Abstract
Biochemical experiments indicate that transcriptional elongation by RNA polymerase II (Pol II) is inhibited by nucleosomes and hence requires chromatin-modifying activities. Here, we examine the fate of histones upon passage of elongating Pol II in vivo. Histone density throughout the entire Saccharomyces cerevisiae GAL10 coding region is inversely correlated with Pol II association and transcriptional activity, suggesting that the elongating Pol II machinery efficiently evicts core histones from the DNA. Furthermore, new histones appear to be deposited onto DNA less than 1 min after passage of Pol II. Transcription-dependent deposition of histones requires the FACT complex that travels with elongating Pol II. Our results suggest that Pol II transcription generates a highly dynamic equilibrium of histone eviction and histone deposition and that there is significant histone exchange throughout most of the yeast genome within a single cell cycle.
The classical view of eukaryotic genomes is that essentially all DNA is stably associated with histone octamers in the form of nucleosome arrays. During S phase, histones are deposited on newly synthesized DNA, but the old histones are presumed to remain associated with DNA. However, recent results indicate that histone-DNA interactions are more dynamic than originally supposed. First, transcriptional activator proteins can cause complete unfolding, and probably dissociation, of histones from promoter regions in Saccharomyces cerevisiae cells (6, 13, 45). Second, the yeast Swr1 complex mediates ATP-dependent exchange of the histone H2AZ variant (24, 26, 36), and this activity protects euchromatin from the spread of heterochromatin (35). Third, in flies and mammals, the histone H3.3 variant is deposited into chromatin in a manner independent of DNA replication (1, 2, 54) but associated with transcription (20, 34).
Biochemical experiments have not revealed a clear understanding for how nucleosomes affect transcriptional elongation by RNA polymerases. Bacterial SP6 and T7 RNA polymerases and yeast RNA polymerase III (Pol III) can mobilize histones and transcribe nucleosomal templates (11, 23, 42, 51-53). Specifically, histone octamers step around a transcribing polymerase without leaving the template, although the enzyme pauses with a pronounced periodicity due to restricted rotation in the intranucleosomal DNA loop. In contrast, although Pol II elongation rates on naked DNA templates are comparable to physiological elongation rates, elongation on chromatin templates is markedly inhibited and produces truncated transcripts (18, 19). In a purified transcription assay lacking chromatin-modifying factors, nucleosomes and histone H3-H4 tetramers nearly completely block Pol II elongation (9). However, under conditions of increased ionic strength, Pol II can elongate through nucleosomes, resulting in the dissociation of a single H2A-H2B dimer but retention of the remaining six subunits of the histone octamer (22).
It is presumed that Pol II elongation in vivo requires modification of chromatin structure. Histone acetylation weakens histone-DNA (16) and nucleosome-nucleosome (30) interactions, and it can increase transcription without affecting nucleosome mobility (56). Nucleosome-remodeling complexes (e.g., Swi/Snf and RSC) can slide or displace histone octamers along the DNA in an ATP-dependent fashion, and they can transfer histone octamers to acceptor DNA molecules (4, 32, 37). The relationship between these biochemical activities and transcriptional elongation through nucleosomal templates is unclear.
Several factors link transcriptional elongation and chromatin structure. The FACT complex facilitates Pol II elongation on chromatin templates by destabilizing nucleosomes and evicting histones H2A and H2B from DNA (5, 43, 44). Additionally, FACT can deposit core histones onto DNA in vitro (5). In yeast cells, FACT travels with elongating Pol II (33), and it plays a role in maintaining normal chromatin structure during transcriptional elongation and prevents inappropriate initiation from cryptic promoters within coding regions (21, 33). Spt6 behaves similarly to FACT in yeast cells, and loss of Spt6 activity causes decreased histone density in actively transcribed mRNA coding regions (21). It is unclear whether the physiological role of Spt6 is related to its biochemical activities of nucleosome assembly (7) and/or transcriptional elongation per se (14). Lastly, Chd1 associates with elongation factors and coding regions of actively transcribed genes (49), and it is part of chromatin-modifying and nucleosome assembly activities in vitro (46, 55).
