Abstract
Previously, we have shown that pinin/DRS (Pnn), a 140-kDa nuclear and cell adhesion-related phosphoprotein, is involved in the regulation of cell adhesion and modulation of the activity of multiple tumor suppressor genes. In the nucleus Pnn is concentrated in the “nuclear speckles,” zones of accumulation of transcriptional and mRNA splicing factors, where Pnn is involved in mRNA processing. Alternatively, other roles of Pnn in gene regulation have not yet been established. By utilizing in vitro pull-down assays, in vivo interaction studies, and immunofluorescence in combination with overexpression and RNA interference experiments, we present evidence that Pnn interacts with the known transcriptional corepressor CtBP1. As a consequence of this interaction Pnn was capable of relieving the CtBP1-mediated repression of E-cadherin promoter activity. Our results suggest that the interaction of Pnn with the corepressor CtBP1 may modulate repression of transcription by CtBP1. This interaction may reflect the existence of coupling factors involved in CtBP-mediated transcriptional regulation and mRNA processing events.
We studied a molecule, pinin (Pnn/DRS/memA), a 140-kDa phosphoprotein associated with the desmosome and localized in the nucleus of various tissues and cultured cell lines (4, 40-42, 52, 53, 62), which seems to play a key role in the establishment and maintenance of epithelia (53, 56). The expression of exogenous Pnn in transformed cells dramatically altered the recipient cells' morphology, driving them to a more epithelial phenotype. Expression of Pnn was linked to the expression of genes such as E-cadherin, p21cip/waf, MIC-1, and Rho-A, which impact epithelial adhesion, proliferation, and cell motility (55). These observations suggest that Pnn may play an integral role in epithelial-cell-specific gene expression.
Transfection experiments revealed that the majority of expressed Pnn is found within the nucleus, exhibiting a diffuse nucleoplasmic and nuclear speckle distribution. However, little, if any, exogenous Pnn was localized to cell-cell adhesion sites. This raises the possibility that Pnn may be exerting its effect predominantly via interaction with nuclear components, perhaps involving mRNA transcription and processing (48, 62). The proposed functions of nuclear speckles include a role as storage compartments for molecular components involved in gene transcription, such as a subpopulation of polymerase II, and mRNA processing machinery, such as the SR family of proteins. SR proteins distinguished by their serine/arginine-rich motifs are required for constitutive mRNA splicing and regulation of alternative splice site selection (6, 30, 36, 37, 47, 58). Previous two-hybrid and colocalization experiments revealed that Pnn indeed binds to, and colocalizes with, the SR protein family members SRp75 and SRm300, known components of the spliceosome machinery, and a novel SR protein (9, 47, 62, 65). In addition, Pnn was demonstrated to contribute to the alternative splice site selection in splicing reporter assays, specifically the regulation of 5′ splice site choice (62). Recently, Mayeda and coworkers described a role for Pnn, along with RNPS1, in alternative pre-mRNA splicing regulation (48).
In addition to its role in RNA processing, expression of Pnn is linked to increased activity of a subset of tumor suppressor genes, including p21 and E-cadherin. In particular, p21 promoter activity was significantly enhanced in response to the exogenously expressed Pnn (55), tempting speculation that Pnn's affect on gene expression may also be conferred through specific protein-protein interactions governing transcriptional regulation. The PEDLS sequence motif found in the proximity of the C terminus of Pnn was shown to interact with the transcriptional corepressor BS69 by two-hybrid analyses (data not shown), raising the possibility that Pnn may bind to transcriptional proteins. Interestingly, the PEDLS motif is known to be involved in the interactions of various transcriptional factors with C-terminal binding protein (CtBP), a transcriptional corepressor with a pronounced function in developmental processes, as well as tumorigenesis. Like BS69, CtBP was first identified through the interaction with the adenoviral E1A protein via E1A's PLDLS motif (3, 17, 26, 33, 49, 50). CtBP has been found to associate with multiple transcriptional repressors, which contain the basic CtBP-binding signature sequence motif PXDLS, although flexibility in the sequence specificity for the CtBP binding has been reported (5, 7, 22, 24, 34, 35, 43-45, 51). Mammalian CtBP family members include CtBP1 and CtBP2 isoforms, which carry diverse functions in embryogenesis and vertebrate development (20). The most well-documented function of CtBP proteins is as short-range transcriptional repressors, when CtBP proteins are recruited to promoters by sequence-specific DNA-binding transcription factors, either through direct physical interaction or indirectly through bridging proteins. CtBP-mediated repression is believed to involve multiprotein associations, which exhibit histone methyltransferase and deacetylase activities, although many details pertaining to the exact mechanism of CtBP-mediated transcriptional silencing remain to be elucidated.
One important target for CtBP-mediated repression is the E-cadherin promoter, in which case CtBP is able to interact with transcriptional repressors and thereby repress E-cadherin gene expression (15, 16, 54, 63). On the basis of these findings, it is possible that CtBP participates in coordinated biochemical and enzymatic events, resulting in the inhibition of E-cadherin transcription, including complex interaction with promoter targeting transcriptional repressors and chromatin remodeling complexes. Given the potential of a Pnn-CtBP interaction through the PEDLS binding motif, it is of interest to explore the possibility of Pnn-CtBP binding and its effect on E-cadherin gene expression. We investigated here the functional interactions between Pnn and CtBP1. We suggest that Pnn interacts with CtBP1 and relieves CtBP1-mediated silencing burden in the context of the E-cadherin promoter by interfering with CtBP1-dependent gene silencing events.
MATERIALS AND METHODS
Cell lines, cell culture, and transfections.
HEK293, MuM-2C, and MDCK cells were cultured in Dulbecco modified Eagle medium (DMEM; BioWhittaker) containing 10% fetal bovine serum (FBS; Cellgro; Mediatech), 2 mM l-glutamine, and 200 U each of streptomycin and penicillin (Cellgro)/ml. Cells were passaged with 0.1% trypsin and 0.04% EDTA in Hanks balanced salt solution. Human corneal epithelial cells (HCETs; RCB1384; K. Araki-Sasaki) were cultured in DMEM-F-12 (BioWhittaker) containing 5% FBS, 5 μg of insulin (Sigma-Aldrich)/ml, 0.1 μg of cholera toxin (Sigma-Aldrich)/ml, 10 ng of human epidermal growth factor (Invitrogen)/ml, and 0.5% dimethyl sulfoxide (Sigma-Aldrich).
Suspensory HeLa cells (s-HeLa) stably expressing Pnn-Flag-HA fusion protein (POZ-N-Pnn-Flag-HA), s-HeLa cells stably expressing CtBP1-Flag-HA fusion protein at a 1:1 ratio to the endogenous CtBP1 (54) (POZ-CtBP1-Flag-HA), and s-HeLa containing empty POZ vector were propagated in suspension at 2 × 105 confluence in Joklik medium (Cambrex) containing 10% FBS (Cellgro) and 200 U each of streptomycin and penicillin/ml.
Cells were transfected at 70 to 90% confluence, utilizing 3 μl of 1 mg of 25-kDa branched polyethylenimine (PEI; Sigma-Aldrich)/ml per 1 μg of DNA. PEI and DNA were incubated in serum-free DMEM for 10 min in separate tubes. After incubation, the contents of two tubes were combined, incubated for additional 10 min, and applied onto cells.
Expression vectors and reporter constructs.
pcI-neo-GFP vector was previously described (61), pcI-neo-PnnGFP and pcI-neo-PnnGFP1-421 deletion vector, which expressed hPnn, were based on pcI-neo-GFP. pCMV-CtBP1-Flag expressing hCtBP1 and pcDNA3.1-Pnn-myc/His expressing hPnn were based on pCMV-Flag (Stratagene) and pcDNA3.1-myc/His (Invitrogen), respectively. Pnn-GALBD vector was based on pBIND construct (Promega) and expressed hPnn and yeast GAL4 DNA-binding domain as a fusion protein. CtBP1-GALBD vector expressing hCtBP1 and yeast GAL4 DNA-binding domain as a fusion protein were a generous gift of Catharina Svensson (Uppsala University). gal4-SV40-luciferase reporter vector carried binding sites for the DNA-binding domain of the yeast GAL4 protein upstream of the constitutively active simian virus 40 (SV40) promoter/enhancer (a generous gift from D. Liao, University of Florida). CtBP1-glutathione S-transferase (GST) fusion construct expressed hCtBP1 as GST fusion protein.
