Abstract
Although the retinoblastoma protein (pRb) has been implicated in the processes of cellular differentiation, there is no compelling genetic or in vivo evidence that such activities contribute to pRb-mediated tumor suppression. Motivated by cell culture studies suggesting that Ras is a downstream effector of pRb in the control of differentiation, we have examined the tumor and developmental phenotypes of Rb and K-ras double-knockout mice. We find that heterozygosity for K-ras (i) rescued a unique subset of developmental defects that characterize Rb-deficient embryos by affecting differentiation but not proliferation and (ii) significantly enhanced the degree of differentiation of pituitary adenocarcinomas arising in Rb heterozygotes, leading to their prolonged survival. These observations suggest that Rb and K-ras function together in vivo, in the contexts of both embryonic and tumor development, and that the ability to affect differentiation is a major facet of the tumor suppressor function of pRb.
Inheritance of a mutated allele of the retinoblastoma gene (Rb) predisposes to familial retinoblastoma, and Rb is also frequently inactivated in sporadic human cancers of diverse histological origins (10). Mice heterozygous for Rb develop pituitary adenocarcinomas and thyroid adenomas (13-15, 44), suggesting that the mouse can be used to understand the pathways through which Rb exerts its tumor suppressor functions. Further, nullizygosity for Rb results in lethality at midgestation, with embryos displaying defects in proliferation and differentiation (5, 15, 21, 47). Thus, the combined analysis of Rb+/− and Rb−/− mice provides an opportunity to dissect the functions of the retinoblastoma protein (pRb) that contribute to its tumor suppressor functions.
pRb is best characterized for its role in controlling proliferation; this is accomplished through its regulated interaction with the E2F family of transcription factors (9). The significance of pRb-mediated inhibition of proliferation through E2F has been analyzed in the mouse. Rb−/− embryos display cell-autonomous deregulated proliferation in tissues such as the central nervous system (CNS) and lens (7, 25, 45), and compound embryos lacking Rb and E2f1, E2f2, or E2f3 show reduced levels of ectopic DNA replication in these tissues (33, 41, 49). Mirroring the effects seen during embryonic development, loss of E2f1 or E2f3 significantly reduced the frequency of grossly detectable pituitary tumors (46, 48), suggesting that deregulated proliferation, mediated by E2f, contributes to tumor formation following loss of Rb.
Cell culture-based experiments suggest that pRb also participates in several differentiation programs such as adipogenesis (3, 6), osteogenesis (34, 40), and myogenesis (11, 27). A role for Rb in skeletal muscle differentiation in the mouse has also been demonstrated (47). The available evidence suggests that pRb regulates differentiation by influencing the activity of a number of differentiation-promoting transcription factors such as MyoD (11, 27, 28), the glucocorticoid receptor (35, 36), C/EBPβ (2-4), and CBFA1 (40). The potential clinical importance of these observations is suggested by the identification of naturally occurring Rb mutants whose protein products fail to interact with E2F, yet which maintain the ability to promote differentiation (19, 34). However, whether the ability of pRb to promote differentiation participates in tumor suppression has not been tested.
Cell culture-based experiments have suggested that Ras is a downstream effector of pRb in the control of cellular differentiation. Nullizygosity for the retinoblastoma gene (Rb) or inactivation of pRb by simian virus 40 T antigen in murine fibroblasts results in aberrantly elevated Ras and mitogen-activated protein kinase (MAPK) activity (22, 31). And the effect of Rb loss on Ras has been linked to a failure in differentiation. Inhibition of Ras in Rb-deficient myoblasts significantly potentiates the activity of MyoD and the expression of a marker of muscle differentiation (22). Similarly, the activity of the glucocorticoid receptor is restored by inhibition of Ras activity in Rb−/− fibroblasts (22). These findings are in keeping with the observation that oncogenic Ras possesses the ability to block the activity of these transcription factors. Additionally, inhibition of the elevated MAPK activity in Rb-deficient mouse embryonic fibroblasts (MEFs) restores their ability to undergo adipogenesis (12). These studies suggest that pRb antagonizes Ras signaling during differentiation.
Here we have investigated the requirement for K-ras function in developmental defects and tumor formation caused by loss of Rb. We find that heterozygosity for K-ras reverses many of the differentiation defects observed in Rb-deficient embryos. In addition, pituitary tumors arising in Rb K-ras heterozygotes are more differentiated than those in Rb+/− mice. These findings link the ability of Rb to positively influence differentiation to its tumor suppressor function.
MATERIALS AND METHODS
Generation of mice and embryos.
Parental Rb+/− (15) and K-ras+/− (16) mice were maintained on a mixed C57BL/6 × 129/Sv genetic background and intercrossed. From the subsequent founders, Rb+/− K-ras+/− females were crossed with Rb+/− K-ras+/− males for both timed pregnancies and the generation of a cohort of adult mice. Timed pregnancies were established by the detection of a plug, taken as embryonic day 0.5 (E0.5). Mice and embryos were genotyped by PCR of genomic DNA extracted from tails and yolk sacs, respectively, as previously described (15, 16). All animal experiments were performed at the Dana-Farber Cancer Institute Animal Resource Facility in accordance with the guidelines of the National Institutes of Health.
Histology and immunohistochemistry.
Embryos were fixed in 4% paraformaldehyde-phosphate buffered saline (PBS, pH 7.4) for 12 h, preserved in 70% ethanol for 5 days with occasional changes of buffer, and embedded in paraffin for sectioning. Sections (thickness, 6 μm) were stained with hematoxylin and eosin (H&E). Unstained sections were incubated with a monoclonal antibody (MY-32; Sigma) to myosin heavy chain (MHC) following deparaffinization and rehydration. To identify proliferating cells, 50 μg of 5-bromodeoxyuridine (BrdU)/g of body weight was injected intraperitoneally into pregnant mice 1 h prior to sacrifice. Sections were treated with 0.05 mM trypsin-2 N HCl in PBS (pH 6.0), blocked with 6% goat serum, and incubated with an anti-BrdU monoclonal antibody (B44; Becton Dickinson) in the presence of 0.5% Tween 20 in PBS (pH 7.4). The bound primary antibody (anti-MHC or anti-BrdU) was detected with the ABC mouse peroxidase detection system (Santa Cruz Biotechnology). For analysis of cell death, embryos were fixed in formalin (3.7% formaldehyde in PBS), from which tissue sections were generated. Apoptosis was measured by a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay using the ApopTag mouse peroxidase plus system (Intergen). Adult animals were fixed in Bouin's solution, and tumor tissue sections were stained with H&E or incubated with a monoclonal antibody to adrenocorticotropin (ACTH) (N1531; Dako) or PCNA (PC10; Sigma), and bound primary antibody was detected with the ENVISION/HRP system (Dako). ACTH-stained sections were pretreated in 0.105 M citrate buffer (pH 6.0) at 120°C for 1 min. All counterstaining was performed with 0.5% methyl blue (Sigma) in 0.1 M sodium acetate buffer (pH 4.0).
Quantifications.
MHC staining and RNase protection assay results were quantified as described elsewhere (38). For quantification of ACTH staining, the intensities of signals in four fields were measured with the aid of the NIH Image program (version 1.61), and the relative level of signal was determined after the level in tumors from Rb+/− K-ras+/+ mice was set to 1.0. For quantification of PCNA staining, the numbers of positive nuclei and total nuclei were counted in four fields. The relative expression ratio was calculated on the basis of the ratio in tumors from Rb+/− K-ras+/+ mice, which was set to 1.0.
RNase protection assay.
RNase protection assays were performed as described elsewhere (38).
Statistics.
Differences in the survival of adult mice and in areas occupied by tumors were analyzed by the log rank test and Fisher's exact test, respectively. A P value of <0.05 was considered to indicate a significant difference.
PCR analysis of tumor DNA.
