Abstract
Btg2 is a primary p53 transcriptional target gene which may function as a coactivator-corepressor and/or an adaptor molecule that modulates the activities of its interacting proteins. We have generated Btg2-null mice to elucidate the in vivo function of Btg2. Btg2-null mice are viable and fertile but exhibit posterior homeotic transformations of the axial vertebrae in a dose-dependent manner. Consistent with its role in vertebral patterning, Btg2 is expressed in the presomitic mesoderm, tail bud, and somites during somitogenesis. We further provide biochemical evidence that Btg2 interacts with bone morphogenetic protein (BMP)-activated Smads and enhances the transcriptional activity of BMP signaling. In view of the genetic evidence that reduced BMP signaling causes posteriorization of the vertebral pattern, we propose that the observed vertebral phenotype in Btg2-null mice is due to attenuated BMP signaling.
B-cell translocation gene 2 (Btg2, also known as Tis21, Pc3, or APRO1) belongs to the antiproliferative (APRO) family, which is classified by the conserved APRO homology boxes A and B separated by 20 to 25 nonconserved amino acids (19, 36). A large body of studies has implicated the role of Btg2 in cell growth, differentiation, survival, and senescence. Overexpression of exogenous Btg2 can induce cell cycle arrest by either Rb-dependent (12) or -independent mechanisms (17). Expression studies have suggested that the antiproliferative property of Btg2 may play a critical role in the procession of precursor neuronal and osteoblast cells from proliferation to differentiation status during development (13, 30). In addition, Btg2 expression was necessary to provide protection against apoptosis in terminally differentiated neuronal cells (6, 8). Other reports have demonstrated that DNA-damaging agents induced Btg2 in a p53-dependent manner (7, 31). Consistent with that finding, it was also shown that the Btg2 promoter contains functional p53-responsive elements (31) and that Btg2 is a primary p53 transcriptional target gene (14).
A growing number of reports have suggested that Btg2 functions as a coactivator-corepressor and/or an adaptor molecule that modulates the activities of its interacting proteins. Both Btg2 and its closest family member, Btg1, have been demonstrated to bind and to positively modulate the activity of the protein arginine methyltransferase 1, Hoxb9 (a homeobox protein), and Caf1 (carbon carbonylate repressor 4 [CCR4]-associated factor 1) (18, 28, 29, 32). It was suggested that the interactions of Btg proteins with a general transcription multisubunit complex CCR4 through Caf1 might be involved in the transcriptional regulation of the genes involved in the control of the cell cycle (22). Tob1, another ARPO member, was shown to interact with bone morphogenetic protein (BMP)-activated Smads and to inhibit BMP/Smad signaling in osteoblasts (42). Targeted disruption of Tob1 resulted in increased osteoblasts and bone mass as well as a predisposition to tumorigenesis (41, 42).
Immunohistological studies have shown that Btg2 is highly expressed in kidney proximal tubules, lung alveolar bronchial epithelia, and prostate acinar cells (21) and that diminished Btg2 expression has been correlated with cancerous states (9, 11, 35). While numerous data have implicated the role of antiproliferative, p53 target Btg2 as a tumor suppressor and a regulator of differentiation of multiple cell lineages, there has been an absence of studies confirming its in vivo function. Canzoniere et al. recently showed that Btg2 overexpression in neuronal tissues of transgenic mice led to a marked increase in the number of postmitotic neurons and to the impairment of cerebellar development (2). This result supports the previously suggested role of Btg2 in controlling the transition of neuronal precursor cells from the proliferative to the differentiative stage in their development and maturation (2, 13).
In this paper, we demonstrate that Btg2 is expressed in the presomitic mesoderm, tail bud, and somites during somitogenesis stages and is necessary for the normal patterning of axial vertebrae. We show that BMP signaling has an antagonistic effect on Gdf11/Acvr2b signaling in vertebral patterning. We further show that Btg2 can interact with BMP-activated Smads and enhance BMP/Smad signaling. We propose the impaired BMP/Smad signaling as a potential mechanism underlying the posterior vertebral transformation in Btg2-deficient mice.
MATERIALS AND METHODS
A Btg2 knockout strain was generated at the University of Florida. Generation of Bmpr2 knockout mice was previously reported (1). Mice were maintained under standard specific-pathogen-free conditions, and all animal procedures performed were reviewed and approved by the University of Florida Institutional Animal Care and Use Committee.
Targeting vector construction and ES cell targeting.
