Abstract
Numerous stressful conditions activate kinases that phosphorylate the alpha subunit of translation initiation factor 2 (eIF2α), thus attenuating mRNA translation and activating a gene expression program known as the integrated stress response. It has been noted that conditions associated with eIF2α phosphorylation, notably accumulation of unfolded proteins in the endoplasmic reticulum (ER), or ER stress, are also associated with activation of nuclear factor kappa B (NF-κB) and that eIF2α phosphorylation is required for NF-κB activation by ER stress. We have used a pharmacologically activable version of pancreatic ER kinase (PERK, an ER stress-responsive eIF2α kinase) to uncouple eIF2α phosphorylation from stress and found that phosphorylation of eIF2α is both necessary and sufficient to activate both NF-κB DNA binding and an NF-κB reporter gene. eIF2α phosphorylation-dependent NF-κB activation correlated with decreased levels of the inhibitor IκBα protein. Unlike canonical signaling pathways that promote IκBα phosphorylation and degradation, eIF2α phosphorylation did not increase phosphorylated IκBα levels or affect the stability of the protein. Pulse-chase labeling experiments indicate instead that repression of IκBα translation plays an important role in NF-κB activation in cells experiencing high levels of eIF2α phosphorylation. These studies suggest a direct role for eIF2α phosphorylation-dependent translational control in activating NF-κB during ER stress.
Diverse stressful conditions lead to the phosphorylation of translation initiation factor 2 on its alpha subunit (eIF2α). Phosphorylated eIF2 inhibits its guanine nucleotide exchange factor, eIF2B, and thereby inhibits the exchange reaction required to generate active GTP-bound eIF2. As a consequence, regulated phosphorylation of eIF2α serves to modulate mRNA translation rates (18, 20). In addition to its negative impact on global protein synthesis, eIF2 phosphorylation also promotes gene-specific upregulation of the translation of certain mRNAs. The two known examples of this involve the yeast transcription factor GCN4 (19) and the mammalian transcription factor ATF4 (12). Regulated gene expression appears to be an important consequence of eIF2α phosphorylation, as mutations that interfere with eIF2α phosphorylation lead to an important defect in stress-induced gene expression (16, 28, 39).
Four known eIF2α kinases couple seemingly unrelated stressful conditions to the aforementioned common translational regulatory event. PKR responds to double-stranded RNA in virally infected cells (23), GCN2 is activated by uncharged tRNAs in amino acid-starved cells (20), HRI is activated by heme depletion in erythroid precursor cells (3), and PERK is activated by unfolded proteins in the endoplasmic reticulum (ER), or ER stress (37). Mutations in each of these four kinases have been produced, and their phenotypes reveal the importance of eIF2α phosphorylation in stressed cells (6).
Nuclear factor kappa B (NF-κB) encompasses a family of stress-induced transcription factors. Like the more ancient eIF2α phosphorylation-dependent signaling, NF-κB signaling is also triggered by diverse stressful conditions, and activated NF-κB has broad effects on gene expression (38). Several studies have suggested cross talk between the eIF2α phosphorylation pathway and NF-κB activation. The double-stranded-RNA-activated eIF2α kinase PKR was noted to phosphorylate the NF-κB inhibitor, IκB (26), and genetic and pharmacological interventions that interfere with PKR activity attenuated NF-κB activation by cytokines (4, 27, 47) or viruses (9, 43). There is some uncertainty regarding the role of eIF2α phosphorylation in NF-κB activation by PKR, as the latter contributes to NF-κB activation by both kinase-dependent (9) and kinase-independent (8) mechanisms.
Conditions that promote accumulation of unfolded proteins in the endoplasmic reticulum lead to high levels of eIF2α phosphorylation (34, 35), which is mediated by the ER-localized kinase PERK (14, 15). These same conditions activate NF-κB (32). A recent study has found that ER stress-mediated NF-κB activation was attenuated both in PERK−/− cells and, importantly, in cells bearing two mutant alleles of EIF2A in which serine 51 (the substrate of the stress-inducible kinases) had been mutated to an alanine. These mutant eIF2αA/A cells were also defective in NF-κB activation by amino acid starvation, as were cells lacking GCN2 (21), the kinase that phosphorylates eIF2α in amino acid-starved cells.
Together these observations point to a nonredundant role for eIF2α phosphorylation in NF-κB activation under various stress conditions. But they provide little insight into the mechanisms involved. One of the best-characterized aspects of NF-κB regulation is the phosphorylation-dependent, proteasome-mediated degradation of its inhibitor, IκB. However, it is not clear if and how eIF2α phosphorylation ties in to IκB levels. Because the stressful conditions used to promote eIF2α phosphorylation have multiple other effects (reviewed in reference 17), it is not even known whether eIF2α phosphorylation plays a permissive role or an instructive role in NF-κB activation, nor is it known whether the phosphorylated form of eIF2α is affecting NF-κB activation as a modified translation initiation factor or by some other means. In an effort to answer some of these questions, we have probed NF-κB activation in an experimental system that uncouples eIF2α phosphorylation from stress signaling and discovered that translational repression of IκB can account for activation of NF-κB under conditions of eIF2α phosphorylation.
MATERIALS AND METHODS
Cell culture, cell transfection, and treatment.
The wild-type cells and EIF2AA/A mutant cells in which the serine at position 51, the regulatory phosphorylation site, had been replaced by an alanine have been previously described (39). They were cultured in Dulbecco's modified Eagle's medium supplemented with glutamine, nonessential amino acids, 55 μM β-mercaptoethanol, and 10% fetal calf serum. The establishment of stable clones of mouse fibroblasts expressing Fv2E-PERK with defined EIF2A genotypes has been previously described (28).
The Fv2E-PERK+ wild-type mouse embryonic fibroblasts described above were transiently transfected using Fugene lipid-based gene transfer reagent (catalog no. 1814443; Roche, Indianapolis, Ind.) with luciferase reporter plasmids containing a minimal rat angiotensinogen promoter driven by four wild-type or mutant NF-κB binding sites from the rat angiotensinogen gene, as previously described (36). One day later the cells were treated for 1 h with the indicated concentration of AP20187 (gift of ARIAD Pharmaceuticals, Cambridge, Mass.), washed free of the activator (to allow translation to recover), and harvested for use in a luciferase assay 24 h later.
