Abstract
Prp2 is an RNA-dependent ATPase that activates the spliceosome before the first transesterification reaction of pre-mRNA splicing. Prp2 has extensive homology throughout the helicase domain characteristic of DEXD/H-box helicases and a conserved carboxyl-terminal domain also found in the spliceosomal helicases Prp16, Prp22, and Prp43. Despite the extensive homology shared by these helicases, each has a distinct, sequential role in splicing; thus, uncovering the determinants of specificity becomes crucial to the understanding of Prp2 and the other DEAH-splicing helicases. Mutations in an 11-mer near the C-terminal end of Prp2 eliminate its spliceosome binding and splicing activity. Here we show that a helicase-associated protein interacts with this domain and that this interaction contributes to the splicing process. First, a genome-wide yeast two-hybrid screen using Prp2 as bait identified Spp2, which contained a motif with glycine residues found in a number of RNA binding proteins. SPP2 was originally isolated as a genetic suppressor of a prp2 mutant. In a reciprocal screen, Spp2 specifically pulled out the C-terminal half of Prp2. Mutations in the Prp2 C-terminal 11-mer that disrupted function or spliceosome binding also disrupted Spp2 interaction. A screen of randomly mutagenized SPP2 clones identified an Spp2 protein with a mutation in the G patch that could restore interaction with Prp2 and enhanced splicing in a prp2 mutant strain. The study identifies a potential mechanism for Prp2 specificity mediated through a unique interaction with Spp2 and elucidates a role for a helicase-associated protein in the binding of a DEXD/H-box protein to the spliceosome.
Pre-mRNA splicing is a dynamic process through which snRNPs and trans-acting proteins interact in an ordered manner in the spliceosome to remove introns from pre-mRNA. The process of splicing can be broken down in vitro to the steps of spliceosome formation, the first transesterification reaction, the second transesterification reaction, the release of mature mRNA, and the recycling of the snRNPs. DEXD/H-box helicases act upon the spliceosome to catalyze assembly, activation, and disassembly of the spliceosome in an ordered, stepwise manner (40). Comprehending the action of these helicases and their effect on the spliceosome is essential to understanding the process of pre-mRNA splicing.
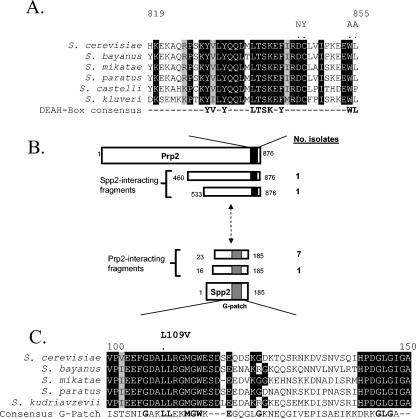
Prp2 is an RNA-dependent ATPase (21) belonging to the DEAH subfamily of DEXD/H-box helicases that includes Prp16, Prp22, and Prp43 (40). The DEAH splicing factors share homology extending from the helicase domain through the carboxyl terminus. Despite the extensive homology shared between the DEAH family splicing helicases, each has a distinct, sequential role in the splicing reaction. Prp2 activates the spliceosome upon ATP hydrolysis before the first transesterification reaction (20, 22), Prp16 acts before the second transesterification reaction (38), Prp22 has roles in the second step of pre-mRNA splicing and mRNA release from the spliceosome (37, 42), and Prp43 is involved in spliceosome disassembly and lariat-intron release (3, 27). The specificity of these helicases could come from their nonconserved N-terminal domain, as was observed with Prp16, which is recruited to the spliceosome primarily through its N terminus (43). However, domain deletion experiments with Prp2 and Prp43 have shown that the bulk of the N-terminal domain is dispensable for their activity in vivo (9, 27). An alternative hypothesis for the case of Prp2 is that the C terminus delivers its specificity. Deletion of the C-terminal 42 amino acids results in a protein that can no longer interact with the spliceosome in vitro or function in vivo (9). Importantly, a double-missense mutation in the C terminus of Prp2, D845N/C846Y (DC/NY), was identified which eliminated the function and spliceosome binding of Prp2 while leaving the ATPase activity intact (9). These and our unpublished observations of a second double mutation (W854A/L855A) led to the conclusion that a discrete 11-mer region of the Prp2 C domain was important for spliceosome binding (Fig. 1A).
FIG. 1.
Spp2 interacts with the C terminus of Prp2. (A) Alignment of the region flanking the D845-W855 11-mer in Prp2 proteins among the Saccharomyces genus. The alignment was created by using the Pretty function in Seqweb 2.0 (Wisconsin GCG package) and was adjusted manually with sequences obtained from the www.yeastgenome.org website. Positions of complete conservation are indicated in black, and positions of strong similarity are outlined in gray. The positions of DC and WL are indicated with dots, and the DC/NY and WL/AA mutations are represented above the dots. A consensus derived from analysis of yeast DEAH-box splicing helicases is also shown. (B) Summary of yeast two-hybrid screening using full-length Prp2 or Spp2. The amino acid positions are indicated in the diagram, while the number of isolates of each prey fragment is listed at the right. The black box in Prp2 represents the D845-W855 11-mer and the flanking region. The gray box in Spp2 represents the G patch. (C) An alignment of the G patch of Spp2 of Saccharomyces spp. The amino acid positions in S. cerevisiae Spp2 are indicated. Positions of complete identity are in black, and positions with strong similarity are outlined in gray. The consensus G patch is shown as identified by the Conserved Protein Domain Database (26). Positions in boldface indicate agreement between Spp2 and the G-patch consensus.
DEXD/H-box helicases often have associated cofactors that help to regulate activity and coordinate function, such as the helicase eukaryotic translation initiation factor 4A (eIF4A) and cofactor eIF4B as well as hepatitis C virus helicase NS3 and cofactor NS4A (39). When eIF4B is present, the otherwise low level of helicase activity measured in eIF4A is stimulated 20-fold (32, 35). Like eIF4A, when Prp2 is highly purified it does not exhibit measurable helicase activity on a test duplex (21). It is not clear whether such helicase-enhancing cofactors exist for Prp2, but we hypothesize that the splicing specificity of Prp2 could be affected by a protein cofactor.