Pituitary tumor regions were marked while in paraffin blocks after observation of corresponding areas in H&E-stained sections and were manually dissected. Subsequently, paraffin blocks were first subjected to a brief heat treatment at 65°C and then suspended in 1× lysis buffer (Applied Biosystems) in the presence of 20 μg of proteinase K (Sigma)/ml at 55°C for 16 h, after which genomic DNA was extracted as described elsewhere (38). PCR was used to determine the genotype for Rb and K-ras as described previously (15, 16).
MEFs and transcriptional transactivation assays.
MEFs were isolated, and MyoD transcriptional transactivation experiments were performed, as described previously (38). pBabe-K-raswt was constructed by PCR amplification using prim-ers 5′-CGCGGATCCGCCACCATGACTGAATATAAACTTGTGGTAGTT-3′ and 5′-CTGCAGAACCAATGCATTGGTTACATAATTACACACTTTGTCTTTGACTT-3′ with pZIPNeoSV(X)I-EE-K-ras4B (17) as the template, followed by digestion with BamHI and BstXI. The sequence was confirmed after subcloning into pBabe-puro. The plasmid encoding H-RasV12 (pCXN2-H-rasV12 [37]) and those encoding pRb (pSG5L-HA-RB), pRb(661W) (pSG5L-HA-RB;661W), pRb(Δex4) (pSG5L-HA-RBΔex4), and pRb(Δex22) (pSG5L-HA-RBΔex22) (34) have been described elsewhere. MyoD expression was detected by using a polyclonal antibody (M-318; Santa Cruz Biotechnology).
MyoD infection and BrdU incorporation assay.
MEFs were infected with a retrovirus encoding MyoD as described elsewhere (38). Forty-eight hours after infection, cells were incubated in differentiation medium (Dulbecco's modified Eagle medium [DMEM] supplemented with 2% horse serum and 10 μg of insulin [Sigma]/ml) for 72 h. In one experiment, cells were further incubated in differentiation medium in the presence of 10 μM BrdU (Becton Dickinson) for 24 h. In another, cells were subsequently restimulated with 20% fetal bovine serum (FBS) in DMEM containing 10 μM BrdU for 24 h. Cells were fixed and stained with an anti-BrdU antibody (B44; Becton Dickinson) as described previously (38). The percentage of cells incorporating BrdU was determined by scoring at least 300 cells.
Cell cycle analysis.
MEFs were maintained in DMEM supplemented with 10% FBS and were harvested with 0.5% trypsin-0.53 mM EDTA. Cells resuspended in PBS were stained with 50 μg of propidium iodide (Molecular Probes)/ml for 30 min in the presence of 0.5 mg of RNase A (Sigma)/ml and were then analyzed by flow cytometry (FACSVantage flow cytometer; Becton Dickinson).
Measurement of K-Ras activity.
K-Ras activity was measured as described elsewhere (22). Five hundred micrograms of whole-cell lysate was used to perform each assay, and 50 μg was used for determination of total K-Ras protein by using a c-K-Ras-specific monoclonal antibody (234-4.2; Sigma).
RESULTS
Prolonged life spans of Rb-deficient embryos lacking a single K-ras allele.
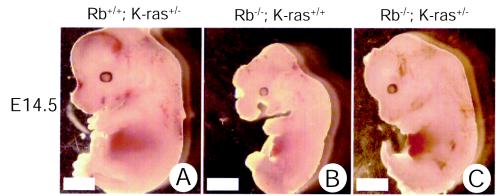
Rb-deficient embryos die at midgestation, displaying a number of developmental abnormalities. Thus, we first tested whether loss of one or both alleles of K-ras prolonged the survival of Rb-deficient embryos. When Rb+/− K-ras+/− mice were intercrossed, no viable Rb−/− K-ras+/− or Rb−/− K-ras−/− pups were recovered. Embryonic viability was then assessed during a time course of gestation (Table 1). Most Rb nullizygous embryos were dead at E14.5 (70%), and no viable embryos were recovered at E15.5 or beyond, in agreement with previous reports (5, 15, 21). K-ras-deficient embryos showed a broad window in the timing of their deaths, as previously reported (16, 18). Remarkably, most Rb-deficient embryos lacking a single allele of K-ras were alive at E14.5 (83%) and E15.5 (64%). Moreover, they displayed a normal appearance, like that of their wild-type littermates (Fig. 1), although the crown-to-rump length, estimated from H&E-stained midsagittal sections, was 11.2% less (at E13.5; n = 12) for Rb+/− K-ras+/− embryos than for their wild-type littermates. Rb−/− animals typically showed highly edematous features, leading to increased crown-to-rump length (7.6% at E12.5; n = 5) compared to that of wild-type littermates. By contrast, loss of both alleles of K-ras and Rb resulted in death as early as E9.5 (Table 1) (data not shown). These results suggest that loss of one but not both alleles of K-ras can significantly prolong the survival of the Rb-deficient embryo.
TABLE 1.
Timing of lethality in progeny arising from Rb+/− × K-ras+/− crossesa
| Stage | No. of live (nonviable) embryos recoveredb
|
|||||
|---|---|---|---|---|---|---|
| Rb+/+ K-ras+/+ | Rb+/+ K-ras+/− | Rb+/+ K-ras−/− | Rb−/− K-ras+/+ | Rb−/− K-ras+/− | Rb−/− K-ras−/− | |
| E9.5 | 2 | 6 | 2 | 3 | 5 | 3 (8) |
| E10.5 | 3 | 7 | 3 (1) | 3 | 6 | (3) |
| E11.5 | 6 | 12 | 5 (2) | 5 | 8 | (2) |
| E12.5 | 4 | 11 | 4 (1) | 5 (3) | 11 | (4) |
| E13.5 | 7 | 13 | 4 (3) | 4 (4) | 12 (1) | (5) |
| E14.5 | 9 | 17 | 4 (5) | 3 (7) | 15 (3) | ND |
| E15.5 | 6 | 10 | 2 (4) | (5) | 7 (4) | ND |
| E16.5 | 7 | 10 | 1 (5) | (6) | (10) | ND |
Embryos with the indicated genotypes were recovered at different embryonic stages and assessed for viability (beating heart).
ND, no data (no embryos were recovered).
FIG. 1.
Effects of heterozygous loss of K-ras on the appearance of Rb-deficient embryos. Live littermate embryos with the indicated genotypes were recovered at E14.5 and immediately photographed in saline. Bars, 1.0 mm.
Improved skeletal muscle development in Rb-deficient embryos following loss of a single K-ras allele.
Given the results noted above, we asked whether heterozygosity for K-ras might reverse some of the developmental abnormalities that characterize Rb-deficient embryos. The most prominent cell-autonomous defects resulting from loss of Rb are those in skeletal muscle development at midgestation (7, 45, 47). Some of these abnormalities become more pronounced and apparent at late gestational stages. Specifically, embryos lacking Rb and E2f1 (41), E2f3 (49), or Id2 (20), or with partial restoration of Rb expression (47), or mutant embryos supplied with a wild-type placenta (7, 45), all of which live longer than Rb-deficient embryos, show similar defects in muscle development. Abnormalities include reduced myotube length, fiber density, and expression of late markers of differentiation, as well as evidence of ectopic S-phase entry and cell death. We determined whether loss of a single allele of K-ras affected these Rb-dependent phenotypes.