To create the E-cadherin promoter reporter construct, the region extending from 427 bases upstream of the E-cadherin transcriptional initiation site down to 53 bases posttranscriptional initiation site was PCR amplified by using adaptor primers coding for KpnI and BglII restriction sites on both ends of the amplified product, with human genomic DNA serving as a template. The PCR was performed at a denaturing temperature of 96°C for 45 s, an annealing temperature of 60°C for 45 s, and an extension temperature of 70°C for 1 min. After the amplification, the PCR product was digested with KpnI and BglII and cloned into the pGL-3 basic luciferase reporter construct (Promega) in BglII and KpnI cloning sites. The promoter sequence was validated by automated DNA sequencing. The cloned region included all of the putative regulatory elements of the E-cadherin promoter including two palindromic elements (E-boxes), CAAT box, and CpG island. E-cadherin promoter truncation mutants were created by PCR by using specific adapter primers, carrying KpnI and BglII restriction sites at the termini and E-cadherin −427+53 pGL-3 basic luciferase reporter vector as a template. Truncations were constructed so that each subsequent truncation step toward the transcriptional start site eliminated a single cis-regulatory element. Mutations included the E-BOX1 deletion; the E-BOX1, CAAT box deletion; the E-BOX1, CAAT box, CpG island deletion; and the E-BOX1, CAAT BOX, CpG island, and E-BOX2 deletion. Truncation mutants were digested with BglII and KpnI and cloned into the pGL-3 basic luciferase reporter vector. Mutations within the promoter region were subjected to automated DNA sequencing and confirmed by sequence analysis. A Pnn-myc/His PEDLS→AADLS mutation of the CtBP1 binding motif was constructed by using a Stratagene QuikChange mutagenesis kit according to the manufacturer's instructions with a wild-type Pnn-myc/His expression vector as a template. Mutations were confirmed by sequence analysis.
In vitro binding assays.
GST and GST-CtBP1 proteins expressed in Escherichia coli BL21-Gold(DE3) (Stratagene) were immobilized on 30 μl of agarose-conjugated glutathione in the presence of HEMGN buffer (25 mM HEPES [pH 7.4], 0.1 mM EDTA, 100 mM KCl, 12.5 mM MgCl2, 0.1% NP-40, 10% glycerol, 2 mM phenylmethylsulfonyl fluoride [PMSF], Complete protease inhibitor cocktail [Roche]). Glutathione-immobilized GST and GST-CtBP1 proteins were then incubated with 50 μl of s-HeLa nuclear extract enriched for Pnn-Flag-HA protein in the presence of 500 μl of binding buffer (20 mM HEPES [pH 7.9], 180 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 2 mM PMSF, Complete protease inhibitor cocktail) for 3 h with rotation at 4°C. After three washes with binding buffer, beads were resuspended in 20 μl of sodium dodecyl sulfate (SDS) loading buffer, and the associated proteins were resolved by SDS-8% polyacrylamide gel electrophoresis (PAGE), followed by Western blotting analysis with mouse anti-Flag (Sigma-Aldrich) at a 1:4,000 dilution and mouse anti-GST (Zymed) at 1:4,000 dilution. For experiments with NADH (Sigma-Aldrich 340-125), the compound was added to the binding buffer before addition of the nuclear lysate.
Pnn and CtBP1-Flag complex isolation.
To determine whether Pnn interacts with nuclear CtBP1, we utilized s-HeLa cells stably expressing human CtBP1 tagged with both Flag and hemagglutinin (HA) epitopes at its amino terminus (POZ-CtBP1-Flag-HA). The expression levels of the tagged CtBP1 were comparable to that of endogenous CtBP1. The detailed procedure for complex isolation was described previously (39). Nuclear extract was made from 4 liters of cells, from which the CtBP1 complex was partially purified by using anti-Flag M2 monoclonal antibody (MAb)-conjugated agarose beads, followed by washes until no CtBP1 was released and elution with SDS loading buffer. For Western blotting, mouse anti-Flag antibody (Covance) at a 1:4,000 dilution and mouse anti-Pnn 143 undiluted were used.
Isolation of endogenous Pnn-CtBP complex.
To determine whether Pnn and CtBP are capable of interaction at the endogenous level, 200 μl of s-HeLa nuclear lysate was brought up to 400 μl by using equilibration buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.2 mM EDTA, 10 mM KCl, 25% [vol/vol] glycerol [Sigma-Aldrich]) and incubated with mouse anti-Pnn 143 (hybridoma supernatant) or control supernatant for 2 h, followed by 1 h of incubation with protein G-Sepharose Fast Flow (Sigma-Aldrich). Beads were then washed with 180 mM NaCl equilibration buffer until no Pnn was released, resuspended in SDS loading buffer, and incubated at 95°C for 5 min. Proteins were resolved by SDS-8% PAGE, followed by Western blotting with anti-Pnn 143 MAb undiluted and anti-CtBP1 and anti-CtBP2 MAbs (Signal Transduction) at a 1:4,000 dilution.
Coimmunoprecipitaton and immunoblotting.
HEK293 cells were grown to confluence in 10-cm dishes and transiently transfected with 10 μg of the expression plasmids Pnn-myc/His or Pnn-AADLS-myc/His and CtBP1-Flag by using Lipofectamine reagent (Life Technologies) according to the manufacturer's instructions. At 24 h posttransfection, the growth medium was aspirated, and cells were washed three times with cold phosphate-buffered saline (PBS). Cells were further lysed in plates by using 1 ml of radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS, 50 mM Tris [pH 8.0], 2 mM PMSF, Complete protease inhibitor cocktail) and then collected in microcentrifuge tubes, where cell lysates were purged 10 times through the 27-gauge syringe, followed by a 30-min incubation with gentle rotation at 4°C. After centrifugation at 4°C for 30 min at 14,000 × g, the supernatant was used for coimmunoprecipitation with 30 μl of anti-myc beads (Sigma-Aldrich). After overnight incubation with rotation, the beads were washed four times with 500 μl of radioimmunoprecipitation assay buffer by using micro-bio spin chromatography columns; immunoprecipitates were eluted with SDS sample buffer, incubated for 5 min at 95°C, and fractionated by SDS-8% PAGE, followed by Western blotting. The antibodies used for immunodetection were anti-myc MAb at a 1:4,000 dilution and anti-Flag MAb at a 1:4,000 dilution.
Cell fixation and immunofluorescence.
Cells grown on coverslips were washed three times for 5 min in PBS (pH 7.4). Cells then were fixed in acetone at −20°C for 2 min, followed by three 5-min washes in PBS. After the washes, coverslips were incubated with primary antibodies for 1 h at 25°C, followed by three 5-min PBS washes. Secondary antibodies conjugated to Cy3 (Chemicon) or Alexa 488 (Chemicon) diluted to 1:2,000 were applied to coverslips for1 h, followed by three 5-min washes in PBS. Coverslips were then mounted on the Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories), and cells were visualized by using a fluorescence microscope (DM IRBE; Leica). For the immunostaining, the following primary antibodies were used: mouse anti-Flag (Sigma-Aldrich) at a 1:500 dilution, rabbit anti-Flag (Sigma-Aldrich) at a 1:500 dilution, mouse anti-SR proteins (Zymed) at a 1:500 dilution, mouse anti-Pnn143 (hybridoma supernatant) undiluted, mouse anti-CtBP1 (Transduction laboratories) at a 1:500 dilution, mouse anti-NuMa (Transduction laboratories) at a 1:500 dilution, mouse anti-PCNA (Transduction laboratories) at a 1:500 dilution, and mouse anti-sm proteins (NeoMarkers) at a 1:500 dilution.
RNAi.
Pnn and CtBP1 RNA interference (RNAi) were carried out by using targeting vectors expressing short hairpin RNAs directed against Pnn or CtBP1 mRNA coding regions as previously described (54). Pnn shRNA expressing vector was constructed by inserting an inverted repeat of Pnn-specific 21-nucleotide sequences into a plasmid BS/U6 harboring PolIII U6 promoter as described previously (59). The sequences corresponding to the shRNA were analyzed by BLAST search to eliminate any possibility of sequence homology to other genes. The first fragment of an inverted repeat was cloned into a BS/U6 vector as an ApaI-HindIII insert. The second fragment, also containing six nucleotides as a loop and five T's as the transcriptional termination signal, was then inserted into the HindIII and EcoRI sites. The coding sequences were GGTAAGGTGGCTCAGCGAGAGA (sense) and AGCTTCTCTCGCTGAGCCACCTTACCCTTTTTG (antisense). CtBP1 shRNA vector was based on shRNA vector harboring the PolIII U6 promoter (54).
Cells were seeded on coverslips at 40% confluence 1 day prior to transfection in six-well dishes. Transfections were carried out by using 3 μl of 1 mg of 25-kDa branched PEI (Sigma-Aldrich)/ml and 1 μg of shRNAi vector per well. At 48 h posttransfection, protein expression and distribution was assayed by immunofluorescence as described in above.
Dual luciferase reporter assays.
The day prior to transfection cells were treated with trypsin and plated at 60 to 70% density in a 24- or 6-well plate with 500 μl or 2 ml of standard DMEM without antibiotics, respectively. Optimal quantities of the reporter DNA were 0.1 μg of E-cadherin and gal4-SV40 reporters per well and the Renilla reporter construct at 0.01 μg per well, which were empirically determined in the pilot experiments. The Renilla construct served as an internal control for transfection efficiency. The concentration of the expression vectors varied depending on the experimental design. The amount of DNA in each transfection reaction was equalized by using an empty expression vector to control for nonspecific effects on the luciferase expression. After transfection, cells were incubated for 24 h and assayed for the luciferase expression by using a dual-luciferase reporter assay system (Promega). All results are shown as the average ± the standard deviation of three independent experiments.