In order to identify cofactors that interact with Prp2, a genome-wide yeast two-hybrid screen was performed utilizing Prp2 as bait. This experiment repeatedly identified a single protein, Spp2, which has previously been shown to interact genetically and biochemically with Prp2 (24, 34). When the reciprocal experiment was performed, Spp2 was found to specifically interact with the C-terminal half of Prp2. We then tested the DC/NY and WL/AA C-domain mutations in both the two-hybrid screen and through a glutathione S-transferase (GST) pulldown and found that these mutations in Prp2 proportionally affect physical interaction with Spp2. To test for splicing relevance of this interaction, we screened for mutations in Spp2 that could restore interaction. We identified a leucine-to-valine change (L109V) within the G patch of Spp2 that greatly enhances the interaction between Spp2 and Prp2 C-terminal mutants. The G-patch motif is a pattern of conserved glycines that is found in many RNA binding proteins (2). Although expression of SPP2L109V did not rescue the growth defect of strains where mutant prp2-DC/NY or prp2-WL/AA provided the sole copy of PRP2, it enhanced the splicing activity of intermediate loss-of-function alleles that map within the D845-L855 11-mer. Taken together, these results are the first to show that the Prp2 C-terminal interaction with Spp2 is essential for activity, that a G-patch motif plays a role in protein-protein interaction, and that the interaction with Spp2 through the unique DC-WL 11-mer of Prp2 could potentially explain its spliceosome specificity.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Protocols for yeast growth conditions (33) and the yeast lithium acetate method for yeast transformation (14, 15) were followed. Strains used were BJ168 (MATa ura3 leu2 trp1 prb pep4 prc), D12 (MATa ura3 leu2 his3 prp2-1), YTY1 (MATa ura3 leu2 trp1 his3 lys2 Δprp2::TRP1 YCp51-PRP2::URA3) (9), JWY36 (MATa his3 leu2 lys2 trp1 ura3 Δspp2::LEU2 YCp50-SPP2) (34), Y187 (MATα ura3 his3 ade2 trp1 leu2 Δgal4 met Δgal80 GAL1UAS-GAL1TATA-lacZ), and CG1945 [MATa ura3 his3 ade2 lys2 trp1 leu2 gal4 gal80 cyhr2 GAL1UAS-GAL1TATA-HIS3::LYS2 GAL417-mers(x3)-CYC1TATA-lacZ::URA3] (Clontech). The YPD medium consists of 1% yeast extract, 2% peptone, and 2% dextrose, and YPGal consists of 1% yeast extract, 2% peptone, and 2% galactose. Benomyl was used at 30 μg/ml.
Plasmid manipulation.
pAS2BJ-PRP2 has the open reading frame (ORF) of PRP2 cloned into a BamHI cloning site, creating a fusion with the GAL4 DNA binding domain under the control of the ADH2 promoter. Site-directed mutagenesis of plasmid DNA was performed with the QuikChange site-directed mutagenesis kit (Stratagene), and mutants were identified by DNA sequencing at the City of Hope sequencing facility. Hydroxylamine mutagenesis of pACTIIst-SPP2 was performed by incubating 10 μg of pACTII-SPP2 in 500 μl of hydroxylamine solution (prepared by dissolving 0.35 g of hydroxylamine-HCl and 0.09 g of NaOH in 5 ml of H2O) for 20 h at 37°C. The reaction was stopped by adding a solution of 10 μl of 5 M NaCl, 50 μl of a 1-mg/ml concentration of bovine serum albumin, and 1 ml of 100% ethanol. DNA was precipitated and resuspended in 100 μl of Tris-EDTA (TE) buffer (pH 8) and was reprecipitated with a mixture of 10 μl of 3 M sodium acetate and 250 μl of 100% ethanol. The pellet was finally resuspended in 100 μl of TE and was used for transformation. A BglII-SalI fragment containing the ORF of SPP2 was cloned into a BamHI-SmaI site of pGEX2T vector (Pharmacia) to create pGEX2T-Spp2. A marker swap was performed to change the marker of pGAL-SPP2L109V(URA3) to pGAL-SPP2L109V(HIS3) as described previously (6). Briefly, a fragment containing an intact HIS3 cassette flanked by a URA3 homology was cut from plasmid pUH7 (6) and was transformed into a strain containing pGAL-SPP2L109V. Plasmids were isolated from His+ transformants by using a yeast plasmid miniprep protocol (31) and were confirmed by DNA analysis. Briefly, yeast plasmid DNA was prepared from 3 ml of overnight culture in selective medium. The pellet was resuspended in 100 μl of STET (8% sucrose, 50 mM Tris [pH 8], 50 mM EDTA, 5% Triton X-100), and then glass beads were added. The mixture was subjected to vigorous vortexing for 5 min, another 100 μl of STET was added, and the mixture was boiled for 3 min. The sample was then spun down for 10 min at 4°C, and the supernatant was transferred to a fresh tube containing 50 μl of 7.5 M ammonium acetate and incubated at −20°C for 1 h. The sample was centrifuged, and the supernatant was precipitated with ethanol.
Plasmid shuffle assay.
Δprp2 strain YTY1 was maintained by a YCp50-PRP2 (URA3 marked) plasmid. Alleles of interest, such as prp2-D845L on a LEU2-marked plasmid, were transformed into YTY1 and then subjected to 5-fluorooritic acid (5-FOA). 5-FOA is a drug that, when utilized by the URA3 gene product in the uracil synthesis pathway, produces an intermediate that is deadly to the cell. Therefore, only cells that can lose the YCp50-PRP2 plasmid can grow on 5-FOA-containing media. Because PRP2 is essential, this means that the strain is forced to rely on the second, LEU2-marked plasmid for cellular Prp2. When this happens, the allele of interest can be assayed for phenotypic effects.
Yeast two-hybrid screens and assays.
To produce the Gal4:Prp2 bait construct, the entire coding sequence of PRP2 was cloned into the BamHI site of pASΔΔ (11) and introduced into yeast strain CG-1945. For the LexA:Spp2 bait, the full coding sequence of SPP2 was cloned into EcoRI-SalI-digested pBTM116 and transformed into L40 cells from Clontech (MATa his3Δ200 trp1-901 leu2-3 leu2-112 ade2 lys2-801am URA3::lexAop8-lacZ LYS2::lexAop4-HIS3). Two-hybrid screens were performed using the FRYL yeast genomic library in Y187 cells as described previously (11). In the Prp2p and Spp2p screens, a total of 5.8 × 107 and 7.3 × 107 diploid cells were tested, respectively. His+ cells were selected and tested for β-galactosidase activity in an 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside agar overlay assay (11), and the prey plasmids in positive clones were analyzed by DNA sequencing. The two-hybrid assays examining the interaction between the Prp2 C-terminal mutants and Spp2 were done as described by van Nues and Beggs (41). The analyses were done with the mated diploid cells shown in Fig. 4, such that the bait would be pretransformed into Y187 and the prey would be pretransformed into CG1945. For spp2 mutant screening, CG1945 with either pASBJ-prp2 (D845N/C846Y) or pASBJ-prp2 (W854A/L855A) was sequentially cotransformed with a hydroxylamine-mutagenized pool of pACTIIst-SPP2. Assays were performed on media selecting for expression of the HIS3 gene. Strength of interaction was ascertained by serial dilution assays and growth on media containing the HIS3 inhibitor 3-aminotriazole (3AT). Because the physical interaction of Prp2 and Spp2 is weak, the low concentration of 1 mM 3AT was sufficient to see differences between wild-type and mutant Prp2 interaction with Spp2.