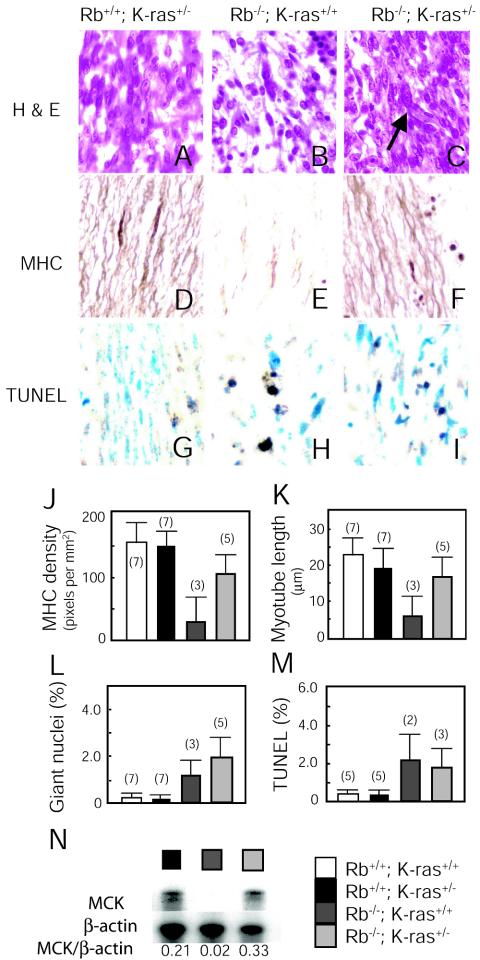
Axial muscles from live Rb+/+ K-ras+/−, Rb−/− K-ras+/+, and Rb−/− K-ras+/− E14.5 littermates were compared. H&E staining of Rb-deficient muscle sections revealed a conspicuous reduction in muscle fiber density compared to Rb+/+ K-ras+/− tissue (Fig. 2A to C). This defect was most pronounced in sections immunostained with an antibody to MHC (Fig. 2D and E). MHC-stained sections also showed reductions in the length and thickness of individual myotubes. In contrast, Rb−/− K-ras+/− axial muscle showed near-normal fiber density (Fig. 2F and J) as well as thickness and length of myotubes (Fig. 2F and K). These features were noted despite evidence of continuing DNA replication (Fig. 2C and L), shown previously to be indicated by abnormally large nuclei (47), and cells undergoing apoptosis (Fig. 2G to I and M). Together, these results suggest that loss of one K-ras allele prevents, although incompletely, the reduction in muscle fiber density and irregular myotube formation that characterize Rb-deficient skeletal muscle, despite having no impact on ectopic DNA synthesis or cell death.
FIG. 2.
Effects of heterozygous loss of K-ras on skeletal muscle development in E14.5 Rb-deficient embryos. (A to F) Longitudinal sections through the fibers of thoracic somite-associated skeletal muscles from sagittal sections of live E14.5 embryos derived from the same litter with the indicated genotypes were stained with H&E (A to C) or immunostained with an antibody to MHC (D to F). A large nucleus (arrow) in myotubes is indicated in panel C. Magnification, ×40. (G to I) Apoptosis (TUNEL) observed in myoblasts of thoracic skeletal muscles of live E14.5 embryos of the indicated genotypes. Magnification, ×40. (J) Density of MHC staining in myotubes. Ten myotubes per embryo were analyzed; average densities of the MHC signal ± standard errors are shown. Numbers of embryos analyzed are given in parentheses. (K) The lengths of myotubes immunostained with an antibody to MHC were quantified. Longitudinal sections of thoracic skeletal muscles from live E14.5 embryos were analyzed by microscopic observation. Twenty myotubes per embryo were measured; average lengths ± standard errors are presented. Numbers of embryos analyzed are given in parentheses. (L) One hundred myotubes in the thoracic skeletal muscle were analyzed for the presence of giant nucleifollowing staining with H&E. Average percentages ± standard errors are presented. Numbers of embryos analyzed are given in parentheses. (M) The level of apoptosis was quantified by counting the frequency of TUNEL-positive cells per 300 nuclei analyzed in the thoracic muscle. Average percentages ± standard errors are presented. Numbers of embryos analyzed are given in parentheses. (N) Expression of MCK in live E14.5 littermates determined by RNase protection assays with RNAs derived from carcasses. Ratios of MCK to β-actin expression are given below the autoradiograph.
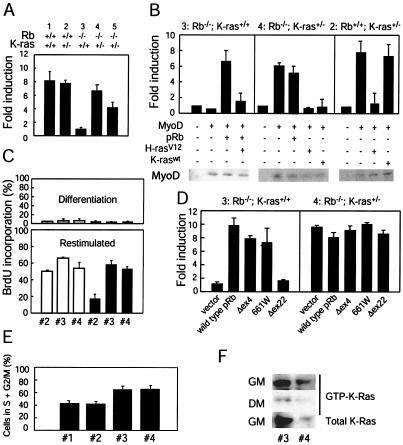
At the molecular level, Rb-deficient muscle is characterized by a failure to induce certain markers of differentiation, e.g., muscle creatine kinase (MCK) (47), which has been linked to a defect in MyoD transcriptional activity (27). Analysis of transcript levels revealed that loss of a single K-ras allele resulted in restoration in MCK expression in Rb-deficient skeletal muscle (Fig. 2N). We also assessed the influence of K-ras heterozygosity in Rb-deficient cells on MyoD function. To this end, Rb+/+ K-ras+/+, Rb+/+ K-ras+/−, Rb−/− K-ras+/+, and Rb−/− K-ras+/− MEFs were generated. MEFs were transfected with a plasmid encoding MyoD and an MCK promoter reporter and were subsequently cultured under conditions known to induce myogenic differentiation. The activity of MyoD was significantly lower in Rb−/− K-ras+/+ myoblasts than in those expressing wild-type Rb (Fig. 3A). In contrast, Rb−/− K-ras+/− myoblasts displayed normal levels of MyoD activity. To rule out the possibility that events secondary to the loss of Rb or K-ras might account for the results described above, a number of experiments were performed. Reintroduction of Rb into Rb-deficient myoblasts restored the activity of MyoD, and oncogenic ras could overcome this effect (Fig. 3B), observations that are consistent with the notion that deregulated Ras signaling following loss of Rb leads to inhibition of MyoD. Further, ectopic expression of either wild-type K-ras or oncogenic H-ras in Rb−/− K-ras+/− myoblasts leads to inhibition of MyoD transcriptional activity (Fig. 3B). By contrast, introduction of K-ras into Rb+/+ K-ras+/− myoblasts has no effect on MyoD activity (Fig. 3B). This suggests that a certain level of endogenous K-Ras activity is required for the negative effect of Rb loss on MyoD activity to be manifest and that the specific loss of K-ras in the context of Rb nullizygosity is responsible for the restoration of MyoD activity in Rb−/− K-ras+/− myoblasts. Together, these in vivo and in vitro analyses suggest that loss of a single K-ras allele can reverse the defect in MyoD function resulting from deficiency in Rb.
FIG. 3.