RESULTS
Pnn associates with CtBP1 in vitro and in vivo.
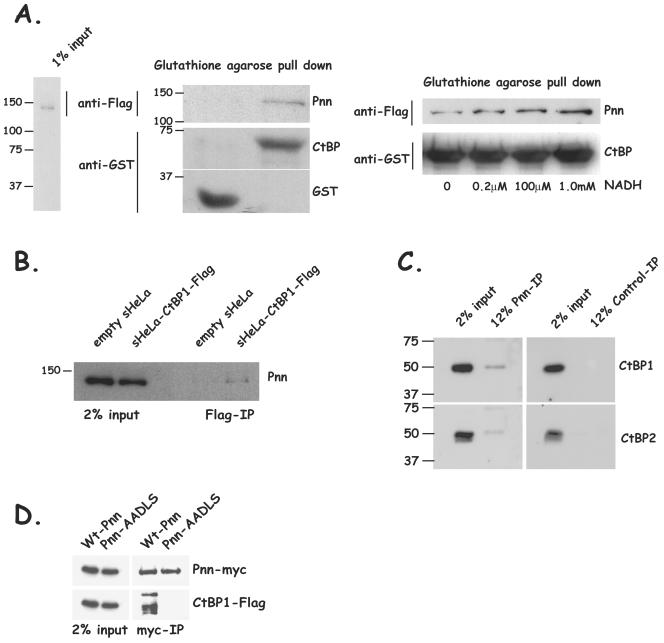
In order to assay the interaction of Pnn with CtBP1, s-HeLa POZ-N-Pnn-Flag-HA nuclear lysates containing epitope-tagged Pnn (Pnn-Flag-HA) were incubated with a CtBP1-GST fusion protein or with GST alone. Immobilized glutathione was utilized to capture CtBP1 and associated proteins. Western blots of pulled-down material revealed the presence of Pnn in the CtBP1-GST containing fractions but not in the fractions containing GST alone (Fig. 1A). Consistent with many other PXDLS motif-containing CtBP1-binding proteins (25, 63), association of Pnn and CtBP1 was noticeably enhanced with increasing concentrations of NADH. These in vitro data suggest that Pnn is indeed capable of binding to CtBP1 and NADH facilitated the binding.
FIG. 1.
Pnn interacts with corepressor CtBP1 in vitro and in vivo. (A) CtBP1-GST fusion protein or GST alone immobilized on glutathione-agarose beads was incubated with the nuclear extracts from s-HeLa cells stably expressing Pnn-Flag-HA fusion protein (s-HeLa-POZ-N-Pnn-Flag-HA) as shown in the nuclear extract input (left panel). The bound material was eluted and evaluated for the presence of Pnn by Western blotting with anti-Flag antibody. The presence of CtBP1-GST or GST was confirmed by using anti-GST antibody. Pnn was found to be associated with CtBP1-GST beads but not beads coupled to GST alone (center panel). CtBP1-GST fusion protein immobilized on glutathione-agarose beads was incubated with s-HeLa-POZ-N-Pnn-Flag-HA nuclear extract in the presence of increasing concentrations of NADH. Bound material from each binding reaction was evaluated for the presence of Pnn with anti-Flag antibody. The presence of CtBP1-GST was confirmed with anti-GST antibody. An increased concentration of NADH resulted in the increased association of CtBP1 with Pnn (right panel). (B) CtBP1-associated complex of proteins was obtained by passing POZ-CtBP1-Flag-HA s-HeLa nuclear extract or the control s-HeLa cell extract over a Flag-M2 column, followed by washes and elution with SDS loading buffer. The bound material was analyzed for the presence of Pnn. Flag-bound material from s-HeLa CtBP1-Flag-HA nuclear extracts but not s-HeLa control nuclear extracts showed the presence of Pnn. (C) Pnn-associated proteins were obtained by coimmunoprecipitation from s-HeLa nuclear lysate with anti-Pnn 143 mouse hybridoma supernatant or control supernatant, followed by washes and elution with SDS loading buffer. Eluted proteins were immunostained for CtBP1 and CtBP2. Both CtBP1 and CtBP2 were found in the Pnn eluate but not in the control eluate. (D) HEK293 cells were transiently cotransfected with CtBP1-Flag expression vector and Pnn-myc/His construct or Pnn-AADLS-myc/His, carrying a PE→AA substitution in the putative CtBP1 interaction motif. Coimmunoprecipitations were performed by using anti-myc agarose affinity gel, followed by Western blotting of the precipitated material with anti-myc and anti-Flag antibodies. Wild type but not Pnn-AADLS was capable of interacting with CtBP1. Wt, wild type.
To assess whether the Pnn-CtBP1 interaction could be detected in vivo, nuclear extracts from either POZ-CtBP1-Flag-HA s-HeLa cells or from s-HeLa cells carrying empty POZ vector were incubated with anti-Flag antibody-conjugated agarose beads. After being washed, the CtBP1-Flag-HA and associated proteins were eluted from the beads. Pnn was found associated with the Flag elutes in s-HeLa-CtBP1-Flag-HA nuclear extracts and not s-HeLa control extracts (Fig. 1B). Furthermore, endogenous CtBP1 and CtBP2 can be coimmunoprecipitated with anti-Pnn antibody from s-HeLa nuclear extracts (Fig. 1C). Together, these data suggest that Pnn interacts with CtBP1 and CtBP2 in vivo.
Next, we sought to determine whether Pnn interacts with CtBP1 through the putative CtBP1 binding motif PEDLS. Cotransfections of HEK293 cells with CtBP1-Flag, along with myc-tagged Pnn or myc-tagged Pnn mutated to contain the sequence AADLS instead of the PEDLS, revealed that the Pnn-CtBP1 interaction was indeed dependent on the PEDLS motif (Fig. 1D). In addition, the Western blots of Pnn-associated material revealed the presence of a higher molecular weight species of CtBP1, perhaps indicative of a posttranslationally modified CtBP1. The higher-molecular-weight CtBP1 has been detected in other pull-down experiments, raising the possibility that the modified CtBP1 may exhibit increased affinity for PXDLS-containing proteins (12, 23, 28, 29).
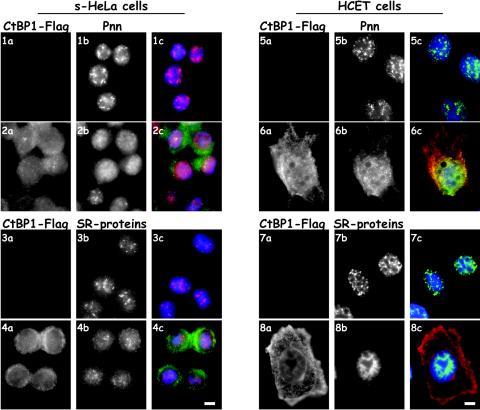
Nuclear distribution of Pnn is altered in response to CtBP1 overexpression.
In the nucleus, Pnn has been shown to concentrate in nuclear speckles and localize more diffusely throughout the interchromatin space (4, 40, 65), whereas CtBP1 is found throughout the nucleoplasm. HCETs transfected with CtBP1 exhibited a more diffuse nuclear distribution of Pnn and increased cytosolic immunostaining for Pnn (Fig. 2). Similar observations were made in s-HeLa cells constructed to express exogenous CtBP1 at 1:1 ratio to the endogenous CtBP1 as described previously (54). Surprisingly, immunostaining with an antibody that reacts with many SR proteins revealed that exogenous CtBP1 expression in HCETs and HeLa cells resulted in a dispersal of SR immunostaining from the nuclear speckles (Fig. 2). These data suggest that the perturbation of the interaction of Pnn with CtBP1 may have significant consequences not only on Pnn but also on the overall organization of SR-containing proteins.
FIG. 2.
Pnn exhibits nuclear redistribution in response to CtBP1 overexpression. (Rows 1 and 2) s-HeLa cells containing empty POZ vector (row 1) or s-HeLa cells stably expressing CtBP1-Flag-HA fusion protein (POZ-CtBP1-Flag-HA) (row 2) were immunostained for CtBP1 with rabbit anti-Flag antibody (panels 1a and 2a) and 143 antibody against the endogenous Pnn (panels 1b and 2b). Panels 1c and 2c show merged DAPI nuclear stain (blue), Pnn (red), and CtBP1 (green) images. (Rows 5 and 6) HCETs were transiently transfected with the CtBP1-Flag expression vector and localization of CtBP1 and Pnn was assessed by immunofluorescence with rabbit anti-Flag antibody (panels 5a and 6a) and 143 antibody to the endogenous Pnn (panels 5b and 6b). Panels 5c and 6c show merged DAPI nuclear stain (blue), Pnn (green), and CtBP1 (red) images. Rows 3 and 4 are the same as rows 1 and 2 and rows 7 and 8 are the same as rows 5 and 6 except, instead of Pnn, cells were immunostained for the endogenous SR proteins with mouse anti-SR proteins antibody. Both Pnn and SR proteins demonstrated nuclear redistribution in response to the CtBP1 overexpression. Bar, 10 μm.
