Abstract
Members of the ternary complex factor (TCF) subfamily of the ETS-domain transcription factors are activated through phosphorylation by mitogen-activated protein kinases (MAPKs) in response to a variety of mitogenic and stress stimuli. The TCFs bind and activate serum response elements (SREs) in the promoters of target genes in a ternary complex with a second transcription factor, serum response factor (SRF). The association of TCFs with SREs within immediate-early gene promoters is suggestive of a role for the ternary TCF-SRF complex in promoting cell cycle entry and proliferation in response to mitogenic signaling. Here we have investigated the downstream gene regulatory and phenotypic effects of inhibiting the activity of genes regulated by TCFs by expressing a dominantly acting repressive form of the TCF, Elk-1. Inhibition of ternary complex activity leads to the downregulation of several immediate-early genes. Furthermore, blocking TCF-mediated gene expression leads to growth arrest and triggers apoptosis. By using mutant Elk-1 alleles, we demonstrated that these effects are via an SRF-dependent mechanism. The antiapoptotic gene Mcl-1 is identified as a key target for the TCF-SRF complex in this system. Thus, our data confirm a role for TCF-SRF-regulated gene activity in regulating proliferation and provide further evidence to indicate a role in protecting cells from apoptotic cell death.
Elk-1 is a member of the ternary complex factor (TCF) subfamily of ETS-domain transcription factors (reviewed in references 46 and 50). In mammals there are two other TCFs, SAP-1 and SAP-2/ERP/Net. These proteins are characterized by their ability to form ternary complexes on target promoters in conjunction with the MADS-box protein serum response factor (SRF). The TCFs share four domains, the ETS DNA-binding domain, the B-box, the D-domain, and the C-domain. SAP-2/Net contains additional regions that impart repressive properties (9, 29). The D- and C-domains constitute the regulatory part of Elk-1 and other TCFs. The D-domain acts as a docking site for mitogen-activated protein kinases (MAPKs) (reviewed in references 15 and 49). These docked kinases can then phosphorylate residues in the C-domain, which constitutes the transcriptional activation domain (TAD). Phosphorylation of the TAD leads to elevation of the transactivation potential of the TCFs and also enhances ternary complex formation (reviewed in references 46, 50, 54, and 58). The TCFs can be phosphorylated by members of all three of the major MAPK pathways present in mammals: ERK, JNK, and p38 (reviewed in references 46, 50, and 58). The ERK MAPK pathway predominantly transmits mitogenic and differentiation stimuli, whereas the JNK and p38 MAPK pathways primarily transduce stress and cytokine stimuli to the nucleus (reviewed in reference 41). The TCFs therefore play a pivotal role in transducing extracellular stimuli into alterations in gene expression in the nucleus.
The B-box of the TCFs is required for ternary complex formation (11, 23, 55) and mediates protein-protein interactions between TCFs and SRF (28, 52). Single point mutations within the B-box in Elk-1 (e.g., leucine 158 to proline) are sufficient to severely disrupt interactions with SRF and hence block ternary complex formation (28). Structural studies of the SRF-SAP-1 complex demonstrate that the B-box binds to a surface-exposed hydrophobic groove on SRF (see Fig. 4A) and consists of two short 310 helices that flank a short β-strand (19). Recently, an additional role for the Elk-1 B-box in inhibiting RhoA signaling to SRF has been uncovered, thereby providing signaling specificity to the TCF-SRF complex (35).