Effects of K-ras heterozygosity on MyoD transcriptional activity in Rb-deficient myoblasts. (A) MEFs of the indicated genotypes were transfected with an MCK promoter reporter construct (MCK-luc; 0.25 μg), pCSA-MyoD (1.25 μg), and pCMV-β-gal (0.25 μg). Twenty-four hours later, the cells were placed in differentiation medium for 48 h. Luciferase and β-galactosidase activities were determined, and normalized fold activations were calculated relative to the corrected luciferase activity in the absence of MyoD. Results are means ± standard errors for three independent experiments performed in triplicate. Numbers above the bar graph represent the particular MEFs used; groups 1 and 5 are derived from matched littermates, as are groups 2, 3, and 4. (B) MEFs 3, 4, and 2 were transfected as described for panel A. Plasmids encoding pRb (0.25 μg), H-rasV12 (0.5 μg), and K-raswt (0.5 μg) were included in the transfections as indicated. Fold activations were calculated relative to the corrected luciferase activity in the absence of MyoD. Results are means ± standard errors for three independent experiments performed in triplicate. Immunoblots for ectopically expressed MyoD are shown. (C) MEFs 2, 3, and 4 (designations as described for panel A) were infected with a retrovirus encoding MyoD (filled bars) or empty vector (open bars) and cultured under differentiation conditions for 72 h. Subsequently, cells were either further cultured in differentiation medium or restimulated with 20% FBS in the presence of BrdU for 24 h. Percentages of cells incorporating BrdU under differentiation conditions (upper panel) and following restimulation (lower panel) were determined. Results are means ± standard errors for two independent experiments. (D) MEFs 3 and 4 were transfected as described for panel A. A plasmid (0.25 μg) encoding pRb, pRb(661W), pRb(Δex4), or pRb(Δex22), or a vector control, was included in the transfection as indicated. Cells were treated as described for panel A, and fold activations were calculated relative to the corrected luciferase activity in the absence of MyoD. Results are means ± standard errors from three independent experiments performed in duplicate. (E) The cell cycle distribution of asynchronous cultures of MEFs 1, 2, 3, and 4 (designations as described for panel A) was determined by fluorescence-activated cell sorting. Results are means ± standard errors for percentages of cells in S and G2/M from four independent experiments. (F) MEFs 3 and 4 (designations as described for panel A) were cultured in 2% horse serum (differentiation medium [DM]) for 72 h. At this time, the level of activated, GTP-bound K-Ras was determined (middle panel). Alternatively, cells cultured in the presence of horse serum (low levels of mitogens) were restimulated with 20% fetal bovine serum (growth medium [GM]) for 6 h; at this time, the level of active K-Ras wasdetermined (top panel). Whole-cell lysates were analyzed for total K-Ras protein levels (bottom panel). Results are representative of three independent experiments.
Though the ability of pRb to influence cell cycle progression and MyoD function are genetically separable (34), it is formally possible that in the absence of Rb, heterozygosity for K-ras influences the ability of myoblasts to withdraw from the cell cycle, which in turn affects MyoD activity. To explore this possibility, the abilities of Rb+/+ K-ras+/−, Rb-deficient, and Rb−/− K-ras+/− myoblasts to withdraw from the cell cycle during myogenic differentiation were compared. MEFs were infected with a retrovirus encoding MyoD and subsequently cultured under conditions known to induce differentiation, and the incorporation of BrdU was measured as an indication of ongoing DNA synthesis. Low levels of BrdU incorporation were noted in Rb+/+ K-ras+/− as well as in Rb−/− and Rb−/− K-ras+/− myoblasts (Fig. 3C), indicating that K-ras heterozygosity did not influence the ability of Rb−/− myoblasts to withdraw from the cell cycle during differentiation. However, upon restimulation of these differentiated myoblasts, both Rb−/− and Rb−/− K-ras+/− myoblasts, unlike those harboring wild-type Rb, showed higher levels of BrdU incorporation (Fig. 3C). These results are in keeping with the previous demonstration that Rb-deficient myoblasts fail to maintain a terminal cell cycle arrest (27), and they indicate that K-ras heterozygosity does not rescue this defect. Further, we determined whether two pRb mutants, 661W and Δex4, previously shown to be deficient in bringing about G1 arrest but to retain the ability to positively affect MyoD activity in an osteosarcoma cell line (34), could restore MyoD activity in our system. These mutants, but not a pRb-null mutant (Δex22), were capable of potentiating MyoD activity in Rb-deficient myoblasts to a level comparable to that observed with the wild-type protein (Fig. 3D). Further, when analyzed in Rb−/− K-ras+/− myoblasts, pRb(661W) and pRb(Δex4) did not further activate MyoD (Fig. 3D), reinforcing the notion that restoration of appropriate cell cycle regulation is not required in a significant way to restore MyoD function in Rb−/− myoblasts.
To analyze the cell cycle issue further, asynchronous populations of MEFs were studied. In agreement with previous observations, Rb-deficient MEFs displayed higher percentages of cells in S and G2/M than wild-type MEFs (Fig. 3E). Rb−/− K-ras+/− MEFs showed a cell cycle distribution similar to that of Rb-deficient cells (Fig. 3E), suggesting that loss of a single K-ras allele does not influence the distribution of cells in each of the cell cycle phases. These findings are consistent with the previous demonstration that Rb-deficient cells, unlike wild-type cells, fail to arrest in G1 following a block to Ras function (23, 26, 30). In addition, we confirmed that heterozygosity for K-ras does reduce the level of Ras activity for this isoform by approximately one-half (Fig. 3F). Together with the observations noted above, these findings suggest that loss of a single K-ras allele in Rb-deficient cells has a more profound impact on the ability of these cells to differentiate than on their cell cycle distribution.
K-ras heterozygosity does not affect the deregulated proliferation that characterizes Rb-deficient embryos.
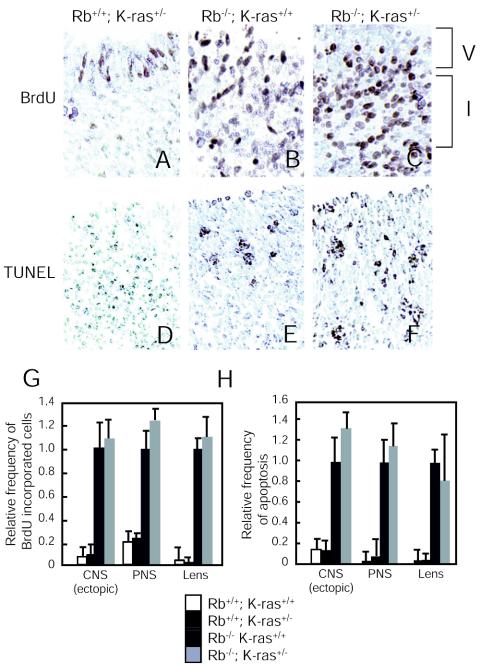
Rb-deficient embryos are characterized by deregulated proliferation and apoptosis in the CNS and peripheral nervous system (PNS) and the developing lens (5, 15, 21), which occur in a cell-autonomous or non-cell-autonomous manner (7, 25, 45). To determine whether loss of one K-ras allele affected these Rb-dependent phenotypes, BrdU was injected into pregnant females at day 13.5 of term, and immunological detection of incorporated BrdU was used as a measure of DNA synthesis. BrdU-positive cells were readily apparent in the intermediate zone of the hindbrain (CNS) in Rb-deficient but not wild-type embryos (Fig. 4A and B). Analysis of Rb−/− K-ras+/− embryos also revealed ectopic DNA synthesis in this tissue (Fig. 4C). Similar findings were made upon inspection of the dorsal root ganglion of the PNS and the fiber cell compartment of the lens (Fig. 4G). TUNEL staining was used to detect apoptosis. Rb−/− K-ras+/− embryos, like Rb-deficient embryos, showed elevated levels of apoptosis in the cortical region around the fourth ventricle of the brain (CNS) (Fig. 4D to F), in the PNS, and in the lens compared to those for wild-type embryos (Fig. 4H). These results suggest that the ectopic proliferation and cell death observed in Rb-deficient embryos are not affected by the loss of a single K-ras allele.
FIG. 4.
Effects of heterozygous K-ras loss on ectopic S-phase entry and cell death. (A-C) Transverse sections of the ventricular (V) and intermediate (I) zones of the hindbrain in the CNS from E13.5 embryos of the indicated genotypes were stained for cells in S phase (BrdU). Magnification, ×20. (D-F) Midsagittal sections of the cortical region around the fourth ventricle from E13.5 embryos of the indicated genotypes were stained for apoptotic cells (TUNEL). Magnification, ×20. (G) The level of ectopic S-phase entry was quantified by counting the frequency of BrdU-positive cells per unit area in tissue sections of the intermediate zone of the hindbrain (CNS [ectopic]), dorsal root ganglia (PNS), and fiber compartment of the lens (Lens). Total cell numbers were determined by counting cells counterstained with methyl blue. Rb−/− samples were set to 1.0, and the relative ratios of BrdU-positive cells are displayed. Values are means ± standard errors for two to four embryos. (H) The level of apoptosis was quantified by counting the frequency of TUNEL-positive cells per unit area of tissues: the cortical region around the fourth ventricle of the CNS, the dorsal root ganglia of the PNS, and the fiber compartment of the lens. Total cell numbers were determined by counting cells counterstained with methyl blue. Rb−/− samples were set to 1.0, and the relative ratios of TUNEL-positive cells are displayed. Values are means ± standard errors for two to four embryos.
