotomous relationship between MHCII and IL‐22 expression was recently reported among intestinal ILC3,60 seemingly in contrast to previous reports that LTi‐like ILC3 are a dominant source of IL‐22 of the intestine56, 67, 97 (Fig. 2). Hence, further investigation is required to determine whether LTi‐like ILC3‐intrinsic expression of MHCII and IL‐22 is mutually exclusive and/or determined by the local tissue environment.
In contrast to canonical antigen‐presenting cells MHCII+ LTi‐like ILC3 lack expression of the classical co‐stimulatory molecules CD80 and CD86 at steady state, in line with an inhibitory role for ILC3‐mediated antigen presentation.55, 76 However, under inflammatory conditions MHCII+ ILC3 can up‐regulate these molecules and act to promote immune responses.95 In addition, ILC3 can augment CD4+ T‐cell responses through expression of auxiliary co‐stimulatory molecules. ILC3 promote the survival of memory CD4+ T cells and aid T cell‐dependent antibody responses by expression of OX40L and CD30L.98, 99, 100 Interactions between ILC3 and adaptive immune populations are probably also bi‐directional as mice lacking T cells demonstrate increased intestinal ILC3 numbers and IL‐22 production.101 Furthermore, a recent study demonstrated CCR6+ LTi‐like ILC3 express the lipid antigen‐presenting molecule CD1d (Fig. 2), and process and present lipids to activate invariant NK T cells, whereas CD1d‐dependent cross‐talk also induced IL‐22 production from the presenting ILC3.102 In addition, ILC3 prevent spontaneous CD8+ T‐cell activation in neonatal lymph nodes, although it remains unclear whether modulation of CD8+ T‐cell responses is also dependent upon ILC3 antigen‐presenting function or cell–cell interactions.103
In addition to cell‐to‐cell interactions with adaptive immune cells, ILC3 have the capacity to modulate adaptive immune responses by the release of soluble mediators in the local tissue microenvironment. In this regard both gut NKp46+ and LTi‐like ILC3 are an important source of granulocyte–macrophage colony stimulating factor (GM‐CSF) under steady‐state conditions. Microbiota‐dependent IL‐1β production by intestinal macrophages activates ILC3 to secrete GM‐CSF, which in turn modulates mononuclear phagocyte function to promote local regulatory T‐cell differentiation and reinforce tolerance.104 ILC3 also express multiple proteins that endow them with the capacity to regulate B‐cell responses. ILC3 resident in Peyer's patches and intestinal lymphoid structures play key roles in the regulation of IgA production in the intestinal tract, via lymphotoxin‐dependent mechanisms.41, 105, 106, 107 LTi‐like ILC3 express both membrane‐bound lymphotoxin heterotrimers (LTα 1 β 2) as well as soluble lymphotoxin α homotrimers (LTα 3), which control T‐independent and T‐dependent IgA responses, respectively.105, 106 Human and mouse splenic ILC3 also stimulate innate‐like B‐cell IgG production through CD40 interactions, production of BAFF/APRIL and via the Notch ligand Delta‐like 1.108, 109 Taken together ILC3 demonstrate increasingly expanding roles in modulating adaptive immune responses.
ILC3 in inflammation and infection
In addition to maintaining tissue homeostasis ILC3 are an important source of cytokines that act to control mucosal infections. In the context of infection and inflammation, IL‐22 production by ILC3 is induced by cytokine signals derived from bacterial‐sensing myeloid cells, such as IL‐23 and IL‐1β.73, 83, 96, 97, 110 CX3CR1 + mononuclear phagocytes are a dominant source of IL‐23 during infection, which stimulates optimal ILC3 IL‐22 production.111 Conversely, ILC3 cytokine production is negatively regulated by epithelially derived cytokines such as thymic stromal lymphopoietin and IL‐25, which act to suppress IL‐22 production.112, 113 Therefore, signals from multiple intestinal resident cells combine to fine‐tune IL‐22 production by ILC3.