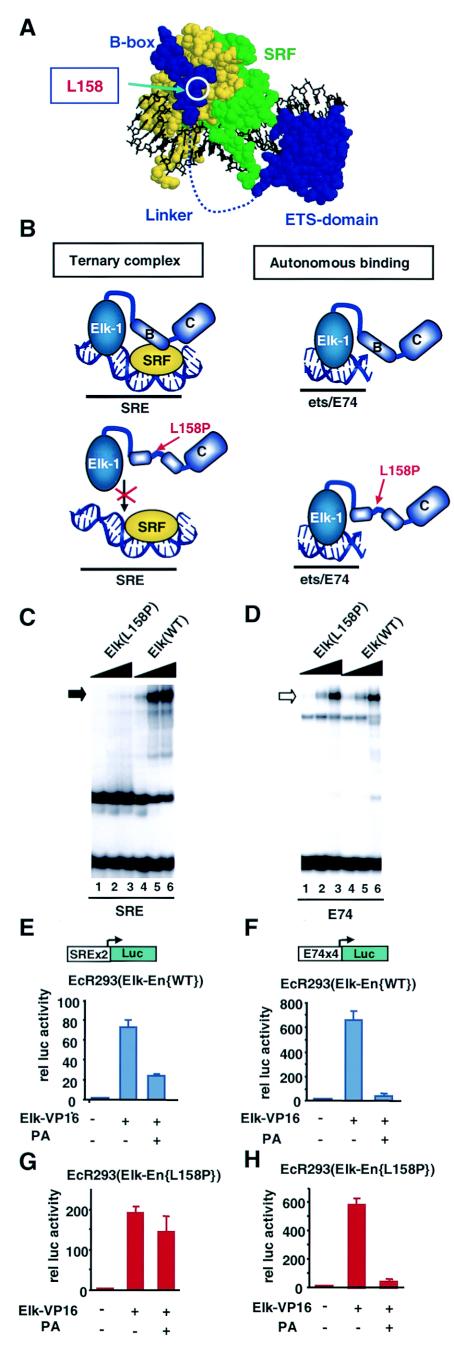
FIG.4.
Elk-En(L158P) mutant is defective in SRE-mediated repression. (A) Structure of the ternary SAP-1-SRF complex (19). SRF monomers are shown in green and yellow, and SAP-1 is shown in blue. The locations of the ETS-domain, linker, B-box region, and the residue corresponding to L158 in Elk-1 are indicated. (B) Diagrammatic representation of ternary Elk-1-SRF-SRE and binary Elk-1ets/E74 complexes in presence of wild-type and L158P mutant Elk-1. Regions corresponding to the B-box (indicated by the letter B) and C-terminal region (indicated by the letter C) are shown. (C and D) Gel retardation analysis of wild-type and L158P mutant Elk-En on the SRE (C) and E74 (D) sites. Increasing amounts of each in vitro-translated protein are included in each set of lanes (ratios: 1 [lanes 1 and 4], 3 [lanes 2 and 5], and 10 [lanes 3 and 6]). (E to H) Reporter gene analysis using SRE-Luc (E and G) and E74-Luc (F and H) reporters in EcR293 lines that inducibly express Elk-En(WT) [#1.3] (E and F) or Elk-En(L158P) [#L8] (G and H). Where indicated, 0.5 μg of Elk-VP16 was transiently transfected to activate the reporters. Elk-En expression is induced by adding 5 μM PA.
The primary action of the TCFs is thought to be the activation of immediate-early genes by forming ternary complexes with SRF at serum response elements (SREs) found in the promoters of these genes. The best-characterized TCF target is the proto-oncogene c-fos (17, 21, 22). c-fos is an immediate-early gene, and c-fos levels are known to increase rapidly upon stimulation of cells with various mitogens. SRF has been found to be constitutively bound to the SRE in the c-fos promoter (21) and is able to recruit Elk-1 despite the suboptimal Elk-1 binding site that makes up part of this promoter element. SREs are also known to be located in the promoters of a variety of other genes, such as egr-1 (early growth response gene-1) and Nur77, the murine homologue of the human TR3 gene. SRE-dependent activation is important for the activation of the Nur77/TR3 gene (26) and the egr-1 gene in response to a variety of signals (1, 6, 12, 33, 38, 39). In vitro studies have implicated the TCFs as an important part of the regulatory complex that forms on these SREs (7, 26). A further target gene identified for the TCFs is Mcl-1, thereby suggesting a potential role in regulating apoptosis (53). Although binding of TCFs to SREs usually requires recruitment by SRF, they can also bind autonomously to high-affinity ets DNA-binding motifs. Additionally, it has been proposed that TCFs are able to recruit SRF to suboptimal SRF-binding sites such as those found in the pip92 and nur77 SREs (26). Indeed, this has been shown to be the case for nur77, where cells deficient in SAP-1 show reduced SRF recruitment to this promoter in vivo (8). The TCF Elk-1 has also been implicated in the regulation of TNF-α (56), 9E3/cCAF (27), and α(1,3)-fucosyltransferase IV (59), where promoter binding apparently occurs independently of SRF.