Cells were treated with thapsigargin (catalog no. T9033; Sigma, St. Louis, Mo.) at a final concentration of 400 nM or cycloheximide (catalog no. C7698; Sigma) at 20 μg/ml. Unless otherwise indicated, AP20187 was used at a concentration of 10 nM. Cells were treated with 20 ng of tumor necrosis factor alpha (TNF-α; catalog no. T7539; Sigma)/ml with or without the proteasome inhibitor MG132 (catalog no. 474790; Calbiochem-Novobiochem, San Diego, Calif.) at 10 μM.
Immunoblotting and immunoprecipitation.
Total IκBα was detected with a purified rabbit immune serum (catalog no. 9242; Cell Signaling, Beverly, Mass.), and IκBα phosphorylated on serine 32 and 36 was detected with an epitope-specific antiserum (catalog no. 9246; Cell Signaling). GADD34 was detected with an antiserum directed to the N terminus of the mouse protein raised in our lab (30). PERK was detected with a 1:1 mixture of two rabbit antisera (NY97, which detects the unphosphorylated form of the protein, and NY201, which detects predominantly the hyperphosphorylated forms of the protein) as described previously (2). Total eIF2α was detected with a monoclonal antibody to human eIF2α, a gift of the late Edward Henshaw (40), and phosphorylated eIF2α was detected with an epitope-specific antiserum (catalog no. RG0001; Research Genetics, Huntsville, Ala.).
Pulse-chase labeling experiments were carried out in the Fv2E-PERK+ wild-type mouse embryonic fibroblasts described above. Cells were switched to Dulbecco's modified Eagle's medium containing 10% of the normal content of methionine and cysteine (these levels of methionine and cysteine are sufficient to suppress activation of the eIF2α kinase GCN2 yet are compatible with high-level incorporation of labeled amino acids into newly synthesized proteins) 15 min before addition of TRANSlabel (MP Biomedical, Irvine, Calif.) 35S-labeled methionine-cysteine mixture at 200 μCi/ml for 10 min. The labeling pulse was terminated by washing the unincorporated label and flooding the cells with complete medium. Following the indicated chase period, during which cells were exposed to AP20187 and/or MG132, the cells were lysed in RIPA buffer (20 mM Tris [pH 8.5], 100 mM NaCl, 0.2% sodium deoxycholate, 0.2% NP-40, 0.2% Triton X-100, 0.1% sodium dodecyl sulfate, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 4 μg of aprotinin/ml, 2 μg of pepstatin/ml), and the lysate was clarified by centrifugation at 14,000 × g for 15 min, precleared on protein A-Sepharose beads (catalog no. 10-1042; Zymed, South San Francisco, Calif.), and subjected to immunoprecipitation with prebound anti-IκBα rabbit immunoglobulin G (catalog no. SC-371 AC; Santa Cruz Biotech, Santa Cruz, Calif.). Radiolabeled proteins found in the immunoprecipitate were resolved by reduced sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the dried gel was exposed to autoradiography using a phosphoimaging cassette (Molecular Dynamics, Sunnyvale, Calif.).
EMSA.
NF-κB DNA binding activity in nuclear extracts was detected by an electrophoretic mobility shift assay (EMSA) performed as previously described (21, 36). The indicated molar excess of unlabeled competitor probe or 1 μl of purified anti-p65 (catalog no. SC-7151; Santa Cruz Biotech) or anti-CHOP antiserum (45) was added to the binding reaction together with the radiolabeled probe.
RESULTS
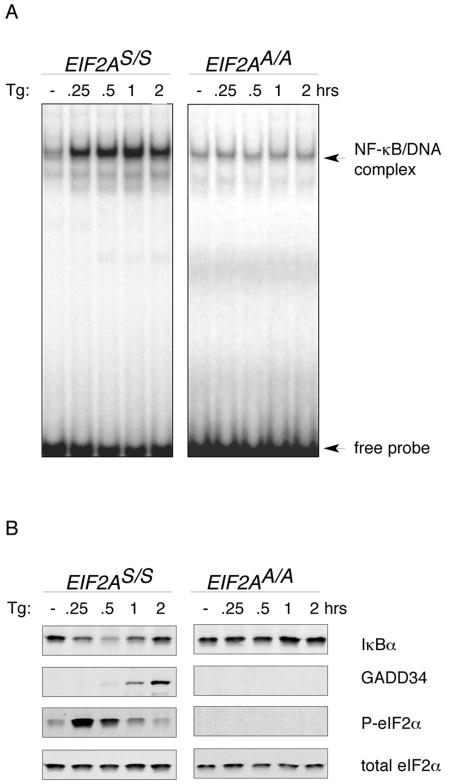
To confirm the previously reported role of eIF2α phosphorylation in NF-κB activation (21), we performed EMSA on nuclear extracts prepared from unstressed cells and cells that had been treated with thapsigargin (Fig. 1A). Thapsigargin-mediated ER calcium depletion leads to rapid onset of ER stress, eIF2α phosphorylation (detected here by immunoblotting with an antiserum specifically reactive with the phosphorylated form), and subsequent ATF4-mediated activation of downstream gene expression, measured here by accumulation of the GADD34 target gene. A protein complex rapidly formed on the NF-κB binding site in nuclear extracts of treated wild-type cells but not in extracts from cells homozygous for the EIF2AA/A mutation that substitutes the serine at position 51 of eIF2α with an alanine and thereby prevents regulatory phosphorylation. Reduced levels of the NF-κB inhibitory protein IκBα, detected by immunoblotting, preceded the induction of NF-κB EMSA activity in thapsigargin-treated cells. The recovery of IκBα levels at longer treatment points correlated with the induction of the GADD34 phosphatase and the dephosphorylation of eIF2α (Fig. 1B).
FIG. 1.