Extensive research has demonstrated protective roles of ILC3 during infection by using the bacterial pathogen Citrobacter rodentium, a model of intestinal inflammation and clinically relevant enteropathic Escherichia coli infection. ILC3‐derived IL‐22 is required for survival and immunity to C. rodentium infection in immunocompromised mice, whereas in the presence of adaptive immunity ILC3‐derived IL‐22 is required to prevent morbidity in the early phase of infection before the generation of a bacteria‐specific T‐cell response.27, 114, 115, 116, 117 As both subsets of ILC3 produce IL‐22 in the intestine the relative contribution of these two subsets to anti‐bacterial immunity has proven controversial. Initial studies proposed that NCR+ ILC3 are important during C. rodentium infection in mice with partial or total immunodeficiency.27, 51, 91, 117 However, NKp46+ ILC3 are dispensable for the control of C. rodentium infection in the presence of T cells.84, 118 In contrast, lack of total ILC3 results in impaired control of infection, suggesting that LTi‐like ILC3 may be sufficient for immunity to C. rodentium.118 In line with this, CD4+ LTi‐like ILC3 have been reported to be a dominant source of IL‐22 during the early stages of C. rodentium infection.97 Nonetheless, NKp46+ ILC3 may have other important non‐redundant roles during C. rodentium infection, as infected mice lacking NKp46+ ILC3 exhibited severe caecal damage.84 In addition, intestinal ILC3‐derived IL‐22 may also contribute to immunity roles in a diverse range of clinically relevant infections in the gut including the nosocomial pathogen Clostridium difficile,119 rotavirus,120 the fungal pathogen Candida albicans,121 as well as gastrointestinal helminths.78 In contrast, some enteric pathogens have evolved to circumvent and exploit the ILC3 response to infection. For example, Salmonella exploit IL‐22‐dependent antimicrobial responses to compete with the commensal microbiota and colonize the inflamed gut.122
ILC3 also contribute to local immunity and inflammation through the production of other cytokines. IL‐17A production by CCR6+ ILC3 is required for Klebsiella pneumoniae clearance and contributes to protection against systemic Candida albicans infection.53, 123 ILC3‐derived IL‐17A has further been associated with pathogenesis of obesity‐associated airway inflammation and psoriasis in mice and humans.124, 125, 126 Although relatively less characterized, ILC3 also secrete IL‐17F and a role for this cytokine in skin inflammation has also been suggested.127
Pro‐inflammatory cytokine production, such as IFN‐γ and tumour necrosis factor‐α, by NCR+ ILC3 and ‘ex‐ILC3’ may also contribute to protective immunity.61 However, chronic activation of ILC3 can result in disease due to uncontrolled inflammation. ILC3 drive colitis in an innate anti‐CD40 driven model of colitis and during Helicobacter hepaticus infection in an IL‐23‐dependent manner, which has been associated with production of GM‐CSF and IFN‐γ by pro‐inflammatory ILC3.71, 118, 128, 129 Neonatal ILC3 can also mediate intestinal pathology in response to transgenic over‐expression of IL‐23 through the release of IL‐17A, IL‐22, IFN‐γ and GM‐CSF.130 In line with a disease‐causing role of ILC3, Crohn's disease patients exhibit increased frequency of NCR− (NKp44− CD56−) CCR6+ ILC3 and have elevated levels of IL‐17A.131 Hence, in contrast to their protective and homeostatic roles, ILC3 may also take on pro‐inflammatory roles and contribute to tissue damage and inflammation during disease. Further studies are required to fully define the signals and environmental cues that regulate the pro‐ versus anti‐inflammatory phenotypes of ILC3 subsets.
Concluding remarks
Group 3 ILC play central roles in lymphoid organogenesis, orchestration of adaptive immunity, regulation of peripheral tolerance and as effector cells in the context of immunity and inflammation. Recent studies have elegantly delineated the developmental and epigenetic networks that dictate ILC3 function and identified novel phenotypes that expand the spectrum of functions ascribed to ILC3 in mucosal barrier tissues. However, challenges remain in resolving the relevance of ILC3 heterogeneity and incorporating recently identified novel transcriptional and metabolic states in the context of current nomenclature. A more nuanced understanding of ILC3 subset‐specific roles in intestinal immunity will aid the development of targeted therapeutic interventions aimed at maintaining beneficial homeostatic ILC3 functions, while neutralizing pro‐inflammatory ILC3 pathways that contribute to the onset or progression of tissue inflammation.
Disclosure
The authors have no competing interests to declare.
Acknowledgements
Research in the Hepworth laboratory is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 105644/Z/14/Z).