Heterozygosity for K-ras affects the differentiation status of pituitary tumors resulting from loss of Rb.
Rb+/− mice are tumor prone, resulting in their shorter life span. Given the results noted above indicating that heterozygosity for K-ras rescues a unique subset of developmental defects associated with Rb nullizygosity, we determined whether loss of a single K-ras allele affects the survival of Rb heterozygotes.
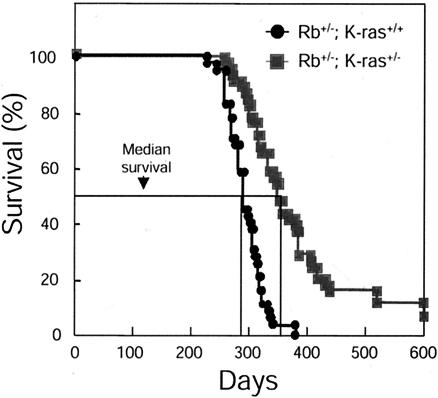
To determine whether loss of a single K-ras allele affects the viability of Rb heterozygotes, a cohort of Rb+/− K-ras+/+ and Rb+/− K-ras+/− mice was observed for 600 days after birth. Loss of a single K-ras allele was found to significantly prolong the survival of Rb heterozygotes (P = 0.0054 by the log rank test) (Fig. 5). The mean age at death of Rb heterozygotes was 270 days (range, 210 to 381 days; standard deviation, ±24 days). In contrast, the mean age at death for Rb+/− K-ras+/− mice was 381 days (range, 241 to 598 days; standard deviation, ±96 days), representing a 41% extension in the life span with loss of a single K-ras allele. Several Rb+/− K-ras+/− mice (6%) survived beyond 598 days, while no Rb+/− K-ras+/+ animals lived past 381 days. The survival curve for Rb+/+ K-ras+/− animals was similar to that of wild-type mice, with more than 90% living beyond 600 days (data not shown). These observations suggest that heterozygosity for K-ras can extend the life spans of Rb+/− mice.
FIG. 5.
Effects of K-ras heterozygosity on the survival of Rb+/− mice. Progeny arising from Rb+/− × K-ras+/− intercrosses were aged together. Shown are survival curves for Rb+/− K-ras+/+ (n = 44) and Rb+/− K-ras+/− (n = 48) mice. Percent survival represents the percentage of the initial starting population surviving at a given age (in days) for the indicated genotype. Median survival is indicated.
Rb heterozygotes develop pituitary adenocarcinomas of the intermediate lobe, resulting in compression of the brain, and medullary (C-cell) thyroid adenomas at high frequencies; the former are responsible for their shorter survival. To examine if loss of a single K-ras allele affects either the incidence or the size of pituitary tumors, we observed the intracranial structures in sick mice after sacrifice and perfusion with a fixative. Frequencies of appearance of macroscopically detectable pituitary tumors were similar in Rb+/− K-ras+/+ (96%) and Rb+/− K-ras+/− (93%) mice, suggesting that loss of a single K-ras allele had no effect on the penetrance of pituitary tumor formation in Rb heterozygotes.
Rb+/− K-ras+/+ and Rb+/− K-ras+/− mice developed tumors of the melanotrophic cells that occupy the intermediate lobe of the pituitary gland. Histological analysis revealed that, at the time of death, tumors arising in Rb+/− K-ras+/− mice tended to be larger than those in Rb+/− K-ras+/+ animals (data not shown), raising the possibility that the prolonged survival of Rb+/− K-ras+/− mice may be a result of their developing less-aggressive pituitary tumors.
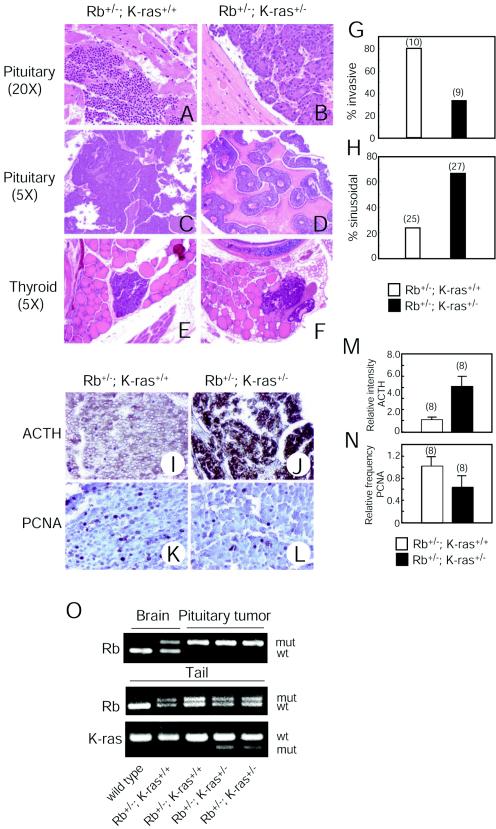
To investigate this issue further, mice were sacrificed at a specific time and then compared. Analysis of animals at 280 days, representing the mean survival of Rb heterozygotes, revealed that the areas of tumors arising in Rb+/− K-ras+/− mice were 1.8-fold smaller than those in Rb+/− K-ras+/+ animals (Table 2), suggesting that pituitary tumors grow more slowly in mice lacking a single K-ras allele. Tumors in Rb+/− K-ras+/− mice analyzed at the age of 360 days (mean survival of Rb+/− K-ras+/− mice) were similar or even larger than those in Rb+/− K-ras+/+ mice at the age of 280 days (Table 2), suggesting that for the same tumor volume in the two genotypes, those arising in Rb+/− K-ras+/− mice might cause less intracranial pressure due to adaptation over time. Further, pituitary tumors arising in Rb+/− K-ras+/+ mice displayed signs of local invasion and were poorly encapsulated; these features were significantly less pronounced in tumors occurring in Rb+/− K-ras+/− mice (Fig. 6A, B, and G).
TABLE 2.
Comparison of pituitary tumor sizesa
| Measurement | Rb+/− K-ras+/+ mice at 280 days (n = 10) | Rb+/− K-ras+/− mice at 280 days (n = 8) | Rb+/− K-ras+/− mice at 360 days (n = 6) |
|---|---|---|---|
| Mean area (mm2) ± SD | 38.9b ± 9.6 | 21.6b ± 11.2 | 53.6 ± 12.0 |
| Range (minimum to maximum) | 12.0-51.2 | 11.2-44.8 | 19.2-61.2 |
Pituitary tumors were observed in mice with the indicated genotypes and ages. Tumor areas were determined under microscopy at the median section of tumors stained with H&E by measuring the longest diameter and its longest perpendicular line.
P = 0.029.
FIG. 6.