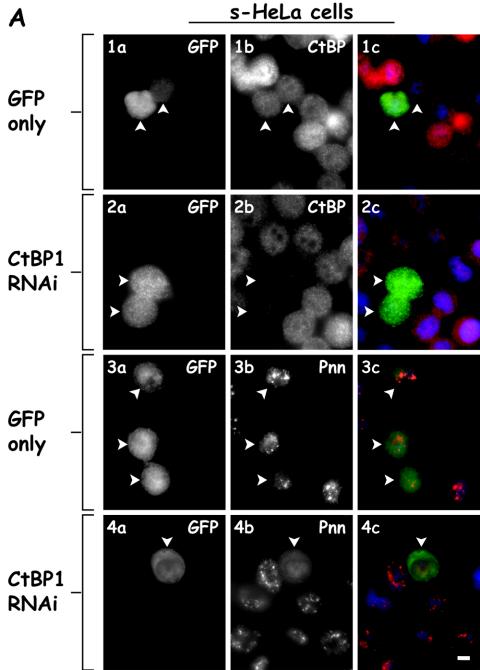
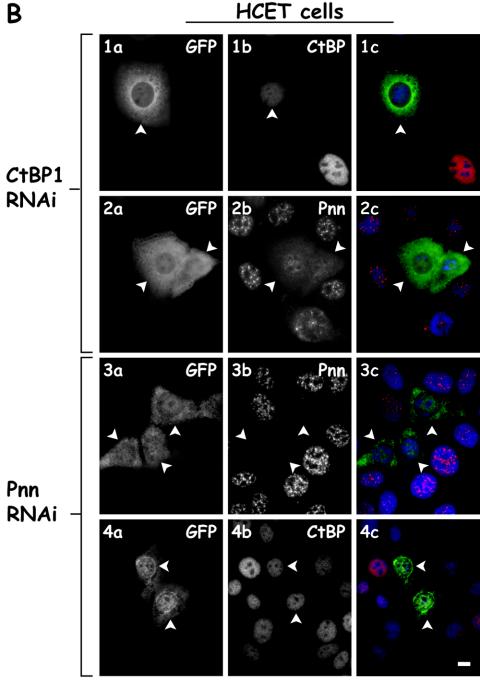
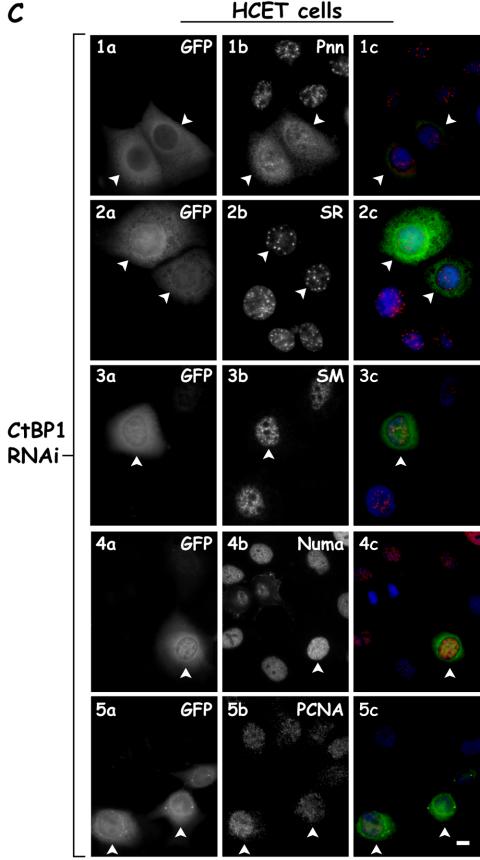
We next sought to determine whether reducing the expression of either Pnn or CtBP1 by utilizing vector based RNAi resulted in a change in the distribution of the other component. Although Pnn RNAi effectively lowered the levels of Pnn, little change was observed in distribution of CtBP1 (Fig. 3B). In contrast, lowering CtBP1 levels resulted in decreased CtBP1 expression and a dramatic change in Pnn's distribution. In CtBP1-knockdown cells and s-HeLa cells (Fig. 3A), as well as HCETs (Fig. 3B), Pnn exhibited a diffuse nucleoplasmic distribution with concurrent increase in cytoplasmic levels. Interestingly, and seemingly paradoxically, both increasing and decreasing CtBP1 expression resulted in a similar distribution change of Pnn, perhaps reflective of the importance of balanced expression of CtBP1 and Pnn. To verify the specificity of the CtBP1 RNAi-dependent Pnn redistribution and to eliminate the possibility of general RNAi-mediated cellular toxicity, which might result in Pnn redistribution, we performed CtBP1 RNAi and immunostained for several nuclear antigens, including SR proteins, sm splicing factors, NuMa, and PCNA antigens. In contrast to its effect on Pnn, CtBP1 RNAi did not exert drastic effect on the distribution of SR, sm, NuMa, and PCNA antigens, supporting the specificity of CtBP1 RNAi on Pnn nuclear redistribution. Interestingly, in some instances we observed accumulation of SR proteins in the nuclear speckles in response to CtBP1 RNAi (unpublished data), raising the possibility that levels of CtBP1 might, directly or indirectly, regulate splicing events.
FIG. 3.
Nuclear distribution of Pnn is altered in response to RNAi-mediated CtBP1 knockdown. (A) s-HeLa cells were transiently cotransfected with a targeting vector expressing short hairpin RNA (shRNA) directed against CtBP1 mRNA coding region and the GFP vector to identify the transfected cells (rows 2 and 4) or the GFP vector alone as a control (rows 1 and 3). Cells were immunostained with mouse anti-CtBP1 antibody (panels 1b and 2b) or 143 antibody against the endogenous Pnn (panels 3b and 4b). Panels 1c to 4c show merged DAPI nuclear stain (blue), GFP (green), and Pnn or CtBP (red) images. Arrowheads indicate the transfected cells. Bar, 10 μm. (B) Rows 1 and 2 show HCETs that were transiently cotransfected with a targeting vector expressing short hairpin RNA (shRNA) directed against CtBP1 mRNA coding region and GFP vector to identify the trasfected cells. Cells were immunostained with mouse anti-CtBP1 antibody (panel 1b) and 143 antibody to the endogenous Pnn (panel 2b). Cells that received CtBP1 shRNAi vector exhibited drastic Pnn redistribution from nuclear speckles. Rows 3 and 4 show HCETs that were transiently cotransfected with a targeting vector expressing short hairpin RNA (shRNA) directed against Pnn mRNA coding region and a GFP vector to identify the transfected cells. Cells were immunostained with 143 antibody to the endogenous Pnn (panel 3b) or mouse anti-CtBP1 antibody (panel 4b). Panels 1c to 4c show merged DAPI nuclear stain (blue), GFP (green), and Pnn or CtBP (red) images. In contrast to the CtBP1 RNAi-dependent Pnn relocalization, RNAi-mediated Pnn knockdown did not result in visible CtBP1 redistribution. Arrowheads indicate the transfected cells. Bar, 10 μm. (C) HCETs were transiently cotransfected with a targeting vector expressing short hairpin RNA (shRNA) directed against CtBP1 mRNA coding region and GFP vector to identify the transfected cells. Cells were immunostained with the following antibodies: anti-Pnn (1b), anti-SR proteins (2b), anti-sm proteins (3b), anti-NuMa (4b), and anti-PCNA (5b). Panels 1c to 5c show merged DAPI nuclear stain (blue), GFP (green), and Pnn, SR, sm, NuMa, and PCNA (red) images. Arrowheads indicate the transfected cells. Bar, 10 μm.
Pnn upregulates the E-cadherin promoter activity.
The effect of Pnn on E-cadherin gene activity was originally described by using EcR293 hPnnGFP-inducible cell lines, in which induction of Pnn expression resulted in upregulation of E-cadherin at both message and protein levels (55). In addition, we demonstrated that RNAi-mediated Pnn knockdown results in the decreased levels of E-cadherin, leading to the disruption of adherence junctions (unpublished data). These findings raised the possibility that Pnn may participate in functional interaction(s) with transcriptional machinery, which contribute to the control of the E-cadherin gene expression, such as interaction with the corepressor CtBP1.
Because the E-cadherin promoter is a known target for the CtBP1-mediated repression (15, 16, 54, 63), we wanted to determine whether Pnn-CtBP1 interaction has functional consequences on the E-cadherin promoter activity.