References
- 1. Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 2016; 17:765–74. [DOI] [PubMed] [Google Scholar]
- 2. Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells – how did we miss them? Nat Rev Immunol 2013; 13:75–87. [DOI] [PubMed] [Google Scholar]
- 3. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G et al Innate lymphoid cells – a proposal for uniform nomenclature. Nat Rev Immunol 2013; 13:145–9. [DOI] [PubMed] [Google Scholar]
- 4. Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity 2014; 41:354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K et al Differentiation of type 1 ILCs from a common progenitor to all helper‐like innate lymphoid cell lineages. Cell 2014; 157:340–56. [DOI] [PubMed] [Google Scholar]
- 6. Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR et al PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci USA 2015; 112:5123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD et al Intraepithelial type 1 innate lymphoid cells are a unique subset of IL‐12‐ and IL‐15‐responsive IFN‐γ‐producing cells. Immunity 2013; 38:769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL‐22. Nat Immunol 2011; 12:383–90. [DOI] [PubMed] [Google Scholar]
- 9. Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol 2015; 15:415–28. [DOI] [PubMed] [Google Scholar]
- 10. Ishizuka IE, Constantinides MG, Gudjonson H, Bendelac A. The innate lymphoid cell precursor. Annu Rev Immunol 2016; 34:299–316. [DOI] [PubMed] [Google Scholar]
- 11. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature 2014; 508:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chea S, Schmutz S, Berthault C, Perchet T, Petit M, Burlen‐Defranoux O et al Single‐cell gene expression analyses reveal heterogeneous responsiveness of fetal innate lymphoid progenitors to notch signaling. Cell Rep 2016; 14:1500–16. [DOI] [PubMed] [Google Scholar]
- 13. Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A et al Single‐cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue‐inducer cell lineage. Nat Immunol 2016; 17:269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med 2012; 209:729–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Satoh‐Takayama N, Lesjean‐Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CA et al IL‐7 and IL‐15 independently program the differentiation of intestinal CD3‐NKp46+ cell subsets from Id2‐dependent precursors. J Exp Med 2010; 207:273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Serafini N, Klein Wolterink RG, Satoh‐Takayama N, Xu W, Vosshenrich CA, Hendriks RW et al Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J Exp Med 2014; 211:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S et al The transcription factor GATA3 is critical for the development of all IL‐7Rα‐expressing innate lymphoid cells. Immunity 2014; 40:378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T et al TCF‐1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J Immunol 2013; 191:4383–91. [DOI] [PubMed] [Google Scholar]
- 19. Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M et al Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med 2014; 211:1733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoyler T, Klose CS, Souabni A, Turqueti‐Neves A, Pfeifer D, Rawlins EL et al The transcription factor GATA‐3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity 2012; 37:634–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J et al T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 2013; 38:694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zook EC, Kee BL. Development of innate lymphoid cells. Nat Immunol 2016; 17:775–82. [DOI] [PubMed] [Google Scholar]
- 23. Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X et al Single‐cell RNA‐seq identifies a PD‐1hi ILC progenitor and defines its developmental pathway. Nature 2016; 539:102–6. [DOI] [PubMed] [Google Scholar]
- 24. Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORγ(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol 2004; 5:64–73. [DOI] [PubMed] [Google Scholar]
- 25. Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L et al Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat Immunol 2009; 10:75–82. [DOI] [PubMed] [Google Scholar]
- 26. Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C et al RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22‐producing NKp46+ cells. Nat Immunol 2009; 10:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Satoh‐Takayama N, Vosshenrich CA, Lesjean‐Pottier S, Sawa S, Lochner M, Rattis F et al Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity 2008; 29:958–70. [DOI] [PubMed] [Google Scholar]
- 28. Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A et al Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat Immunol 2011; 12:949–58. [DOI] [PubMed] [Google Scholar]
- 29. Montaldo E, Teixeira‐Alves LG, Glatzer T, Durek P, Stervbo U, Hamann W et al Human RORγt+CD34+ cells are lineage‐specified progenitors of group 3 RORγt+ innate lymphoid cells. Immunity 2014; 41:988–1000. [DOI] [PubMed] [Google Scholar]
