Abstract
Mice with a homozygous knockout of the jumonji (jmj) gene showed abnormal heart development and defective regulation of cardiac-specific genes, including the atrial natriuretic factor (ANF). ANF is one of the earliest markers of cardiac differentiation and a hallmark for cardiac hypertrophy. Here, we show that JMJ represses ANF gene expression by inhibiting transcriptional activities of Nkx2.5 and GATA4. JMJ represses the Nkx2.5- or GATA4-dependent activation of the reporter genes containing the ANF promoter-enhancer or containing the Nkx2.5 or GATA4-binding consensus sequence. JMJ physically associates with Nkx2.5 and GATA4 in vitro and in vivo as determined by glutathione S-transferase pull-down and immunoprecipitation assays. Using mutational analyses, we mapped the protein-protein interaction domains in JMJ, Nkx2.5, and GATA4. We identified two DNA-binding sites of JMJ in the ANF enhancer by gel mobility shift assays. However, these JMJ-binding sites do not seem to mediate ANF repression by JMJ. Mutational analysis of JMJ indicates that the protein-protein interaction domain of JMJ mediates the repression of ANF gene expression. Therefore, JMJ may play important roles in the down-regulation of ANF gene expression and in heart development.
Cardiac-specific gene regulations appear to be dependent on combinatorial associations between cardiac-specific transcription factors and ubiquitous factors. The precise regulation of temporal and spatial expression of tissue-specific genes may require interactions among trans-activators and repressors. Likewise, specification and differentiation of the cardiac muscle lineage appear to require a combinatorial network of many trans-acting factors. For example, the cardiac-specific α-myosin heavy chain (MHC) expression is synergistically activated by myocyte-specific enhancer factor 2 (MEF2) and the thyroid hormone receptor. This activation depends on the binding of each factor to the DNA target sequences (25). The ventricular chamber-specific homeobox protein Irx4 is essential for activation and repression of the cardiac ventricular and atrial MHC expression, respectively (2). The cardiac α-actin expression is activated by serum response factor (SRF), which recruits the murine cardiac-restricted homeobox protein, Nkx2.5 (4) or myocardin (44) for synergistic activation. The GATA4 transcription factor transactivates the cardiac muscle-specific troponin C promoter in nonmuscle cells (18).
Recent evidence suggests that transcriptional repressors play important roles in regulating cardiac muscle gene expression. MEF2, whose activity is essential for early normal cardiac development and muscle differentiation (28), is inhibited by Twist, a basic helix-loop-helix protein that is specifically expressed in the mesoderm (38). Activated Notch (45) and histone deacetylase, a general transcriptional repressor (30, 49), have been shown to directly interact with MEF2 to inhibit the MEF2 transcriptional activities. The thyroid hormone receptor plays a critical role in activating cardiac α-MHC (20, 25). The transcriptional activity of the thyroid hormone receptor can be repressed by interacting with a nuclear receptor corepressor, SMART/NcoR, which results in silencing or repressing the target gene activation (8, 35). An unusual homeodomain protein, HOP, inhibits the activation of atrial natriuretic factor (ANF), cardiac α-actin, and SM22 (37) by interacting with SRF to inhibit SRF-dependent activation. A GATA4-interacting factor, Friend of GATA-2 (FOG-2), represses GATA4-mediated transcriptional activation of several cardiac gene promoters, including the ANF, BNP, and cardiac muscle-specific troponin C (40, 42).
The molecular mechanism of the regulation of ANF expression has been studied extensively because of its early onset of expression and its cardiac-specific pattern of expression in development. Near the time of birth, the expression of ANF in the ventricle of the normal heart decreases significantly. However, the ANF gene is reexpressed in response to hypertrophic stimulation (7, 34). Expression of the ANF gene is activated by either Nkx2.5 or GATA4 or both with synergism (9, 26). The T box-containing transcription factor Tbx5 associates with Nkx2.5 and synergistically activates ANF expression in vitro (3, 17). The ANF promoter is also a target of PITX2 homeobox protein and one of the PITX2 isoforms, PITX2C, can synergistically activate the ANF promoter with Nkx2.5 (12).
Jumonji (JMJ) is a member of the jumonji family of transcription factors (for reviews, see references 1 and 6). JMJ is a nuclear factor (27) that contains a domain homologous to a DNA-binding domain of the A/T-rich interaction domain (ARID) transcription factors based on amino acid sequence analysis (13, 16, 19, 24). We and others have previously demonstrated that the jmj gene plays a critical role in the development of the heart (27) and other organs (23, 33, 41). Cardiac defects in jmj mutant mice embryos include ventricular septal defects, double outlet right ventricles, and thin ventricular walls (27). Examination of the jmj homozygote mutants revealed that regulation of cardiac-specific gene expression was defective in the hearts (27). It was of interest that down-regulation of the ANF gene expression, which occurs in the normal ventricle near birth, was defective in the hearts of the mutants compared to those of the wild-type littermates (27). These data suggest that JMJ suppresses expression of the ANF gene as the heart develops to maturity. The recent report regarding the structural and functional analyses of JMJ showed that JMJ contains domains for the powerful transcriptional repression, DNA binding, and nuclear localization signal (22). Together, these data led us to investigate the molecular mechanism of JMJ on regulating ANF gene expression.
The present study demonstrates that JMJ inhibited ANF activation by Nkx2.5 and GATA4. JMJ interacts physically with cardiac-restricted transcription factors Nkx2.5 and GATA4 and represses their abilities to activate target gene expression. Although we identified two JMJ-binding sites in the ANF enhancer, these JMJ-binding sites were dispensable in ANF repression by JMJ. This study provides molecular mechanisms of cardiac-specific gene regulation and, therefore, of cardiac development, through the repression function of JMJ in ANF gene expression.
MATERIALS AND METHODS
Plasmid constructs.
Various jmj constructs in the pcDNA3.1 vector and in pGEX2T encoding glutathione S-transferase (GST) have been described earlier (22, 27). Nkx2.5/pcDNA3 (26, 32), GATA4/pMT2 (26), various deletion ANF reporter genes (26, 32), the Nkx2.5 reporter gene A20-TATA-Luc (10), and the GATA4 reporter gene GATA-TK-Luc (46) were described elsewhere. To generate Myc-tagged Nkx2.5 and GATA4, the corresponding PCR product was subcloned in frame into the pcDNA3.1-myc containing a C-terminal Myc tag sequence (Invitrogen). The plasmids encoding GST-Nkx2.5 and GST-GATA4 were constructed by subcloning the full-length cDNAs into the pGEX2T vector (Amersham). All new constructs were confirmed by restriction digestion followed by sequencing.
Transfection and reporter gene assay.
Neonatal primary rat cardiomyocytes were prepared and transfected by Lipofectamine (Invitrogen) as described elsewhere (32). Transient transfection assays were done by the calcium phosphate precipitation method as described previously (22, 26). Briefly, mouse embryo fibroblast (10T1/2) cells in 60-mm dish plates were transfected with 1 to 2 μg of reporter gene and/or various transcription factors (Nkx2.5, GATA4, and/or JMJ) in mammalian expression vectors and 0.5 μg of pCMV-βgal. Two days after glycerol shock, cell lysates were assayed (Promega) for luciferase activity by using a luminometer according to the manufacturer's recommendations. Reporter gene activity was normalized to β-galactosidase activity to correct for variations in transfection efficiency. Data are given as means ± standard errors. Statistical analyses were performed by using the analysis of variance. The posttest comparison was performed by paired t test. Results were accepted as significant when P was <0.05.
To examine whether cotransfection of JMJ affects the expression levels of Nkx2.5 or GATA4, Western blot analysis was performed as described previously (22).
Protein-protein interaction.