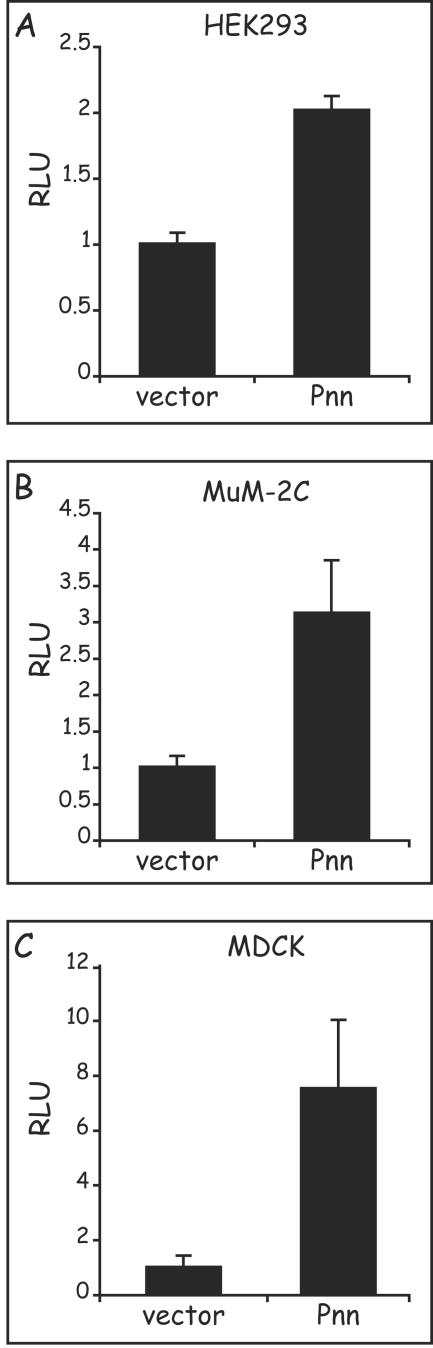
E-cadherin luciferace promoter (−427+53) reporter vector was constructed and assayed in the presence of overexpressed Pnn in the HEK293 cell line. Cotransfection of the Pnn expression vector resulted in upregulation of the E-cadherin promoter reporter activity, suggesting that Pnn can activate the E-cadherin promoter (Fig. 4A). To explore the effect of Pnn expression on the E-cadherin promoter activity in different cellular contexts, we cotransfected Pnn expression vector and E-cadherin reporter construct into invasive uveal melanoma cells, MuM-2C (Fig. 4B), and the epithelial cell line MDCK (Fig. 4C). Pnn was able to upregulate E-cadherin promoter reporter activity in all of the cell lines, thus providing evidence that Pnn possesses a general positive regulatory function. Interestingly, the greatest relative increase was seen in MDCK cells, which exhibit more robust morphological and physiological characteristics of epithelial cells than do the epithelium-derived HEK293 cells or the MuM-2C cells, a finding suggestive of a context-dependent degree of response.
FIG. 4.
Pnn is capable of upregulation of the E-cadherin promoter activity, independent of cell type origin. MDCK, uveal melanoma MuM-2C, and HEK293 cells were transiently cotransfected with 100 ng of E-cadherin luciferase reporter construct with either 500 ng of the Pnn-GFP expression vector or the GFP vector alone. The numerical data obtained from the luciferase readings was normalized to the values of the GFP vector alone control transfections, which represented a value of 1. In HEK293 and MuM-2C cells E-cadherin promoter exhibited 2- and 2.5-fold increases in luciferase activity in response to Pnn expression, respectively, whereas in MDCK cells a fourfold increase was observed.
Full-length E-cadherin promoter is required for Pnn-dependent upregulation of transcription.
In order to assess the requirement of the immediate region of the E-cadherin promoter for the Pnn-induced activation, we created E-cadherin promoter truncations and utilized them in the luciferase reporter assays in context of exogenous Pnn overexpression (Fig. 5).
FIG. 5.
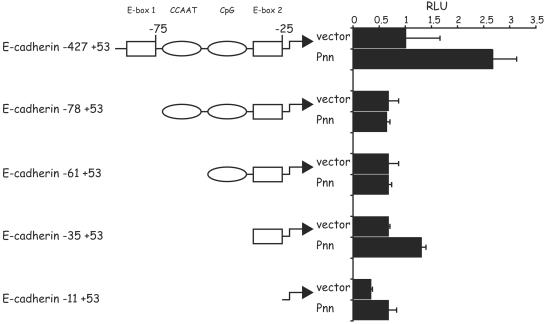
Identification of the Pnn-responsive regions in the E-cadherin promoter. Each truncation of the E-cadherin promoter luciferase reporter is represented in the diagram, with the indicated distances from the transcriptional initiation site. Squares and ovals demarcate known regulatory elements within the promoter region. Bars on the right represent luciferase activity of each of the truncation construct in response to Pnn-myc/His expression (Pnn) or expression of the myc/His vector alone (vector). A total of 100 ng of each promoter construct was transiently cotransfected into HEK293 cells, along with 500 ng of the Pnn-myc/His expression vector or myc/His vector alone. The numerical data obtained from the luciferase readings was normalized to the values of the full-length E-cadherin promoter (−427+53) activity when cotransfected with the myc/His vector alone, which represented a value of 1. E-box 1 and E-box 2 regions, as well as the region adjacent to the transcriptional start site, demonstrated an increased activity in the presence of Pnn.
E-cadherin promoter contains several key cis-regulatory elements implicated in modulation and cell type-specific patterns of E-cadherin gene expression. They include E-boxes 1 (CAGGTG; −78) and 2 (CACCTG; −28), the conserved CCAAT box (−65), and the CpG island (−52 −32), which contribute to the epithelial specific E-cadherin gene expression (8, 11, 14, 18, 19, 21). The modular structure of the E-cadherin gene control region allows interdependent regulation by trans-acting transcriptional factors, which utilize common or adjacent cis-regulatory regions of the promoter.
When cotransfected with the Pnn expression vector, −427+53 E-cadherin promoter was the most responsive to the Pnn-induced transcriptional activation, suggesting that the increase in the transcriptional output triggered by Pnn requires an intact, immediate E-cadherin promoter region (Fig. 5). In particular, deletion of the distal promoter region, which includes E-box 1, abolished Pnn-dependent upregulation of the E-cadherin promoter activity, indicating that this region may be involved in the enhancement of the E-cadherin promoter transcriptional output by Pnn. We are currently investigating the possibility that Pnn is a part of transcriptional complexes occupying specific sites on the E-cadherin promoter.
Pnn derepresses CtBP1-mediated silencing of the E-cadherin promoter.
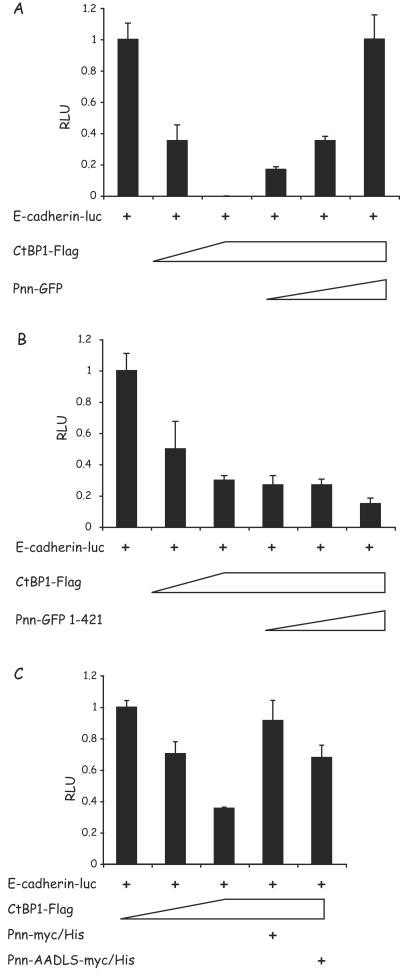
In order to establish the functional relationship between CtBP1 and Pnn at the E-cadherin promoter region, HEK293 cells were cotransfected with the E-cadherin reporter construct and the CtBP1 vector, with or without the wild-type Pnn (wtPnn) construct. As predicted (15, 16, 54, 63), CtBP1 expression resulted in dose-dependent repression of E-cadherin promoter activity, suggesting that CtBP1 serves as a transcriptional corepressor in the E-cadherin promoter context (Fig. 6A). Although CtBP1 was capable of repressing E-cadherin expression, increasing concentrations of the Pnn expression vector resulted in a dose-dependent reversal of CtBP1-mediated repression of the reporter activity. These data are consistent with the proposal that Pnn may function as a transcriptional activator indirectly by binding to nuclear CtBP1, resulting in transactivation of E-cadherin transcription. Pnn1-421, a Pnn truncation that expresses Pnn lacking its C-terminal portion, which includes the region containing the PEDLS motif, did not display reversal of CtBP1-mediated repression (Fig. 6B). Furthermore, cells cotransfected with Pnn-AADLS also exhibited moderate relief of CtBP1-mediated repression. However, the effect of Pnn-AADLS was less dramatic than the full deletion of the C terminus of Pnn, raising the possibility that other regions of Pnn may have effects on transcriptional regulation of E-cadherin.