- 30. Sawa S, Cherrier M, Lochner M, Satoh‐Takayama N, Fehling HJ, Langa F et al Lineage relationship analysis of RORγt+ innate lymphoid cells. Science 2010; 330:665–9. [DOI] [PubMed] [Google Scholar]
- 31. Scoville SD, Mundy‐Bosse BL, Zhang MH, Chen L, Zhang X, Keller KA et al A progenitor cell expressing transcription factor RORγt generates all human innate lymphoid cell subsets. Immunity 2016; 44:1140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Onder L, Danuser R, Scandella E, Firner S, Chai Q, Hehlgans T et al Endothelial cell‐specific lymphotoxin‐beta receptor signaling is critical for lymph node and high endothelial venule formation. J Exp Med 2013; 210:465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3– LTβ + cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity 1997; 7:493–504. [DOI] [PubMed] [Google Scholar]
- 34. van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol 2010; 10:664–74. [DOI] [PubMed] [Google Scholar]
- 35. Kim MY, McConnell FM, Gaspal FM, White A, Glanville SH, Bekiaris V et al Function of CD4+CD3– cells in relation to B‐ and T‐zone stroma in spleen. Blood 2007; 109:1602–10. [DOI] [PubMed] [Google Scholar]
- 36. Eberl G. Inducible lymphoid tissues in the adult gut: recapitulation of a fetal developmental pathway? Nat Rev Immunol 2005; 5:413–20. [DOI] [PubMed] [Google Scholar]
- 37. Lindemans CA, Calafiore M, Mertelsmann AM, O'Connor MH, Dudakov JA, Jenq RR et al Interleukin‐22 promotes intestinal‐stem‐cell‐mediated epithelial regeneration. Nature 2015; 528:560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aparicio‐Domingo P, Romera‐Hernandez M, Karrich JJ, Cornelissen F, Papazian N, Lindenbergh‐Kortleve DJ et al Type 3 innate lymphoid cells maintain intestinal epithelial stem cells after tissue damage. J Exp Med 2015; 212:1783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hanash AM, Dudakov JA, Hua G, O'Connor MH, Young LF, Singer NV et al Interleukin‐22 protects intestinal stem cells from immune‐mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity 2012; 37:339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG et al Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 2008; 456:507–10. [DOI] [PubMed] [Google Scholar]
- 41. Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II et al Requirement for lymphoid tissue‐inducer cells in isolated follicle formation and T cell‐independent immunoglobulin A generation in the gut. Immunity 2008; 29:261–71. [DOI] [PubMed] [Google Scholar]
- 42. Gomez de Aguero M, Ganal‐Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H et al The maternal microbiota drives early postnatal innate immune development. Science 2016; 351:1296–302. [DOI] [PubMed] [Google Scholar]
- 43. van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira‐Santos L et al Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature 2014; 508:123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goverse G, Labao‐Almeida C, Ferreira M, Molenaar R, Wahlen S, Konijn T et al Vitamin A controls the presence of RORγ + innate lymphoid cells and lymphoid tissue in the small intestine. J Immunol 2016; 196:5148–55. [DOI] [PubMed] [Google Scholar]
- 45. Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, Boyd A et al Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science 2014; 343:432–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C et al Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 2011; 334:1561–5. [DOI] [PubMed] [Google Scholar]
- 47. Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX et al The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 2012; 36:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M et al AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol 2012; 13:144–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D et al Group 3 innate lymphoid cells inhibit T‐cell‐mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 2013; 39:386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chea S, Perchet T, Petit M, Verrier T, Guy‐Grand D, Banchi EG et al Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci Signal 2016; 9:ra45. [DOI] [PubMed] [Google Scholar]
- 51. Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA et al The transcription factor T‐bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol 2013; 14:389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 2015; 350:981–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Withers DR, Hepworth MR, Wang X, Mackley EC, Halford EE, Dutton EE et al Transient inhibition of ROR‐gammat therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med 2016; 22:319–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kloverpris HN, Kazer SW, Mjosberg J, Mabuka JM, Wellmann A, Ndhlovu Z et al Innate lymphoid cells are depleted irreversibly during acute HIV‐1 infection in the absence of viral suppression. Immunity 2016; 44:391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J et al Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria‐specific CD4+ T cells. Science 2015; 348:1031–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK et al Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol 2015; 16:306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bjorklund AK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D et al The heterogeneity of human CD127+ innate lymphoid cells revealed by single‐cell RNA sequencing. Nat Immunol 2016; 17:451–60. [DOI] [PubMed] [Google Scholar]