Recently, mouse knockout studies on individual TCFs have been completed, but minimal phenotypes were observed, suggesting that functional redundancy might exist. SAP-2/Net has a role in egr-1 repression and vascular development (2). SAP-1 is important in the positive selection of thymocytes (8), whereas Elk-1 plays a role in the neuronal expression of immediate-early genes like c-fos in the brain (5). Due to the possible functional redundancy of the TCFs, we have used a repressive form of Elk-1 to probe the potential role of TCFs in regulating cell growth and gene expression. We demonstrate that inhibition of TCF-regulated gene expression leads to growth arrest and apoptosis. These apoptotic effects are dependent on the downregulation of the key antiapoptotic regulator Mcl-1, which has been previously implicated as a TCF target gene (53). By using mutant Elk-1 alleles, we further demonstrate that these effects are via an SRF-dependent mechanism. A large body of literature links the activation of the Ras/ERK pathway in controlling cell proliferation and survival. Our data demonstrate that members of one class of nuclear target of this pathway, the TCFs, act through ternary TCF-SRF-SRE complexes to regulate genes whose activity is required for proliferation and also for protecting cells from apoptotic death.
MATERIALS AND METHODS
Plasmid constructs.
pSRE-Luc (pAS821) contains two copies of the c-fos SRE (nucleotides −357 to −275, containing both an SRF binding site and an adjacent ets motif) upstream of a minimal tk promoter and the luciferase gene (44). pE74-Luc (pNC101) contains four E74 ets binding sites upstream of a minimal tk promoter and the firefly luciferase gene (kindly provided by Erik Jansen). pMcl-1-Luc (pAS2156) contains the human Mcl-1 promoter (−3893 to +25) upstream from the firefly luciferase gene (kindly provided by Steve Edwards), pCH110 (Pharmacia) contains a simian virus 40-driven β-galactosidase (LacZ) gene and is used to monitor transfection efficiency. pRSV-Elk-1-VP16 (pAS348), pRSV-SAP-1-VP16, and pRSV-SAP-2-VP16 are Rous sarcoma virus (RSV) promoter-driven vectors encoding full-length wild-type human Elk-1, SAP-1, and SAP-2 fused to residues 410 to 490 of a VP16 C-terminal sequence (37). pCMV-MEK1 encodes constitutively active MEK-1(ΔNS218E-S222D) (30). pAS515 encodes full-length zFli-1 fused to the engrailed (En) repressor domain (provided by Dave Hickleton). pAS728 (encoding full-length His-FLAG-tagged Elk-1) was created by ligating a NcoI-BamHI fragment from pAS278 (60) into the same sites in pAS38 (47). pEGFP-N1 (Clontech) encodes enhanced green fluorescent protein (EGFP).
The following plasmids were constructed for use in in vitro transcription-translation of proteins. pAS1407, encoding full-length Elk-1 fused to the En repressor domain and the FLAG epitope (Elk-En), was produced by ligating the HindIII-XbaI fragment from pAS1406 (described below) into pAS728. pAS1409, encoding Elk-1(L158P) fused to the En repressor domain and FLAG epitope, [Elk-En(L158P)], was constructed by ligating an NcoI-StuI fragment from pAS853 (61) into the same sites in pAS1407.
The following vectors were used for in vivo expression in mammalian cells. pAS1402 (encoding RSV-driven Elk-VP16{L158P})was created by ligating an NcoI/HindIII fragment from pAS1403 into the same sites in pAS348. pAS1403 was created by inserting a StuI/NcoI fragment from pAS853 into the same sites in pRSETB-Elk-VP16. pRSETB-Elk-VP16 was created by inserting an NcoI/HindIII fragment from pRSV-Elk-VP16 into pRSETB. To create stable cell lines which can be induced with ponasterone A (PA) to express FLAG-tagged Elk-En and Elk-En(L158P), pAS1406 and pAS1410 were constructed. pAS1406, encoding Elk-En driven by ecdysone response elements, was constructed by ligating a HindIII/XhoI fragment from pAS1404 into the same sites of pAS728. pAS1404, encoding the En repressor domain (amino acids 2 to 298) fused to the FLAG epitope, was constructed by cloning an XhoI/XbaI-cleaved PCR fragment (primer pair ADS731-ADS732; template pAS515) into the same sites in pIND(SP1) (Invitrogen). The FLAG epitope was created through the use of primer ADS738, which was designed to include the FLAG sequence. pAS1410, encoding Elk-1(L158P)-En-FLAG, was created by ligating the HindIII/XbaI fragment of pAS1409 into the same sites in pAS1406. pAS1408 and pAS1411, encoding Elk-En-FLAG wild type (WT) and L158P, were created by ligating HindIII/XbaI fragments of pAS1406 and pAS1409 into the same sites in pCDNA3. pexpMcl-1 (pAS2157) encodes full-length FLAG-tagged human Mcl-1 in a cytomegalovirus (CMV)-driven vector (kindly provided by X. Wang) (36).