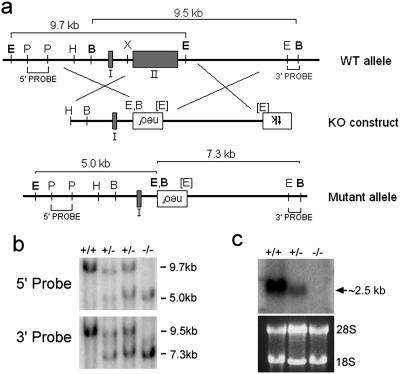
A bacterial artificial chromosome (BAC) clone containing the entire Btg2 gene was isolated by screening the mouse 129Sv BAC library (Incyte Genomics, St. Louis, Mo.) with PCR primers generated from exons I and II of the Btg2 gene. Upon cloning, restriction enzyme mapping, and sequencing, we identified exons I and II of the BAC subclones (Fig. 1a). The targeting vector was generated by sequential subclonings of the 5′ and 3′ homology arms into pPNT vector (38). A 3.0-kb HindIII-XbaI fragment containing exon I and a 4.7-kb EcoRI-EcoRI fragment were sequentially inserted into the EcoRI and XhoI sites, respectively, of the pPNT vector. The targeting vector resulted in the replacement of the entire exon II, including the region containing APRO boxes A and B, with the neomycin resistance cassette. Linearized Btg2 targeting vector was then electroporated into 4 × 107 J1 embryonic stem (ES) cells, and G418- and fialuridine-resistant colonies were selected. Approximately 400 G418- and fialuridine-resistant colonies were randomly picked for homologous recombination screening by genomic Southern blot analysis. Upon initial screening with a 3′ probe followed by confirmation with a 5′ probe and Southern blot analysis (Fig. 1b), one ES clone was identified to contain the correctly targeted Btg2 locus. ES cells from the positive clone were injected into blastocysts on the C57BL/6 (B6) strain. Chimeric male mice were mated with females on the B6 background.
FIG. 1.
Gene targeting of the Btg2 locus. (a) Diagram showing the Btg2 WT genetic locus, the corresponding targeting construct, and the resulting targeted mutant allele. Two exons are indicated by shaded boxes. Both 5′ and 3′ probe regions for genomic Southern screening are indicated, as are the expected sizes of fragments from WT and mutant Btg2 loci. Restriction enzyme sites for BamHI (sites labeled B), EcoRI (E), HindIII (H), PstI (P), and XbaI (X) are indicated. Restriction enzyme sites in brackets indicate the nullification of the recognition sequence. Neomycin resistance (neor) and thymidine kinase (tk) cassettes are in the opposite direction of the Btg2 gene. KO, knockout. (b) Genomic Southern analysis of Btg2+/− crossing. Genomic DNA from WT and Btg2+/− and Btg2−/− mice was digested with EcoRI and BamHI and subsequently hybridized with 5′ and 3′ probes. (c) Northern blot analysis of Btg2+/− crossing. Note the absence of the Btg2 transcript in the kidney of the Btg−/− mutant.
Skeleton preparation and whole-mount in situ hybridization.
Newborn pups either from Btg2+/− females mated with Btg2+/− or Btg2−/− males or from Acvr2b+/− Bmpr2+/− females mated with Acvr2b−/− males were subjected to skeleton preparations as previously described (24). For studying the expression patterns of Btg2 in developing mouse embryos, wild-type (WT) embryos, ranging from embryonic day 9.0 (E9.0) to E14.5, were collected. Antisense Btg2 RNA probe was generated by using a digoxigenin-UTP labeling kit (Roche Diagnostics, Indianapolis, Ind.), and whole-mount in situ hybridization was performed as described previously (40).
Transfection and reporter assays.
C2C12 cells were transiently transfected with SBE4-, ARE-, or BRE-luc and the internal control pCMV-β-gal with or without the Btg2 expression vector (1 μg/well) in six-well plates by using Lipofectin (Invitrogen, Rockville, Md.) according to the manufacturer's instructions. Twelve hours after transfection, cells were treated with medium containing 50 ng of BMP-2/ml for 18 h. Luciferase activity was quantified by using an enhanced luciferase assay kit (BD Biosciences, Franklin Lakes, N.J.). Values were normalized with β-galactosidase activity. All assays were performed in triplicate, and results are given as the means (± standard errors) of three independent transfections.
Immunoblotting and immunoprecipitation.
293T cells were used for the detection of protein-protein interactions in vivo. 293T cells were transiently transfected with the indicated plasmids. Forty-eight hours after transfection, cells were lysed in a buffer containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM EDTA, and a protease inhibitor cocktail (Roche Diagnostics). Extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred to polyvinylidene difluoride membranes; they were then probed with polyclonal or monoclonal antisera, followed by horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG, respectively, and visualized by chemiluminescence (Pierce Biotechnology, Rockford, Ill.) according to the manufacturer's instructions. The GST pull-down assay was performed by incubating glutathione-Sepharose 4B beads (Amersham Biosciences, Piscataway, N.J.) with each extract for 1 h. After the beads were washed four times with the buffer used for cell solubilization, immunoblots assays were performed.