FIG. 4.
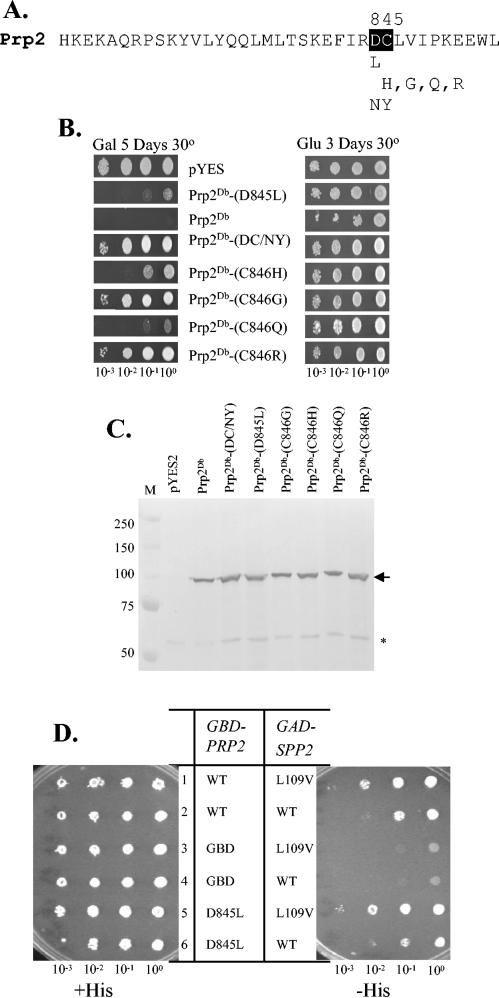
Random mutations in D845 and C846 dipeptide of Prp2 reveal a variety of loss-of-function phenotypes. (A) Amino acid sequence at the C terminus of S. cerevisiae Prp2 flanking the DC dipeptide (in black). The identities of the mutations depicted in this figure are shown underneath the sequence. (B) Growth phenotype on galactose medium of cells harboring GAL-PRP2 plasmids. Cells were grown overnight in glucose medium, diluted, and spotted on plates as indicated. (C) Western blotting of yeast lysates upon induction of GAL-PRP2 expression in galactose (Gal) medium. Glass bead extract from galactose-induced strains bearing GAL-PRP2 constructs was separated in an SDS-9% PAGE gel and probed with anti-Prp2 antibody as described in the legend to Fig. 2B. The arrow indicates Prp2, and the asterisk indicates the cross-reacting band. (D) Yeast two-hybrid assay of prp2-D845L with wild-type and L109V SPP2. Cells carrying two plasmids (one GBD derivative and one GAD derivative) were spotted and grown in the presence (+His) or absence (−His) of histidine for 3 days as indicated.
Yeast glass bead extracts.
For yeast glass bead extracts, an overnight culture was grown in minimal raffinose medium and then was diluted 1:10 in the same medium and grown for 2 h. Galactose was then added to 2% and the culture was grown for an additional 6 h. After induction, 1.5 ml of cells was harvested. The two-hybrid constructs were grown overnight in YPD, and 1.5 ml of cells was directly harvested. For Western blotting cell pellets were resuspended in 30 μl of ESB (80 mM Tris [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 1.5% dithiothreitol [DTT], 0.1 mg of bromphenol blue/ml) and boiled for 3 min. Glass beads were then added until the suspension was filled, and then the suspensions were subjected to a 2-min vortexing. Seventy microliters of ESB was added, and the solution was mixed and boiled for 1 min. After centrifugation, 12 μl of the supernatant was used for Western blot analysis. For the GST-pulldown assay, extracts were prepared as described previously (9).
GST pulldown.
To produce GST-tagged Spp2, BL21(DE3)pLysS Escherichia coli carrying pGEX2T-SPP2 culture was grown overnight and then was diluted 1:100 in 500 ml of Luria-Bertani-carbenicillin. The culture was grown until the optical density at 600 nm (OD600) reached 0.6, at which point isopropyl-β-d-thiogalactopyranoside was added to 1 mM. The culture was grown for 3 h and then was harvested. The pellet was washed in 10 ml of buffer A (10 mM Tris-HCl, 100 mM NaCl, 1 mM DTT) and resuspended in 15 ml of buffer B (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5% glycerol, 2 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.1% NP-40). Cells were then broken through freeze/thaw and sonication. After centrifugation for 20 min, the supernatant was collected. The supernatant was adjusted to contain 300 mM NaCl and 0.1% Triton-X, and 100 μl of the supernatant was added to 50 μl of glutathione agarose and bound at 4°C with mechanical rotation. The glutathione-agarose was then washed three times with 10 bed volumes of 1× Tris-buffered saline (TBS) containing 500 mM NaCl-0.5% Triton-X and then was washed three times with 10 bed volumes of 1× TBS containing 300 mM NaCl-0.1% NP-40. Two-hundred microliters of glass bead extracts from yeast was then added, and the supernatant was incubated for 1 h at 4°C. The unbound fraction was retrieved, and the agarose fraction was washed as described above. The bound fraction was then separated by directly loading 12 μl of protein-bound beads on an SDS-10% polyacrylamide gel electrophoresis (PAGE) gel.
Western analysis.
Proteins separated by SDS-PAGE were transferred onto a polyvinylidene difluoride membrane (Millipore) by using a Bio-Rad Trans-Blot SD semidry transfer cell per the manufacturer's instructions. The membrane was then incubated in Blotto (5% nonfat dry milk) for 1 h and was processed as described previously (9, 20, 21). N-terminal-specific anti-Prp2 polyclonal immunoglobulin G (IgG) was used at a 1:2,000 dilution, and the blots were developed (20) with a goat anti-rabbit IgG alkaline phosphatase-conjugated antibody (Bio-Rad).
Splicing reporter assay.