Effects of K-ras heterozygosity on characteristics of tumors arising in Rb+/− mice. (A-D) H&E-stained sections of pituitary adenocarcinomas from mice of the indicated genotypes. Magnifications, ×18 (A and B) and ×4.5 (C and D). Original magnifications are given in parentheses. (E and F) H&E-stained sections of C-cell adenomas from mice of the indicated genotypes. Magnification, ×4.5. Original magnification is given in parentheses. (G) Frequency of appearance of invasive tumors. Genotypes and numbers of mice analyzed are given. (H) Frequencyof appearance of sinusoidal (versus diffuse) pattern of growth. Genotypes and numbers of mice analyzed are given. (I and J) Anti-ACTH immunostaining of pituitary tumors arising in mice of the indicated genotypes. Magnification, ×18. (K and L) Anti-PCNA immunostaining of pituitary tumors arising in mice of the indicated genotypes. Magnification, ×36. (M) Quantification of ACTH immunostaining. Genotypes and numbers of mice analyzed are given. Bars represent means ± standard errors. (N) Quantification of PCNA immunostaining. Genotypes and numbers of mice analyzed are given. Bars represent means ± standard errors. (O) Genotyping of Rb and K-ras in normal brain tissue, pituitary tumors, and tails from mice of the indicated genotypes by PCR. The weak band for the wild-type Rb allele that appeared in one of the pituitary tumors is likely derived from nontumor cells residing in tumors. wt, wild type; mut, mutant.
Morphological studies of pituitary tumors were performed, restricting the analysis to the window of time when 80 to 20% of the mice were still alive for the respective genotypes. The majority of tumors arising in Rb+/− K-ras+/+ mice showed a poorly differentiated, diffuse pattern of growth (Fig. 6C and H). In striking contrast, most tumors analyzed in Rb+/− K-ras+/− animals displayed a well-differentiated sinusoidal pattern of growth (Fig. 6D and H). Indeed, a sinusoidal morphology is one of the features used to distinguish pituitary adenomas from pituitary adenocarcinomas (1). This suggests that loss of a single K-ras allele results in the formation of less-aggressive pituitary tumors owing to their more-differentiated phenotype.
To explore the differentiation status of the pituitary tumors further, ACTH expression was studied. Secretion of ACTH is often associated with melanotrophic tumors of the intermediate lobe (1), and it has been reported that there is an inverse correlation between ACTH levels and the aggressiveness of pituitary tumors (43). Analysis of several mice at the approximate median survival for the cohort revealed that pituitary tumors arising in Rb+/− K-ras+/− mice express significantly higher levels of ACTH than those in Rb+/− K-ras+/+ animals (Fig. 6I, J, and M), consistent with the morphological studies described above. We also analyzed the growth fraction of the tumors by staining the same sections used in the ACTH studies for PCNA. Per unit area, pituitary tumors arising in Rb+/− K-ras+/− mice showed a 30% reduction in the frequency of PCNA-positive cells compared to those in Rb+/− K-ras+/+ animals (Fig. 6K, L, and N). Loss of the remaining wild-type Rb allele was observed in pituitary tumors from both Rb+/− K-ras+/+ and Rb+/− K-ras+/− mice, suggesting that K-ras heterozygosity does not influence this genetic event (Fig. 6O). Together these observations suggest that the reduced volume of the tumors arising in Rb+/− K-ras+/− mice is due to their more-differentiated characteristics and somewhat lower proliferative activity rather than to delayed tumor onset.
C-cell tumors resulting from loss of Rb cannot be detected macroscopically. Histological analysis of multiple sections revealed that 100% of the Rb+/− K-ras+/+ mice developed C-cell adenomas (n = 26). Similar findings were noted for Rb+/− K-ras+/− mice (n = 26). Rb+/− K-ras+/+ and Rb+/− K-ras+/− mice sacrificed at 280 days after birth showed the presence of histologically similar tumors (Fig. 6E and F). These data suggest that, in contrast to pituitary adenocarcinomas, loss of a K-ras allele does not influence the initiation or progression of medullary thyroid adenomas resulting from loss of Rb.
DISCUSSION
A block to differentiation is thought to contribute to tumor formation. However, there is little understanding of the genetic pathways that contribute to tumorigenesis by disrupting the processes of differentiation (39). Part of this difficulty stems from the observation that aberrant alterations in cell cycle control can hinder the normal execution of various differentiation programs. This can complicate efforts to dissect the relative contributions of specific pathways governing cell cycle progression and differentiation to tumorigenesis. pRb is a case in point, as it is involved in both processes. Genetic alterations affecting components of the pRb pathway (Rb, cyclin D, cdk4, p16) leading to deregulation of E2F activity are thought to provide tumors with a selective advantage by increasing their proliferative potential. It is likewise well established that pRb can also positively regulate differentiation through mechanisms distinct from its regulation of E2F and proliferation. And in this study we provide genetic evidence that the ability of pRb to promote differentiation is linked to its tumor suppressor function.
Several defects characterize Rb-deficient embryos, most notably a failure in fetal liver erythropoiesis, ectopic proliferation and apoptosis of the central and peripheral nervous systems and lens, and aberrant differentiation of skeletal muscle. Recently, it has been demonstrated that the origin of the defect in erythropoiesis and ectopic apoptosis in the central nervous system is the Rb-deficient placenta and not the embryo proper, while the other abnormalities noted above still persist when Rb-deficient embryos are supplied with a wild-type placenta (7, 45). Our finding that loss of a single K-ras allele can extend the life spans of Rb-deficient embryos suggests that fetal liver erythropoiesis is improved in these animals. Preliminary studies indicate that Rb+/− K-ras+/− embryos do display an increased hematocrit compared to that of Rb heterozygotes, likely reflecting at least a partial rescue of erythropoiesis (C. Takahashi, M. Socolovsky, and M. E. Ewen, unpublished data). Further, heterozygosity for K-ras rescues many of the skeletal muscle defects that characterize Rb-deficient embryos. By contrast, we find no effect of K-ras heterozygosity on the ectopic proliferation and apoptosis observed in the CNS, PNS, and lens resulting from Rb deficiency. Thus, loss of K-ras appears to affect a unique subset of developmental defects that characterize Rb-deficient embryos.
The differentiation function of pRb has been best characterized for myogenesis; the requirement for Rb during skeletal muscle differentiation has been documented both in vivo and in vitro. Cell culture studies indicate that pRb controls myogenesis in an E2F-independent manner (34). In agreement with this observation, Rb E2f compound embryos, while living longer than Rb-deficient embryos, continue to display abnormalities in muscle differentiation (41, 49). Our findings suggest that the genetic interaction between Rb and ras has a significant role in skeletal muscle development. Loss of a single K-ras allele rescued many of the defects that characterize Rb-deficient skeletal muscle, including fiber density and myotube length. Importantly, expression of MCK, a late marker of muscle differentiation and a transcriptional target of MyoD, was also rescued. Consistent with this observation made in vivo, heterozygosity for K-ras restored MyoD transcriptional activity in myoblasts lacking Rb, in the apparent absence of an influence on the cell cycle. Previously, similar observations were made in an analysis of Rb−/− N-ras−/− embryos and myoblasts (38), suggesting that a reduced level of total ras is required in order to observe an amelioration of defects brought about by Rb deficiency. In contrast to the observed effects on differentiation and MyoD function, loss of a single K-ras allele did not suppress ectopic proliferation in developing Rb-deficient skeletal muscle. Thus, in a given tissue, skeletal muscle, pRb appears to affect proliferation and differentiation through distinct mechanisms.
Understanding how tumor suppressors such as Rb affect processes involved in tumorigenesis is a goal of cancer research. The contribution of the proliferation function of Rb to tumor suppression has been established through analysis of Rb E2f compound mutant mice (see the introduction). By contrast, no role in tumor suppression has been found for Rb's function in promoting differentiation. However, genetic analysis of Rb suggests the possibility that the positive effect of Rb on differentiation does contribute to its tumor suppression function. In classical familial retinoblastoma, bilateral tumors are noted in more than 90% of affected individuals. By contrast, Rb mutations have been identified where carriers show no signs of disease, develop unilateral retinoblastoma, or suffer benign retinomas (8, 19, 24, 29), suggesting that the protein products encoded by these partially penetrant alleles do possess tumor suppressor activity. Characterization of these pRb mutants in vitro indicates that they fail to interact with E2F (19, 34, 42) yet maintain the ability to promote differentiation (34) and restore the aberrantly high levels of Ras activity observed in Rb-deficient cells to normal levels (22). Together, these studies have raised the possibility that Rb might function together with ras to affect tumor suppression by influencing differentiation.