NF-κB activation during ER stress depends on eIF2α phosphorylation and is associated with declining levels of the NF-κB inhibitor IκBα. (A) Autoradiogram of an NF-κB EMSA performed with nuclear extracts of thapsigargin-treated mouse fibroblasts (Tg) with wild-type (EIF2AS/S) or mutant (EIF2AA/A) EIF2A genotypes. The free radiolabeled probe and the labeled NF-κB/DNA complex are indicated. (B) Immunoblots of IκBα, GADD34, phosphorylated eIF2α, and total eIF2α from extracts of the cells shown in panel A, detected with specific antibodies.
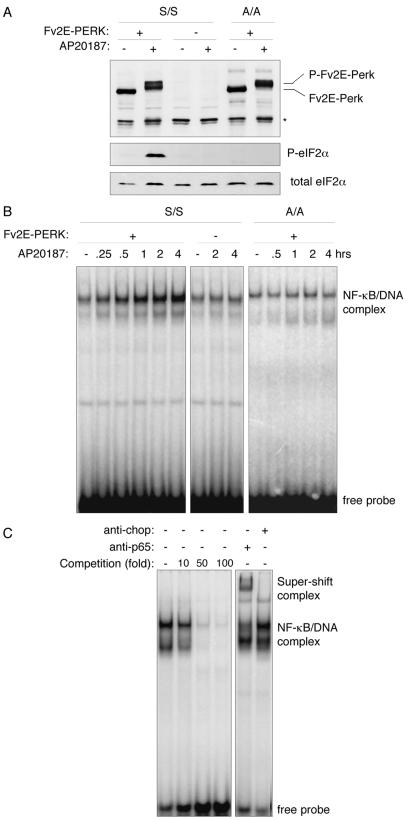
To more closely examine the role of eIF2α phosphorylation in NF-κB activation, we made use of an experimental system that uncouples eIF2α phosphorylation from stress signaling. PERK, the ER stress-inducible eIF2α kinase, is normally activated by oligomerization in the plane of the ER membrane (2). We fused PERK's eIF2α kinase domain to a protein module with two high-affinity binding sites for the otherwise inert bivalent compound AP20187. When expressed in cells, this artificial kinase, Fv2E-PERK, is subordinate to AP20187 treatment (28) and is activated independently of any stress signaling. AP20187 treatment led to high-level eIF2α phosphorylation in Fv2E-PERK+ cells but had no effect on the parental cells lacking the artificial kinase (Fig. 2A). Fv2E-PERK was readily activated in mutant EIF2AA/A cells, but this predictably failed to induce eIF2α phosphorylation. EMSA of nuclear extracts showed that AP20187 induced NF-κB activity in Fv2E-PERK+ wild-type (EIF2AS/S) cells but not in the mutant EIF2AA/A cells (Fig. 2B). Homologous competition binding assays and antibody supershift experiments confirmed the identity of the NF-κB protein-DNA complex detected in the assay (Fig. 2C).
FIG. 2.
Phosphorylation of eIF2α on serine 51 is sufficient to activate NF-κB DNA binding activity in vivo. (A) Immunoblots of ligand-activable Fv2E-PERK (upper panel), phosphorylated eIF2α (P-eIF2α; middle panel), and total eIF2α (lower panel) in extracts of mouse fibroblasts of wild-type (S/S) and EIF2AA/A mutant (A/A) genotypes that do and do not stably express the chimeric eIF2α kinase, Fv2E-PERK. Where indicated, the cells had been treated with the Fv2E ligand, AP20187. Endogenous PERK is not detected at this exposure. The asterisk marks the position of a nonspecific band reactive with the anti-PERK sera. (B) Autoradiogram of an NF-κB EMSA performed with nuclear extracts of cells treated as described for panel A. (C) Autoradiogram of an NF-κB EMSA with nuclear extract obtained from AP20187-treated Fv2E-PERK+ cells performed in the presence of the indicated excess of an unlabeled homologous competitor oligonucleotide (left panel) or in the presence of antisera to CHOP (a negative control) or p65 (a component of the NF-κB DNA binding complex) (right panel). The positions of the free radiolabeled probe, the NF-κB/DNA complex, and antiserum supershifted complex are indicated.
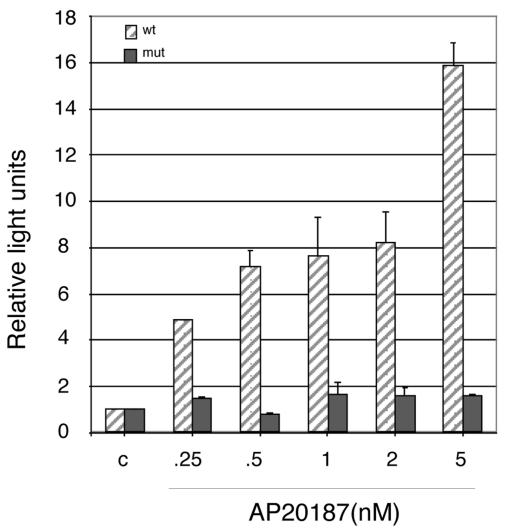
To gauge the functional significance of Fv2E-PERK-mediated eIF2α phosphorylation and activation of NF-κB DNA binding activity, we measured the activity of a transfected reporter gene driven by four copies of a wild-type NF-κB binding site. A brief (60-min) pulse of AP20187 induced marked activation of the wild-type reporter gene (measured 24 h later [Fig. 3]). No activation of a reporter gene driven by mutant NF-κB sites was observed. In addition, endogenous NF-κB target genes, such as those encoding the major histocompatibility complex heavy chains (H2-Q8, H2-2KF, H2-K2, and H2-D1) and β2 microglobulin (Qb-1), were induced in the Fv2E-PERK+ cells by AP20187 treatments and in wild-type mouse fibroblasts by exposure to tunicamycin (National Center for Biotechnology Information Gene Expression Omnibue [GEO] data set GDS405).