- 58. Koues OI, Collins PL, Cella M, Robinette ML, Porter SI, Pyfrom SC et al Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell 2016; 165:1134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR et al Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell 2016; 165:1120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gury‐BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara‐Astiaso D et al The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 2016; 166:1231–46 e13. [DOI] [PubMed] [Google Scholar]
- 61. Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y et al A T‐bet gradient controls the fate and function of CCR6‐RORγt+ innate lymphoid cells. Nature 2013; 494:261–5. [DOI] [PubMed] [Google Scholar]
- 62. Vitenshtein A, Charpak‐Amikam Y, Yamin R, Bauman Y, Isaacson B, Stein N et al NK cell recognition of Candida glabrata through binding of NKp46 and NCR1 to fungal ligands Epa1, Epa6, and Epa7. Cell Host Microbe 2016; 20:527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Glatzer T, Killig M, Meisig J, Ommert I, Luetke‐Eversloh M, Babic M et al RORγt+ innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity 2013; 38:1223–35. [DOI] [PubMed] [Google Scholar]
- 64. Johnson H, Scorrano L, Korsmeyer SJ, Ley TJ. Cell death induced by granzyme C. Blood 2003; 101:3093–101. [DOI] [PubMed] [Google Scholar]
- 65. Cortez VS, Colonna M. Diversity and function of group 1 innate lymphoid cells. Immunol Lett 2016; 179:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Veiga‐Fernandes H, Coles MC, Foster KE, Patel A, Williams A, Natarajan D et al Tyrosine kinase receptor RET is a key regulator of Peyer's patch organogenesis. Nature 2007; 446:547–51. [DOI] [PubMed] [Google Scholar]
- 67. Ibiza S, Garcia‐Cassani B, Ribeiro H, Carvalho T, Almeida L, Marques R et al Glial‐cell‐derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature 2016; 535:440–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Olivier BJ, Cailotto C, van der Vliet J, Knippenberg M, Greuter MJ, Hilbers FW et al Vagal innervation is required for the formation of tertiary lymphoid tissue in colitis. Eur J Immunol 2016; 46:2467–80. [DOI] [PubMed] [Google Scholar]
- 69. Weiss JM, Bilate AM, Gobert M, Ding Y, Curotto de Lafaille MA, Parkhurst CN et al Neuropilin 1 is expressed on thymus‐derived natural regulatory T cells, but not mucosa‐generated induced Foxp3+ T reg cells. J Exp Med 2012; 209:1723–42, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yadav M, Louvet C, Davini D, Gardner JM, Martinez‐Llordella M, Bailey‐Bucktrout S et al Neuropilin‐1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo . J Exp Med 2012; 209:1713–22, S1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T et al Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor‐expressing RORγt+ innate lymphocytes. Immunity 2010; 33:736–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K et al Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol 2013; 14:221–9. [DOI] [PubMed] [Google Scholar]
- 73. Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed‐Nielsen M et al Interleukin‐12 and ‐23 control plasticity of CD127+ Group 1 and Group 3 innate lymphoid cells in the intestinal lamina propria. Immunity 2015; 43:146–60. [DOI] [PubMed] [Google Scholar]
- 74. Verrier T, Satoh‐Takayama N, Serafini N, Marie S, Di Santo JP, Vosshenrich CA. Phenotypic and functional plasticity of murine intestinal NKp46+ Group 3 innate lymphoid cells. J Immunol 2016; 196:4731–8. [DOI] [PubMed] [Google Scholar]
- 75. Viant C, Rankin LC, Girard‐Madoux MJ, Seillet C, Shi W, Smyth MJ et al Transforming growth factor‐β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci Signal 2016; 9:ra46. [DOI] [PubMed] [Google Scholar]
- 76. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R et al Innate lymphoid cells regulate CD4+ T‐cell responses to intestinal commensal bacteria. Nature 2013; 498:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity 2015; 42:28–39. [DOI] [PubMed] [Google Scholar]
- 78. Turner JE, Stockinger B, Helmby H. IL‐22 mediates goblet cell hyperplasia and worm expulsion in intestinal helminth infection. PLoS Pathog 2013; 9:e1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gulhane M, Murray L, Lourie R, Tong H, Sheng YH, Wang R et al High fat diets induce colonic epithelial cell stress and inflammation that is reversed by IL‐22. Sci Rep 2016; 6:28990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shih VF, Cox J, Kljavin NM, Dengler HS, Reichelt M, Kumar P et al Homeostatic IL‐23 receptor signaling limits Th17 response through IL‐22‐mediated containment of commensal microbiota. Proc Natl Acad Sci USA 2014; 111:13942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L et al IL‐22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol 2013; 