To test this hypothesis and to extend the observations described above to an in vivo setting, we asked whether loss of a ras allele, which should theoretically lower the total level of Ras activity, might affect tumor development in Rb heterozygotes. Our studies reveal that this is the case. Loss of a single K-ras allele resulted in less-aggressive pituitary tumors and correspondingly increased the life spans of Rb heterozygous mice. Rb K-ras heterozygotes developed smaller pituitary tumors than their age-matched Rb+/− littermates. Importantly, and in consonance with our embryological studies, histological and molecular analyses revealed that loss of a K-ras allele resulted in more-differentiated tumors. The modest reduction in the growth fraction of these tumors is likely secondary to their more-differentiated phenotype. These findings are in contrast to those obtained in the analysis of Rb heterozygotes lacking E2f1 or E2f3, where it was found that while the sizes of the pituitary tumors were reduced, the tumors were histologically indistinguishable from those arising in Rb+/− animals (46, 48), suggesting that downstream effectors of Rb in the control of proliferation do not influence the differentiation status of pituitary tumors resulting from Rb loss. Together these observations suggest that loss of Rb contributes to pituitary tumorigenesis by affecting differentiation (through K-ras) and proliferation (through E2f). Consistent with this interpretation, treatment of pituitary tumors arising in Rb+/− mice with an adenovirus expressing wild-type pRb prolonged their survival, and this correlated with a decrease in proliferation and a more-differentiated phenotype (32).
Our analysis of C-cell tumors revealed no effect of K-ras deficiency, suggesting that the signaling output from the genetic interaction between Rb and K-ras influencing the differentiation status during tumor development is cell type specific. Thus far, our embryological studies identify MyoD as a key molecular target of the antagonism between Rb and K-ras signaling during skeletal muscle development. Only when we identify the repertoire of the effectors of Rb/K-ras signaling will we understand the tissue-specific nature of the genetic interaction between Rb and K-ras.
Virtually all studies linking ras proto-oncogenes to tumorigenesis pertain to the oncogenic constitutively active forms. By contrast, the work presented here identifies wild-type K-ras as a participant in pituitary tumor progression following loss of Rb. Thus, there is the possibility that, as a function of the particular genetic lesions driving tumorigenesis and perhaps the tissue type, wild-type ras may contribute materially to the malignant behavior of certain cancers. Current strategies that target oncogenic Ras in human tumors (e.g., farnesyltransferase inhibitors) do not distinguish between the mutant constitutively active and wild-type forms. This suggests that such therapeutic strategies may be applied in the context of cancers that harbor wild-type Ras.
Acknowledgments
We express special thanks to K. Y. Lee, M. Noda, C. McMahon, J. Lamb, D. Livingston, A. Lassar, G. Dranoff, S. Lux, P. Sicinski, M. Ciemerych, W. Kaelin, J. DeCaprio, K. Tsai, R. Takahashi, and T. Arai for help, advice, and encouragement. We thank T. Jacks for kindly providing Rb and K-ras mutant mice, Y. Itokazu for FACS analysis, T. Kawai and Z. Lee for technical help, J. Jackson, A. Lassar, W. Sellers, J. Miyazaki, and W. Wright for plasmids, and J. Lamb, C. McMahon, W. Sellers, and J. Lee for critical review of the manuscript.
This work was supported by National Institutes of Health grants R01CA65842 (to M.E.E.) and P01CA89021 (to M.L.) and by a Massachusetts Prostate Cancer Research Grant and a grant from the Japanese Ministry of Education and Science to C.T. C.T. was supported in part by the NCI-JFCR Scientist Exchange Program. During this work M.E.E. was a Leukemia and Lymphoma Society Scholar.
REFERENCES
- 1.Capen, C. C., E. Karbe, U. Deschl, C. George, P.-G. Germann, C. Gopinath, J. F. Hardisty, J. Kanno, W. Kaufmann, G. Krinke, K. Kuttler, B. Kulwich, C. Landes, B. Lenz, L. Longeart, I. Paulson, E. Sander, and K. Tuch. 2001. Endocrine system, p. 269-322. In U. Mohr (ed.), International classification of rodent tumors: the mouse. Springer-Verlag, Berlin, Germany.
- 2.Charles, A., X. Tang, E. Crouch, J. S. Brody, and Z. X. Xiao. 2001. Retinoblastoma protein complexes with C/EBP proteins and activates C/EBP-mediated transcription. J. Cell. Biochem. 83:414-425. [DOI] [PubMed] [Google Scholar]
- 3.Chen, P.-L., D. J. Riley, Y. Chen, and W.-H. Lee. 1996. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 10:2794-2804. [DOI] [PubMed] [Google Scholar]
- 4.Chen, P.-L., D. J. Riley, S. Chen-Kiang, and W.-H. Lee. 1996. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 93:465-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, A. R., E. R. Maandag, M. van Roon, N. M. T. van der Lugt, M. van der Valk, M. L. Hooper, A. Berns, and H. te Riele. 1992. Requirement for a functional Rb-1 gene in murine development. Nature 359:328-330. [DOI] [PubMed] [Google Scholar]
- 6.Classon, M., B. K. Kennedy, R. Mulloy, and E. Harlow. 2000. Opposing roles of pRB and p107 in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 97:10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bruin, A., L. Wu, H. I. Saavedra, P. Wilson, W. Yang, T. J. Rosol, M. Weinstein, M. L. Rosinson, and G. Leone. 2003. Rb function in extraembryonic lineages suppresses apoptosis in CNS of Rb-deficient mice. Proc. Natl. Acad. Sci. USA 100:6546-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dryja, T. P., J. Rapaport, T. L. McGee, T. M. Nork, and T. L. Schwartz. 1993. Molecular etiology of low-penetrance retinoblastoma in two pedigrees. Am. J. Genet. 52:1122-1128. [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 10.Goodrich, D. W., and W. H. Lee. 1993. Molecular characterization of the retinoblastoma susceptibility gene. Biochim. Biophys. Acta 1155:43-61. [DOI] [PubMed] [Google Scholar]
- 11.Gu, W., J. W. Schneider, G. Condorelli, S. Kaushal, V. Mahdavi, and B. Nadal-Ginard. 1993. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell 72:309-324. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, J. B., R. K. Petersen, C. Jorgensen, and K. Kristiansen. 2002. Deregulated MAPK activity prevents adipocyte differentiation in fibroblasts lacking the retinoblastoma protein. J. Biol. Chem. 277:26335-26339. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, D. J., M. L. Hooper, J. F. Armstrong, and A. R. Clarke. 1995. Effects of heterozygosity for the Rb-1t19neo allele in the mouse. Oncogene 20:1615-1620. [PubMed] [Google Scholar]
- 14.Hu, N., A. Gutsmann, D. C. Herbert, A. Bradley, W.-H. Lee, and E. Y. Lee. 1994. Heterozygous Rb-1 delta 20/+ mice are predisposed to tumors of the pituitary gland with a nearly complete penetrance. Oncogene 9:1021-1027. [PubMed] [Google Scholar]
- 15.Jacks, T., A. Fazeli, E. M. Schmitt, R. T. Bronson, M. A. Goodell, and R. A. Weinberg. 1992. Effects of an Rb mutation in the mouse. Nature 359:295-300. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, L., D. Greenbaum, K. Cichowski, K. Mercer, E. Murphy, E. Schmitt, R. T. Bronson, H. Umanoff, E. Windfried, R. Kucherlapati, and T. Jacks. 1997. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 11:2468-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, M. K., and J. H. Jackson. 1998. Ras-GRF activates Ha-Ras, but not N-Ras or K-Ras 4B, protein in vivo. J. Biol. Chem. 273:1782-1787. [DOI] [PubMed] [Google Scholar]