FIG. 3.
eIF2α phosphorylation is sufficient to activate an NF-κB reporter gene. The activity of a transiently transfected reporter gene consisting of a minimal promoter driven by four wild-type (wt) or mutant (mut) NF-κB binding sites in mouse fibroblasts stably expressing Fv2E-PERK is shown following treatment with the indicated concentration of the activating ligand AP20187. The results are expressed as relative light units, and the activity of the reporter in untreated cells is arbitrarily set at 1. Shown are means and standard errors of the means of results from an experiment performed in triplicate and reproduced twice.
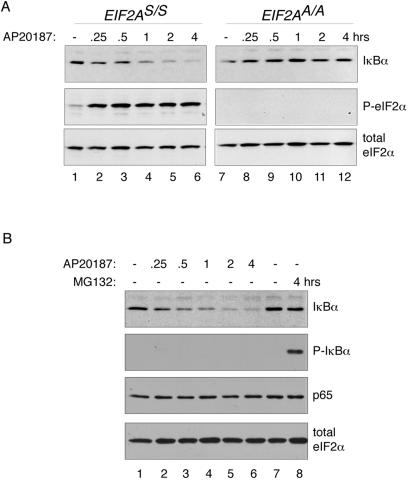
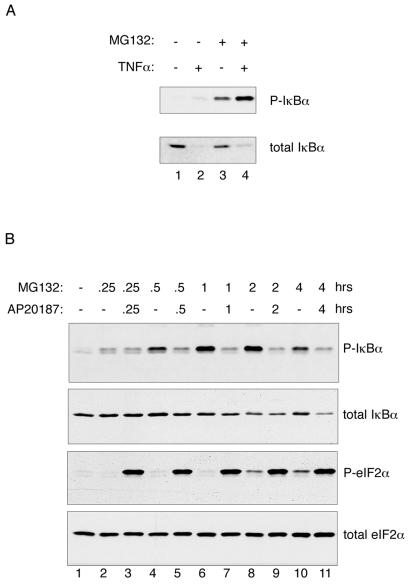
Fv2E-PERK-mediated eIF2α phosphorylation and NF-κB activation correlated with a time-dependent decrease in IκBα levels that was not observed in the mutant EIF2AA/A cells (Fig. 4A). Interestingly, Fv2E-PERK activation had no measurable effect on levels of the p65 NF-κB subunit, which is consistent with the known stability of that protein (24) and with the induction of NF-κB binding activity that we observe. eIF2α levels were similarly stable, attesting to the effect's specificity to IκBα (Fig. 4B). Canonical activators of NF-κB access signal transduction pathways that promote phosphorylation of the inhibitor IκBα on serines 32 and 36 (38). A ubiquitin ligase complex recognizes the phosphorylated form of IκBα, and polyubiquitinated IκBα is degraded by the proteasome. Fv2E-PERK activation by AP20187 did not promote a measurable increase in levels of phosphorylated IκBα, which remained undetectable. However, phosphorylated IκBα was readily detectable in lysates of cells treated with the proteasome inhibitor, MG132, which stabilizes the phosphorylated form of the protein (Fig. 4B).
FIG. 4.
eIF2α phosphorylation reduces cellular levels of IκBα. (A) Immunoblots of total IκBα (upper panel), phosphorylated eIF2α (P-eIF2α; middle panel), and total eIF2α (lower panel) in extracts of wild-type (EIF2AS/S) and mutant (EIF2AA/A) Fv2E-PERK+ mouse fibroblasts following treatment with the activating ligand AP20187 for the indicated periods of time. (B) Immunoblots of total IκBα, phosphorylated IκBα (P-IκBα), p65 NF-κB subunit, and total eIF2α in extracts of wild-type (EIF2AS/S) Fv2E-PERK+ mouse fibroblasts following treatment with the activating ligand AP20187 or the proteasome inhibitor (MG132) for the indicated periods of time.
Because it is rapidly degraded, signal-dependent accumulation of phosphorylated IκBα is difficult to detect, rendering an Fv2E-PERK-mediated increase in IκBα phosphorylation potentially easy to miss. Therefore, to determine if the eIF2α phosphorylation-dependent decline in IκBα levels correlated with any increased phosphorylation on serines 32 and 36, we exposed the AP20187-treated cells to the proteasome inhibitor MG132. As expected, proteasome inhibition markedly increased the levels of phosphorylated IκBα in tumor necrosis factor alpha-treated cells (Fig. 5A). Interestingly, proteasome inhibition led to only modest stabilization of total IκBα, an observation that is consistent with the existence of proteasome-independent mechanisms for IκBα degradation (5, 11).
FIG. 5.
Reduction in levels of IκBα in cells with elevated eIF2α phosphorylation occurs independently of IκBα phosphorylation. (A) Immunoblots of IκBα phosphorylated on serines 32 and 36 (P-IκBα; upper panel) and total IκBα (lower panel) in extracts of mouse fibroblasts treated with TNF-α and/or the proteasome inhibitor MG132. (B) Immunoblots of phosphorylated IκBα (P-IκBα), total IκBα, phosphorylated eIF2α (P-eIF2α), and total eIF2α in extracts of Fv2E-PERK+ mouse fibroblasts treated with the activating ligand AP20187 and/or the proteasome inhibitor MG132 for the indicated periods of time.
MG132 treatment led to a progressive increase in phosphorylated IκBα levels in cells that were otherwise unperturbed (Fig. 5A, compare lanes 1 and 3, and B, compare lane 1 with lanes 2, 4, 6, 8, and 10). This observation is consistent with a relatively high basal phosphorylation-dependent turnover of IκBα in these cells. The decline in IκBα levels effected by Fv2E-PERK was only slightly attenuated by proteasome inhibition (compare Fig. 4, lanes 4 to 6, with 5B, lanes 7, 9, and 11). Furthermore, proteasome inhibition promoted some eIF2α phosphorylation (Fig. 5, lanes 8 and 10), presumably mediated by proteotoxic stress. Remarkably, however, Fv2E-PERK activation and eIF2α phosphorylation not only failed to increase IκBα phosphorylation but also significantly attenuated the accumulation of phosphorylated IκBα in proteasome-inhibited cells (Fig. 5B, compare odd- and even-numbered lanes). These observations indicate that eIF2α phosphorylation does not activate NF-κB by accessing one of the canonical IκBα phosphorylation-promoting pathways and must use a different mechanism.