190:5306–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ et al An IL‐23R/IL‐22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 2015; 163:381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rankin LC, Girard‐Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A et al Complementarity and redundancy of IL‐22‐producing innate lymphoid cells. Nat Immunol 2016; 17:179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pham TA, Clare S, Goulding D, Arasteh JM, Stares MD, Browne HP et al Epithelial IL‐22RA1‐mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe 2014; 16:504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV et al Rapid fucosylation of intestinal epithelium sustains host‐commensal symbiosis in sickness. Nature 2014; 514:638–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A et al Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014; 345:1254009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J et al Innate lymphoid cells promote anatomical containment of lymphoid‐resident commensal bacteria. Science 2012; 336:1321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D et al Lymphoid‐tissue‐resident commensal bacteria promote members of the IL‐10 cytokine family to establish mutualism. Immunity 2016; 44:634–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Obata T, Goto Y, Kunisawa J, Sato S, Sakamoto M, Setoyama H et al Indigenous opportunistic bacteria inhabit mammalian gut‐associated lymphoid tissues and share a mucosal antibody‐mediated symbiosis. Proc Natl Acad Sci USA 2010; 107:7419–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Satoh‐Takayama N, Serafini N, Verrier T, Rekiki A, Renauld JC, Frankel G et al The chemokine receptor CXCR6 controls the functional topography of interleukin‐22 producing intestinal innate lymphoid cells. Immunity 2014; 41:776–88. [DOI] [PubMed] [Google Scholar]
- 92. Chea S, Possot C, Perchet T, Petit M, Cumano A, Golub R et al CXCR6 expression is important for retention and circulation of ILC precursors. Mediators Inflamm 2015; 2015:368427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hepworth MR, Sonnenberg GF. Regulation of the adaptive immune system by innate lymphoid cells. Curr Opin Immunol 2014; 27:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mackley EC, Houston S, Marriott CL, Halford EE, Lucas B, Cerovic V et al CCR7‐dependent trafficking of RORγ + ILCs creates a unique microenvironment within mucosal draining lymph nodes. Nat Commun 2015; 6:5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. von Burg N, Chappaz S, Baerenwaldt A, Horvath E, Bose Dasgupta S, Ashok D et al Activated group 3 innate lymphoid cells promote T‐cell‐mediated immune responses. Proc Natl Acad Sci USA 2014; 111:12835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG et al Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 2014; 40:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D et al CD4+ lymphoid tissue‐inducer cells promote innate immunity in the gut. Immunity 2011; 34:122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Withers DR, Gaspal FM, Mackley EC, Marriott CL, Ross EA, Desanti GE et al Cutting edge: Lymphoid tissue inducer cells maintain memory CD4 T cells within secondary lymphoid tissue. J Immunol 2012; 189:2094–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gaspal FM, Kim MY, McConnell FM, Raykundalia C, Bekiaris V, Lane PJ. Mice deficient in OX40 and CD30 signals lack memory antibody responses because of deficient CD4 T cell memory. J Immunol 2005; 174:3891–6. [DOI] [PubMed] [Google Scholar]
- 100. Kim MY, Gaspal FM, Wiggett HE, McConnell FM, Gulbranson‐Judge A, Raykundalia C et al CD4+CD3– accessory cells costimulate primed CD4 T cells through OX40 and CD30 at sites where T cells collaborate with B cells. Immunity 2003; 18:643–54. [DOI] [PubMed] [Google Scholar]
- 101. Korn LL, Thomas HL, Hubbeling HG, Spencer SP, Sinha R, Simkins HM et al Conventional CD4+ T cells regulate IL‐22‐producing intestinal innate lymphoid cells. Mucosal Immunol 2014; 7:1045–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Saez de Guinoa J, Jimeno R, Farhadi N, Jervis PJ, Cox LR, Besra GS et al CD1d‐mediated activation of group 3 innate lymphoid cells drives IL‐22 production. EMBO Rep 2016; doi: 10.15252/embr.201642412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bank U, Deiser K, Finke D, Hammerling GJ, Arnold B, Schuler T. Cutting edge: innate lymphoid cells suppress homeostatic T cell expansion in neonatal mice. J Immunol 2016; 196:3532–6. [DOI] [PubMed] [Google Scholar]
- 104. Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y et al Microbiota‐dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 2014; 343:1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, Cyster JG. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer's patches. Science 2016; 352:aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kruglov AA, Grivennikov SI, Kuprash DV, Winsauer C, Prepens S, Seleznik GM et al Nonredundant function of soluble LTα3 produced by innate lymphoid cells in intestinal homeostasis. Science 2013; 342:1243–6. [DOI] [PubMed] [Google Scholar]