RESULTS AND DISCUSSION
Generation of Btg2-deficient mice.
In order to investigate the in vivo function of Btg2 in mice, we have performed gene targeting experiments. Btg2 transcript encodes 158 amino acids (aa) in two exons separated by a small intron (Fig. 1a) (10). The first exon contains sequence encoding the translation start site and 48 aa, while the second exon contains sequence for the remaining 110 aa. To generate the Btg2-null mutation, we designed a targeting vector in which the entire second exon, containing APRO homology boxes A and B, was replaced with a neomycin resistance cassette (Fig. 1a). Homologous recombination was confirmed by genomic Southern blot analysis using both 5′ and 3′ probes (Fig. 1b), and the absence of Btg2 transcript in homozygous Btg2 mutants (Btg2−/−) was confirmed by Northern blot analysis (Fig. 1c).
Btg2 is not essential for the viability of mice.
Btg2 appeared dispensable for the viability of embryonic development and postnatal life, as we observed the Mendelian ratio of viable offspring at the weaning age from Btg2+/− crosses. Both male and female Btg2−/− mice were normal in appearance and fertile. Furthermore, female Btg2−/− mice nurtured their offspring normally, indicating that Btg2 is unessential for mammary gland development. Flow cytometry analysis of lymphoid development in thymus and spleen with antibodies specific for CD4, CD8, CD45R, and CD19 showed no obvious differences between Btg2−/− and wild-type mice (data not shown). In addition, genotoxic agent-induced growth arrest was not significantly different in murine embryonic fibroblasts isolated from Btg2−/− and WT embryos (data not shown).
To investigate whether Btg2 played an essential role in cerebellar development, we examined the cerebella of Btg2−/− mice at postnatal day 4 (PN4) and at two months of age. The morphologies, sizes, and cell densities of cerebella of Btg2−/− were grossly indistinguishable from those of their littermate controls (data available on request). In PN4 cerebella, a comparable number and thickness of granule cell progenitors in the external granular layer were observed for Btg2−/− mice and for their littermate controls (unpublished data). The lengths and sizes of the cerebellar lobules, as well as the cellular morphologies and densities of the cerebella of adult Btg2−/− mice were also normal (unpublished data). A detailed molecular and immunological analysis of cerebellar development may further address whether Btg2 is essential in regulating genes, such as cyclin D1 or Math1, involved in the proliferation and differentiation of granule cell progenitors during cerebellar development. Histological examinations of various organs and tissues of 9-week-old Btg2−/− mice (n = 3) found no obvious abnormalities, with the exception of the vertebral columns as detailed below.
Btg2−/− mice exhibit posterior homeotic transformation of axial vertebrae.
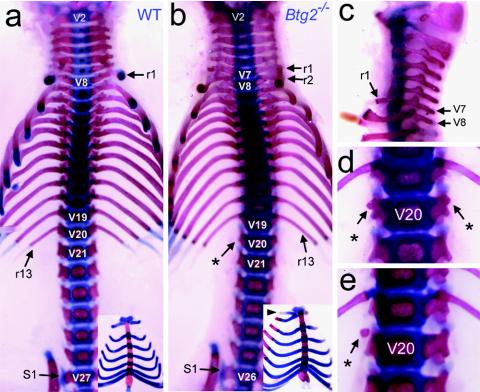
During the expansion of the Btg2 mutant line, one Btg2+/− mouse displayed paralysis of the hind limbs at weaning and the necropsy result showed an obliquely positioned lumbar vertebra. This finding, together with the previous report of the interaction of BTG2 with Hoxb9 (29), which is involved in the specification of axial vertebrae, led us to investigate the developmental defects of axial skeletons for Btg2−/− mice. The axial vertebrae are categorized as cervical, thoracic, lumbar, sacral, or caudal depending on their position along the anteroposterior axis and on their morphological characteristics. For example, the thoracic vertebrae are characterized by the attachment of the ribs, whereas the sacral vertebrae are fused to each other to form a structure called the sacrum. The numbers of vertebrae in each category are mostly invariable within a species. Mice have 7 cervical (C), 13 thoracic (T), and 6 lumbar (L) vertebrae, represented by the C7-T13-L6 vertebral pattern (Fig. 2a). We found that the majority of Btg2−/− mice exhibited abnormal vertebral patterns.
FIG. 2.
Posterior transformation of axial vertebrae in Btg2−/− mice. (a to e) Axial skeletons and vertebrosternal ribs (insets) of wild-type (a) and Btg2−/− (b to e) newborn pups. In wild-type mice, the thoracic, lumbar, and sacral vertebrae begin at V8, V21, and V27, respectively (a). In Btg2−/− mice, the first rib (r1) is attached to V7 instead of V8 (b and c) and is joined to the second rib ventrally at the rostral end of the first sternebra (b, inset). The asterisks indicate the ribs attached in V20, which are either absent (b) or in the form of minute rudiments (d and e). The sacral vertebrae begin at V26 instead of V27 (b).