Despite the above connections between Pol II elongation and chromatin structure, little is known about the fate of nucleosomes during transcriptional elongation in vivo. We report here that, in yeast cells, histone density throughout the entire coding region is inversely related to transcriptional activity. This observation has been made on a genome-wide basis in a report submitted contemporaneously with the present paper (29). Furthermore, we show that low histone density at actively transcribed genes is reversed in less than 1 min after inhibition of transcription. These observations suggest that histones are efficiently evicted throughout the entire coding region upon Pol II elongation, with new histones being rapidly deposited following Pol II clearance. We also show that, like Spt6, FACT is required for transcription-association deposition of histones. Our results suggest that Pol II transcription generates a highly dynamic equilibrium of histone eviction and histone deposition and that there is significant histone exchange throughout most of the yeast genome within a single cell cycle.
MATERIALS AND METHODS
Yeast strains and media.
Occupancy of histones H2B and H4 was monitored, respectively, with strains expressing FLAG-tagged H2B (41) or Myc-tagged histone H4 (38). Isogenic wild-type and kin28-ts16 strains (10) were grown overnight at room temperature and then shifted to 37°C for 3 h, prior to cross-linking. A similar protocol was employed for isogenic FY56 (wild type) and L577 (spt16-197) strains (31). The GAL1-YLR454 strain expresses hemagglutinin-Rpb3, and its use for monitoring the kinetics of transcriptional elongation has been described previously (33). All strains were grown in yeast extract-peptone (YP) containing 2% carbon source, except for the GAL1-YLR454 strain, which was grown in medium containing 2% carbon source and Casamino Acids. For galactose-to-glucose shifts, cells were removed from medium containing 2% galactose by rapid centrifugation and then quickly resuspended in new medium containing 2% glucose.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was carried out with a modified version of a procedure described previously (3, 28). Cells (A600 = 0.7) were fixed in 1% formaldehyde for 20 min at room temperature, quenched for 5 min with glycine, and lysed with zirconia-silica beads (BioSpec Products) in a mini-bead beater (BioSpec Products). Chromatin was first pelleted by centrifugation and then solubilized by sonication (Branson Sonifier 350, three times, 100% duty, power 5, 30 s for each cycle). Cross-linked chromatin was immunoprecipitated with monoclonal antibodies to hemagglutinin (F7) and Myc (9e10; both from Santa Cruz Biotechnology) epitopes and Rbp1 (8WG16; Covance) and polyclonal antibodies to histone H3 (AbCam). Quantitative PCR analyses were performed in real time with an Applied Biosystems 7700 sequence detector. The PCR primers (coordinates defined relative to the transcription start site) correspond to the GAL7 promoter (−230 to +57), GAL10 promoter (−152 to +56), GAL10 5′ coding region (+174 to +292), and GAL10 3′ coding region (+1575 to +1788). Relative occupancy values were calculated by determining the apparent immunoprecipitation efficiency (amount of PCR product in the immunoprecipitated sample divided by the amount of PCR product in the input sample) and normalized to the level observed at an intergenic region of chromosome I, which was usually defined as 1.0. Congruency of data was also verified by normalizing the data to a gene expressed at low levels that appears to be unaffected by carbon source shifts (Rad53). For all data shown, each experiment was performed independently and replicated at least twice. Error bars shown reflect the standard deviations of the means.
RESULTS
An inverse correlation between histone and Pol II density.
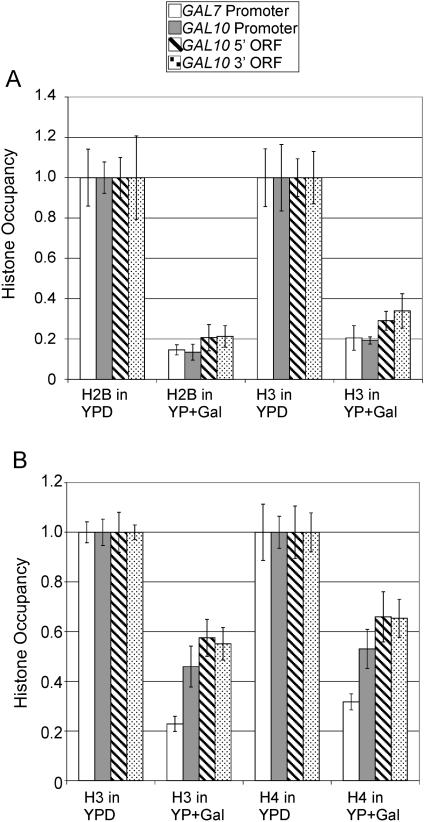
We examined the relationship between Pol II occupancy and histone density at multiple positions within the GAL genes in cells grown under conditions where the genes are activated (galactose medium) or repressed (glucose medium). In a strain expressing FLAG-tagged H2B, the densities of H2B and H3 are about fivefold lower under activating conditions than under repressing conditions in both promoter and coding regions of genes (Fig. 1A). In a different strain expressing Myc-tagged H4, the densities of H3 and H4 are about twofold lower under activating conditions (Fig. 1B). Although GAL activation reduces histone density to different extents in the two strains, the effect on H3 is quantitatively similar to that on both H2B and H4. The quantitatively similar behavior of all three histones strongly suggests that both the promoter and coding regions of the GAL genes are deficient in nucleosomes.