β-Galactosidase activity expressed from the pJYHZ splicing reporter was assayed by using o-nitrophenyl-β-d-galactopyranoside (ONPG) (25). Briefly, cells were grown as a starter culture overnight in selective media at a permissive temperature. The culture were then split to an OD600 of 0.25, and one tube was grown for 4 h at a restrictive temperature and one tube was grown for 4 h at a permissive temperature before harvest. After harvest, the OD600 was recorded and the cells were pelleted. Two-hundred-fifty microliters of breaking buffer (100 mM Tris-Cl [pH 8], 1 mM DTT, 20% glycerol) and 12.5 μl of phenylmethylsulfonyl fluoride stock (40 mM in isopropanol) was added to each pellet, and the cells were resuspended. Glass beads were then added until the suspension was filled, and the mixture was vortexed six times for 15 s each, alternating with 15-s incubations on ice to prevent overheating. After this step, 200 μl of breaking buffer was added and the supernatant was rescued to another tube. This supernatant was clarified by spinning at top speed in a microcentrifuge at 4°C. One-hundred microliters of extract was added to 900 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, pH 7.0) and preincubated for 5 min at 26°C in a water bath. Two-hundred microliters of a 4-mg/ml concentration of ONPG (in Z buffer) was then added, and the mixture was incubated at 26°C in a water bath. The reaction was timed and stopped by the addition of 0.5 ml of 1 M Na2CO3 solution once it turned a light yellow. At that point the OD420 was recorded. Assays were carried out three times per sample, and at least three independent samples from each strain were tested. Adjusted units were determined by the following formula: adjusted units = OD420/(OD600 × time of reaction). Ratios of 4-h shifted units/no shift units are, in some cases, multiplied by 100 to give a percentage.
RESULTS
The carboxyl terminus of Prp2 interacts with Spp2.
To identify potential cofactors of Prp2, we utilized the yeast two-hybrid system. Full-length PRP2 was fused to a GAL4 DNA binding domain (GBD) and was used as bait to screen the Fromont-Racine yeast library (11) fused to the GAL4 transactivation domain (GAD). One factor, SPP2, was pulled out eight times (Fig. 1B). The shortest fragment of SPP2 that was isolated from the screen encoded all but the 22 amino acids of the N terminus (Fig. 1B). Spp2 is a high-copy-number suppressor of a prp2-1 temperature-sensitive mutant (24) and has been shown to interact with Prp2 and promote splicing (34). To evaluate the uniqueness of the Prp2-Spp2 interaction, full-length SPP2 was used to screen the GAD-fused two-hybrid library. Two fragments of PRP2 were identified; both included most of the C-terminal half of Prp2. The shortest fragment included amino acids 533 to 876, encompassing the end portion of the H domain and the entire C-terminal domain (Fig. 1B). These results indicate that Spp2 interacts with the C terminus of Prp2.
Point mutations in the C domain of Prp2 eliminate the interaction between Prp2 and Spp2.
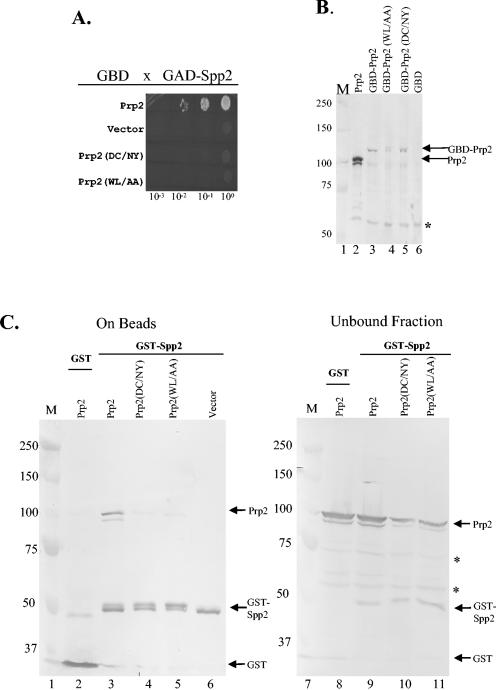
To determine whether mutations in the C-terminal DC-WL 11-mer in Prp2 that affect spliceosome binding (Fig. 1A) could affect the interaction with Spp2, we constructed a GBD-PRP2 fusion bearing the desired alleles and assayed their ability to interact with GAD-SPP2 in a two-hybrid assay. We found that both the D845N/C846Y mutations and the W854A/L855A mutations eliminated the two-hybrid interaction (Fig. 2A). The lack of interaction is not due to the lack of protein, because each of the fusion proteins could be detected by immunoblot analysis (Fig. 2B, lanes 3 to 5). Thus, it appeared that these mutant Prp2 proteins failed to interact with Spp2.
FIG. 2.
Interaction between Prp2 C-terminal mutations and Spp2 as determined by two-hybrid and GST-pulldown assays. (A) Yeast two-hybrid spot assay. Y187 cells containing the indicated GBD-Prp2 constructs were mated with CG1945 cells containing GAD-Spp2, and the diploid colonies were selected and grown overnight in liquid medium containing histidine. The overnight cultures were then serially diluted as indicated. Three microliters of each dilution was spotted onto a medium plate omitting histidine, leucine, and tryptophan. Incubation was at 30°C for 3 days. Cell growth indicates interaction between the two-hybrid proteins. (B) Western blotting of lysates from Y187 cells containing the indicated plasmids. Twelve microliters of lysate from an overnight culture was analyzed by using anti-Prp2 antibody (lanes 3 to 6). Protein markers (lane 1, in kilodaltons) and a cell lysate from a Prp2-overproducing strain (8) (lane 2) were included. The GBD-Prp2 fusion and the overexpressed Prp2 are marked with arrows. The asterisk indicates a protein that cross-reacted to the antibody. (C) GST-pulldown assay followed by Western blotting shows that Prp2-D845N/C846Y (DC/NY) and Prp2-W854A/L855A (WL/AA) lose the ability to interact with GST-Spp2. GST-Spp2 was bound to glutathione-agarose beads. Extracts from yeast strains overexpressing PRP2, prp2-D845N/C846Y (DC/NY), or prp2-W854A/L855A (WL/AA) were mixed with the beads and were allowed to bind at 4°C for 1 h. The bound (lanes 2 to 6) and the unbound (lanes 8 to 11) samples were separated and loaded onto a gel with protein markers (lanes 1 and 7, in kilodaltons). The blot was probed with an antibody raised against GST-Prp2 (N domain) (20), which could recognize GST, GST-Spp2, and Prp2 (marked with arrows). Asterisks indicate nonspecific cross-reacting bands in the unbound fraction.
To further investigate the nature of the Prp2-Spp2 interaction, we fused Spp2 with GST and asked if recombinant Spp2 could interact with wild-type or mutant Prp2 proteins in a GST-pulldown assay. We generated PRP2 constructs under control of the GAL promoter and induced them to overexpress PRP2, prp2-D845N/C846Y, or prp2-W854A/L855A. We added extracts from these cells to GST-Spp2 bound to glutathione beads. The proteins that bound were subjected to Western blotting with an anti-Prp2 antibody (Fig. 2C). Consistent with results of a previous report (34), Prp2 could bind GST-Spp2 (Fig. 2C, lane 3) but not GST (Fig. 2C, lane 2). Neither Prp2-D845N/C846Y nor Prp2-W854A/L855A bound to GST-Spp2 (Fig. 2C, lanes 4 and 5). Western blotting of the unbound fractions showed that wild-type and mutant Prp2 proteins were all present in the extracts (Fig. 2C, lanes 8 to 11). Taken together, both D845N/C846Y and W854A/L855A mutations eliminate the interaction of Prp2 with Spp2. Because Prp2 protein with the D845N/C846Y mutation does not bind to the spliceosome (9), and because Spp2 is necessary for Prp2 to associate with the spliceosome (34), the loss of spliceosome binding exhibited by the D845N/C846Y mutation may be a direct result of the inability to bind Spp2.