Specifically, Btg2−/− mice displayed a series of posterior vertebral transformations (Table 1). As shown in Fig. 2b and c, the first set of ribs of Btg2−/− mice were attached to the seventh vertebra (V7), indicating the transformation of C7 to T1. The T13-to-L1 transformation was also apparent, as the rib rudiments in the 20th vertebra (V20) of Btg2−/− mice were minute or completely absent (Fig. 2b to e). In addition, V26, which forms L6 in wild-type mice, was transformed to S1 in Btg2−/− mice (Fig. 2b). Interestingly, a significant number of the Btg2+/− mice also exhibited similar, but less intense, vertebral abnormalities compared to Btg2−/− mice, indicating a haploinsufficiency effect (Table 1).
TABLE 1.
Posterior vertebral transformation in Btg2−/− mice
| Vertebral transformation | No. of mice
|
||
|---|---|---|---|
| Btg2+/+ (n = 9) | Btg2+/− (n = 21) | Btg2−/− (n = 22) | |
| V7 to C7 | 9 | 20 | 16 |
| V7 to T1 | 0 | 1 | 6 |
| V20 to T13 | 9 | 19 | 4 |
| V20 to T13(s)a | 0 | 2 | 11 |
| V20 to L1 | 0 | 0 | 7 |
| V26 to L6 | 9 | 10 | 0 |
| V26 to S1 | 0 | 11 | 22 |
(s), a vertebra exhibiting small rib attachments in the form of minute rudiment.
Dynamic Btg2 expression in somitogenesis stage embryos.
Each vertebra is derived from two adjacent somites (5), which form bilaterally on either side of the neural tube. The first somite appears shortly after gastrulation. Thereafter, new somites are added at a relatively constant rate from the anterior end of the presomitic mesoderm (PSM), while new mesoderm is being added concomitantly at the posterior end of PSM from the primitive streak and then from the tail bud (27). It has been shown that the positional information of a vertebra is determined in the somitomeres of the PSM even before they become mature somites (23, 26).
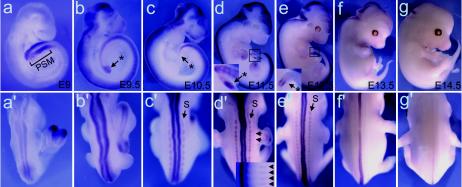
Btg2 expression was detected in the PSM region of E9 embryos (Fig. 3a). PSM and tail bud-specific expression of Btg2 continued until E12.5 (Fig. 3b to e) and faded away when somitogenesis was completed (Fig. 3f and g). Btg2 transcripts were also detected in the somites of E10.5 to E12.5 embryos (Fig. 3c′ to e′). Btg2 transcripts on the dorsal side of the neural tube emerged at E9 and faded away starting at E13.5 (Fig. 3a′ to g′). Btg2 expression in the PSM-tail bud region and somites of embryos during active somitogenesis is consistent with its role in vertebral patterning.
FIG. 3.
Whole-mount in situ hybridization showing a dynamic Btg2 expression pattern in PSM-tail bud regions, somites, and the neural tubes. Lateral (a to b) and dorsal (a′ to g′) views of E9 to E14.5 embryos hybridized with Btg2 antisense probe are shown. Btg2 transcripts were specifically detected in the PSM region at E9 (a) and in the tail buds (indicated by asterisks) at subsequent stages (b to e). Tail bud-specific expression disappeared from E13.5 embryos (f and g). Btg2 transcripts were detected also on the dorsal side of the neural tubes from E9 to E13.5. This neural tube expression peaked at E11.5 and faded away by E13.5. Expressions in the somite (S) were also visible in E10.5 to E12.5 embryos. The inset in panel d′ shows Btg2 expression in developing rib-like streaks in the trunk at E11.5.
It has been shown that Btg2 interacts with and increases the transcriptional activity of Hoxb9 (29). If the loss of interaction between Btg2 and Hoxb9 were the cause of vertebral defects in Btg2−/− mice, we should have observed an anterior vertebral transformation consistent with most Hox gene-deficient mutants (4, 39). In this regard, the posterior vertebral transformation in Btg2−/− is unlikely due to the attenuated transcriptional activities of Hox genes.
BMP type II receptor (Bmpr2) haploinsufficiency attenuates vertebral defects in Acvr2b−/− mice.