To examine the in vivo association of JMJ with Nkx2.5 or GATA4, coimmunoprecipitation was performed as described previously (26) with minor modifications. Briefly, human embryonic kidney 293 cells (3 × 106 cells/10-cm culture dish) were transfected with 6 μg of JMJ/pFLAG-CMV2 and/or 3 μg of Nkx2.5/pcDNA3.1-myc or 3 μg of GATA4/pcDNA3.1-myc by using Lipofectamine and Plus reagents (Invitrogen). The cells were briefly sonicated in 1.5 ml of a lysis buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.5% NP-40, 10% glycerol, 10 mM Na2HPO4, and 1 mM dithiothreitol). The cell extracts (500 μg) were diluted with the lysis buffer containing 0.1% NP-40 and precleared by incubation with 5 μg of rabbit immunoglobulin G (IgG) and 50 μl of protein A-agarose at 4°C for 2 h. The precleared cell extracts were incubated with 3 μg of polyclonal anti-Flag antibodies (Ab) (Sigma) at 4°C for 3 h, followed by incubation with protein A-agarose at 4°C for 2 h. After washing five times with NETN buffer (100 mM NaCl, 1 mM EDTA, 1 mM Tris-HCl [pH 8.0], 0.5% NP-40, and 1 mM dithiothreitol) at room temperature for 5 min, bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting with the anti-Myc Ab (Santa Cruz Biotechnology). To perform reciprocal experiments, the cell extracts were precleared by incubation with 5 μg of mouse IgG and protein A-agarose, followed by coimmunoprecipitation with 3 μg of monoclonal anti-Myc Ab and protein A-agarose. After extensive washes with NETN buffer, the bound proteins were analyzed by Western blotting with the anti-JMJ Ab that was characterized previously (22).
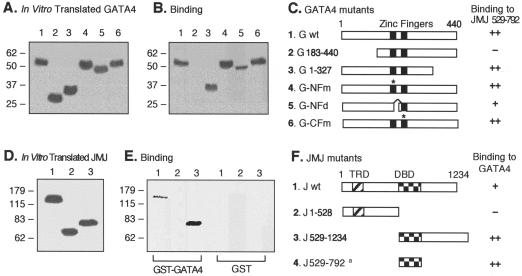
To map the protein-protein interaction domains between JMJ and Nkx2.5 or GATA4, GST pull-down assays were performed as described previously (26). The GATA4 mutant constructs (see Fig. 6C) are as follows. In the N-terminal zinc finger point mutant, cysteine residues at 236 and 239 were replaced by serine residues (C236S and C239S). The N-terminal zinc finger deletion mutant consists of amino acids 1 to 213 and 242 to 440. The C-terminal zinc finger point mutant contains a C290S mutation (26).
FIG. 6.
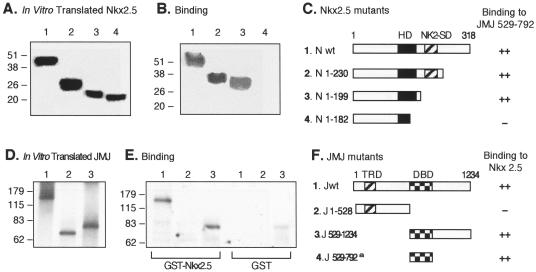
Mapping of the protein-protein interaction domains in GATA4 and JMJ. Various 35S-labeled GATA4 mutants (A) were incubated with approximately 1 μg of GST-JMJ 529-792 fusion proteins coupled to agarose beads and were then subjected to SDS-PAGE (B). A diagram of protein structures for the GATA4 mutants and a summary of the physical interaction are shown (C). The asterisks in panel C indicate the positions of the point mutations (26). For converse experiments, various 35S-labeled JMJ mutants (D) were incubated with approximately 1 μg of GST-GATA4 beads or GST beads, as indicated, followed by SDS-PAGE (E). A diagram of protein structures for JMJ mutants and the summary are shown (F); the a indicates the binding activity of the GST-JMJ 529-792 to 35S-GATA4 as shown in panels B and C. Association affinity: ++, strong interaction; +, moderate interaction; −, no interaction. G, GATA4; J, JMJ; wt, wild type.
To test whether JMJ blocks the interaction between Nkx2.5 and GATA4 (see Fig. 8), equal amounts of in vitro-translated 35S-GATA4 were diluted in 500 μl of NETN buffer and incubated with 80 or 400 μg of 293 cell extract containing the JMJ protein at 4°C for 1 h. Expression of JMJ in 293 cells transfected with a plasmid encoding jmj cDNA was confirmed by Western blot analysis (see Fig. 4B, lane 7). For control experiments, 400 μg of control 293 cell extract was incubated. The solution was then incubated with equal protein amounts (0.6 μg) of GST-Nkx2.5-agarose beads or GST-agarose beads at 4°C for 2 h. After washing, bound proteins were resolved by SDS-PAGE and detected by autoradiography. A reciprocal experiment was performed with 35S-Nkx2.5-4 and GST-GATA4-agarose beads as described above.
FIG. 8.
JMJ does not interfere with the interaction between Nkx2.5 and GATA4. 35S-GATA4 (lane 1) was incubated with increasing amounts of 293 cell extract containing overexpressed JMJ proteins (lanes 2 and 3) or control 293 cell extract (lane 4) as described in Materials and Methods. To these incubation mixtures, GST-Nkx2.5 agarose beads (lanes 2 to 4) or GST agarose beads (lane 5) were added. As a reciprocal experiment, 35S-Nkx2.5 (lane 6) was incubated with increasing amounts of 293 cell extract containing JMJ (lanes 7 and 8) or control 293 cell extract (lane 9), followed by incubation with GST-GATA4 agarose beads (lanes 7 to 9) or GST agarose beads (lane 10). Molecular mass markers (in kilodaltons) are indicated on the left.
FIG. 4.
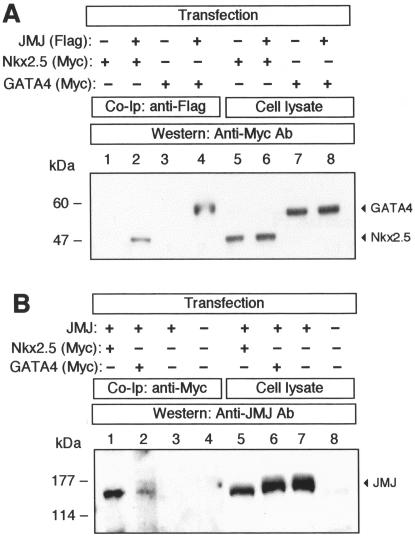
Association of JMJ with Nkx2.5 and GATA4 in vivo. (A) 293 cells were cotransfected with the expression vectors encoding Flag-tagged JMJ and Myc-tagged Nkx2.5 or Myc-tagged GATA4, as indicated. After the cell lysates were precleared by incubation with rabbit IgG and protein A-agarose, JMJ proteins were immunoprecipitated with anti-Flag tag Ab followed by immunoblotting with anti-Myc Ab to detect Nkx2.5 (lane 2) or GATA4 (lane 4). The cell lysates (12 μg/lane; 4% input) were loaded in the same gel to confirm the expression of Nkx2.5 (lanes 5 and 6) and GATA4 (lanes 7 and 8). (B) For reciprocal experiments, the precleared cell lysates were subjected to immunoprecipitation with a monoclonal anti-Myc Ab. The JMJ proteins coimmunoprecipitated with Nkx2.5 (lane 1) or GATA4 (lane 2) were detected with anti-JMJ polyclonal Ab. The expression of JMJ was confirmed by direct Western blot analyses (lanes 5 to 7). Molecular mass markers (in kilodaltons) are indicated on the left of each panel.
GMSA.