A mutation in Spp2 can restore the two-hybrid interaction of Prp2 and Spp2.
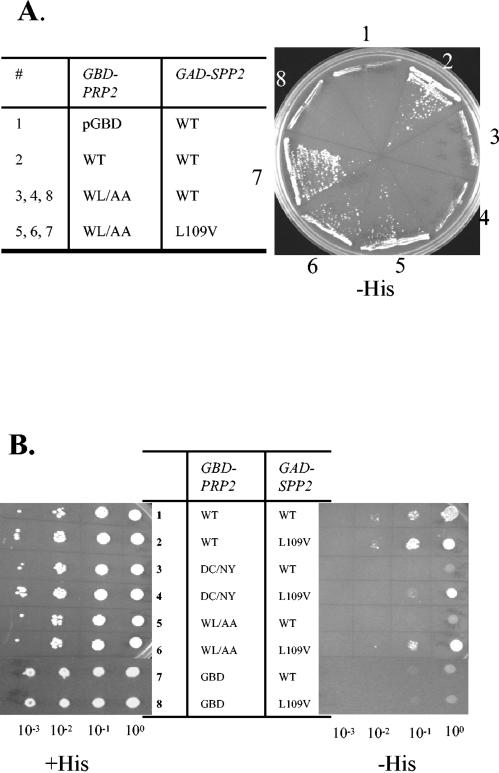
We wanted to look for Spp2 mutants that restore the interaction with the D845N/C846Y or W854A/L844A mutants in the hopes that they would also restore Prp2 function. We screened for spontaneous and hydroxylamine-mutagenized mutants in GAD-SPP2 constructs that would activate the expression of the two-hybrid reporter in the W854A/L855A mutant. A leucine-to-valine change at amino acid 109 (SPP2L109V) enabled GBD-prp2-W854A/L855A to grow in the absence of histidine (Fig. 3A, numbers 5, 6, and 7). This leucine residue was conserved in Spp2 proteins belonging to the Saccharomyces genus and was found in the consensus G-patch domain (Fig. 1C) (2). The T-to-G transversion mutation that caused the L109V change in SPP2 was specifically remade via site-directed mutagenesis and was further assayed for the ability to interact with GBD-prp2-W854A/L855A, GBD-prp2-D845N/C846Y, and GBD-PRP2. The remade GAD-SPP2L109V restored interaction with GBD-prp2-W854A/L855A (Fig. 3B, rows 5 and 6) and with GBD-prp2-D845N/C846Y, although to a lesser extent (Fig. 3B, rows 3 and 4). Interestingly, GAD-SPP2L109V interacted slightly better with GBD-PRP2 than did wild-type GAD-Spp2 (Fig. 3B, rows 1 and 2). It is not clear whether Spp2L109V functions either to make Spp2 associate with higher affinity to Prp2, to increase the amount of Spp2, or to structurally relax Spp2, enabling it to accommodate mutant Prp2. Nonetheless, the L109V mutation did restore Spp2 interaction with mutants mapping to the DC-WL 11-mer in Prp2.
FIG. 3.
Yeast two-hybrid assay showing that a mutation in SPP2 can restore the interaction with mutant prp2. (A) Retransformation of strain CG1945 carrying the indicated pGBD-derived plasmid with the parental GAD-SPP2 or the isolated GAD-SPP2L109V plasmid. The transformants were streaked and grown on a plate omitting histidine at 30°C for 4 days. (B) The GAD-SPP2L109V plasmid was recreated via site-directed mutagenesis and transformed into CG1945 strains containing GBD derivatives. The transformants were grown, diluted as indicated, and spotted onto a plate containing histidine (+His) and a plate without histidine (−His). WT, wild type; DC/NY, D845N/C846Y; WL/AA, W854A/L855A.
To further test the involvement of L109 of Spp2 in Prp2 interaction, we created L109S, L109N, L109R, L109E, L109F, and L109Y mutations in GAD-SPP2. None of these mutations could interact in a yeast two-hybrid assay with GBD-prp2-D845N/C846Y or GBD-prp2-W854A/L855A, and the L109R and L109E mutations further lost interaction with wild-type GBD-PRP2 (Table 1). This provides further evidence that L109 is likely in a region critical for Prp2-Spp2 interaction.
TABLE 1.
Yeast two-hybrid interaction between Prp2 mutants and Spp2 mutants
| SPP2a |
PRP2b
|
||
|---|---|---|---|
| WT | DC/NY | WL/AA | |
| WT | ++c | − | − |
| L109V | +++ | + | +++ |
| L109S | ++ | − | − |
| L109N | + | − | − |
| L109R | − | − | − |
| L109E | − | − | − |
| L109F | + | − | − |
| L109Y | + | − | − |
The allele of SPP2 in the GAD-SPP2 construct. WT, wild type.
The alleles of PRP2 in the GBD-PRP2 construct; DC/NY is D845N/C846Y; WL/AA is W854A/L855A.
The assay was performed as described in the legend to Fig. 3, with +++ indicating very good growth, ++ indicating healthy growth, + indicating slightly impaired growth, and − indicating little growth.
In order to test the ability of SPP2L109V to restore Prp2 function, an SPP2 ORF driven by a GAL1 promoter (34) was altered to L109V via site-directed mutagenesis. pGAL-SPP2L109V was able to complement a Δspp2 strain as the sole copy of SPP2, and like wild-type SPP2 it was also able to suppress the temperature-sensitive phenotype of prp2-1 when SPP2L109V was overexpressed (data not shown). Thus, Spp2L109V retains most, if not all, SPP2 function. pGAL-SPP2L109V was transformed into a Δprp2 strain carrying two plasmids, one carrying wild-type PRP2 and one with prp2-W854A/L855A or prp2-D845N/C846Y. Plasmid shuffle assays showed that SPP2L109V failed to allow growth in a Δprp2 strain expressing only prp2-W854A/L855A or prp2-D845N/C846Y (data not shown). Thus, it appeared that SPP2L109V could restore interaction but not the function of the mutant Prp2 proteins that is needed for cell viability. We also carried out a plasmid shuffle screen in an attempt to isolate any Spp2 mutant that could allow a prp2-W854A/L855A or prp2-D845N/C846Y strain to grow. The screen failed to identify such SPP2 mutants. This result raised the possibility that the dysfunction of these Prp2 DC-WL 11-mer mutants extends beyond the interaction with Spp2 and suggests an additional role of the WL and DC residues.