FIG. 6.
Wild-type Pnn, but not the truncated or mutant form of Pnn, is capable of relieving CtBP1-mediated repression of the E-cadherin promoter. (A) HEK293 cells were transiently cotransfected with the full-length E-cadherin luciferase reporter vector and increasing amounts of the CtBP1-Flag expression vector (0.1 and 1 μg). Parallel transfection reactions contained increasing concentrations of Pnn-GFP (0.01, 0.1, and 1 μg) in the presence of 1 μg of the CtBP1-Flag vector. (B) The same experiment was performed with Pnn-GFP1-421, a Pnntruncation mutant that lacks CtBP1 binding site. (C) HEK293 cells were transiently cotransfected with the full-length E-cadherin luciferase reporter vector and increasing amounts of the CtBP1-Flag expression vector (0.1 and 1 μg). Parallel transfection reactions contained 1 μg of the CtBP1-Flag vector and 1 μg of the Pnn-myc/His or Pnn-AADLS-myc/His vector carrying PE→AA point mutations in the Pnn's PEDLS CtBP1 binding motif. The numerical data obtained from the luciferase readings was normalized to the values of full-length E-cadherin reporter in the absence of CtBP1 or Pnn constructs, which represented a value of 1. In contrast to wild-type Pnn, Pnn1-421 deletion construct was not capable of relieving CtBP1-mediated repression of the E-cadherin promoter activity. The Pnn PE→AA mutant only partially relieved the promoter activity.
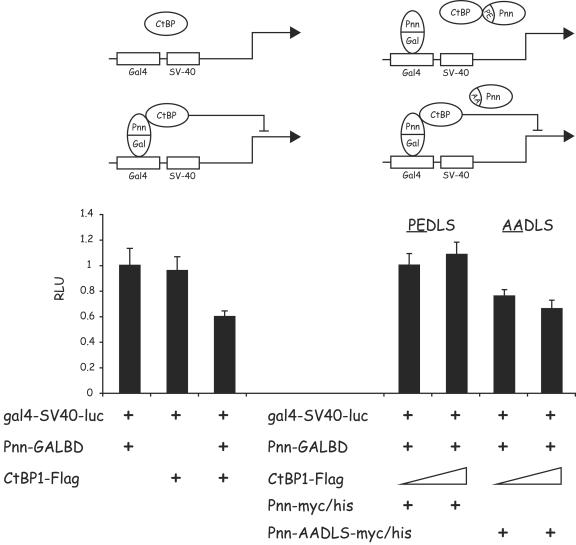
Further evidence of Pnn functioning by interacting with CtBP1 was obtained in the reporter assays in which Pnn-GALBD was tethered to Gal4 binding sites immediately upstream of the constitutively expressed viral SV40 promoter/enhancer. Only in the presence of Pnn-GALBD could CtBP1 repress SV40 promoter activity (Fig. 7, left panel). However, coexpression of wtPnn, but not Pnn-AADLS, effectively blocked the Pnn-GALBD/CtBP1 repression (Fig. 7, right panel).
FIG. 7.
Pnn can recruit CtBP1 to a heterologous promoter, leading to a CtBP1-mediated repression of the promoter activity. (Left panel) HEK293 cells were transiently cotransfected with the luciferase reporter construct gal4-SV40-luc carrying GAL4 binding sites upstream of the SV40 constitutive promoter-enhancer and the following constructs: 100 ng of the Pnn-GALBD fusion construct alone and 1 μg of the CtBP1-Flag expression vector in the presence or absence of the Pnn-GALBD fusion construct (100 ng). gal4-SV40-luc activity was subject to CtBP1-mediated repression only in the presence of the Pnn-GALBD fusion construct, indicating that Pnn-CtBP1 interaction was responsible for this effect. (Right panel) HEK293 cells were transiently cotransfected with the gal4-SV40-luc reporter, carrying GAL4 biding sites upstream of the SV40 constitutive promoter/enhancer, 100 ng of the Pnn-GALBD fusion construct, and increasing concentrations of CtBP1-Flag (100 ng and 1 μg). In addition, transfection reactions contained 1 μg of the wild-type Pnn-myc/His or mutant form of this vector Pnn-AADLS-myc/His, carrying PE→AA substitution in the CtBP1 binding motif, as depicted in the diagram. The numerical data obtained from the luciferase readings was normalized to the value of gal4-SV40-luc reporter in the presence of Pnn-GALBD only, which represented a value of 1. The wild-type, but not mutant, form of Pnn was capable of competition with Pnn-GALBD, thereby relieving CtBP1-mediated repression of the reporter activity.
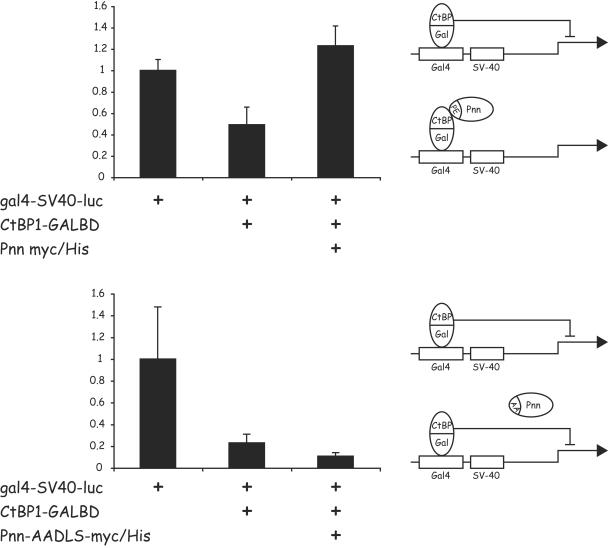
In a reciprocal experiment CtBP1-GALBD, when tethered upstream of the SV40 promoter, was able to repress the activity of the SV40 promoter (Fig. 8). However, in this scenario, only the coexpression of wtPnn, but not Pnn-AADLS, was capable of blocking CtBP1-mediated repression. We speculate that wtPnn may bind to the tethered CtBP1 and occupy or block a site on CtBP1 that is required for the recruitment of additional protein(s) that enable CtBP1 to repress transcription.
FIG. 8.
Pnn, but not the Pnn mutant carrying a mutation in the CtBP1 binding motif, is capable of relieving CtBP1-mediated repression of a heterologous promoter activity. HCETs were transiently transfected with the gal4-SV40-luc reporter construct carrying GAL4 binding sites upstream of the SV40 promoter-enhancer. Transfection reactions also included 1 μg of CtBP1-GALBD fusion vector alone, and in the presence of 1 μg of Pnn-myc/His construct (upper panel) or its mutant Pnn-AADLS-myc/His, carrying the PE→AA substitution in the PEDLS CtBP1 binding motif (lower panel). The numerical data obtained from the luciferase readings was normalized to the value of the gal4-SV40-luc reporter alone, which represented a value of 1. Wild type, but not mutant form of Pnn, was capable of relieving CtBP1-mediated repression of the promoter activity, as depicted in the diagram.
DISCUSSION
In our experiments we demonstrated that Pnn interacted with transcriptional corepressor CtBP1 in vitro and in vivo. As a consequence of their interaction, exogenously expressed Pnn was capable of reversing CtBP1-mediated repression of E-cadherin promoter, providing some mechanistic insights into the functional interaction of Pnn and CtBP1, as well as its effect on gene transcription.
Our data regarding Pnn-CtBP1 interaction places Pnn as a promising candidate modifier of the CtBP1 silencing function. This is in accord with similar reports describing adenoviral E1A protein, responsible for the acquisition of the epithelial phenotype by invasive tumors by binding CtBP and preventing its repressive influence on the target genes (10, 49, 63, 64). It is tempting to speculate that by binding to CtBP E1A exploits preexisting functional protein-protein interactions of the host cellular transcriptional machinery.
CtBP family members interact with, and modify, the activity of a large number of proteins with key roles in development, differentiation, oncogenesis, cell cycle control, and apoptosis. Interaction of CtBP with the C terminus of E1A antagonizes a transcriptional activation mediated by the N terminus of E1A (7, 50, 60). In addition, corepressor CtBP has been shown to associate with the DNA-binding transcriptional repressors ZEB1, and SIP1 (ZEB2), histone deacetylase 1 (HDAC1) and HDAC2, related histone methyltransferases G9a and Eu-HMTase1 (EuHMT), two chromodomain-containing proteins HPC2 and CDYL, transcriptional repressor CoREST, and its related protein KIAA1343 (8, 45, 54). Taken together, these data suggest that CtBP-mediated repression encompasses combinatorial events involving DNA binding repressors and coordinated activity of the HDACs and methyltransferases. Consistent with the fact that CtBP binds a diverse array of transcriptional regulators, CtBP deficiency dramatically compromised mouse embryo-wide development as a result of genetic and biochemical perturbations (20). Therefore, by binding to CtBP1, Pnn is likely to alter CtBP1-dependent silencing functions of transcriptional repression machinery, resulting in activation of the CtBP1 target genes.