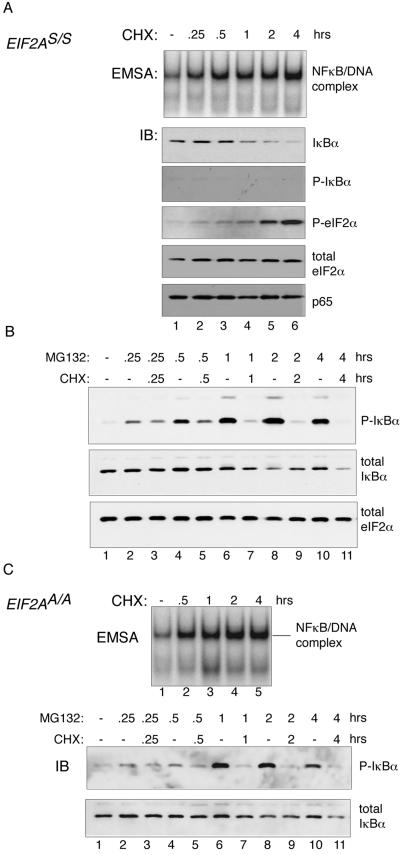
The original descriptions of IκB emphasized the lability of the factor, as translational inhibitors were noted to promote NF-κB DNA binding activity (1, 42). Given that eIF2α phosphorylation also inhibits protein synthesis, we decided to explore this facet of NF-κB activation in more detail. NF-κB DNA binding activity was increased by cycloheximide treatment of wild-type cells, as previously reported (42), and this correlated with reduced levels of the inhibitor, IκBα (Fig. 6A). Cycloheximide treatment led to no measurable decrease in p65 or eIF2α protein levels, attesting to the stability of these proteins. The effects of cycloheximide on levels of phosphorylated IκBα also resembled those of Fv2E-PERK activation (Fig. 4B) in that no increase in the phosphorylated protein was observed in cells treated with cycloheximide alone. Proteasome inhibitor, by itself, led to a progressive increase in levels of phosphorylated IκBα, whereas the addition of cycloheximide strongly attenuated this increase (Fig. 6B).
FIG. 6.
Reduction in levels of IκBα in cells treated with the protein synthesis inhibitor cycloheximide occurs independently of IκBα phosphorylation or eIF2α phosphorylation. (A) The top panel is an autoradiogram of an NF-κB EMSA from nuclear extracts of untreated and cycloheximide (CHX)-treated mouse fibroblasts. The lower panels are immunoblots (IB) of total IκBα, phosphorylated IκBα (P-IκBα), phosphorylated eIF2α (P-eIF2α), total eIF2α, and the p65 NF-κB subunit from the same cells. (B) Immunoblots of phosphorylated IκBα (P-IκBα), total IκBα, and total eIF2α in extracts of wild-type (EIF2AS/S) Fv2E-PERK+ mouse fibroblasts treated with cycloheximide and/or the proteasome inhibitor MG132 for the indicated periods of time are shown. (C) The same assays as shown in panels A and B were conducted with mutant (EIF2AA/A) Fv2E-PERK+ cells.
As previously noted, cycloheximide treatment induced eIF2α phosphorylation (21, 22) (Fig. 6A), an effect that might be attributed to loss of the labile eIF2α phosphatase CReP (22). To study the role of eIF2α phosphorylation in cycloheximide-mediated activation of NF-κB, we treated mutant EIF2AA/A cells with the protein synthesis inhibitor and studied NF-κB activation by EMSA and IκBα levels by immunoblotting. The EIF2AA/A genotype, which inhibits regulatory phosphorylation of eIF2α, had no measurable effect on NF-κB activation, IκBα phosphorylation, or total IκBα levels in cycloheximide-treated cells (Fig. 6C). These observations suggest that inhibition of new protein synthesis can adequately explain the effects of cycloheximide on NF-κB activity without evoking an additional role for eIF2α phosphorylation.
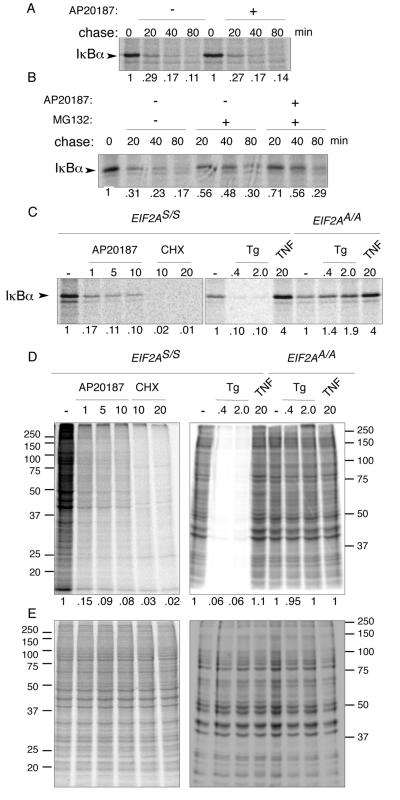
Induced degradation of IκBα plays an important role in canonical activation of NF-κB. To address the possibility that eIF2α phosphorylation might affect this aspect of IκBα metabolism (independently of IκBα phosphorylation), we performed pulse-chase labeling experiments, tracking the fate of newly synthesized IκBα. The basal turnover of IκBα in murine fibroblasts proved very high. Less than 30% of the signal measured at the end of the 10-min labeling pulse was present after a 20-min chase. Furthermore, activation of Fv2E-PERK during the chase had no measurable effect on the decay of the IκBα signal (Fig. 7A). Addition of proteasome inhibitor during the chase stabilized IκBα somewhat; however, in that context, too, activation of Fv2E-PERK during the chase did not accelerate IκBα degradation and may have even contributed modestly to its stability (Fig. 7B). We conclude that IκBα turns over rapidly in murine fibroblasts and that eIF2α phosphorylation does not exert its effects on the levels of the inhibitor by further enhancing its degradation.