Identification of Prp2 with an intermediate phenotype.
If the increased interaction observed between Prp2 and Spp2L109V is beneficial, it should enhance the ability of Prp2 to function in splicing. Because SPP2L109V could not rescue growth in a Δprp2 strain expressing only the prp2-W854A/L855A or prp2-D845N/C846Y allele, we hypothesized that the L109V effect might be measured in C-terminal 11-mer mutants of Prp2 with an intermediate phenotype. To generate these types of mutants, we performed random site-directed mutagenesis on residues D845 and C846 in a dominant-negative prp2DB mutant, which bears double mutations in the helicase domain of Prp2 at positions H349D and Q548H (8). These H-domain mutations prevent mutant prp2 from hydrolyzing ATP and releasing from the spliceosome. When this type of allele is under control of the GAL1 promoter and is overexpressed on galactose, spliceosomes are sequestered and cells cease to grow, giving a strong Gal mutant phenotype (8, 29). Plasmids with mutations allowing Gal+ colonies to form were recovered, sequenced, and retransformed back into the yeast strain to confirm the phenotype. One mutation at residue 845, prp2Db-D845L, conferred an intermediate Gal+ phenotype, while prp2Db-C846H, prp2Db-C846G, prp2Db-C846Q, and prp2Db-C846R also had various degrees of Gal+ phenotypes (Fig. 4A and B). Each of these mutants produced full-length protein at comparable levels, as measured by probing the extracts with anti-Prp2 antibodies (Fig. 4C). Thus, the weak Gal+ phenotype in D845L, C846H, and C846Q mutants suggested defects in spliceosome and perhaps Spp2 binding.
To test the effect on Spp2 interaction, the D845L mutation was created in the two-hybrid construct and was assayed for its interaction with Spp2 or Spp2L109V. In the presence of wild-type GAD-SPP2, there was a slight decrease in two-hybrid reporter activity in D845L compared to that of wild-type Prp2 (Fig. 4D, rows 2 and 6), which is consistent with the intermediate GAL+ phenotype seen in Fig. 4B. The interaction between the D845L mutant and Spp2 was restored when the L109V mutation was present in Spp2 (Fig. 4D, rows 5 and 6).
We next tested whether the D845L mutation would affect Prp2's function in splicing or cell growth. The D845L mutation was introduced into pRS415-PRP2 (9) to create construct 415-prp2-D845L. The D845L mutation conferred a slight temperature-sensitive phenotype when 415-prp2-D845L was shuffled into a Δprp2 strain (Fig. 5A, top panels). To test whether the mutation had a defect in splicing, we measured benomyl sensitivity as a splicing readout. Benomyl sensitivity is a measure of microtubule instability caused by an insufficiency in the amount of α-tubulin (30). Because α-tubulin genes TUB1 and TUB3 and microtubule-promoting factor GIM5 all contain introns, perturbations in splicing lead to benomyl sensitivity (4, 5, 28). In the Δprp2 strain, the growth defect of prp2-D845L is lethal at restrictive temperature on benomyl plates, consistent with having a splicing defect (Fig. 5A, bottom panels). To confirm this, a splicing reporter consisting of a Kluyveromyces lactis actin intron fused to β-galactosidase (7, 25) was transformed into a Δprp2 strain containing either 415-PRP2 or 415-prp2-D845L. The β-galactosidase activity in prp2-D845L was 61% of the activity in a PRP2 strain at the restrictive temperature of 37°C (Fig. 5B). Thus, the D845L mutation decreased the dominant-negative effect, the Spp2 interaction, cell growth, and splicing at elevated temperatures.
FIG. 5.
SPP2L109V enhances splicing reporter activity of a prp2-D845L strain compared to activity of SPP2. (A) Temperature sensitivity of prp2-D845L. After plasmid shuffle, cells are maintained by a plasmid containing either PRP2 (pRS415-PRP2) or prp2-D845L (pRS415-prp2-D845L). These strains were tested at 30 and 37°C on YPD and YPD-containing benomyl plates. Serial dilution spot assays were performed as described in the legend to Fig. 1, and cells were grown for 3 days. (B) β-Galactosidase (β-gal) splicing reporter activity of Δprp2 strains containing only prp2-D845L or PRP2. Cells were grown overnight at 30°C and diluted, and fractions of the culture were grown for 4 h at the indicated temperature. The β-galactosidase activity in the cell lysate was measured and normalized. Numbers indicate the average normalized units of β-galactosidase activity from three independent trials. Error bars represent one standard deviation. (C) SPP2L109V and SPP2 can rescue the benomyl-induced temperature-sensitive growth defect of prp2-D845L. Cultures were diluted, spotted on plates as indicated, and grown for 3 days (YPD) or a week (YPD-benomyl and YPGal-benomyl). (D) SPP2L109V and SPP2 can rescue the splicing defect exhibited by D845L at restrictive temperature. Assays were carried out as described in the legend to panel B after the transformation of HIS3-marked YCp50, pGAL-SPP2, or pGAL-SPP2L109V constructs. Numbers indicate the average normalized units of activity from three independent trials. Error bars represent one standard deviation.
Effect of SPP2L109V on intermediate loss-of-function allele prp2-D845L.
SPP2L109V restored interaction with Prp2-D845L (Fig. 4D), and here we tested whether L109V could rescue the temperature-sensitive splicing defect in prp2-D845L. The effect of extra SPP2 or SPP2L109V on benomyl sensitivity of the D845L mutant was tested. Both SPP2 and SPP2L109V were able to rescue the growth defect of prp2-D845L on benomyl plates at semirestrictive temperatures of 34 and 35°C (Fig. 5C, compare row 10 to rows 11 and 12). Overexpression of SPP2 (Fig. 5C, row 17) or SPP2L109V (row 18) as measured on YPGal-benomyl plates also showed rescue (lane 16). Growth of wild-type PRP2 on benomyl plates was not affected by SPP2 alleles under the assay conditions (Fig. 5C, rows 7 to 9 and 13 to 15). We further tested β-galactosidase activity in the splicing reporter assay. Ectopically expressed SPP2 or SPP2L109V had no effect on the splicing reporter activity of a prp2-D845L or PRP2 strain at the permissive temperature (Fig. 5D, 30°C). In contrast, expression of SPP2 and SPP2L109V rescued the splicing reporter activity of a Δprp2 strain expressing prp2-D845L, with SPP2L109V restoring the activity to a slightly higher level than SPP2 (Fig. 5D, the bottom graph, 37°C). Thus, SPP2L109V appeared to rescue the splicing defect as well as restore the interaction between Spp2 and Prp2-D845L.