In our experiments, Pnn interacted with CtBP1 in the nucleus and exhibited drastic nuclear redistribution in response to CtBP1 overexpression or RNAi-mediated CtBP1 knockdown, findings supportive of Pnn/CtBP1 functional association. Although the data presented here pertain to CtBP1, we expect the same effect on Pnn nuclear distribution in response to perturbations of the CtBP2 levels. Both CtBP1 and CtBP2 are found to be associated with Pnn in the nucleus and, therefore, can possibly exert similar effects on Pnn's nuclear localization. Interestingly, CtBP1 and CtBP2 can be SUMOylated (23, 29) and, as in case of CtBP1, exhibit SUMOylation-dependent subcellular redistribution. However, interaction of CtBP1 with PXDLS-containing proteins is not affected by the CtBP1 SUMOylation status (29), suggesting that SUMOylation does not affect CtBP1 association with the transcriptional proteins, although it can possibly dictate the amount of CtBP1 present in the nucleus.
As with the majority of the interactions between CtBP and its interacting partners, Pnn's interaction with CtBP1 is mediated through the PXDLS motif within the C terminus of Pnn. In addition, NADH facilitates Pnn-CtBP1 binding. It has been demonstrated that NAD+/NADH-induced conformational change in CtBP family members results in an increase in their affinity for PXDLS motif-containing proteins (7, 25, 31, 63). It is possible that the ability of NADH to stimulate CtBP oligomerization could contribute to the enhanced binding to PXDLS-containing proteins such as Pnn. Indeed, Nardini et al. recently determined the structural basis of the increased affinity of CtBP for NADH (38). These authors proposed that nucleotide binding induces a conformational change that promotes CtBP dimerization, which is essential for corepressor activity. Our finding that NADH stimulated Pnn and CtBP1 binding implies that the physiological state of the cell can modulate Pnn-CtBP1 interaction, an idea that is consistent with the previously reported data describing CtBP interactions with E1A and the transcriptional repressor ZEB1 (25, 63).
We show here that Pnn is capable of the reversal of CtBP1-mediated repression of the E-cadherin promoter activity. According to pull-down experiments it appears that approximately 1 to 2% of nuclear CtBP is bound to Pnn, suggesting that change in Pnn levels is unlikely to drastically affect available nuclear CtBP pool. Therefore, we suggest that Pnn, rather than acting as a CtBP sink, may be recruited to transcriptional regulatory complexes and, through CtBP, alter CtBP-mediated repression, contributing in a positive manner to E-cadherin transcriptional regulation. Thus, it is tempting to speculate that Pnn-CtBP interaction reflects the existence of bridging machinery, a framework for coupling of transcription and mRNA processing. It remains to be established how Pnn-CtBP interaction with the transcriptional machinery will affect Pnn-dependent splicing events.
The E-cadherin promoter is a well-characterized target for CtBP-mediated transcriptional repression, which may be manifested in the course of epithelial-mesenchymal transitions associated with the malignant transformation of tumors and cell migration during embryogenesis. Recent attempts to elucidate the nature of CtBP's repressive functions resulted in a series of crucial findings linking CtBP-mediated E-cadherin silencing to the local chromatin state at the promoter region, as well as to the intranuclear NAD+/NADH ratio, establishing CtBP's role as an important regulator of E-cadherin activity in context of the local metabolic and biochemical environment (54, 63). Therefore, it has been speculated that because CtBP appears to play important roles in normal development and in oncogenesis, expression of CtBP target genes such as E-cadherin may be influenced by the cellular redox status. In this context, NADH-dependent transcriptional properties of CtBP1, as well as its Pnn-binding specificities, imposes significant consequences of physiological changes on the nature of Pnn-CtBP1 interaction as related to transcriptional regulation. Significantly, Pnn was first characterized as a component of the complex containing dehydrogenase activity (2), which may provide the foundation for the underlying mechanism governing the Pnn and CtBP1 functional relationship. Given the assumption that Pnn and CtBP1 can be responsive to the common and unique factor, which defines patterns of cellular metabolism, such as NAD+/NADH, it is plausible to speculate that the general metabolic spectrum of enzymatic activities can evoke opposite responses depending on the innate characteristics of transcriptional regulator.
Although the role of Pnn in mRNA splicing activities has been described (27, 62, 65), its function as a transcriptional regulator has not been explored in detail. Our data identified Pnn as a transcriptional regulator, which functionally antagonizes the effect of corepressor-mediated transcriptional silencing. Interestingly, Dellaire et al. have reported that PRP4 kinase, which binds to Pnn, is part of an N-CoR-containing deacetylase complex (9), again potentially linking Pnn to transcriptional regulation. We suggest that Pnn may belong to an emerging family of proteins involved in nuclear functions of transcription and mRNA processing. Taken together, these data raise an interesting possibility in connection with the transcriptional regulation and splicing activity. It is known that splicing activities are coupled to transcription through the interaction of transcriptional factors and constituents of the splicing complexes (12, 13, 46). As a result of the functional interplay between the factors responsible for orchestrated gene activation, such as chromatin remodeling enzymes, transcriptional regulators, and splicing machinery, gene expression acquires the necessary flexibility and synchronization of regulatory events due to the possibility of multiple control points (9, 32, 57). It remains to be established whether Pnn-associated modulation of transcriptional output and splicing activities represents two distinct mechanisms, which Pnn is a part of, or is the effect of a single functional regulatory unit. In any case, Pnn-CtBP1 interaction at the E-cadherin promoter provides a novel insight into the nature of gene regulation through the combinatorial association with repression factors and mRNA splicing machinery, which supplements transcriptional mechanisms with the necessary means for a flexible, multilayered control of gene expression.
Acknowledgments
We thank D. Liao (University of Florida) for the gal4-SV40-luc reporter and Catharina Svensson (Uppsala University, Uppsala, Sweden) for the CtBP1-GALBD expression vector. We also thank Todd Barnash and Lynda Hanssen for graphics support.
This study was supported by NIH award EY07883 to S.P.S.
REFERENCES
- 1.Barnes, C. J., R. K. Vadlamudi, S. K. Mishra, R. H. Jacobson, F. Li, and R. Kumar. 2003. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat. Struct. Biol. 10:622-628. [DOI] [PubMed] [Google Scholar]
- 2.Bellavite, P., F. Bazzoni, M. A. Cassatella, K. J. Hunter, and J. V. Bannister. 1990. Isolation and characterization of a cDNA clone for a novel serine-rich neutrophil protein. Biochem. Biophys. Res. Commun. 170:915-922. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis, and metastasis. EMBO J. 12:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandner, J. M., S. Reidenbach, and W. W. Franke. 1997. Evidence that “pinin,” reportedly a differentiation-specific desmosomal protein, is actually a widespread nuclear protein. Differentiation 62:119-127. [DOI] [PubMed] [Google Scholar]
- 5.Burke, L. J., and A. Bahianmad. 2000. Corepressors 2000. FASEB J. 14:1876-1888. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F., T. Misteli, G. R. Screaton, D. L. Spector, and A. R. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 8.Comijn, J., G. Berx, P. Vermassen, K. Verschueren, L. van Grunsven, E. Bruyneel, M. Mareel, D. Huylebroeck, and F. van Roy. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7:1267-1278. [DOI] [PubMed] [Google Scholar]
- 9.Dellaire, G., E. M. Makarov, J. J. Cowger, D. Longman, H. G. Sutherland, R. Luhrmann, J. Torchia, and W. A. Bickmore. 2002. Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and the N-CoR deacetylase complexes. Mol. Cell. Biol. 22:5141-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frisch, S. M. 1994. E1a induces the expression of epithelial characteristics. J. Cell Biol. 127:1085-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giroldi, L. A., P. P. Bringuier, M. de Weijert, C. Jansen, A. van Bokhoven, and J. A. Schalken. 1997. Role of E boxes in the repression of E-cadherin expression. Biochem. Biophys. Res. Commun. 241:453-458. [DOI] [PubMed] [Google Scholar]
- 12.Goldstrohm, A. C., T. R. Albrecht, C. Sune, M. T. Bedford, and M. A. Garcia-Blanco. 2001. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21:7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstrohm, A. C., A. L. Greenleaf, and M. A. Garcia-Blanco. 2001. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277:31-47. [DOI] [PubMed] [Google Scholar]