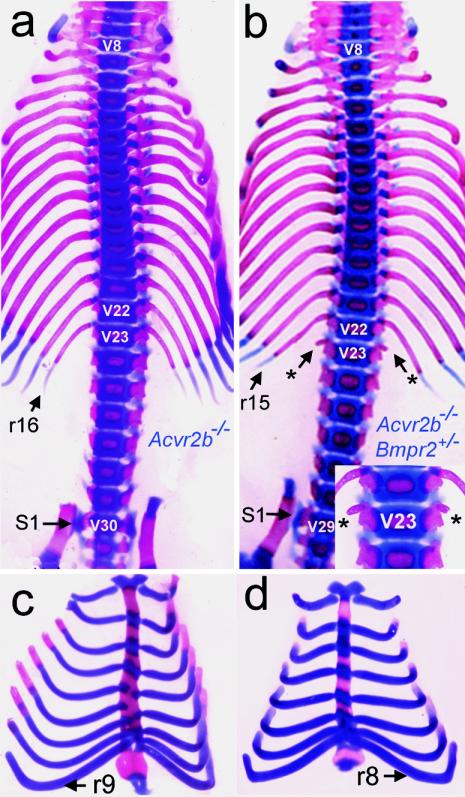
It was previously demonstrated that activin type IIB receptor (Acvr2b)-null (Acvr2b−/−) mice exhibited an anterior vertebral transformation resulting in the C7-T16-L6 vertebral pattern, with nine vertebrosternal (VS) ribs (24) (Fig. 4a and c). We have also shown that additional heterozygous deletion of its subfamily receptor Acvr2 gene in the Acvr2b−/− background intensified the Acvr2b−/− phenotype of vertebral transformation, exhibiting the C7-T17-L7 pattern, with 10 VS ribs, and that Gdf11 is the likely ligand for Acvr2 and Acvr2b (20, 25). In the course of studying the genetic interactions between Acvr2b and BMP type II receptor (Bmpr2), we discovered that Acvr2b−/− Bmpr2+/− mice showed alleviated vertebral defects compared to Acvr2b−/− mice. As shown in Fig. 4b and Table 2, the transformation of V23 to T16 was incomplete and V29 was transformed to S1 instead of L6 in the majority of Acvr2b−/− Bmpr2+/− mice. Furthermore, eight ribs, instead of nine, were attached to the sternum (Fig. 4c and d; Table 2). These data indicate that there are antagonistic roles for BMP and Gdf11 signaling in vertebral development and that attenuated BMP signals cause posteriorization of the vertebral patterns.
FIG. 4.
Representative axial skeletons of Acvr2b−/− Bmpr2+/− newborn pups. (a and b) Axial skeletons of Acvr2b−/− (a) and Acvr2b−/− Bmpr2+/− (b) mice. Transformation of V23 to T16 in Acvr2b−/− mice (a) was incomplete in Acvr2b−/− Bmpr2+/− mice (b). Asterisks and inset in panel b show small rib rudiments, indicating the incomplete transformation of V23 to the thoracic rib. V29, which mostly forms L6 in Acvr2b−/− (a), formed S1 in Acvr2b−/− Bmpr2+/− (b). (c and d) Sternum and vertebrosternal ribs of Acvr2b−/− (c) and Acvr2b−/− Bmpr2+/− (d) mice, showing eight VS ribs in Acvr2b−/− Bmpr2+/− mice, instead of nine.
TABLE 2.
Vertebral transformation in Acvr2b−/− and Acvr2b−/− Bmpr2+/− mice
| Vertebral transformation | No. of mice
|
||
|---|---|---|---|
| WT (n = 9) | Acvr2b−/−Bmpr2+/− (n = 13) | Acvr2b−/− (n = 15) | |
| V23 to L3 | 9 | 0 | 0 |
| V23 to L1 | 0 | 4 | 0 |
| V23 to T16(s)a | 0 | 7 | 0 |
| V23 to T16 | 0 | 2 | 15 |
| V29 to S3 | 9 | 0 | 0 |
| V29 to S1 | 0 | 13 | 0 |
| V29 to L6 | 0 | 0 | 15 |
(s), a vertebra exhibiting small rib attachments in the form of minute rudiment.
Btg2 enhances BMP/Smad-mediated transcriptional activation and interacts with Smad1 and Smad8.