DISCUSSION
This study identifies an interaction between a helicase (Prp2) and a helicase-associated protein (Spp2) that is required for splicing and cell viability. The interaction is likely to ensure the binding of these extrinsic splicing factors to the spliceosome, as has been suggested by Roy et al. (34). We demonstrated that point mutations localized to a defined region of either protein resulted in loss of interaction and loss of splicing function. We further suggest that the Spp2 interaction specifies the binding of Prp2 to the spliceosome and that this type of protein-protein interaction is an example of a general scheme for the specificity of related spliceosomal helicases.
Spp2 is a basic protein (pI of 8.8) of 185 amino acid residues with one conserved domain known as a G patch, which is named after a pattern of conserved glycine residues (2). The G patch is present in many RNA binding proteins, but the function of the domain remains unclear. In one study, it was shown that the G patch is necessary for the function of Gno1, an rRNA processing factor (16). Our study provides the first evidence that a mutation in a G patch can affect protein-protein binding. The leucine at position 109 within the G patch of Spp2 is critical for Prp2 interaction, because an arginine or glutamate substitution eliminated the interaction while a valine substitution enhanced the interaction (Table 1). Leucine and valine are very similar, differing by only one extra carbon in the side chain. It is interesting that such an apparently small change can affect a protein significantly. There are examples that an L-to-V change can be detrimental; leucine-to-valine changes in Bacillus thuringiensis Cry4B disrupt structure (12), and the pathogenic L392V mutation in presenilin 1 reduces affinity to glycogen synthase kinase 3β (12, 23).
We identified several intermediate loss-of-function single mutations in the DC dipeptide of Prp2. For example, prp2-D845L is slightly temperature sensitive, and it has a growth defect when grown on benomyl at restrictive temperature as well as defective splicing reporter activity. The fact that single changes at each residue could give a mutant phenotype illustrates that both residues D845 and C846 are important for Prp2 function. Amino acid residues 845 to 853 of Prp2, including the DC dipeptide, diverge considerably from the corresponding region in other spliceosomal DEAH-box proteins, but this region is quite conserved among Prp2 orthologues in the Saccharomyces genus (Fig. 1A). The Saccharomyces genus also has highly conserved Spp2 (Fig. 1C), which is interesting because an obvious homologue of Spp2 has yet to be identified outside the genus or in mammals. The notion that the major role of the DC region in Prp2 is to coordinate with a helicase-associated protein is quite intriguing.
Because the DC region of Prp2 is unique among DEAH-box spliceosomal helicases, it suggests that the interaction between the DC region of Prp2 and the G patch of Spp2 may be responsible for the specificity observed for Prp2 entry into the spliceosome. Unlike Prp2, other DEAH-box splicing factors, such as Prp16, Prp22, and Prp43, all have a tail domain extending beyond the C-terminal domain homology. These tail domains are rich in positively charged amino acids (19 of 76 [25%] in Prp16, 12 of 40 [30%] in Prp22, and 11 of 29 [38%] in Prp43). Although a deletion of the tail domain is generally tolerated, in the case of Prp16 there is a detrimental effect when its tail domain is truncated in conjunction with N-terminal deletions (43). Note that the N-terminal domains of Prp16 (17, 43) and Prp22 (36) are sensitive to deletion, whereas deletion of the N terminus of either Prp2 (9) or Prp43 (27) has no measurable effect in vivo. Because Spp2 is a highly charged spliceosomal protein, it may perform some function for Prp2 that the N terminus and tail domains perform for Prp16 and Prp22. This function would involve coupling Prp2 with the spliceosome via interaction between the G patch and the C-terminal domain of Prp2 and between the N-terminal charged domain of Spp2 and the spliceosome.
One reason the Prp2-Spp2 relationship is so essential appears to be because spliceosome binding of Prp2 is dependent on interaction with Spp2. Antibodies against Spp2 can immunoprecipitate spliceosomes prepared from a heat-treated prp2-1 extract (34); in addition, anti-Prp2 antibodies cannot immunoprecipitate spp2-1 spliceosomes unless recombinant Spp2 is added (34; our unpublished data). These data indicate that the presence of Spp2 is required for Prp2 interaction with the spliceosome. When Prp2-D845N/C846Y is added to prp2-1 splicing extract, immunoprecipitation of the spliceosome by an anti-Prp2 antibody fails, indicating that the DC motif is essential for Prp2 interaction with the spliceosome (9). We have further observed that although recombinant wild-type Spp2 protein could rescue splicing in spp2-1 temperature-sensitive extracts, Spp2 with the L109E mutation that eliminates Prp2 interaction could not. Interestingly, the antibody against Spp2 could immunoprecipitate this inactive spliceosome, while the antibody against Prp2 could not (our unpublished results). The lack of splicing and the absence of Prp2 in this Spp2L109E spliceosome further demonstrated the importance of the association between the two proteins, mediated by the G patch of Spp2 and the DC-WL region of Prp2, in spliceosome function.
The finding that the interaction between Prp2 and Spp2 is critical for splicing could have important implications for the understanding of the function of DEXD/H-box helicases. Typically, regions important for a helicase-cofactor interaction can be mapped to within a defined area in a protein. This demonstrates that the information that governs a DEXD/H-box helicase interaction with another protein can be encoded within, but not necessarily limited to, a small, discrete area in each protein. When short stretches and combinations of amino acids can mediate a helicase-cofactor interaction, specificity can be achieved even in DEAH-box splicing factors that are highly conserved. The study of how the interaction with Spp2 is essential for Prp2 activity could provide a roadmap for other helicase-accessory factor interactions. Accessory factors can interact with helicases to affect activity in several ways (39). Among these, an accessory factor can assist a helicase through physical modulation of activity or through recruitment to its RNP target complex. There are several examples of cofactors that interact with DEXD/H-box helicases in a temporally and spatially important manner in pre-mRNA splicing (39). These cofactors can interact in a cooperative manner with or antagonistically to the partner helicase. For Prp2, interaction with Spp2 enables Prp2 activity on the spliceosome. A similar example of cooperation in splicing is seen in humans with U2AF65 and DEXD/H-box helicase UAP56. U2AF65 has RNA binding activity and is involved in the association of the U2 snRNP to the branchpoint sequence. UAP56 is an RNA helicase that is essential for U2 snRNP addition. U2AF65 physically interacts with UAP56; when this interaction is eliminated, U2 snRNP addition catalyzed by UAP56 is deficient (10). Another example includes Slu7, which is a protein required for Prp22 interaction with the spliceosome (19) and which interacts with Prp22 in a yeast two-hybrid assay (41). Slu7 could also be important for the role Prp16 plays in the second step of pre-mRNA splicing (1). Perhaps similar factors could fill the roles of Spp2 or Slu7 for other spliceosomal DEAH-box helicases.