FIG. 7.
eIF2α phosphorylation inhibits synthesis of IκBα but does not destabilize the preexisting protein. (A) Autoradiogram of IκBα immunoprecipitated from wild-type (EIF2AS/S) Fv2E-PERK+ mouse fibroblasts following a brief, 10-min [35S]methionine- and cysteine-labeling pulse and cold chase of the indicated duration. The chase was conducted in the presence or absence of the activating ligand AP20187. The IκBα signal intensity is expressed as a fraction of that present at the end of the labeling pulse and is depicted beneath each lane. (B) Same assay as shown in panel A except that the proteasome inhibitor, MG132, was included during the chase where indicated. (C) Autoradiogram of the radiolabeled IκBα present at the end of the 10-min labeling pulse in wild-type (EIF2AS/S) or mutant (EIF2AA/A) Fv2E-PERK+ mouse fibroblasts treated with the indicated concentration of AP20187 ligand (in nM), cycloheximide (in μg/ml), thapsigargin (in μM), or TNF-α (in ng/ml) starting 30 min before and continuing throughout the pulse. (D) Autoradiogram ([35S]methionine) of equal fractions of the cell lysates used in panel C. The right panel is of a gel that was run longer than the left panel, accounting for differences in appearance of the two. (E) Coomassie stain of the gels shown in panel D.
Next we compared the rates of synthesis of IκBα in untreated cells with those in cells treated with AP20187, cycloheximide, the ER stress-promoting agent thapsigargin, and the canonical NF-κB activator TNF-α. The amount of radiolabeled IκBα immunoprecipitated with a specific antibody following a short labeling pulse was markedly diminished by activation of the eIF2α kinase Fv2E-PERK by AP20187, by treatment with cycloheximide, or by exposure to conditions that cause ER stress (thapsigargin) (Fig. 7C). The effect of thapsigargin on IκBα synthesis depended on eIF2α phosphorylation, since it was abolished in the EIF2AA/A mutant cells (Fig. 7C), and the decline in IκBα synthesis paralleled the global inhibition in protein synthesis in the cells exposed to conditions promoting eIF2α phosphorylation (Fig. 7D). By contrast, exposure to the canonical NF-κB activator, TNF-α, increased IκBα synthesis, suggesting a completely different mechanism of action. These observations are consistent with a role for inhibited synthesis of IκBα in mediating the effects of eIF2α phosphorylation on NF-κB activation both in ER-stressed cells and following activation of Fv2E-PERK.
DISCUSSION
Signaling through stress-induced phosphorylation of eIF2α is conserved among the eukaryotes and represents one of the oldest pathways for stress-induced gene expression. Furthermore, eIF2α phosphorylation is concerned mostly with autonomous cell adaptations to stress. NF-κB signaling, on the other hand, is found in metazoans, and canonical activators of NF-κB signaling, such as cytokines, are intercellular signaling molecules. However, over the years evidence that autonomous cell phenomena, such as ER stress, are also associated with NF-κB activation has accrued, with the suggestion that ancient, autonomous cell signaling pathways might be linked to NF-κB activation.
This study confirms the established role of eIF2α phosphorylation in NF-κB activation by ER stress (21). Using an inducible system that uncouples eIF2α phosphorylation from other stress signals, we find that eIF2α phosphorylation can have an instructive role in NF-κB activation. In other words, activation of an eIF2α kinase provides a signal sufficient for NF-κB activation in cultured mouse fibroblasts. Our study also reveals significant differences between the mechanism used by canonical inducers of NF-κB and the consequences of eIF2α phosphorylation. Unlike canonical inducers of NF-κB, eIF2α phosphorylation promoted neither phosphorylation nor degradation of IκBα. Instead, our data argue that the major impact of eIF2α phosphorylation on NF-κB activation is inhibition of the synthesis of the labile inhibitor IκBα.
The mechanism uncovered in this study suggests that the link between eIF2α phosphorylation and NF-κB activation depends on the lability of the inhibitor, which, in turn, likely depends on basal levels of signaling through the canonical pathway(s) that activates NF-κB. Indeed, the rapid accumulation of phosphorylated IκBα in mouse fibroblasts treated with proteasome inhibitor is consistent with high basal levels of IκBα kinase activity in these cells. It is worth noting that both eIF2α phosphorylation and cycloheximide treatment disproportionately reduced the levels of phosphorylated IκBα, compared with their effect on the levels of total IκBα. Inhibited protein synthesis may attenuate basal activity of an IκBα kinase and account for some of this effect. Alternatively, newly synthesized IκBα might constitute a preferred substrate for its kinases. The plausibility of the latter explanation is supported by evidence for the existence of multiple pools of IκBα in cells (25, 33, 41). The existence of more than one pool of IκBα might also explain the discrepancy between the short half-life of newly synthesized IκBα (measured by the pulse-chase method [Fig. 7A and B]) and the much longer half-life inferred from the gradual decline in total IκBα protein levels in the cycloheximide-treated and Fv2E-PERK-activated cells (Fig. 4, 5B, and 6A and B). However, these potential complexities of IκBα metabolism do not weaken our conclusion that attenuated synthesis of the inhibitor plays a major role in mediating activation of NF-κB by eIF2α phosphorylation in mouse fibroblasts.
Our findings are at odds with those reported by Jiang and colleagues, who found no decrease in steady-state IκBα levels in thapsigargin-treated cells and instead uncovered evidence for dissociation of the IκBα-NF-κB complex under those conditions (see Fig. 6 in reference 21). We have no explanation for these differences; however, we do note that since the submission of the present study Wu and colleagues have reported that induction of NF-κB DNA binding activity in cells exposed to UV light is also associated with eIF2α phosphorylation-dependent repression of IκBα synthesis (46).