- 14.Graff, J. R., E. Gabrielson, H. Fujii, S. B. Baylin, and J. G. Herman. 2000. Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J. Biol. Chem. 275:2727-2732. [DOI] [PubMed] [Google Scholar]
- 15.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. USA 100:4568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grooteclaes, M. L., and S. M. Frisch. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823-3828. [DOI] [PubMed] [Google Scholar]
- 17.Hateboer, G., A. Gennissen, Y. F. Ramos, R. M. Kerkhoven, V. Sonntag-Buck, H. G. Stunnenberg, and R. Bernards. 1995. BS69, a novel adenovirus E1A-associated protein that inhibits E1A transactivation. EMBO J. 14:3159-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennig, G., J. Behrens, M. Truss, S. Frisch, E. Reichmann, and W. Birchmeier. 1995. Progression of carcinoma cells is associated with alterations in chromatin structure and factor binding at the E-cadherin promoter in vivo. Oncogene 11:475-484. [PubMed] [Google Scholar]
- 19.Hennig, G., O. Lowrick, W. Birchmeier, and J. Behrens. 1996. Mechanisms identified in the transcriptional control of epithelial gene expression. J. Biol. Chem. 271:595-602. [DOI] [PubMed] [Google Scholar]
- 20.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22:5296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosono, S., I. Gross, M. A. English, K. M. Hajra, E. R. Fearon, and J. D. Licht. 2000. E-cadherin is a WT1 target gene. J. Biol. Chem. 275:10943-10953. [DOI] [PubMed] [Google Scholar]
- 22.Izutsu, K., M. Kurokawa, Y. Imai, K. Maki, K. Mitani, and H. Hirai. 2001. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood 97:2815-2822. [DOI] [PubMed] [Google Scholar]
- 23.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 24.Koipally, J., and K. Georgopoulos. 2000. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 275:19594-19602. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, V., J. E. Carlson, K. A. Ohgi, T. A. Edwards, D. W. Rose, C. R. Escalante, M. G. Rosenfeld, and A. K. Aggarwal. 2002. Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol. Cell 10:857-869. [DOI] [PubMed] [Google Scholar]
- 26.Ladendorff, N. E., S. Wu, and J. S. Lipsick. 2001. BS69, an adenovirus E1A-associated protein, inhibits the transcriptional activity of c-Myb. Oncogene 20:125-132. [DOI] [PubMed] [Google Scholar]
- 27.Li, C., R. I. Lin, M. C. Lai, P. Ouyang, and W. Y. Tarn. 2003. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol. Cell. Biol. 23:7363-7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, X., Y. Y. Liang, B. Sun, M. Liang, Y. Shi, F. C. Brunicardi, and X. H. Feng. 2003. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol. Cell. Biol. 23:9081-9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, X., B. Sun, M. Liang, Y. Y. Liang, A. Gast, J. Hildebrand, F. C. Brunicardi, F. Melchior, and X. H. Feng. 2003. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell 11:1389-1396. [DOI] [PubMed] [Google Scholar]
- 30.Longman, D., I. L. Johnstone, and J. F. Caceres. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marmorstein, R. 2002. Dehydrogenases, NAD, and transcription: what's the connection? Structure 10:1465-1466. [DOI] [PubMed] [Google Scholar]
- 32.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masselink, H., and R. Bernards. 2000. The adenovirus E1A binding protein BS69 is a corepressor of transcription through recruitment of N-CoR. Oncogene 19:1538-1546. [DOI] [PubMed] [Google Scholar]
- 34.Melhuish, T. A., and D. Wotton. 2000. The interaction of C-terminal binding protein with the Smad corepressor TG-interacting factor is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem. 275:39762-39766. [DOI] [PubMed] [Google Scholar]
- 35.Meloni, A. R., E. J. Smith, and J. R. Nevins. 1999. A mechanism for Rb/p130-mediated transcription repression involving recruitment of the CtBP corepressor. Proc. Natl. Acad. Sci. USA 96:9574-9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mintz, P. J., and D. L. Spector. 2000. Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 129:241-251. [DOI] [PubMed] [Google Scholar]
- 37.Misteli, T., J. F. Caceres, and D. L. Spector. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387:523-527. [DOI] [PubMed] [Google Scholar]
- 38.Nardini, M., S. Spano, C. Cericola, A. Pesce, A. Massaro, E. Millo, A. Luini, D. Corda, and M. Bolognesi. 2003. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 22:3122-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 40.Ouyang, P. 1999. Antibodies differentiate desmosome-form and nucleus-form pinin: evidence that pinin is a moonlighting protein with dual location at the desmosome and within the nucleus. Biochem. Biophys. Res. Commun. 263:192-200. [DOI] [PubMed] [Google Scholar]
- 41.Ouyang, P., and S. P. Sugrue. 1996. Characterization of pinin, a novel protein associated with the desmosome-intermediate filament complex. J. Cell Biol. 135:1027-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouyang, P., and S. P. Sugrue. 1992. Identification of an epithelial protein related to the desmosome and intermediate filament network. J. Cell Biol. 118:1477-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phippen, T. M., A. L. Sweigart, M. Moniwa, A. Krumm, J. R. Davie, and S. M. Parkhurst. 2000. Drosophila C-terminal binding protein functions as a context-dependent transcriptional cofactor and interferes with both mad and groucho transcriptional repression. J. Biol. Chem. 275:37628-37637. [DOI] [PubMed] [Google Scholar]
- 44.Postigo, A. A., and D. C. Dean. 2000. Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors. Proc. Natl. Acad. Sci. USA 97:6391-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Postigo, A. A., and D. C. Dean. 1999. ZEB represses transcription through interaction with the corepressor CtBP. Proc. Natl. Acad. Sci. USA 96:6683-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed, R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15:326-331. [DOI] [PubMed] [Google Scholar]
- 47.Sacco-Bubulya, P., and D. L. Spector. 2002. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakashita, E., S. Tatsumi, D. Werner, H. Endo, and A. Mayeda. 2004. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 24:1174-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaeper, U., T. Subramanian, L. Lim, J. M. Boyd, and G. Chinnadurai. 1998. Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J. Biol. Chem. 273:8549-8552. [DOI] [PubMed] [Google Scholar]
- 51.Sewalt, R. G., M. J. Gunster, J. van der Vlag, D. P. Satijn, and A. P. Otte. 1999. C-Terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi, J., and S. P. Sugrue. 2000. Dissection of protein linkage between keratins and pinin, a protein with dual location at desmosome-intermediate filament complex and in the nucleus. J. Biol. Chem. 275:14910-14915. [DOI] [PubMed] [Google Scholar]
- 53.Shi, Y., P. Ouyang, and S. P. Sugrue. 2000. Characterization of the gene encoding pinin/DRS/memA and evidence for its potential tumor suppressor function. Oncogene 19:289-297. [DOI] [PubMed] [Google Scholar]
- 54.Shi, Y., J. Sawada, G. Sui, B. Affar el, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, and Y. Nakatani. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 55.Shi, Y., M. N. Simmons, T. Seki, S. P. Oh, and S. P. Sugrue. 2001. Change in gene expression subsequent to induction of Pnn/DRS/memA: increase in p21cip1/waf1. Oncogene 20:4007-4018. [DOI] [PubMed] [Google Scholar]
- 56.Shi, Y., M. Tabesh, and S. P. Sugrue. 2000. Role of cell adhesion-associated protein, pinin (DRS/memA), in corneal epithelial migration. Investig. Ophthalmol. Vis. Sci. 41:1337-1345. [PubMed] [Google Scholar]
- 57.Shin, C., and J. L. Manley. 2002. The SR protein SRp38 represses splicing in M phase cells. Cell 111:407-417. [DOI] [PubMed] [Google Scholar]
- 58.Spector, D. L. 2001. Nuclear domains. J. Cell Sci. 114:2891-2893. [DOI] [PubMed] [Google Scholar]
- 59.Sui, G., C. Soohoo, B. Affar el, F. Gay, Y. Shi, and W. C. Forrester. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sundqvist, A., E. Bajak, S. D. Kurup, K. Sollerbrant, and C. Svensson. 2001. Functional knockout of the corepressor CtBP by the second exon of adenovirus E1a relieves repression of transcription. Exp. Cell Res. 268:284-293. [DOI] [PubMed] [Google Scholar]
- 61.Wang, D., R. Miller, R. Shaw, and S. G. 1996. The Pleckstrin homology Domain of Human βIΣII Spectrin is targeted to the plasma membrane in vivo. Biochem. Biophys. Res. Commun. 225:420-426. [DOI] [PubMed] [Google Scholar]
- 62.Wang, P., P. J. Lou, S. Leu, and P. Ouyang. 2002. Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem. Biophys. Res. Commun. 294:448-455. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of corepressor function by nuclear NADH. Science 295:1895-1897. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, Q., H. Yao, N. Vo, and R. H. Goodman. 2000. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc. Natl. Acad. Sci. USA 97:14323-14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimowska, G., J. Shi, G. Munguba, M. R. Jackson, R. Alpatov, M. N. Simmons, Y. Shi, and S. P. Sugrue. 2003. Pinin/DRS/memA interacts with SRp75, SRm300, and SRrp130 in corneal epithelial cells. Investig. Ophthalmol. Vis. Sci. 44:4715-4723. [DOI] [PubMed] [Google Scholar]