FIG. 1.
GAL activation causes reduced histone density at promoters and coding regions. (A) Densities of histones H2B and H3 at the indicated promoter and coding regions in a strain expressing FLAG-tagged H2B grown in YP medium containing either 2% galactose or glucose were normalized to an open reading frame (ORF)-free internal control region, and the highest level was arbitrarily set to 1. (B) Densities of histones H4 and H3 in a strain expressing Myc-tagged H4 were analyzed in a similar manner.
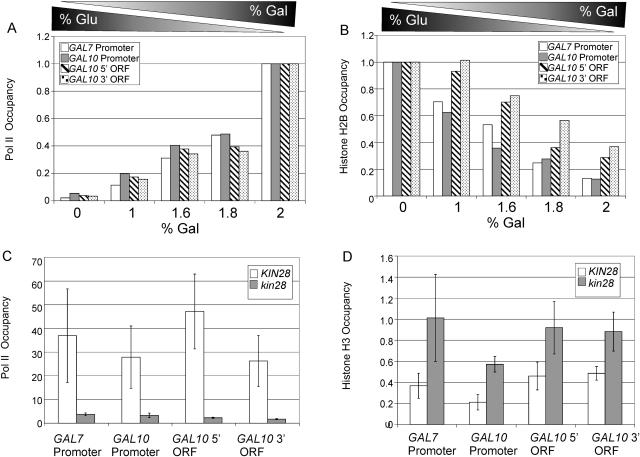
We further addressed the relationship between Pol II occupancy and H2B density by growing cells in media containing various mixtures of glucose and galactose (the final concentration of carbon source was fixed at 2%). As galactose concentrations increase relative to glucose concentrations, Pol II occupancy progressively increases in both promoters and coding regions, as expected (Fig. 2A). Conversely, H2B occupancy progressively decreases as galactose concentrations increase in both the promoters and coding region analyzed (Fig. 2B). These results clearly demonstrate an inverse correlation between histone H2B occupancy and transcriptional activity.
FIG. 2.
Inverse correlation between histone and Pol II occupancy. (A) Pol II and (B) H2B density at the indicated promoter and coding regions in cells expressing FLAG-tagged H2B and grown in YP medium containing the indicated mixtures of glucose and galactose. (C) Pol II and (D) H3 density in wild-type and kin28-ts16 cells grown in YP medium containing 2% galactose and shifted to 37°C for 2 h. ORF, open reading frame.
To directly demonstrate that histone density is inversely related to transcriptional activity, we analyzed a strain containing a temperature-sensitive allele of Kin28, a subunit of transcription factor IIH that phosphorylates the Pol II C-terminal domain at serine 5 in the vicinity of the promoter (25, 48). When the kin28-ts16 strain is shifted to the restrictive temperature, transcription (10) as well as Pol II (48) and transcription factor IIH (33) occupancy at the promoter is drastically reduced, whereas TATA binding protein (TBP) occupancy is unaffected (28). As expected, Pol II occupancy at promoter and coding regions is reduced in the kin28 mutant, compared to the isogenic wild-type strain (Fig. 2C). In contrast, the kin28 mutant exhibits an increase in histone H3 occupancy in all regions examined (Fig. 2D), and similar results were observed upon thermal inactivation of Pol II (rpb1-1 strain; data not shown). Thus, Pol II occupancy causes an inverse change in histone occupancy.
Rapid increase in histone density after passage of elongating Pol II.