- 18.Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-Ras is essential for the development of the mouse embryo. Oncogene 15:1151-1159. [DOI] [PubMed] [Google Scholar]
- 19.Kratzke, R. A., G. A. Otterson, A. Hogg, A. B. Coxon, J. Geradts, J. K. Cowell, and F. J. Kaye. 1994. Partial inactivation of RB product in a family with incomplete penetrance of familial retinoblastoma and benign retinal tumors. Oncogene 9:1321-1326. [PubMed] [Google Scholar]
- 20.Lasorella, A., M. Noseda, M. Beyna, and A. Iavarone. 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407:592-598. [DOI] [PubMed] [Google Scholar]
- 21.Lee, E. Y.-H. P., C.-Y. Chang, N. Hu, Y.-C. J. Wang, C.-C. Lai, K. Herrup, W.-H. Lee, and A. Bradley. 1992. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 359:288-294. [DOI] [PubMed] [Google Scholar]
- 22.Lee, K. Y., M. H. Ladha, C. McMahon, and M. E. Ewen. 1999. The retinoblastoma protein is linked to the activation of Ras. Mol. Cell. Biol. 19:7724-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leone, G., J. DeGregori, R. Sears, L. Jakoi, and J. R. Nevins. 1997. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 387:422-426. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann, D. R., B. Brandt, W. Hopping, E. Passarge, and B. Horsthemke. 1994. Distinct RB1 gene mutations with low penetrance in hereditary retinoblastoma. Hum. Genet. 94:349-354. [DOI] [PubMed] [Google Scholar]
- 25.MacPherson, D., J. Sage, D. Crowley, A. Trumpp, R. T. Bronson, and T. Jacks. 2003. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol. Cell. Biol. 23:1044-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittnacht, S., H. Paterson, M. F. Olson, and C. J. Marshall. 1997. Ras signalling is required for inactivation of tumour suppressor pRb cell-cycle control protein. Curr. Biol. 7:219-221. [DOI] [PubMed] [Google Scholar]
- 27.Novitch, B. G., G. J. Mulligan, T. Jacks, and A. B. Lassar. 1996. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J. Cell Biol. 135:441-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novitch, B. G., D. B. Spicer, P. S. Kim, W. L. Cheung, and A. B. Lassar. 1999. pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 9:449-459. [DOI] [PubMed] [Google Scholar]
- 29.Onadim, Z., A. Hogg, P. N. Baird, and J. K. Cowell. 1992. Oncogenic point mutations in exon 20 of the RB1 gene in families showing incomplete penetrance and mild expression of the retinoblastoma phenotype. Proc. Natl. Acad. Sci. USA 89:6177-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peeper, D. S., T. M. Upton, M. H. Ladha, E. Neuman, J. Zalvide, R. Bernards, J. A. DeCaprio, and M. E. Ewen. 1997. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386:177-181. [DOI] [PubMed] [Google Scholar]
- 31.Raptis, L., H. L. Brownell, M. J. Corbley, K. W. Wood, D. Wang, and T. Haliotis. 1997. Cellular ras gene activity is required for full neoplastic transformation by the large tumor antigen of SV40. Cell Growth Differ. 8:891-901. [PubMed] [Google Scholar]
- 32.Riley, D. J., A. Y. Nikitin, and W.-H. Lee. 1996. Adenovirus-mediated retinoblastoma gene therapy suppresses spontaneous pituitary melanotroph tumors in Rb+/− mice. Nat. Med. 2:1316-1321. [DOI] [PubMed] [Google Scholar]
- 33.Saavendra, H. I., L. Wu, A. de Bruin, C. Timmers, T. J. Rosol, M. Weinstein, M. L. Robinson, and G. Leone. 2002. Specificity of E2F1, E2F2 and E2F3 in mediating phenotypes induced by loss of Rb. Cell Growth Differ. 13:215-225. [PubMed] [Google Scholar]
- 34.Sellers, W. R., B. G. Novitch, S. Miyake, A. Heith, G. A. Otterson, F. J. Kaye, A. B. Lassar, and W. G. J. Kaelin. 1998. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor growth. Genes Dev. 12:96-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, P., S. W. Chan, and W. Hong. 2001. Retinoblastoma protein is functionally distinct from its homologues in affecting glucocorticoid receptor-mediated transcription and apoptosis. J. Biol. Chem. 276:13762-13770. [DOI] [PubMed] [Google Scholar]
- 36.Singh, P., J. Coe, and W. Hong. 1995. A role for the retinoblastoma protein in potentiating transcriptional activation of the glucocorticoid receptor. Nature 374:562-565. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi, C., N. Akiyama, T. Matsuzaki, S. Takai, H. Kitayama, and M. Noda. 1996. Characterization of a human MSX-2 cDNA and its fragment isolated as a transformation suppressor gene against v-Ki-ras oncogene. Oncogene 12:2137-2146. [PubMed] [Google Scholar]
- 38.Takahashi, C., R. T. Bronson, M. Socolovsky, B. Contreras, K. Y. Lee, T. Jacks, M. Noda, R. Kucherlapati, and M. E. Ewen. 2003. Rb and N-ras function together to control differentiation in the mouse. Mol. Cell. Biol. 23:5256-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenen, D. G. 2003. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3:89-101. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, D. M., S. A. Carty, D. M. Piscopo, J.-S. Lee, W.-F. Wang, W. C. Forrester, and P. W. Hinds. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 8:303-316. [DOI] [PubMed] [Google Scholar]
- 41.Tsai, K. Y., Y. Hu, K. F. Macleod, D. Crowley, L. Yamasaki, and T. Jacks. 1998. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 2:293-304. [DOI] [PubMed] [Google Scholar]
- 42.Whitaker, L. L., H. Su, R. Baskaran, E. S. Knudsen, and J. Y. J. Wang. 1998. Growth suppression by an E2F-binding-defective retinoblastoma protein (RB): contribution from the RB C pocket. Mol. Cell. Biol. 18:4032-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White, A., and S. Gibson. 1998. ACTH precursors: biological significance and clinical relevance. Clin. Endocrinol. (Oxford) 48:251-255. [DOI] [PubMed] [Google Scholar]
- 44.Williams, B. O., L. Remington, D. M. Albert, S. Mukai, R. T. Bronson, and T. Jacks. 1994. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat. Genet. 7:480-484. [DOI] [PubMed] [Google Scholar]
- 45.Wu, L., A. de Bruin, H. I. Saavedra, M. Starovic, A. Trimboli, Y. Yang, J. Opavska, P. Wilson, J. C. Thompson, M. C. Ostrowski, T. J. Rosol, L. A. Woollett, M. Weinstein, J. C. Cross, M. L. Rosinson, and G. Leone. 2003. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature 421:942-947. [DOI] [PubMed] [Google Scholar]
- 46.Yamasaki, L., R. Bronson, B. O. Williams, N. J. Dyson, E. Harlow, and T. Jacks. 1998. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1+/− mice. Nat. Genet. 18:360-364. [DOI] [PubMed] [Google Scholar]
- 47.Zacksenhaus, E., Z. Jiang, D. Chung, J. D. Marth, R. A. Phillips, and B. L. Gallie. 1996. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 10:3051-3064. [DOI] [PubMed] [Google Scholar]
- 48.Ziebold, U., E. Y. Lee, R. T. Bronson, and J. A. Lees. 2003. E2F3 loss has opposite effects on different pRB-deficient tumors, resulting in suppression of pituitary tumors but metastasis of medullary thyroid carcinomas. Mol. Cell. Biol. 23:6542-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziebold, U., T. Reza, A. Caron, and J. A. Lees. 2001. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 15:386-391. [DOI] [PMC free article] [PubMed] [Google Scholar]