A double-stranded oligonucleotide probe for the Nkx2.5-binding element (NKE) in the ANF promoter (from bp −92 to −71 of the transcription initiation site) was produced by annealing a sense strand (5′-CGCCGCAAGTGACAGAATGGGG) and an antisense strand (5′-CTCCCCATTCTGTCACTTGCGG) (9, 10). The GATA4-binding site in the ANF promoter (from bp −136 to −115) was probed with 5′-AGCTTCGCTGGACTGATAACTT and 5′-TAAAGTTATCAGTCCAGCGAAG (9, 26). The various probes for the A/T-rich sequence positioned at bp −580 of the ANF promoter-enhancer were as follows: −580A/T, 5′-ACTCTAAAAAAATATAATAGC and 5′-AGCTATTATATTTTTTTAGA; −425A/T, 5′-CAGCTGCCTGTATTGCCTCTCC and 5′-GAGGAGAGGCAATACAGGCAGC; −405A/T, 5′-CTCCCGCCCTTATTTGGAGCCC and 5′-AGGGGCTCCAAATAAGGGCGGG; −265A/T, 5′-CCAAGGACTATTTTCTGCTCTT and 5′-AGAAGAGCAGAAAATAGTCCTT; −220A/T, 5′-CTCTTGAGGCAAATCATCAAGA and 5′-ATTCTTGATGATTTGCCTCAAG; −110A/T, 5′-ATAACTTTAAAAGGGCATCTTC and 5′-GAGAAGATGCCCTTTTAAAGTT. The Nkx2.5, GATA4-binding site, and A/T-rich sequences are underlined. Gel mobility shift assays (GMSA) were performed as described elsewhere (22, 25).
RESULTS
JMJ represses ANF gene expression.
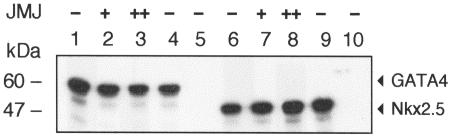
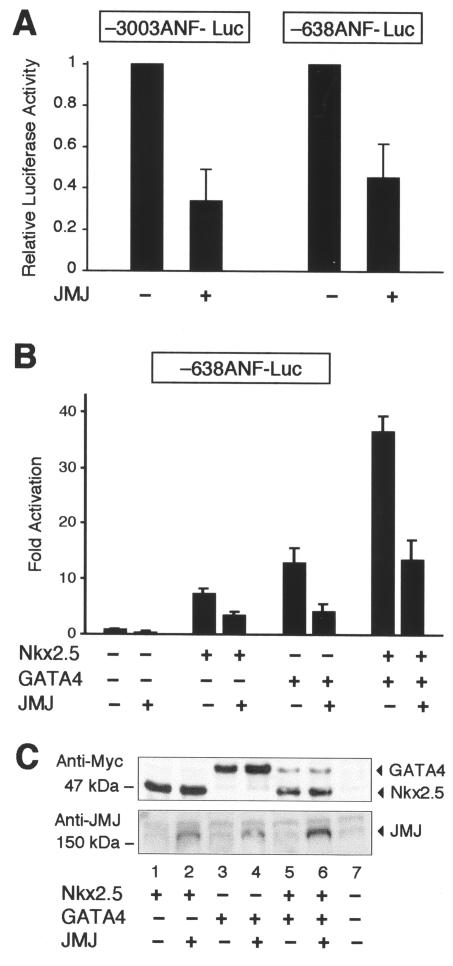
To search for endogenous target genes of JMJ, we performed transient transfection assays using the reporter genes containing either bp −3003 or −638 of the ANF promoter-enhancer linked to a luciferase gene in primary neonatal rat cardiomyocytes. As shown in Fig. 1A, JMJ repressed both ANF reporter genes by more than 50% compared to the activity of either reporter gene alone, indicating that JMJ represses ANF gene expression and that the −638 ANF promoter-enhancer region is sufficient to mediate repression by JMJ. We then began to examine molecular mechanisms of the repression function of JMJ. The cardiac-specific ANF promoter-enhancer is a transcriptional target for both Nkx2.5 and GATA4 (10, 26). It has been shown that either Nkx2.5 or GATA4 alone activates, or both together synergistically activate, ANF expression in cultured fibroblasts (9, 26). Therefore, it was of interest to examine whether JMJ represses ANF activation by Nkx2.5 or GATA4 (Fig. 1B). Nkx2.5 and GATA4 activated the −638 ANF reporter gene in 10T1/2 cells as reported previously (9, 26). Interestingly, JMJ repressed activation of the ANF reporter gene by Nkx2.5 or GATA4 or synergistic activation by both, suggesting that JMJ inhibits the transcriptional activation function of Nkx2.5 and GATA4. These data correlate well with in situ analyses of jmj-knockout mouse heart, which show that down-regulation of the ANF gene is defective in the mutant embryonic heart near birth (27). To exclude the possibility that cotransfection of JMJ decreases the expression of Nkx2.5 or GATA4, Western blot analyses were performed by using the same cell extracts as described for Fig. 1B. As shown in Fig. 1C (upper panel), the cotransfection of JMJ with Nkx2.5 or GATA4 (lane 2 or 4) did not decrease the expression level of Nkx2.5 or GATA4 compared to that of Nkx2.5 or GATA4 alone (lane 1 or 3, respectively). Similarly, cotransfection of JMJ did not affect the expression levels of Nkx2.5 and GATA4 when all three plasmids were expressed (compare lanes 5 and 6). Therefore, the reduced activation of the ANF reporter gene by JMJ is not due to decreases in expression of Nkx2.5 or GATA4.
FIG. 1.
JMJ represses ANF gene expression. (A) JMJ inhibits ANF gene expression in cardiomyocytes. The ANF reporter gene −3003ANF-Luc or −638ANF-Luc (2 μg) was cotransfected with 0.1 μg of the JMJ expression vector into rat neonatal primary cardiomyocytes. Relative luciferase activity was calculated when luciferase activity with the reporter gene alone was set at 1. (B) JMJ inhibits activation of the ANF gene by Nkx2.5 or GATA4 or both. The ANF reporter gene −638ANF-Luc (2 μg) was cotransfected with 2 μg of various transcription factors in the expression vectors into 10T1/2 cells grown on 60-mm plates by calcium phosphate precipitation methods. Luciferase activity was normalized with β-galactosidase activity to correct transfection efficiency. Relative luciferase activity was expressed as the activation level (n-fold) above that of the reporter gene alone. Filled bars indicate the means and T bars indicate standard errors of the means for four separate transfection assays with duplicate plates. (C) Cotransfection of JMJ does not affect the expression level of Nkx2.5 or GATA4. Western blots show expression levels of exogenous genes in the same 10T1/2 cell extracts transfected for the reporter gene assays described for panel B. After cotransfection of the Myc-tagged Nkx2.5 and/or GATA4 expression vector with JMJ as indicated, equivalent amounts of the cell extract (100 μg/lane) were subjected to immunoblotting for Nkx2.5 and GATA4 or JMJ.
Identification of JMJ-binding sites in the ANF enhancer.
It has been reported that JMJ is a DNA-binding protein (22). Results from DNA-binding site selection experiments indicated that JMJ prefers binding to A/T-rich sequences (22). Interestingly, the ANF promoter-enhancer region within −638 nucleic acids contains six A/T-rich sequences, which are the putative DNA-binding sites of JMJ. Therefore, there are several possible mechanisms by which JMJ represses ANF activation. JMJ may bind directly to the ANF enhancer region and repress ANF expression since JMJ contains the transcriptional repression domain (22). Alternatively, JMJ may physically interact with Nkx2.5 or GATA4, which interferes with their transcriptional activation function, or a combination of both mechanisms may lead to repression of ANF expression.