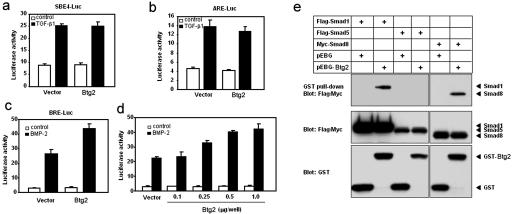
Another APRO family member, Tob1, has been shown to antagonize BMP/Smad signaling. Tob1 deficiency leads to enhanced BMP/Smad-dependent osteoblast function and to an increased incidence of spontaneous tumor formation (41, 42). We have investigated whether Btg2 also has a similar modulating effect on BMP or transforming growth factor β (TGF-β) signaling as Tob1. Based on the posterior transformation phenotype, we anticipated that Btg2 would either enhance BMP signaling, which is mediated by Smad1, Smad5, and/or Smad8, or inhibit Gdf11 signaling, which is mediated by Smad2 and/or Smad3 (25). To test this hypothesis, we analyzed the effect of Btg2 on the transcriptional activities of TGF-β/activin/Gdf11- or BMP-responsive promoters. Using the artificial SBE4-Luc reporter, which contains four tandem repeats of Smad-binding elements and measures the Smad3- and Smad4-specific response (43), we showed that overexpression of Btg2 has no effect on TGF-β-induced transcriptional activity (Fig. 5a). To address the effect of Btg2 on a Smad2-dependent promoter, we examined induction of the activin response element (ARE) from the Xenopus Mix.2 gene (16). Cotransfection of Btg2 with the ARE-Luc reporter and FAST1 on C2C12 cells showed that Btg2 has no effect on the TGF-β-induced activity of ARE-Luc (Fig. 5b). To address whether Btg2 has an effect on BMP-induced transcriptional activity, we tested BMP-responsive element (BRE), which is found in the mouse Id1 gene and responds to BMP signals but not to TGF-β or to activin (15). When the BMP-responsive element BRE-Luc was used, Btg2 enhanced BMP-2-induced transcriptional activity in a dose-dependent manner (Fig. 5c and d).
FIG. 5.
BTG2 activates BMP-dependent transcription and associates with Smad1 or Smad8 in vivo. (a and b) Effect of Btg2 on TGF-β1-induced transcription of SBE4-Luc (a) and ARE-Luc reporters (b). C2C12 cells were transiently transfected with a control vector (pCMV-β-gal) or Btg2 (1 μg/well) along with either SBE4-Luc (a) or ARE reporter plus FAST1 (b). (c and d) Effect of Btg2 on BMP-induced transcription of BRE-Luc reporter. (c) C2C12 cells were transiently transfected with the control vector or Btg2 (1 μg/well) along with BRE-Luc. The cells were treated with 50 ng of BMP-2/ml (black bars) or left untreated (white bars). (d) C2C12 cells were transiently transfected with the control vector or increasing amounts of Btg2 along with BRE-Luc. Luciferase activity was normalized to β-galactosidase activity and plotted as the mean and standard deviation for triplicates from a representative experiment. (e) GST-BTG2 was transfected into 293T cells with the Flag-tagged Smad1 or Smad5 or Myc-tagged Smad8 construct. Cell extracts were subjected to GST pull-down assay using glutathione-Sepharose 4B beads, followed by immunoblotting with anti-Flag or anti-Myc antibody. Expression of GST, GST-BTG2, and Smads was monitored as indicated.
To examine the possibility that Btg2 interacts directly with Smad proteins in vivo, 293T cells were transfected with Flag-tagged Smad1 or Smad5 or Myc-tagged Smad8 along with GST-tagged Btg2. The transfected cells were then subjected to pull-down with glutathione-Sepharose 4B beads, followed by immunoblotting with Flag or Myc antibodies. As shown in Fig. 5e, Btg2 showed strong interactions with Smad1 or Smad8 but not with Smad5. The interaction between Btg2 and Smad1 or Smad8 was not increased in the presence of BMP-2 (data not shown).
These results suggest that the posterior vertebral transformation phenotype observed with Btg2−/− mice might be due to the attenuation of BMP signaling. To test whether additional heterozygous mutation of the Bmpr2 gene in Btg2−/− mice increases the severity of the posterior transformation phenotype, we compared the vertebral pattern of Btg2−/− Bmpr2+/− (n = 11) with Btg2−/− (n = 7) littermates. However, no difference in the severities of the phenotypes between the two groups was found.
In this report, we have shown that Btg2 is expressed in the PSM-tail bud region and in developing somites with dynamic expression patterns and that Btg2 is involved in vertebral patterning via activating BMP signaling. Our results support the hypothesis that Btg2 may function as a coactivator-repressor and/or adaptor molecule in regulating the transcription status of various target genes. The posterior homeotic transformation phenotype of Btg2−/− mice is reminiscent of those found in several knockout mice deficient in Polycomb group (PcG) genes, which are required in order to maintain repression of Hox genes (33). A remarkably similar posterior transformation phenotype was also found in mice deficient in E2f6, a transcriptional repressor, which was found to associate with a number of PcG proteins (34, 37). Interactions between TGF-β-activated Smad proteins with E2f family proteins have been demonstrated (3). We speculate that Btg2 may play an adaptor function in the interaction of BMP-activated Smads and E2f6 in the PcG complex. Our results also suggest the intriguing possibility that two APRO family members, Btg2 and Tob1, may have a balancing role in BMP signaling: Btg2 activates BMP signaling, whereas Tob1 inhibits it (42).