Additional proteins other than Spp2 have been reported to interact with Prp2 under certain experimental conditions. Prp2 has been found in complexes containing Cef1 (13) and Cin2 (18) in genome-wide proteomic screens. Interestingly, DEXD/H-box factor Brr2/Prp44 interacts with Prp2, Prp16, and Prp22 in a yeast two-hybrid assay, suggesting a potential role for Brr2/Prp44 in spliceosome coordination (41). Therefore, the recruitment of a transient or extrinsic DEAH-box helicase to the spliceosome is likely influenced by multiple protein-protein and perhaps additional protein-RNA interactions.
Acknowledgments
We are grateful to P. Legrain and M. Fromont-Racine in the European Commission-funded TAPIR Network for providing the FRYL yeast two-hybrid library. We thank J. Woolford for his generous gift of SPP2 strains and plasmids and John Rossi for the splicing reporter plasmid. We express our gratitude to Glenn Manthey and Gretchen Edwalds-Gilbert for suggestions and comments on the manuscript.
The work was supported by a Wellcome Trust grant to J.D.B. and NIH grant GM40639 to R.J.L. A.M. was partially supported by a postdoctoral fellowship from the International Agency for Research on Cancer.
REFERENCES
- 1.Ansari, A., and B. Schwer. 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 14:4001-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1999. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24:342-344. [DOI] [PubMed] [Google Scholar]
- 3.Arenas, J. E., and J. N. Abelson. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. USA 94:11798-11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns, C. G., R. Ohi, S. Mehta, E. T. O'Toole, M. Winey, T. A. Clark, C. W. Sugnet, M. Ares, Jr., and K. L. Gould. 2002. Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla, G., A. K. Sapra, U. Surana, and U. Vijayraghavan. 2003. Dependence of pre-mRNA introns on PRP17, a non-essential splicing factor: implications for efficient progression through cell cycle transitions. Nucleic Acids Res. 31:2333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross, F. R. 1997. ‘Marker swap' plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647-653. [DOI] [PubMed] [Google Scholar]
- 7.Deshler, J. O., and J. J. Rossi. 1991. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 5:1252-1263. [DOI] [PubMed] [Google Scholar]
- 8.Edwalds-Gilbert, G., D. H. Kim, S. H. Kim, Y. H. Tseng, Y. Yu, and R. J. Lin. 2000. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DEXD/H-box proteins. RNA 6:1106-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwalds-Gilbert, G., D. H. Kim, E. Silverman, and R. J. Lin. 2004. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA 10:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 11.Fromont-Racine, M., J. C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277-282. [DOI] [PubMed] [Google Scholar]
- 12.Gantier, R., D. Gilbert, C. Dumanchin, D. Campion, D. Davoust, F. Toma, and T. Frebourg. 2000. The pathogenic L392V mutation of presenilin 1 decreases the affinity to glycogen synthase kinase-3 beta. Neurosci. Lett. 283:217-220. [DOI] [PubMed] [Google Scholar]
- 13.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 14.Gietz, R. D., and R. A. Woods. 2001. Genetic transformation of yeast. BioTechniques 30:816-828. [DOI] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and R. A. Woods. 1994. High efficiency transformation in yeast, p. 121-134. In J. A. Johnston (ed.), Molecular genetics of yeast: practical approaches. Oxford University Press, Oxford, United Kingdom.
- 16.Guglielmi, B., and M. Werner. 2002. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J. Biol. Chem. 277:35712-35719. [DOI] [PubMed] [Google Scholar]
- 17.Hotz, H. R., and B. Schwer. 1998. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics 149:807-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, S. A., W. Turner, and B. Schwer. 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8:1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S. H., and R. J. Lin. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16:6810-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. H., J. Smith, A. Claude, and R. J. Lin. 1992. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 11:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King, D. S., and J. D. Beggs. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 18:6559-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krittanai, C., A. Bourchookarn, W. Pathaichindachote, and S. Panyim. 2003. Mutation of the hydrophobic residue on helix alpha5 of the Bacillus thuringiensis Cry4B affects structural stability. Protein Peptide Lett. 10:361-368. [DOI] [PubMed] [Google Scholar]
- 24.Last, R. L., J. R. Maddock, and J. L. Woolford, Jr. 1987. Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics 117:619-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, J., and J. J. Rossi. 1996. Identification and characterization of yeast mutants that overcome an experimentally introduced block to splicing at the 3′ splice site. RNA 2:835-848. [PMC free article] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, A., S. Schneider, and B. Schwer. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 277:17743-17750. [DOI] [PubMed] [Google Scholar]
- 28.O'Keefe, R. T. 2002. Mutations in U5 snRNA loop 1 influence the splicing of different genes in vivo. Nucleic Acids Res. 30:5476-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plumpton, M., M. McGarvey, and J. D. Beggs. 1994. A dominant negative mutation in the conserved RNA helicase motif ′SAT' causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 13:879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards, K. L., K. R. Anders, E. Nogales, K. Schwartz, K. H. Downing, and D. Botstein. 2000. Structure-function relationships in yeast tubulins. Mol. Biol. Cell 11:1887-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robzyk, K., and Y. Kassir. 1992. A simple and highly efficient procedure for rescuing autonomous plasmids from yeast. Nucleic Acids Res. 20:3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rogers, G. W., Jr., N. J. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914-30922. [DOI] [PubMed] [Google Scholar]
- 33.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Roy, J., K. Kim, J. R. Maddock, J. G. Anthony, and J. L. Woolford, Jr. 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1:375-390. [PMC free article] [PubMed] [Google Scholar]
- 35.Rozen, F., I. Edery, K. Meerovitch, T. E. Dever, W. C. Merrick, and N. Sonenberg. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider, S., and B. Schwer. 2001. Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem. 276:21184-21191. [DOI] [PubMed] [Google Scholar]
- 37.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwer, B., and C. Guthrie. 1992. A dominant negative mutation in a spliceosomal ATPase affects ATP hydrolysis but not binding to the spliceosome. Mol. Cell. Biol. 12:3540-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverman, E., G. Edwalds-Gilbert, and R. J. Lin. 2003. DEXD/H-box proteins and their partners: helping RNA helicases unwind. Gene 312:1-16. [DOI] [PubMed] [Google Scholar]
- 40.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 41.van Nues, R. W., and J. D. Beggs. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157:1451-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner, J. D., E. Jankowsky, M. Company, A. M. Pyle, and J. N. Abelson. 1998. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 17:2926-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y., and C. Guthrie. 1998. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA 4:1216-1229. [PMC free article] [PubMed] [Google Scholar]