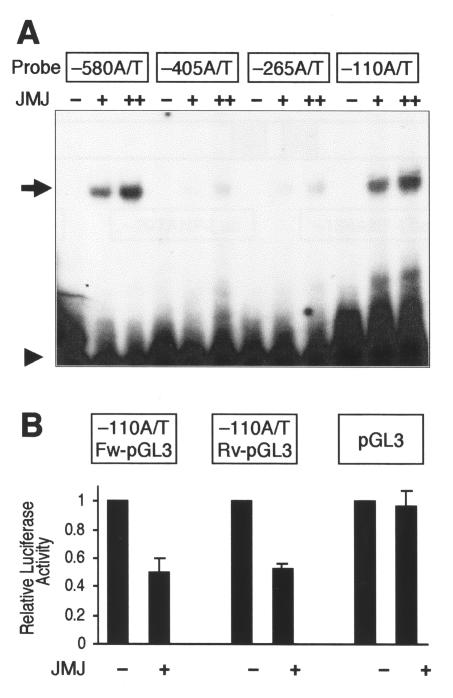
To investigate whether JMJ binds to any of the A/T-rich sequences in the ANF enhancer-promoter region, GMSA were performed (Fig. 2A). Each of six A/T-rich oligonucleotides was labeled with 32P and incubated with GST-JMJ 529-792, the DNA-binding domain (DBD) of JMJ (22). JMJ showed stronger binding to two of the sequences, located at bp −580 and −110, in a dose-dependent manner, than to the A/T-rich sequences located at bp −405 and −265. To determine whether these two A/T-rich sequences mediate the repression function of JMJ in vivo, reporter plasmids were constructed by inserting the A/T-rich oligonucleotides upstream of the SV40 promoter linked to luciferase in the pGL3-promoter vector (Promega). The reporter plasmids, containing an A/T-rich sequence at bp −110 in the ANF promoter in the forward and reverse directions, were designated −110A/T Fw-pGL3 and −110A/T Rv-pGL3, respectively. When cotransfected (Fig. 2B), JMJ repressed both −110A/T Fw-pGL3 and −110A/T Rv-pGL3 by about 50%. Regardless of its orientation, the −110A/T sequence could mediate the transcriptional repression activity of JMJ. The enhancer mediates transcriptional activity regardless of its orientation. The repression function of JMJ was specific to the DNA binding of JMJ, because JMJ did not have any effect on transcriptional activity of the pGL3-promoter vector. JMJ repressed expression of the reporter genes containing −580 A/T-rich sequence in the forward and reverse directions (−580A/T Fw-pGL3 and −580A/T Rv-pGL3) at a level similar to −110A/T-pGL3 (data not shown). JMJ also repressed the reporter gene containing the weaker binding site of JMJ, located at bp −405 or −265, by about 30% (data not shown). These data suggest that JMJ may mediate the repression of ANF expression via these binding sites.
FIG. 2.
JMJ binds to DNA motifs in the ANF enhancer, which mediate repression by JMJ. (A) Representative GMSA with the six A/T-rich sequences in the −638 ANF promoter-enhancer. The 32P-end-labeled probe (20 fmol; 50,000 cpm/lane) was incubated with 20 (+) or 100 ng (++) of GST-JMJ 529-792, as indicated. The reaction mixtures were loaded onto 5% nondenatured PAGE and autoradiographed. The arrow and arrowhead indicate the probe bound to JMJ and free probe, respectively. JMJ showed stronger binding to two of the sequences (bp −580 and −110) (arrow), in a dose-dependent manner, than to other A/T-rich sequences. Sequences of the six oligonucleotides are presented in Materials and Methods. (B) JMJ represses the reporter genes containing the JMJ-binding site in the ANF enhancer. The reporter plasmids containing the −110 A/T-rich sequence selected by GMSA were constructed by subcloning the oligonucleotide into the pGL3-promoter vector in the forward (−110A/T Fw-pGL3) or reverse (−110A/T Rv-pGL3) direction. Transient transfection assays were performed by using 10T1/2 cells as described for Fig. 1B. The reporter genes (1 μg) were cotransfected with 0.5 μg of JMJ in the expression vector. Filled bars represent the means and T bars indicate the standard errors of the means for three separate transfection assays with duplicate plates.
Mutational analyses of the ANF enhancer.
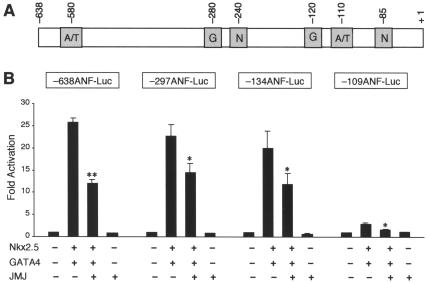
To determine whether these A/T sequences that JMJ binds to indeed mediate the repression function of JMJ, serial mutational analyses of the ANF reporter genes were performed (Fig. 3). The ANF reporter genes were cotransfected with Nkx2.5 and GATA4 in the presence or absence of JMJ. Significant repression of −638 ANF-Luc by JMJ was observed as before (Fig. 1B). The −297ANF-Luc reporter gene, for which the A/T sequences located at bp −580, −425, and −405 are deleted, was significantly repressed by JMJ. The −134ANF-Luc deleted further upstream of the ANF promoter showed a repression pattern similar to that of −297ANF-Luc with JMJ. JMJ also repressed the −109 ANF-Luc reporter gene expression by 44% compared to that by both Nkx2.5 and GATA4. It should be noted that JMJ significantly repressed −109ANF-Luc, which does not contain the A/T-rich sequences but contains an NKE. These results suggest that there are alternative mechanisms for JMJ to repress ANF gene expression that do not require the binding of JMJ to its DNA-binding site.
FIG. 3.
Mutational analyses of the rat ANF promoter-enhancer. (A) Schematic diagram of the ANF enhancer-promoter showing the positions of the two A/T-rich sequences (A/T) that JMJ binds to, Nkx2.5-binding sites (N), and GATA4-binding sites (G). (B) Mutational analyses of the ANF reporter genes. The mutant ANF reporter genes (2 μg) were cotransfected with Nkx2.5 (1 μg) and GATA4 (1 μg) and/or JMJ (1 μg) expression vector into 10T1/2 cells as described for Fig. 1B. Filled bars represent the means and T bars indicate the standard errors of the means for three separate transfection assays with duplicate plates. *, P < 0.05; **, P < 0.01 compared to respective control activation.
Intracellular associations of JMJ with Nkx2.5 and GATA4.
Since the DNA-binding sites for JMJ do not seem to mediate the repression function of JMJ, we explored the possibility that JMJ physically interacts with Nkx2.5 or GATA4 in vivo, resulting in decreased transcriptional activities of these activators. Coimmunoprecipitation was performed by using HEK 293 cells with overexpressed JMJ and Nkx2.5 or GATA4 (Fig. 4). Before immunoprecipitation, the cell extracts were precleared by incubation with rabbit or mouse IgG to remove proteins that may bind nonspecifically to IgG or protein A-agarose. As shown in Fig. 4A, the Myc-tagged Nkx 2.5 or GATA4 protein was detected (lane 2 or 4, respectively) when cell extracts coexpressing Flag-tagged JMJ and Nkx2.5 or GATA4 were immunoprecipitated with the rabbit anti-Flag Ab, followed by Western blotting with the mouse anti-Myc Ab. In the control cell extract expressing only Nkx2.5 or GATA4 (lane 1 or 3, respectively), neither Nkx2.5 nor GATA4 protein was detected, indicating the specific interaction of JMJ with Nkx2.5 or GATA4. Nkx2.5 or GATA4 protein migrated as an approximately 45-kDa (Fig. 4A, lanes 5 and 6) or 52-kDa (lanes 7 and 8) band, respectively, upon direct Western blotting of the transfected cells. Nkx2.5 and GATA4 are not expressed in all cell lines employed in this study, and the endogenous JMJ could not be detected in cell extracts by Western blot analyses (Fig. 4B, lane 8). The converse coimmunoprecipitation experiment also revealed the specific interactions of JMJ with Nkx2.5 or GATA4 in vivo (Fig. 4B). When the cell extract containing JMJ and Myc-tagged Nkx2.5 (lane 1) or GATA4 (lane 2) was immunoprecipitated with anti-Myc Ab, Nkx2.5, and GATA4 were able to pull down JMJ. In contrast, JMJ was not detected when coimmunoprecipitation was performed with anti-Myc Ab in the cell extract containing only JMJ (lane 3) or nontransfected control cell extract (lane 4). These data clearly demonstrate that JMJ physically associates with Nkx2.5 and GATA4 in vivo when the proteins are coexpressed.