Acknowledgments
We thank Jennifer Embury and Naime Akdede for the necropsy analysis of Btg2−/− mice.
This study was supported in part by NIH grant HL64024 to S.P.O. and by Korea Heath 21 R&D project grant 02-PJ10-PG8 EC01-0028 to I.K.L.
REFERENCES
- 1.Beppu, H., M. Kawabata, T. Hamamoto, A. Chytil, O. Minowa, T. Noda, and K. Miyazono. 2000. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 221:249-258. [DOI] [PubMed] [Google Scholar]
- 2.Canzoniere, D., S. Farioli-Vecchioli, F. Conti, M. T. Ciotti, A. M. Tata, G. Augusti-Tocco, E. Mattei, M. K. Lakshmana, V. Krizhanovsky, S. A. Reeves, R. Giovannoni, F. Castano, A. Servadio, N. Ben-Arie, and F. Tirone. 2004. Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1. J. Neurosci. 24:3355-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 4.Chen, F., and M. R. Capecchi. 1997. Targeted mutations in hoxa-9 and hoxb-9 reveal synergistic interactions. Dev. Biol. 181:186-196. [DOI] [PubMed] [Google Scholar]
- 5.Christ, B., C. Schmidt, R. Huang, J. Wilting, and B. Brand-Saberi. 1998. Segmentation of the vertebrate body. Anat. Embryol. (Berlin) 197:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Corrente, G., D. Guardavaccaro, and F. Tirone. 2002. PC3 potentiates NGF-induced differentiation and protects neurons from apoptosis. Neuroreport 13:417-422. [DOI] [PubMed] [Google Scholar]
- 7.Cortes, U., C. Moyret-Lalle, N. Falette, C. Duriez, F. E. Ghissassi, C. Barnas, A. P. Morel, P. Hainaut, J. P. Magaud, and A. Puisieux. 2000. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol. Carcinog. 27:57-64. [PubMed] [Google Scholar]
- 8.el Ghissassi, F., S. Valsesia-Wittmann, N. Falette, C. Duriez, P. D. Walden, and A. Puisieux. 2002. BTG2(TIS21/PC3) induces neuronal differentiation and prevents apoptosis of terminally differentiated PC12 cells. Oncogene 21:6772-6778. [DOI] [PubMed] [Google Scholar]
- 9.Ficazzola, M. A., M. Fraiman, J. Gitlin, K. Woo, J. Melamed, M. A. Rubin, and P. D. Walden. 2001. Antiproliferative B cell translocation gene 2 protein is down-regulated post-transcriptionally as an early event in prostate carcinogenesis. Carcinogenesis 22:1271-1279. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher, B. S., R. W. Lim, B. C. Varnum, D. A. Kujubu, R. A. Koski, and H. R. Herschman. 1991. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J. Biol. Chem. 266:14511-14518. [PubMed] [Google Scholar]
- 11.Guan, H., D. A. Smirnov, and R. P. Ricciardi. 2003. Identification of genes associated with adenovirus 12 tumorigenesis by microarray. Virology 309:114-124. [DOI] [PubMed] [Google Scholar]
- 12.Guardavaccaro, D., G. Corrente, F. Covone, L. Micheli, I. D'Agnano, G. Starace, M. Caruso, and F. Tirone. 2000. Arrest of G1-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol. Cell. Biol. 20:1797-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iacopetti, P., M. Michelini, I. Stuckmann, B. Oback, E. Aaku-Saraste, and W. B. Huttner. 1999. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc. Natl. Acad. Sci. USA 96:4639-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kannan, K., N. Amariglio, G. Rechavi, J. Jakob-Hirsch, I. Kela, N. Kaminski, G. Getz, E. Domany, and D. Givol. 2001. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20:2225-2234. [DOI] [PubMed] [Google Scholar]
- 15.Korchynskyi, O., and P. ten Dijke. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277:4883-4891. [DOI] [PubMed] [Google Scholar]
- 16.Labbe, E., C. Silvestri, P. A. Hoodless, J. L. Wrana, and L. Attisano. 1998. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 2:109-120. [DOI] [PubMed] [Google Scholar]
- 17.Lim, I. K., M. S. Lee, M. S. Ryu, T. J. Park, H. Fujiki, H. Eguchi, and W. K. Paik. 1998. Induction of growth inhibition of 293 cells by downregulation of the cyclin E and cyclin-dependent kinase 4 proteins due to overexpression of TIS21. Mol. Carcinog. 23:25-35. [DOI] [PubMed] [Google Scholar]
- 18.Lin, W. J., J. D. Gary, M. C. Yang, S. Clarke, and H. R. Herschman. 1996. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271:15034-15044. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda, S., J. Rouault, J. Magaud, and C. Berthet. 2001. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 497:67-72. [DOI] [PubMed] [Google Scholar]