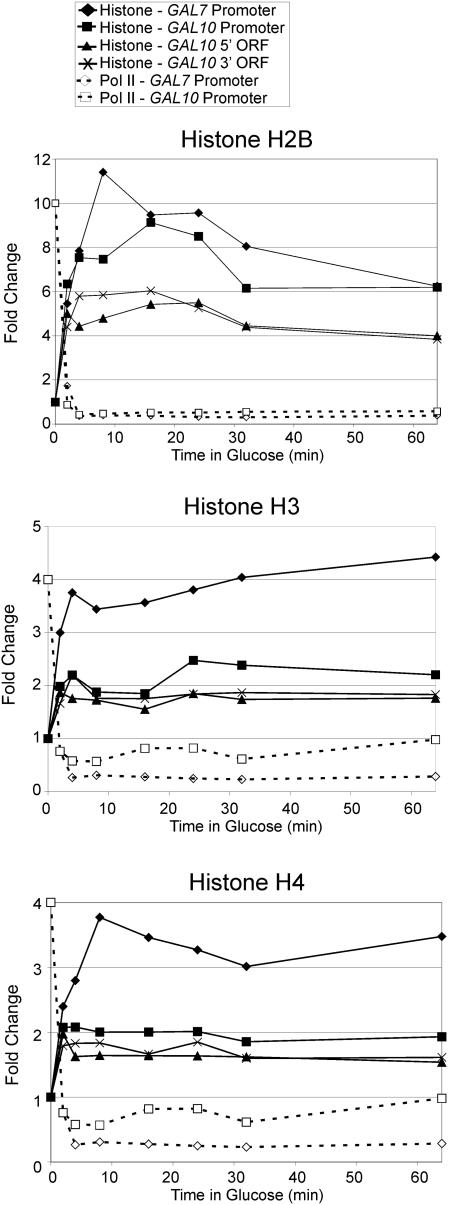
The fact that actively transcribed genes have low histone density provides an opportunity to investigate the kinetics of transcription-dependent deposition of histones in vivo. Specifically, we shifted galactose-grown cells into medium containing glucose and measured histone density at the GAL genes at various times after transcriptional initiation (Fig. 3). In accord with previous work (33, 40, 50), Pol II occupancy decreases 20- to 30-fold within 4 min after the shift to glucose medium. Concomitant with this decrease in transcription, occupancy of histones H2B, H3, and H4 increases rapidly at both promoter and coding regions. The behaviors of all three histones tested are kinetically similar, and histone densities increase to the level observed in glucose medium within 2 min after the shift.
FIG. 3.
Core histones are deposited upon cessation of transcription. Shown are densities of Pol II and histones H2B, H3, and H4 at the indicated promoter and coding regions in galactose-grown cells (zero time point) shifted into glucose medium for the indicated time (in minutes). Histone densities were normalized to an open reading frame (ORF)-free control region, and the zero time point was arbitrarily defined. Standard deviations (not shown) were generally less than 15% of the means.
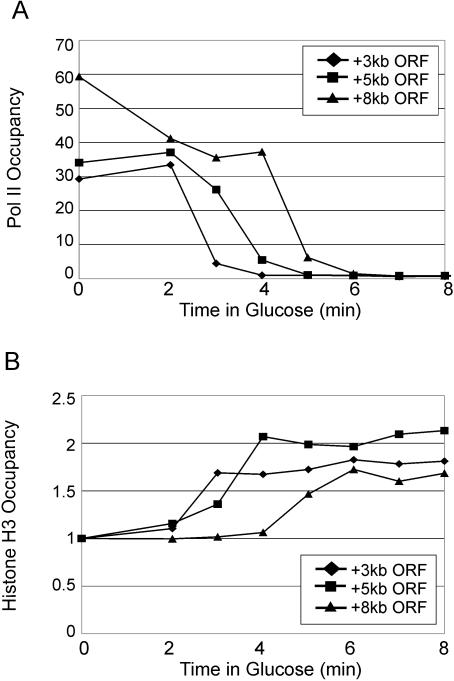
To determine more precisely when histones are deposited as Pol II travels across a gene, we examined a gene containing a GAL1 promoter upstream of the 8-kb YLR454 coding region, which kinetically monitors the last wave of Pol II transcription upon a shift to glucose medium (33, 50). As expected, Pol II is rapidly cleared from the gene upon the shift into glucose medium, with elongation occurring at a rate of approximately 2 kb/min (Fig. 4). Strikingly, histone H3 deposition across the gene occurs at a comparable rate, with H3 deposition lagging less than 1 min after clearance of Pol II. Thus, it appears that new histones are deposited almost immediately onto DNA as Pol II travels across a gene.
FIG. 4.