It has been shown that JMJ is expressed in the heart (27, 43), as are Nkx2.5 and GATA4. To confirm these protein-protein interactions in cardiomyocytes, we also performed coimmunoprecipitation experiments using mouse heart extracts. However, the protein-protein interaction between JMJ and Nkx2.5 or GATA4 could not be confirmed by the conditions we employed, possibly due to the low expression level of each factor.
The C-terminal part of homeodomain in Nkx2.5 physically interacts with the DBD of JMJ.
To map the protein-protein interaction domain between Nkx2.5 and JMJ, GST pull-down assays were performed (Fig. 5). Various 35S-labeled Nkx2.5 wild type and mutants were translated in vitro (Fig. 5A) and incubated with GST-wild-type JMJ (Jwt) or GST-JMJ 529-792 that contains the DBD of JMJ (Fig. 5B). A schematic diagram of Nkx2.5 mutants and summary of the binding results are shown in Fig. 5C. The Nkx2.5 wild type and the C-terminal deletion mutants, Nkx2.5 with a deletion of amino acids (aa) 1 to 230 or 1 to 199 (N 1-230 or N 1-199, respectively), interacted well with JMJ (lane 1 in Fig. 5B). When the C-terminal part of the homeodomain was deleted (Nkx2.5 1-182), the interaction was abolished (lane 4 in Fig. 5B). The reciprocal experiments were performed to map the interaction domain of JMJ. The GST-Nkx2.5 protein coupled to agarose beads was incubated with the in vitro-translated 35S-JMJ (Fig. 5D). Jwt (Fig. 5E, lane 1) and the C-terminal-containing mutant, JMJ 529-1234 (lane 3), interacted well with GST-Nkx2.5, but JMJ 1-528 did not (lane 2). The binding results are summarized in Fig. 5F. These data indicate that the DBD of JMJ (aa 529 to 792) mediates the interaction with Nkx2.5.
FIG. 5.
Mapping of the protein-protein interaction domains in Nkx2.5 and JMJ. Various 35S-labeled Nkx2.5 mutants were prepared by using an in vitro transcription and translation kit (Promega) and confirmed by SDS-PAGE and autoradiography (A). Equal amounts of 35S-Nkx2.5 were incubated with approximately 1 μg of GST-JMJ 529-792 fusion proteins coupled to agarose beads followed by extensive washing and SDS-PAGE (B). A diagram of protein structures for the Nkx2.5 mutants and a summary of the physical interaction with JMJ are shown (C). To perform reciprocal experiments, various 35S-labeled JMJ mutants (D) were incubated with approximately 1 μg of GST-Nkx2.5 beads or GST beads, as indicated, followed by SDS-PAGE (E). A diagram of protein structures for JMJ mutants and a summary of binding to Nkx2.5 are shown (F); the a indicates the binding activity of GST-JMJ 529-792 to 35S-Nkx2.5 as shown in panels B and C. The homeodomain of Nkx2.5 (HD) and the Nkx2-specific domain (NK2-SD) consist of aa 137 to 196 and 210 to 226, respectively (27). The transcriptional repression domain (TRD) and DBD of JMJ consist of aa 131 to 222 and 529 to 792, respectively. Association affinity: ++, strong interaction; +, moderate interaction; −, no interaction. N, Nkx2.5; J, JMJ; wt, wild type.
The N-terminal part of GATA4 physically associates with the DBD of JMJ.
Similar GST pull-down assays were performed to map the interaction domains between GATA4 and JMJ (Fig. 6). Various in vitro-translated GATA4 mutants (Fig. 6A) were incubated with GST-JMJ, and the bound proteins were subjected to SDS-PAGE (Fig. 6B). All deletion or point mutants of GATA4 bound to JMJ (lanes 1 and 3 to 6), while the N-terminal deletion mutant GATA4 containing aa 183 to 440 (G 183-440) did not interact with JMJ (lane 2), indicating that the N terminus of GATA4 (aa 1 to 182) is necessary for interaction with JMJ. The reciprocal experiments (Fig. 6D to F) indicated that the DBD of JMJ (aa 528 to 792) was also necessary for interaction with GATA4 (panel E). The full-length JMJ bound to GATA4 more weakly than did JMJ 529-1234 (panel E). These data so far show that the DBD of JMJ mediates physical interactions with the N-terminal end of GATA4 and the C-terminal part of the homeodomain in Nkx2.5.
JMJ represses the reporter gene containing the Nkx2.5- or GATA4-binding consensus sequence.
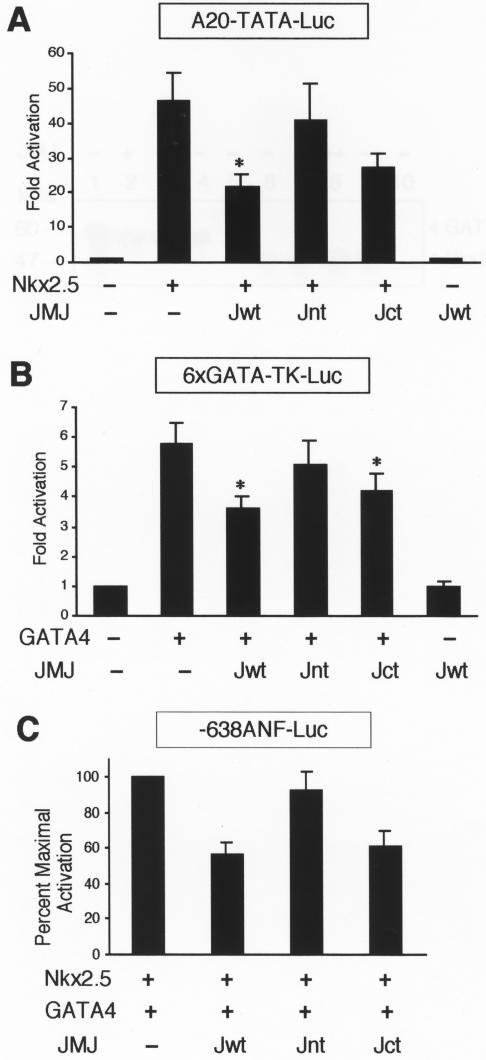
We then investigated whether JMJ represses the Nkx2.5- or GATA4-dependent activation via a DNA-binding-independent mechanism. Cotransfection assays were performed by using the reporter genes containing either the Nkx2.5-binding consensus site, A20-TATA-Luc (10), or the GATA4-binding site, GATA4-TK-Luc (46). These reporter genes do not contain the DNA-binding site of JMJ. As shown in Fig. 7A, the A20 reporter gene was activated when cotransfected with Nkx2.5. Wild-type JMJ (Jwt) significantly repressed this Nkx2.5-dependent activation by 53% (46.2- versus 21.5-fold activation). The N-terminal part of JMJ (Jnt; aa 1 to 528) did not seem to repress the A20 reporter gene. Jnt does not contain the protein-protein interaction domain with Nkx2.5 and GATA4, but it contains the transcriptional repression domain. In contrast, the activation of the reporter gene by Nkx2.5 was markedly repressed with the coexpression of Jct, where the JMJ C-terminal aa 529 to 1234 is fused to the nuclear localization signal aa 1 to 130 (22). Jct contains the protein-protein interaction domain for Nkx2.5 (Fig. 5) and GATA4 (Fig. 6), but does not contain the transcriptional repression domain of JMJ. These data suggest that the protein-protein interaction domain of JMJ is required for most of the repression activity.