Tissue culture, cell transfection, and reporter gene assays.
EcR293 cells (Invitrogen) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL) and 400 μg of Zeocin (Invitrogen)/ml. HeLa cells were grown in DMEM-10% FBS. Where indicated, the caspase inhibitor Z-VAD-fmk (Calbiochem) was added together with PA. Transient transfection experiments were carried out using Superfect transfection reagent (QIAGEN). Stable cell lines exhibiting inducible expression of Elk-En (57) or Elk-En(L158P) were created by transfecting EcR293 cells with SspI-linearized pAS1406 or pAS1410 to create EcR293(Elk-En) and EcR293(Elk-En{L158P}) lines. Stably transfected clones were selected as described previously (57). Western blotting and reporter assays were used to look for inducible expression of FLAG-tagged Elk-En or Elk-En(L158P) and to check for leaky transcription from the vectors. A total of 30 separate clones were expanded for each cell line and assayed by Western blotting; 5 clones were found to be positive for inducible Elk-En expression and 9 were found to be positive for inducible Elk-En(L158P). These were then used in reporter gene analysis experiments.
For reporter gene assays, SRE- and E74-driven vectors containing two tandem SREs and four E74 response elements, respectively, upstream of a minimal promoter element or a Mcl-1 promoter fragment, followed by the firefly luciferase gene, were transfected into EcR293 cells or into the EcR293(Elk-En) stable cell lines. The cells were also transfected with pCH110 to monitor transfection efficiency and where indicated, with either pCMV-MEK or pRSV-Elk-VP16 to stimulate reporter activity. Following transfection, cells were left in DMEM containing 0.05% FBS for 24 h. In some experiments the cells were pretreated with 5 μM PA (Invitrogen) for 6 h prior to transfection to stimulate Elk-En expression and then maintained in 0.05% FBS containing PA following transfection for a further 24 h. Cells were then solubilized, and luciferase assays were carried out using a dual light reporter gene assay system (Tropix). Light emissions were measured for 10 s with a Turner TD-20/20 luminometer.
Western blot analysis.
Cells were harvested by direct lysis or lysed following trypsinization in lysis buffer (Tropix) containing phenylmethylsulfonyl fluoride and aprotinin. Anti-p53, p21cip1 (Santa Cruz), and a mouse monoclonal anti-M2 FLAG antibody (Sigma) were used along with horseradish peroxidase-conjugated secondary antibodies followed by detection using Supersignal West Dura Extended Duration substrate (Pierce) and visualization using a Bio-Rad Fluor-S MultiImager. Quantification of proteins was carried out using Quantity One software (Bio-Rad).
Gel retardation assays.
Gel retardation assays were carried out with a 32P-labeled 49-bp fragment of the c-fos promoter, containing the SRE, the E74 site, or an 88-bp fragment of the Mcl-1 promoter (nucleotides −79 to −127; ADS1226 [5′-AGCAACCCTCCGGAAGCTGCCGCCCCTTTCCCCTTTTATGGGAA-3′] and ADS1225 [5′-AGTATTCCCATAAAAGGGGAAAGGGGCGGCAGCTTCCGGAGGGT-3′]) as described previously (48, 52). Elk-1 was produced as a His-tagged protein in bacteria (see Fig. 7) (61) or as FLAG-tagged derivatives by in vitro transcription and translation using a TNT-coupled reticulocyte lysate system (Promega) (see Fig. 4). 35S-labeled proteins were analyzed by electrophoresis through use of a 0.1% sodium dodecyl sulfate (SDS)-12% polyacrylamide gel before visualization and quantification using a PhosphorImager and Quantity One software (Bio-Rad). Proteins were incubated with labeled probe and coreSRF purified from bacteria where indicated (52). Protein-DNA complexes were analyzed on nondenaturing 5% polyacrylamide gels cast in 0.5× Tris-borate-EDTA and visualized by autoradiography and phosphorimaging.