Histone H3 deposition tracks with clearance of Pol II. Galactose-grown cells containing GAL1-YLR454 were shifted into glucose medium for the indicated time (in minutes) and examined for (A) Pol II density (8-min time point arbitrarily set to 1) and (B) H3 density (0-min time point arbitrarily set to 1) at the indicated regions within the 8-kb YLR454 coding region. Standard deviations (not shown) were generally less than 15% of the means. ORF, open reading frame.
FACT is important for transcription-dependent histone deposition.
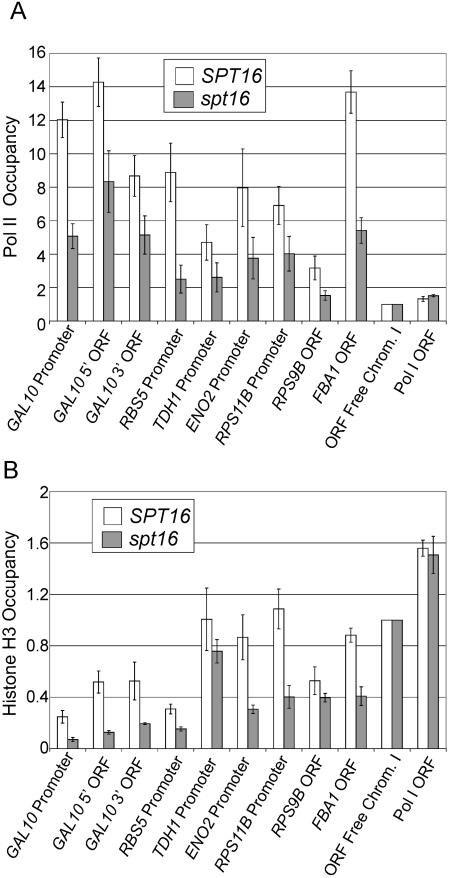
Given the role of FACT in the eviction and deposition of H2B in vitro (5) and suppression of inappropriate initiation within coding regions in vivo (33), we examined Pol II and H3 density in wild-type and spt16-197 strains grown in galactose medium. As expected (33), the spt16 mutant strain exhibits reduced Pol II occupancy in promoter and coding regions of the GAL10 and several other highly transcribed genes (Fig. 5A). However, in contrast to the inverse relationship between Pol II occupancy and histone density (Fig. 2), association of H3 at the GAL10 promoter and coding region decreases in the spt16 mutant strain (Fig. 5B). A similar reduction in H3 density is observed in the promoter and coding regions of several other highly transcribed genes (Fig. 5B). As controls, H3 density is unaffected at an intergenic region of chromosome I and at the POL1 coding region, which is poorly transcribed. These observations indicate that FACT is important for transcription-dependent histone deposition, and it suggests that FACT may not be involved in eviction of H3. In this regard, FACT evicts H2A-H2B dimers, but not H3-H4 tetramers, from DNA in vitro, but it can mediate deposition of all four core histones under certain conditions (5). The effect of FACT on histone density is similar to that of Spt6, another factor that travels with elongating Pol II (21), and this property is likely to explain why both FACT and Spt6 are important for suppression of initiation within coding regions (21, 33).
FIG. 5.
Spt16 is important for transcription-associated histone deposition. (A) Pol II and (B) H3 density at the indicated promoter and coding regions in wild-type and spt16-197 cells grown in YP medium containing 2% galactose and shifted to 37°C for 1 h. ORF, open reading frame.
DISCUSSION
While it is well recognized that nucleosomes inhibit Pol II elongation, the fate of histones upon passage of elongating Pol II in vivo has never been addressed. Here, we present evidence that the elongating Pol II machinery (with associated factors) evicts histones throughout the entire mRNA coding region and that new core histones are rapidly deposited on DNA after passage of Pol II. We cannot exclude the formal possibility that the apparent changes in histone density might reflect conformational alterations in histone-DNA interactions that affect cross-linking efficiency. However, this formal possibility does not easily explain why the three histones tested behave in a quantitatively and kinetically indistinguishable manner, i.e., it seems unlikely that each histone undergoes a reversible conformational change that has a similar effect on cross-linking to DNA. In addition, H2B is evicted from chromatin under various conditions in vitro (5, 22), yet it behaves similarly to H3 and H4 in vivo. Our results are also in general accord with transcriptional-associated deposition of the histone H3.3 (20).