- 107. Macpherson AJ, Koller Y, McCoy KD. The bilateral responsiveness between intestinal microbes and IgA. Trends Immunol 2015; 36:460–70. [DOI] [PubMed] [Google Scholar]
- 108. Cella M, Otero K, Colonna M. Expansion of human NK‐22 cells with IL‐7, IL‐2, and IL‐1β reveals intrinsic functional plasticity. Proc Natl Acad Sci USA 2010; 107:10961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Magri G, Miyajima M, Bascones S, Mortha A, Puga I, Cassis L et al Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol 2014; 15:354–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Seo SU, Kuffa P, Kitamoto S, Nagao‐Kitamoto H, Rousseau J, Kim YG et al Intestinal macrophages arising from CCR2+ monocytes control pathogen infection by activating innate lymphoid cells. Nat Commun 2015; 6:8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER et al CX(3)CR1+ mononuclear phagocytes support colitis‐associated innate lymphoid cell production of IL‐22. J Exp Med 2014; 211:1571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Giacomin PR, Moy RH, Noti M, Osborne LC, Siracusa MC, Alenghat T et al Epithelial‐intrinsic IKKα expression regulates group 3 innate lymphoid cell responses and antibacterial immunity. J Exp Med 2015; 212:1513–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sawa S, Lochner M, Satoh‐Takayama N, Dulauroy S, Berard M, Kleinschek M et al RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol 2011; 12:320–6. [DOI] [PubMed] [Google Scholar]
- 114. Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q et al Interleukin‐22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008; 14:282–9. [DOI] [PubMed] [Google Scholar]
- 115. Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA et al Induction of innate lymphoid cell‐derived interleukin‐22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity 2014; 40:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Basu R, O'Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W et al Th22 cells are an important source of IL‐22 for host protection against enteropathogenic bacteria. Immunity 2012; 37:1061–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK et al A human natural killer cell subset provides an innate source of IL‐22 for mucosal immunity. Nature 2009; 457:722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappenbeck TS et al Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J Exp Med 2015; 212:1869–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Abt MC, Lewis BB, Caballero S, Xiong H, Carter RA, Susac B et al Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe 2015; 18:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F et al Interferon‐λ and interleukin 22 act synergistically for the induction of interferon‐stimulated genes and control of rotavirus infection. Nat Immunol 2015; 16:698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G et al Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin‐22. Immunity 2013; 39:372–85. [DOI] [PubMed] [Google Scholar]
- 122. Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W et al The cytokine IL‐22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 2014; 40:262–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Xiong H, Keith JW, Samilo DW, Carter RA, Leiner IM, Pamer EG. Innate Lymphocyte/Ly6Chi monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell 2016; 165:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kim HY, Lee HJ, Chang YJ, Pichavant M, Shore SA, Fitzgerald KA et al Interleukin‐17‐producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity‐associated airway hyperreactivity. Nat Med 2014; 20:54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK et al Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol 2014; 134:984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Dyring‐Andersen B, Geisler C, Agerbeck C, Lauritsen JP, Gudjonsdottir SD, Skov L et al Increased number and frequency of group 3 innate lymphoid cells in nonlesional psoriatic skin. Br J Dermatol 2014; 170:609–16. [DOI] [PubMed] [Google Scholar]
- 127. Li Z, Hodgkinson T, Gothard EJ, Boroumand S, Lamb R, Cummins I et al Epidermal Notch1 recruits RORγ + group 3 innate lymphoid cells to orchestrate normal skin repair. Nat Commun 2016; 7:11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ et al Innate lymphoid cells drive interleukin‐23‐dependent innate intestinal pathology. Nature 2010; 464:1371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Pearson C, Thornton EE, McKenzie B, Schaupp AL, Huskens N, Griseri T et al ILC3 GM‐CSF production and mobilisation orchestrate acute intestinal inflammation. Elife 2016; 5:e10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chen L, He Z, Slinger E, Bongers G, Lapenda TL, Pacer ME et al IL‐23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol 2015; 8:390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Geremia A, Arancibia‐Carcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ et al IL‐23‐responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med 2011; 208:1127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]