FIG.7.
JMJ inhibits transcriptional activation of Nkx2.5 and GATA4. (A) JMJ inhibits transcriptional activation by Nkx2.5. A reporter plasmid (1 μg) containing the Nkx2.5 DNA-binding consensus sequence A20 (AGTTAATTG) linked to the cardiac α-actin TATA minimal promoter, A20-TATA-Luc, was cotransfected with 0.1 μg of Nkx2.5 and/or 0.5 μg of JMJ wild-type (Jwt), Jnt (aa 1 to 528), or Jct (aa 1 to 130 fused to aa 529 to 1234) into 10T1/2 cells as indicated. Filled bars indicate the means and T bars indicate the standard errors of the means for three separate experiments with duplicate plates. *, P < 0.05 compared to control activation. (B) JMJ inhibits transcriptional activation by GATA4. Transient transfection assays were performed as described for panel A with a reporter plasmid containing six GATA4-binding sequence linked to the thymidine kinase (TK) promoter and/or GATA4 expression vector. (C) JMJ inhibits ANF activation by Nkx2.5 and GATA4. The −638ANF reporter plasmid (1 μg) was cotransfected with 0.2 μg of Nkx2.5 and GATA4 and 0.5 μg of JMJ in expression vectors. The percentage of maximal repression was calculated by setting the maximum activation of the ANF gene by Nkx2.5 and GATA4 at 100%.
Similarly, Jwt also significantly inhibited the GATA4-dependent activation of the reporter gene, which contains only the GATA4-binding site, by 37% (Fig. 7B). While Jct repressed the GATA4-dependent activation significantly, Jnt showed very weak repression activity, if any. Similar results were observed when Jwt and its mutants were cotransfected with the ANF reporter (Fig. 7C). Jwt and Jct repressed the ANF activation by 42 and 39%, respectively, while Jnt produced only 8% repression. JMJ alone did not inhibit the basal reporter gene activity, indicating the specific repression by JMJ on ANF activation. These results indicate that the major repression function of JMJ requires the protein-protein interaction with Nkx2.5 and GATA4, and the transcriptional repression domain seems to contribute to the minor repression activity of JMJ, if any.
It is possible that JMJ may compete with Nkx2.5 or GATA4 at their DNA-binding sites. However, our GMSA experiments (data not shown) indicated that JMJ bound to neither the NKE- nor the GATA-binding sequence identified in the ANF promoter (9, 26). The oligonucleotide sequences of the probes are described in Materials and Methods. In addition, the JMJ protein did not decrease the DNA-binding activity of Nkx2.5 or GATA4 to its own binding site when JMJ was coincubated with Nkx2.5 or GATA4 (data not shown). Therefore, it is unlikely that JMJ competes with Nkx2.5 or GATA4 at their DNA-binding sites, resulting in repression of the target gene expression.
JMJ does not interfere with the interaction between Nkx2.5 and GATA4.
It has been reported that Nkx2.5 and GATA4 physically associate and cooperatively activate transcription of the ANF gene (9, 26). Because our data show that JMJ can interact with both Nkx2.5 and GATA4, we examined whether JMJ interferes with the physical interaction between Nkx2.5 and GATA4 (Fig. 8), which may lead to a decrease in their synergistic cooperation for ANF activation. 35S-labeled GATA4 preincubated with an excess amount of JMJ was then pulled down with GST-Nkx2.5, followed by SDS-PAGE and autoradiography. If JMJ interferes with the interaction between GATA4 and Nkx2.5, GST-Nkx2.5 would not efficiently pull down 35S-GATA4. When 35S-GATA4 (Fig. 8, lane 1) was preincubated with increasing amounts of cell extracts with JMJ overexpressed (lanes 2 and 3), GST-Nkx2.5 interacted with 35S-GATA4 as efficiently as with the control cell extract (lane 4). Similarly, 35S-Nkx2.5 (lane 6) preincubated with increasing amounts of JMJ (lanes 7 and 8) retained its ability to interact with GST-GATA4 as well as with control cell extracts (lane 9). These interactions were specific since neither 35S-GATA4 nor Nkx2.5 was pulled down by beads of GST alone (lanes 5 and 10). Although it is not possible to calculate the exact amount of each protein in the reaction, we preincubated minimal amounts of 35S-GATA4 or 35S-Nkx2.5 with large excess amounts of JMJ extracts as evidenced by Western blotting (data not shown) to saturate 35S-GATA4 and 35S-Nkx2.5. Therefore, it is unlikely that JMJ inhibits the physical interaction between Nkx2.5 and GATA4.
DISCUSSION
Transcriptional repression plays a critical role in precisely controlling gene expression in a spatial and temporal manner. Many gene-specific repressors in bacteria and eukaryotes have been identified (21, 29). Transcriptional repressors either bind directly to the cis-acting DNA element of a target gene promoter or interact with other transcriptional activators. The target gene expression can be inhibited subsequently by recruiting transcriptional corepressors or by masking a transcriptional activation function of an activator. We have previously reported that JMJ contains a transcriptional repression domain and can bind to DNA in vitro (22). The present study demonstrates that JMJ, a nuclear factor that is critical for normal heart development (27), represses ANF gene expression by physical interaction with the cardiac-restricted transcription factors Nkx2.5 and GATA4. Although JMJ can bind to the two cis elements of the ANF gene promoter, these binding sequences did not seem to mediate the repression activity of JMJ.
Atrial natriuretic factor (ANF) is an important hormonal mediator of body fluid and electrolyte balance in both the normal and hypertensive states of mammalian species (7, 34). During normal development, ANF is expressed in both the atrial and ventricular chambers of the embryonic heart (48). Near birth, expression of ANF in the ventricular compartment decreases significantly, leading to the atrial-specific expression of the ANF gene in the normal adult heart. However, in response to increased demands for cardiac muscle work, cardiac muscle is activated by hypertrophic stimulation, which results in reactivation of the ANF gene expression in the ventricle. This reactivation of ANF is a hallmark for inducible gene expression in hypertrophied cardiac muscle cells (7, 34). An understanding of induction or repression of ANF expression will also provide valuable insights into the mechanism of hypertrophic changes of ventricle cells in the heart.
Regulation of ANF expression is controlled primarily at the transcriptional level. The transcription factors involved in activation of ANF expression include Nkx2.5, GATA4, MEF2, PitX2, and Tbx5 (3, 10, 12, 26, 31), which bind to their cis elements in the ANF enhancer region. In contrast, molecular mechanisms that repress ANF expression have not been well characterized. Several repressors of the ANF gene have been recently reported. For example, Tbx2, a Tbx isoform, inhibits ANF expression in the atrioventricular canal (14, 47). HOP and FOG2 inhibit ANF expression by interacting with SRF (5, 37) and GATA4 (39, 42), respectively. Our present data, together with the observation that JMJ-knockout mutant mice showed elevated levels of ANF expression in the late-stage embryonic ventricle compared to that of wild type (27), demonstrate that JMJ mediates down-regulation of ANF expression at the late embryonic stage of the ventricle. Since the expression level of JMJ is increased in the ventricle as it develops (27, 43), it is plausible that an increased amount of JMJ in the ventricle of the late-stage embryo results in repression of ANF expression.