Our results differ from in vitro analyses indicating that Pol II elongation evicts a single H2A-H2B dimer without affecting the remaining histone hexamer (5, 22). Further, they suggest that transcription-dependent eviction of histones is very efficient, perhaps occurring with every passage of elongating Pol II. Reduced histone density is clearly observed under conditions where Pol II association with the GAL genes is as low as 10% of maximum (1% galactose). Under conditions of full activation, TBP occupancy at GAL promoters is approximately half-maximal (28), which corresponds to initiation approximately every 10 s (17). Thus, reduced histone density is observed when Pol II passage through a given region occurs approximately once per minute, even though it takes less than 1 min to deposit new histones on DNA after cessation of transcription. Our data are most consistent with the idea that each passage of Pol II evicts histones, but we cannot exclude the possibility that eviction is slightly less efficient (for example, eviction every two or three passages of Pol II).
How might histones be evicted by the elongating Pol II machinery in vivo? Based on biochemical experiments, it seems unlikely that elongating Pol II per se can mediate eviction (9, 18, 19), although Pol II can dissociate a single H2A-H2B dimer under artificial conditions of high ionic strength (22). Similarly, the biochemical properties of the FACT complex (5, 43, 44) suggest that it may play a role in eviction of H2A-H2B dimers but not in eviction of H3 or H4. ATP-dependent nucleosome remodeling complexes are likely to be important for histone eviction in vivo, because some of them can transfer histone octamers between different DNA templates in vitro (4, 32, 37). However, a failure to evict histones upon Pol II elongation is likely to be a lethal event, and RSC is the only nucleosome-remodeling complex essential for yeast cell growth. RSC is the most abundant nucleosome-remodeling complex in yeast (8), but it does not specifically associate with active Pol II genes (12, 39). We suspect that transcription-dependent eviction of histones in vivo might depend on the combined activities of multiple nucleosome-remodeling complexes. In any event, our results suggest that the various in vitro assays used to analyze Pol II elongation on nucleosomal template may be lacking factors that are crucial for the process in vivo.
The transcription-associated increase in histone density follows closely in the wake of elongating Pol II, occurring less than 1 min after Pol II clearance from coding regions. The low density of histones in the spt16-197 mutant suggests that FACT is important for such histone deposition, consistent with its ability to travel with elongating Pol II in vivo (33, 47) and to deposit all core histones on DNA in vitro (5). However, FACT is not sufficient for transcription-associated deposition of histones, because Spt6 also can mediate nucleosome assembly in vitro (7), travels with elongating Pol II in vivo (27), and is required for normal chromatin structure in actively transcribed regions (21). Furthermore, FACT and Spt6 are both required to suppress inappropriate Pol II initiation within coding regions (21, 33). Thus, in yeast cells both FACT and Spt6 are important for transcription-associated deposition of histones, and other factors may also be involved.
Our results suggest that Pol II transcription generates a highly dynamic equilibrium of histone eviction and deposition. Such histone exchange may occur at each passage (or perhaps every few passages) of Pol II through a given region, and any evicted histone appears to be rapidly replaced (less than 1 min). Although our experiments involve situations in which transcription is rapidly activated or repressed, it is highly likely that the same dynamic situation occurs at all genes. Specifically, the rate of Pol II initiation determines how often an elongating Pol II molecule passes through a given region and hence the relative amount of time that nucleosomes in this region are present or absent. Estimates of Pol II initiation rates in yeast cells based on absolute levels and half-lives of RNA indicate that initiation of most yeast genes occurs multiple times within a single cell cycle (15, 17). In this regard, we observe reduced histone density at the GAL10 gene under conditions where transcription occurs at a level that is only 3- to 10-fold higher than that for the vast majority of yeast genes. Consequently, our results suggest that there is significant histone exchange throughout most of the yeast genome within a single cell cycle.
Acknowledgments
We thank Paul Mason for invaluable discussions and reagents. We are grateful to Joseph Wade, Joseph Geisberg, Zarmik Moqtaderi, Edward Sekinger, and Yael Katan-Khaykovich for insightful discussions, analyses, and technical assistance. We thank Fred Winston and Mary Cismowski for yeast strains.
M.A.S. is supported by an NIH Pharmacological Sciences Training Grant. This work was supported by a grant to K.S. from the National Institutes of Health (GM30186).