Our study does not address the physiological significance of the link between eIF2α phosphorylation and NF-κB activation. It is worth noting that we have but an incomplete understanding of the relative significance of regulated protein synthesis versus activation of gene expression programs as readouts of eIF2α phosphorylation. In yeast it is fairly clear that mutations in the transcription factor GCN4 phenocopy mutations in the upstream kinase GCN2 or in the gene encoding its substrate SUI2 (yeast eIF2α) (6, 7). In mammalian cells too, some of the phenotypes of loss of PERK gene function or the EIF2AA/A genotype are mimicked by mutations in the gene encoding the downstream transcription factor ATF4 (16, 29, 39). Furthermore, in both yeast and mammalian cells, translation activation of the transcription factors GCN4 and ATF4 occurs at levels of eIF2α phosphorylation that have only a modest impact on global protein synthesis (7, 44; Lu et al., unpublished observation). By contrast, our proposed mechanism of cross talk between eIF2α phosphorylation and NF-κB signaling is proportional to the repression of IκBα translation. Such levels of repression are easily attained in thapsigargin-treated cells (14) or in Fv2E-PERK+ cells activated by AP20187 (28) and are clearly sufficient to activate NF-κB in cultured mouse fibroblasts (Fig. 1A and 2B) (21).
The extent to which translational repression contributes to NF-κB activation in more physiological contexts in which eIF2α kinases are activated is not known. However, we note that endogenous proinflammatory NF-κB target genes, such as those encoding the major histocompatibility complex heavy and light chains, the interleukin 17 receptor, and a complement receptor-related protein, were all induced in the Fv2E-PERK+ cells by AP20187 treatment and in wild-type mouse fibroblasts by exposure to tunicamycin (National Center for Biotechnology Information GEO data set GDS405). The PERK-dependent induction of NF-κB target genes by tunicamycin is potentially significant, as global repression of mRNA translation is relatively modest under those conditions (14), mimicking physiological stress situations. Furthermore, loss-of-function mutations in the eIF2α kinase PERK or HRI or the EIF2AA/A genotype all predispose cells to programmed cell death under physiologically stressful conditions (10, 13, 14, 39, 48); however, the role of defective activation of NF-κB in this phenotype, if any, remains to be explored.
Translational repression in response to activation of eIF2α kinases tends to be transient (34, 35). Translational recovery is mediated in part by activation of GADD34, an eIF2α-specific regulatory subunit of a holophosphatase complex (30, 31), which is itself a target of the eIF2α phosphorylation-dependent gene expression program, the integrated stress response (16, 29, 30). GADD34-mediated translational recovery is therefore likely to reestablish IκBα translation and reverse the effects of eIF2α phosphorylation on NF-κB activity, since the stress response is attenuated (Fig. 1B). Furthermore, while activation of NF-κB proceeds through utilization of preformed components, the response in terms of target gene expression depends on new protein synthesis. Thus, the biphasic nature of the inhibition of protein synthesis, which is inherent to stressful conditions that promote eIF2α phosphorylation, is also predicted to contribute to the expression of NF-κB target genes.
In conclusion, our study indicates that the pathways promoting eIF2α phosphorylation and those that activate NF-κB interact through translational repression of the inhibitor IκBα. Our study also suggests that the importance of this link is likely to be influenced by signaling through canonical NF-κB activation pathways that define the turnover rate of IκBα. As such, eIF2α phosphorylation and the consequent inhibition of eIF2B might modulate NF-κB signaling by parallel pathways active in stressed cells.
Acknowledgments
We thank Yinon Ben Neriah and Haoyuan Jiang for scientific advice and the ARIAD Corporation for the gift of the inducible dimerization system.
This work was supported by NIH grants ES08681 and DK47119 (to D.R.) and DK42394 (to R.J.K.). D.R. is a Scholar of the Ellison Medical Foundation.
REFERENCES
- 1.Baeuerle, P. A., and D. Baltimore. 1988. I kappa B: a specific inhibitor of the NF-kappa B transcription factor. Science 242:540-546. [DOI] [PubMed] [Google Scholar]
- 2.Bertolotti, A., Y. Zhang, L. Hendershot, H. Harding, and D. Ron. 2000. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J. 2000. Heme-regulated eIF2α kinase, p. 529-546. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. CSHL Press, Cold Spring Harbor, N.Y.
- 4.Cheshire, J. L., B. R. Williams, and A. S. Baldwin, Jr. 1999. Involvement of double-stranded RNA-activated protein kinase in the synergistic activation of nuclear factor-kappaB by tumor necrosis factor-alpha and gamma-interferon in preneuronal cells. J. Biol. Chem. 274:4801-4806. [DOI] [PubMed] [Google Scholar]
- 5.Cuervo, A. M., W. Hu, B. Lim, and J. F. Dice. 1998. IkappaB is a substrate for a selective pathway of lysosomal proteolysis. Mol. Biol. Cell 9:1995-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 7.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 8.Donze, O., J. Deng, J. Curran, R. Sladek, D. Picard, and N. Sonenberg. 2004. The protein kinase PKR: a molecular clock that sequentially activates survival and death programs. EMBO J. 23:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gil, J., J. Rullas, M. A. Garcia, J. Alcami, and M. Esteban. 2001. The catalytic activity of dsRNA-dependent protein kinase, PKR, is required for NF-kappaB activation. Oncogene 20:385-394. [DOI] [PubMed] [Google Scholar]
- 10.Han, A. P., C. Yu, L. Lu, Y. Fujiwara, C. Browne, G. Chin, M. Fleming, P. Leboulch, S. H. Orkin, and J. J. Chen. 2001. Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20:6909-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han, Y., S. Weinman, I. Boldogh, R. K. Walker, and A. R. Brasier. 1999. Tumor necrosis factor-alpha-inducible IkappaBalpha proteolysis mediated by cytosolic m-calpain. A mechanism parallel to the ubiquitin-proteasome pathway for nuclear factor-kappaB activation. J. Biol. Chem. 274:787-794. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H., I. Novoa, Y. Zhang, H. Zeng, R. C. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 13.Harding, H., H. Zeng, Y. Zhang, R. Jungreis, P. Chung, H. Plesken, D. Sabatini, and D. Ron. 2001. Diabetes Mellitus and exocrine pancreatic dysfunction in Perk−/− mice reveals a role for translational control in survival of secretory cells. Mol. Cell 7:1153-1163. [DOI] [PubMed] [Google Scholar]
- 14.Harding, H., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5:897-904. [DOI] [PubMed] [Google Scholar]
- 15.Harding, H., Y. Zhang, and D. Ron. 1999. Translation and protein folding are coupled by an endoplasmic reticulum resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 16.Harding, H., Y. Zhang, H. Zeng, I. Novoa, P. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. Stojdl, J. Bell, T. Hettmann, J. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 17.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 18.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. CSHL Press, Cold Spring Harbor, N.Y.