GMSA indicates that JMJ binds to at least two A/T-rich cis elements in the ANF enhancer in vitro (Fig. 2A). JMJ inhibits expression of the reporter genes that contain these A/T-rich sequences (Fig. 2B). However, these cis elements do not seem to play a major role in JMJ-dependent repression of ANF expression, since JMJ inhibits activities of the ANF reporter genes where all or some of these A/T-rich sequences are deleted (Fig. 3B). The JMJ binding site located at bp −580 is particularly interesting, because this element has been demonstrated to mediate the up-regulation of ANF expression during cardiac myocyte hypertrophy (15) and also binds weakly to MEF2 (31). The transacting factor that binds to this site during cardiac hypertrophy has not been identified. Therefore, JMJ may be a putative therapeutic factor that prevents the reexpression of ANF in hypertrophic hearts by binding to the A/T-rich sequence at bp −580 in the ANF enhancer.
Our data suggest that the protein-protein interactions of JMJ with Nkx2.5 and GATA4 are essential in mediating the repression function of JMJ. Specifically, we mapped the protein-protein interaction domain of JMJ to a region between aa 529 and 792 (Fig. 5 and 6), which overlaps with the previously identified DBD of JMJ (22). It is interesting that JMJ interacts with the N-terminal region of GATA4 between aa 1 and 181, which is the transactivation domain of GATA4. Therefore, the physical association of JMJ with GATA4 may lead to the inhibition of the transcriptional activity of GATA4. The interaction site of GATA4 with JMJ is rather unusual, because the zinc finger region of GATA4 is known to serve as a protein interaction domain. For instance, the C-terminal and N-terminal zinc finger regions of GATA4 interact with Nkx2.5 (9, 26) and FOG-2 (39), respectively. The DBD region of JMJ (aa 529 to 792) is essential for interacting with Nkx2.5; therefore, this region seems to serve as a protein-protein interaction domain of JMJ. The C-terminal part of the homeodomain in Nkx2.5 mediates the interaction with JMJ and also mediates interaction with GATA4 to synergistically activate ANF expression (9, 26). Because JMJ interacts with Nkx2.5 and GATA4 interacts with Nkx2.5 via the same interaction domain, JMJ may interfere with the interaction between Nkx2.5 and GATA4. Therefore, we investigated by GST pull-down assay whether JMJ inhibits the physical association of Nkx2.5 with GATA4. As shown in Fig. 8, JMJ does not seem to interfere with the physical association between Nkx2.5 and GATA4 under the conditions we employed.
JMJ appears to repress ANF activation via protein-protein interactions with Nkx2.5 and GATA4, because Jnt, the JMJ mutant that does not contain the protein-protein interaction domain, lost most of its repression activity as shown in Fig. 7. In addition, Jct, which contains the protein-protein interaction domain, retains most of its repression activity. To exclude the possibility that Jnt contains an additional low-affinity protein interaction domain(s), several short JMJ mutants (such as aa 1 to 130, 1 to 220, 1 to 377, and 130 to 220) were subjected to a GST pull-down assay using GST-Nkx2.5 or GST-GATA4. However, none of the short fragments of JMJ bound to Nkx2.5 or GATA4 in vitro (data not shown).
JMJ contains a homologous region to the DNA-binding domain of the ARID transcription factor family. The members of this gene family include the B-cell-specific transactivator Bright (16) and the dead ringer gene product of Drosophila (13). JMJ has other motifs homologous to yeast SWI1, which mediates transcriptional activation (36), and two retinoblastoma-binding proteins (11), which bind to a cell cycle regulator, pRb. Thus, JMJ may also be involved in cell proliferation and differentiation via interaction with the cell cycle regulator pRb or with pRb-related factors, such as p107 (50). It would also be interesting to identify the novel factors that may mediate the physiological and transcriptional regulatory functions of JMJ.
JMJ may regulate expression of other genes, since jmj knockout caused embryonic lethality in mice and JMJ is expressed widely throughout development (27, 33, 41). The ANF gene may be one of the endogenous target genes of JMJ in the ventricles of last-stage embryos, and there may be other target genes of JMJ in different tissues and at different developmental stages. Indeed, our study of transcriptional repression function of JMJ in the heart is supported by a recent paper, which was published while the present study was in preparation, describing how JMJ represses cyclin D1 expression in the heart (43). Taken together, these studies demonstrate that JMJ is a DNA-binding trans-acting factor involved in a transcription factor cascade in heart development.
Acknowledgments
We thank Robert Schwartz, Kenneth Chien, Seigo Izumo, Bruce Markham, Hisamaru Hirai, and Jeffery Molkentin for providing valuable plasmids and Alice J. Song for excellent technical support.
This work was supported in part by grants from NIH (HL67050) and the American Heart Association (0030002N) to Y. Lee and a grant from NIH (HL67724) to J. Sadoshima.
REFERENCES
- 1.Balciunas, D., and H. Ronne. 2000. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 25:274-276. [DOI] [PubMed] [Google Scholar]
- 2.Bao, Z. Z., B. G. Bruneau, J. G. Seidman, C. E. Seidman, and C. L. Cepko. 1999. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science 283:1161-1164. [DOI] [PubMed] [Google Scholar]
- 3.Bruneau, B. G., G. Nemer, J. P. Schmitt, F. Charron, L. Robitaille, S. Caron, D. A. Conner, M. Gessler, M. Nemer, C. E. Seidman, and J. G. Seidman. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709-721. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Y., and R. J. Schwartz. 1996. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol. Cell. Biol. 16:6372-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, F., H. Kook, R. Milewski, A. D. Gitler, M. M. Lu, J. Li, R. Nazarian, R. Schnepp, K. Jen, C. Biben, G. Runke, J. P. Mackay, J. Novotny, R. J. Schwartz, R. P. Harvey, M. C. Mullins, and J. A. Epstein. 2002. Hop is an unusual homeobox gene that modulates cardiac development. Cell 110:713-723. [DOI] [PubMed] [Google Scholar]
- 6.Clissold, P. M., and C. P. Ponting. 2001. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem. Sci. 26:7-9. [DOI] [PubMed] [Google Scholar]
- 7.Dickstein, K., T. Aarsland, and C. Hall. 1997. Plasma N-terminal atrial natriuretic factor: a predictor of survival in patients with congestive heart failure. J. Card. Fail. 3:83-89. [DOI] [PubMed] [Google Scholar]
- 8.Dilworth, F. J., and P. Chambon. 2001. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene 20:3047-3054. [DOI] [PubMed] [Google Scholar]
- 9.Durocher, D., F. Charron, R. Warren, R. J. Schwartz, and M. Nemer. 1997. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 16:5687-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durocher, D., C. Y. Chen, A. Ardati, R. J. Schwartz, and M. Nemer. 1996. The atrial natriuretic factor promoter is a downstream target for Nkx-2.5 in the myocardium. Mol. Cell. Biol. 16:4648-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fattaey, A. R., K. Helin, M. S. Dembski, N. Dyson, E. Harlow, G. A. Vuocolo, M. G. Hanobik, K. M. Haskell, A. Oliff, D. Defeo-Jones, and R. E. Jones. 1993. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 8:3149-3156. [PubMed] [Google Scholar]
- 12.Ganga, M., H. M. Espinoza, C. J. Cox, L. Morton, T. A. Hjalt, Y. Lee, and B. A. Amendt. 2003. PITX2 isoform-specific regulation of atrial natriuretic factor expression: synergism and repression with Nkx2.5. J. Biol. Chem. 278:22437-22445. [DOI] [PubMed] [Google Scholar]