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2002. Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99:16477-16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., J. V. Geisberg, and K. Struhl. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, p. 21.3.1-21.3.17. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 5.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1093. [DOI] [PubMed] [Google Scholar]
- 6.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 7.Bortvin, A., and F. Winston. 1996. Evidence that Spt6 controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C. H., and D. S. Luse. 1997. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem. 272:23427-23434. [DOI] [PubMed] [Google Scholar]
- 10.Cismowski, M. J., G. M. Laff, M. J. Solomon, and S. I. Reed. 1995. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, D. J., and G. Felsenfeld. 1992. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell 71:11-22. [DOI] [PubMed] [Google Scholar]
- 12.Damelin, M., I. Simon, T. I. Moy, B. Wilson, S. Komili, P. Tempst, F. P. Roth, R. A. Young, B. R. Cairns, and P. A. Silver. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563-573. [DOI] [PubMed] [Google Scholar]
- 13.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endoh, M., W. Zhu, J. Hasegawa, H. Watanabe, D. K. Kim, M. Aida, N. Inukai, T. Narita, T. Yamada, A. Furuya, H. Sato, Y. Yamaguchi, S. S. Mandal, D. Reinberg, T. Wada, and H. Handa. 2004. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24:3324-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 16.Hong, L., G. P. Schroth, H. R. Matthews, P. Yau, and E. M. Bradbury. 1993. Studies of the DNA binding properties of histone H4 amino terminus: thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268:305-314. [PubMed] [Google Scholar]
- 17.Iyer, V., and K. Struhl. 1996. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5208-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izban, M. G., and D. S. Luse. 1992. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J. Biol. Chem. 267:13647-13655. [PubMed] [Google Scholar]
- 19.Izban, M. G., and D. S. Luse. 1991. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 5:683-686. [DOI] [PubMed] [Google Scholar]
- 20.Janicki, S. M., T. Tsukamoto, S. E. Salghetti, W. P. Tansey, R. Sachidanandam, K. V. Prasanth, T. Ried, Y. Shav-Tai, E. Bertrand, R. H. Singer, and D. L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 22.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9:541-552. [DOI] [PubMed] [Google Scholar]
- 23.Kirov, N., I. Tsaneva, E. Einbinder, and R. Tsanev. 1992. In vitro transcription through nucleosomes by T7 RNA polymerase. EMBO J. 11:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komarnitsky, P., E.-J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 27.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomic approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-612. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regulation genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 30.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 31.Malone, E. A., C. D. Clark, A. Chiang, and F. Winston. 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5710-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 33.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKittrick, E., P. R. Gafken, K. Ahmad, and S. Henikoff. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. USA 101:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 36.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 37.Narlikar, G. J., H.-Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 38.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 41.Ng, H. H., R. M. Xu, Y. Zhang, and K. Struhl. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3-lysine 79. J. Biol. Chem. 277:34655-34657. [DOI] [PubMed] [Google Scholar]
- 42.O'Neill, T. E., J. G. Smith, and E. M. Bradbury. 1993. Histone octamer dissociation is not required for transcript elongation through arrays of nucleosome cores by phage T7 RNA polymerase in vitro. Proc. Natl. Acad. Sci. USA 90:6203-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 44.Orphanides, G., W.-H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human Spt16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 45.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 46.Robinson, K. M., and M. C. Schultz. 2003. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol. Cell. Biol. 23:7937-7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J. T. Lis. 2003. Tracking FACT and RNA polymerase II elongation complex through chromatin in vivo. Science 301:1094-1096. [DOI] [PubMed] [Google Scholar]
- 48.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with mRNA export. Nature 417:304-307. [DOI] [PubMed] [Google Scholar]
- 51.Studitsky, V. M., D. J. Clark, and G. Felsenfeld. 1994. A histone octamer can step around a transcribing polymerase without leaving the template. Cell 76:371-382. [DOI] [PubMed] [Google Scholar]
- 52.Studitsky, V. M., D. J. Clark, and G. Felsenfeld. 1995. Overcoming a nucleosomal barrier to transcription. Cell 83:19-27. [DOI] [PubMed] [Google Scholar]
- 53.Studitsky, V. M., G. A. Kassavetis, E. P. Geiduschek, and G. Felsenfeld. 1997. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science 278:1960-1963. [DOI] [PubMed] [Google Scholar]
- 54.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 55.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromodomain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 19:2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ura, K., H. Kurumizaka, S. Dimitrov, G. Almouzni, and A. P. Wolffe. 1997. Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J. 16:2096-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]