- 19.Hinnebusch, A. 1996. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2, p. 199-244. In J. Hershey, M. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Hinnebusch, A. G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. CSHL Press, Cold Spring Harbor, N.Y.
- 21.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Scheuner, R. J. Kaufman, D. R. Cavener, and R. C. Wek. 2003. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol. Cell. Biol. 23:5651-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jousse, C., S. Oyadomari, I. Novoa, P. D. Lu, Y. Zhang, H. P. Harding, and D. Ron. 2003. Inhibition of a constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman, R. J. 2000. The double-stranded RNA-activated protein kinase PKR, p. 503-527. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. CSHL Press, Cold Spring Harbor, N.Y.
- 24.Krappmann, D., and C. Scheidereit. 1997. Regulation of NF-kappa B activity by I kappa B alpha and I kappa B beta stability. Immunobiology 198:3-13. [DOI] [PubMed] [Google Scholar]
- 25.Krappmann, D., F. G. Wulczyn, and C. Scheidereit. 1996. Different mechanisms control signal-induced degradation and basal turnover of the NF-kappaB inhibitor IkappaB alpha in vivo. EMBO J. 15:6716-6726. [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, A., J. Haque, J. Lacoste, J. Hiscott, and B. R. Williams. 1994. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc. Natl. Acad. Sci. USA 91:6288-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, P. D., C. Jousse, S. J. Marciniak, Y. Zhang, I. Novoa, D. Scheuner, R. J. Kaufman, D. Ron, and H. P. Harding. 2004. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma, Y., and L. M. Hendershot. 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 278:34864-34873. [DOI] [PubMed] [Google Scholar]
- 30.Novoa, I., H. Zeng, H. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novoa, I., Y. Zhang, H. Zeng, R. Jungreis, H. P. Harding, and D. Ron. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22:1180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pahl, H., and P. Baeuerle. 1995. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by the transcription factor NF-κB. EMBO J. 14:2580-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pando, M. P., and I. M. Verma. 2000. Signal-dependent and -independent degradation of free and NF-kappa B-bound IkappaBalpha. J. Biol. Chem. 275:21278-21286. [DOI] [PubMed] [Google Scholar]
- 34.Prostko, C. R., M. A. Brostrom, and C. O. Brostrom. 1993. Reversible phosphorylation of eukaryotic initiation factor 2 alpha in response to endoplasmic reticular signaling. Mol. Cell. Biochem. 127-128:255-265. [DOI] [PubMed] [Google Scholar]
- 35.Prostko, C. R., M. A. Brostrom, E. M. Malara, and C. O. Brostrom. 1992. Phosphorylation of eukaryotic initiation factor (eIF) 2 alpha and inhibition of eIF-2B in GH3 pituitary cells by perturbants of early protein processing that induce GRP78. J. Biol. Chem. 267:16751-16754. [PubMed] [Google Scholar]
- 36.Ron, D., A. R. Brasier, K. A. Wright, J. E. Tate, and J. F. Habener. 1990. An inducible 50-kilodalton NFkB-like protein and a constitutive protein both bind the acute-phase response element of the angiotensinogen gene. Mol. Cell. Biol. 10:1023-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ron, D., and H. Harding. 2000. PERK and translational control by stress in the endoplasmic reticulum, p. 547-560. In J. Hershey, M. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Rothwarf, D. M., and M. Karin. 1999. The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Sci. STKE 1999:RE1. [DOI] [PubMed] [Google Scholar]
- 39.Scheuner, D., B. Song, E. McEwen, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in-vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 40.Scorsone, K. A., R. Panniers, A. G. Rowlands, and E. C. Henshaw. 1987. Phosphorylation of eukaryotic initiation factor 2 during physiological stresses which affect protein synthesis. J. Biol. Chem. 262:14538-14543. [PubMed] [Google Scholar]
- 41.Scott, M. L., T. Fujita, H. C. Liou, G. P. Nolan, and D. Baltimore. 1993. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 7:1266-1276. [DOI] [PubMed] [Google Scholar]
- 42.Sen, R., and D. Baltimore. 1986. Inducibility of kappa immunoglobulin enhancer-binding protein Nf-kappa B by a posttranslational mechanism. Cell 47:921-928. [DOI] [PubMed] [Google Scholar]
- 43.Taddeo, B., T. R. Luo, W. Zhang, and B. Roizman. 2003. Activation of NF-kappaB in cells productively infected with HSV-1 depends on activated protein kinase R and plays no apparent role in blocking apoptosis. Proc. Natl. Acad. Sci. USA 100:12408-12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tzamarias, D., I. Roussou, and G. Thireos. 1989. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell 57:947-954. [DOI] [PubMed] [Google Scholar]
- 45.Ubeda, M., X.-Z. Wang, H. Zinszner, I. Wu, J. Habener, and D. Ron. 1996. Stress-induced binding of transcription factor CHOP to a novel DNA-control element. Mol. Cell. Biol. 16:1479-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, S., M. Tan, Y. Hu, J. L. Wang, D. Scheuner, and R. J. Kaufman. 2004. Ultraviolet light activates NFκB through translational inhibition of IκBα synthesis. J. Biol. Chem. 279:34898-34902. [DOI] [PubMed] [Google Scholar]
- 47.Zamanian-Daryoush, M., T. H. Mogensen, J. A. DiDonato, and B. R. G. Williams. 2000. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol. Cell. Biol. 20:1278-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, P., B. McGrath, S. Li, A. Frank, F. Zambito, J. Reinert, M. Gannon, K. Ma, K. McNaughton, and D. R. Cavener. 2002. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22:3864-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]