- 20.McPherron, A. C., A. M. Lawler, and S. J. Lee. 1999. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22:260-264. [DOI] [PubMed] [Google Scholar]
- 21.Melamed, J., S. Kernizan, and P. D. Walden. 2002. Expression of B-cell translocation gene 2 protein in normal human tissues. Tissue Cell 34:28-32. [DOI] [PubMed] [Google Scholar]
- 22.Morel, A. P., S. Sentis, C. Bianchin, M. Le Romancer, L. Jonard, M. C. Rostan, R. Rimokh, and L. Corbo. 2003. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J. Cell Sci. 116:2929-2936. [DOI] [PubMed] [Google Scholar]
- 23.Nowicki, J. L., and A. C. Burke. 2000. Hox genes and morphological identity: axial versus lateral patterning in the vertebrate mesoderm. Development 127:4265-4275. [DOI] [PubMed] [Google Scholar]
- 24.Oh, S. P., and E. Li. 1997. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 11:1812-1826. [DOI] [PubMed] [Google Scholar]
- 25.Oh, S. P., C. Y. Yeo, Y. Lee, H. Schrewe, M. Whitman, and E. Li. 2002. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev. 16:2749-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Packard, D. S., Jr. 1978. Chick somite determination: the role of factors in young somites and the segmental plate. J. Exp. Zool. 203:295-306. [DOI] [PubMed] [Google Scholar]
- 27.Pourquie, O. 2001. Vertebrate somitogenesis. Annu. Rev. Cell Dev. Biol. 17:311-350. [DOI] [PubMed] [Google Scholar]
- 28.Prevot, D., A. P. Morel, T. Voeltzel, M. C. Rostan, R. Rimokh, J. P. Magaud, and L. Corbo. 2001. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J. Biol. Chem. 276:9640-9648. [DOI] [PubMed] [Google Scholar]
- 29.Prevot, D., T. Voeltzel, A. M. Birot, A. P. Morel, M. C. Rostan, J. P. Magaud, and L. Corbo. 2000. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J. Biol. Chem. 275:147-153. [DOI] [PubMed] [Google Scholar]
- 30.Raouf, A., and A. Seth. 2002. Discovery of osteoblast-associated genes using cDNA microarrays. Bone 30:463-471. [DOI] [PubMed] [Google Scholar]
- 31.Rouault, J. P., N. Falette, F. Guehenneux, C. Guillot, R. Rimokh, Q. Wang, C. Berthet, C. Moyret-Lalle, P. Savatier, B. Pain, P. Shaw, R. Berger, J. Samarut, J. P. Magaud, M. Ozturk, C. Samarut, and A. Puisieux. 1996. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat. Genet. 14:482-486. [DOI] [PubMed] [Google Scholar]
- 32.Rouault, J. P., D. Prevot, C. Berthet, A. M. Birot, M. Billaud, J. P. Magaud, and L. Corbo. 1998. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J. Biol. Chem. 273:22563-22569. [DOI] [PubMed] [Google Scholar]
- 33.Schumacher, A., and T. Magnuson. 1997. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 13:167-170. [PubMed] [Google Scholar]
- 34.Storre, J., H. P. Elsasser, M. Fuchs, D. Ullmann, D. M. Livingston, and S. Gaubatz. 2002. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep. 3:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struckmann, K., P. Schraml, R. Simon, K. Elmenhorst, M. Mirlacher, J. Kononen, and H. Moch. 2004. Impaired expression of the cell cycle regulator BTG2 is common in clear cell renal cell carcinoma. Cancer Res. 64:1632-1638. [DOI] [PubMed] [Google Scholar]
- 36.Tirone, F. 2001. The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J. Cell. Physiol. 187:155-165. [DOI] [PubMed] [Google Scholar]
- 37.Trimarchi, J. M., B. Fairchild, J. Wen, and J. A. Lees. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98:1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 39.Wellik, D. M., and M. R. Capecchi. 2003. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301:363-367. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson, D. G. 1992. In situ hybridization: a practical approach. Oxford University Press, London, United Kingdom.
- 41.Yoshida, Y., T. Nakamura, M. Komoda, H. Satoh, T. Suzuki, J. K. Tsuzuku, T. Miyasaka, E. H. Yoshida, H. Umemori, R. K. Kunisaki, K. Tani, S. Ishii, S. Mori, M. Suganuma, T. Noda, and T. Yamamoto. 2003. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 17:1201-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida, Y., S. Tanaka, H. Umemori, O. Minowa, M. Usui, N. Ikematsu, E. Hosoda, T. Imamura, J. Kuno, T. Yamashita, K. Miyazono, M. Noda, T. Noda, and T. Yamamoto. 2000. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103:1085-1097. [DOI] [PubMed] [Google Scholar]
- 43.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1:611-617. [DOI] [PubMed] [Google Scholar]