- 13.Gregory, S. L., R. D. Kortschak, B. Kalionis, and R. Saint. 1996. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 16:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habets, P. E., A. F. Moorman, D. E. Clout, M. A. van Roon, M. Lingbeek, M. van Lohuizen, M. Campione, and V. M. Christoffels. 2002. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 16:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, A. N., P. Ruiz-Lozano, Y. F. Chen, P. Sionit, Y. T. Yu, B. Lilly, E. N. Olson, and K. R. Chien. 1997. A novel A/T-rich element mediates ANF gene expression during cardiac myocyte hypertrophy. J. Mol. Cell. Cardiol. 29:515-525. [DOI] [PubMed] [Google Scholar]
- 16.Herrscher, R. F., M. H. Kaplan, D. L. Lelsz, C. Das, R. Scheuermann, and P. W. Tucker. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9:3067-3082. [DOI] [PubMed] [Google Scholar]
- 17.Hiroi, Y., S. Kudoh, K. Monzen, Y. Ikeda, Y. Yazaki, R. Nagai, and I. Komuro. 2001. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 28:276-280. [DOI] [PubMed] [Google Scholar]
- 18.Ip, H. S., D. B. Wilson, M. Heikinheimo, Z. Tang, C. N. Ting, M. C. Simon, J. M. Leiden, and M. S. Parmacek. 1994. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol. Cell. Biol. 14:7517-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwahara, J., and R. T. Clubb. 1999. Solution structure of the DNA binding domain from Dead ringer, a sequence-specific AT-rich interaction domain (ARID). EMBO J. 18:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumo, S., and V. Mahdavi. 1988. Thyroid hormone receptor α isoforms generated by alternative splicing differentially activate myosin HC gene transcription. Nature 334:539-542. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, A. D. 1995. The price of repression. Cell 81:655-658. [DOI] [PubMed] [Google Scholar]
- 22.Kim, T. G., J. C. Kraus, J. Chen, and Y. Lee. 2003. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J. Biol. Chem. 278:42247-42255. [DOI] [PubMed] [Google Scholar]
- 23.Kitajima, K., M. Kojima, K. Nakajima, S. Kondo, T. Hara, A. Miyajima, and T. Takeuchi. 1999. Definitive but not primitive hematopoiesis is impaired in jumonji mutant mice. Blood 93:87-95. [PubMed] [Google Scholar]
- 24.Kortschak, R. D., P. W. Tucker, and R. Saint. 2000. ARID proteins come in from the desert. Trends Biochem. Sci. 25:294-299. [DOI] [PubMed] [Google Scholar]
- 25.Lee, Y., B. Nadal-Ginard, V. Mahdavi, and S. Izumo. 1997. Myocyte-specific enhancer factor 2 and thyroid hormone receptor associate and synergistically activate the α-cardiac myosin heavy-chain gene. Mol. Cell. Biol. 17:2745-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, Y., T. Shioi, H. Kasahara, S. M. Jobe, R. J. Wiese, B. E. Markham, and S. Izumo. 1998. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol. Cell. Biol. 18:3120-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, Y., A. J. Song, R. Baker, B. Micales, S. J. Conway, and G. E. Lyons. 2000. Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res. 86:932-938. [DOI] [PubMed] [Google Scholar]
- 28.Lin, Q., J. Schwarz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maldonado, E., M. Hampsey, and D. Reinberg. 1999. Repression: targeting the heart of the matter. Cell 99:455-458. [DOI] [PubMed] [Google Scholar]
- 30.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2000. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 97:14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin, S., F. Charron, L. Robitaille, and M. Nemer. 2000. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 19:2046-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morisco, C., K. Seta, S. E. Hardt, Y. Lee, S. F. Vatner, and J. Sadoshima. 2001. Glycogen synthase kinase 3β regulates GATA4 in cardiac myocytes. J. Biol. Chem. 276:28586-28597. [DOI] [PubMed] [Google Scholar]
- 33.Motoyama, J., K. Kitajima, M. Kojima, S. Kondo, and T. Takeuchi. 1997. Organogenesis of the liver, thymus and spleen is affected in jumonji mutant mice. Mech. Dev. 66:27-37. [DOI] [PubMed] [Google Scholar]
- 34.Needleman, P., S. P. Adams, B. R. Cole, M. G. Currie, D. M. Geller, M. L. Michener, C. B. Saper, D. Schwartz, and D. G. Standaert. 1985. Atriopeptins as cardiac hormones. Hypertension 7:469-482. [DOI] [PubMed] [Google Scholar]
- 35.Ordentlich, P., M. Downes, and R. M. Evans. 2001. Corepressors and nuclear hormone receptor function. Curr. Top. Microbiol. Immunol. 254:101-116. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C. L., and I. Herskowitz. 1992. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68:573-583. [DOI] [PubMed] [Google Scholar]
- 37.Shin, C. H., Z. P. Liu, R. Passier, C. L. Zhang, D. Z. Wang, T. M. Harris, H. Yamagishi, J. A. Richardson, G. Childs, and E. N. Olson. 2002. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110:725-735. [DOI] [PubMed] [Google Scholar]
- 38.Spicer, D. B., J. Rhee, W. L. Cheung, and A. B. Lassar. 1996. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272:1476-1480. [DOI] [PubMed] [Google Scholar]
- 39.Svensson, E. C., G. S. Huggins, F. B. Dardik, C. E. Polk, and J. M. Leiden. 2000. A functionally conserved N-terminal domain of the friend of GATA-2 (FOG-2) protein represses GATA4-dependent transcription. J. Biol. Chem. 275:20762-20769. [DOI] [PubMed] [Google Scholar]
- 40.Svensson, E. C., R. L. Tufts, C. E. Polk, and J. M. Leiden. 1999. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl. Acad. Sci. USA 96:956-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi, T., Y. Yamazaki, Y. Katoh-Fukui, R. Tsuchiya, S. Kondo, J. Motoyama, and T. Higashinakagawa. 1995. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 9:1211-1222. [DOI] [PubMed] [Google Scholar]
- 42.Tevosian, S. G., A. E. Deconinck, A. B. Cantor, H. I. Rieff, Y. Fujiwara, G. Corfas, and S. H. Orkin. 1999. FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl. Acad. Sci. USA 96:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyoda, M., H. Shirato, K. Nakajima, M. Kojima, M. Takahashi, M. Kubota, R. Susuki-Migishima, Y. Mogegi, M. Yokoyama, and T. Takeuchi. 2003. Jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev. Cell 5:85-97. [DOI] [PubMed] [Google Scholar]
- 44.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 45.Wilson-Rawls, J., J. D. Molkentin, B. L. Black, and E. N. Olson. 1999. Activated Notch inhibits myogenic activity of the MADS-box transcription factor myocyte enhancer factor 2C. Mol. Cell. Biol. 19:2853-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamagata, T., J. Nishida, R. Sakai, T. Tanaka, H. Honda, N. Hirano, H. Mano, Y. Yazaki, and H. Hirai. 1995. Of the GATA-binding proteins, only GATA-4 selectively regulates the human interleukin-5 gene promoter in interleukin-5-producing cells which express multiple GATA-binding proteins. Mol. Cell. Biol. 15:3830-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamagishi, H., C. Yamagishi, O. Nakagawa, R. P. Harvey, E. N. Olson, and D. Srivastava. 2001. The combinatorial activities of Nkx2.5 and dHAND are essential for cardiac ventricle formation. Dev. Biol. 239:190-203. [DOI] [PubMed] [Google Scholar]
- 48.Zeller, R., K. D. Bloch, B. S. Williams, R. J. Arceci, and C. E. Seidman. 1987. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes Dev. 1:693-698. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, L., S. van den Heuvel, K. Helin, A. Fattaey, M. Ewen, D. Livingston, N. Dyson, and E. Harlow. 1993. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 7:1111-1125. [DOI] [PubMed] [Google Scholar]