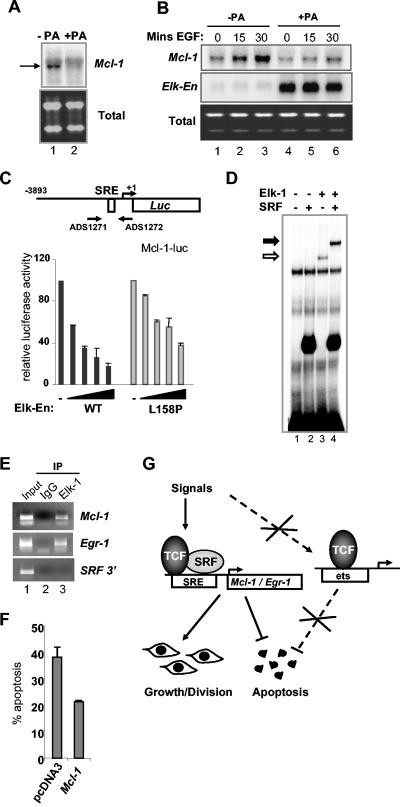
FIG. 7.
Mcl-1 is an antiapoptotic Elk-1 target gene. (A and B) Northern blot analysis of Mcl-1 expression in serum-starved EcR293(Elk-En)#1.3 cells in the presence and absence of treatment with 5 μM PA for 18 h (A) or 2.5 h (B). (B) Cells were stimulated with EGFP for the indicated times. Lower panels show total mRNA. (C) Mcl-1 promoter analysis. 293 cells were transiently transfected with a luciferase construct containing the Mcl-1 promoter and with increasing concentrations of Elk-En(WT) and Elk-En(L158P) (0, 1, 5, 10, 25 ng for each of the WT and L158P bars from left to right, respectively). Luciferase activity was measured 24 h after transfection. The results were normalized to β-galactosidase activity. (D) Gel retardation assay with a fragment of Mcl-1 promoter (−79 to −127). The DNA was incubated with the indicated combinations of purified coreSRF and full-length Elk-1 in the presence of unprogrammed rabbit reticulocyte lysate. Binary and ternary complexes containing Elk-1 are shown by open and closed arrows, respectively. (E) Chromatin IP of Elk-1 bound to the Mcl-1 and Egr-1 promoters. HeLa cells were starved in serum-free DMEM for 48 h and then stimulated with 10 nM PMA for 15 min. Sonicated chromatin was immunoprecipitated with either anti-Elk-1antibody or nonspecific IgG. PCR analysis of eluted DNA was performed using oligonucleotides specific for Mcl-1 and Egr-1 promoters and an SRF intronic sequence. Lane 1, 2% of input DNA. (F) EcR293(Elk-En) cells were transfected with 1.5 μg of a CMV-driven Mcl-1 expression vector or with pCDNA3. At 24 h after transfection, the cells were stimulated with 5 μM PA. At 48 h later, cells were subjected to Hoechst staining and apoptotic cells were counted. Data are representative of two independent experiments. (G) Model showing that ternary TCF-SRF complex-specific gene expression is required to promote cellular proliferation and inhibit cell death. Genes such as Mcl-1 that are regulated by the ternary TCF-SRF-SRE complex must be active to permit cell growth and division and to inhibit apoptotic cell death. Blocking the activity of this complex with Elk-En causes cell cycle arrest and apoptotic cell death. Experiments using mutant forms of Elk-En (L158P) demonstrated that the major route of action is not via binary TCF-DNA complexes.
RT-PCR, Northern and microarray analysis.
Cells were maintained in 0.05% FBS for 24 h and then treated with 5 μM PA in ethanol or with ethanol alone for 24 h. Epidermal growth factor (EGF) (Sigma) was then added to the cells to a final concentration of 50 nM for 15, 30, or 60 min where indicated. Cells were then harvested, and total RNA was extracted using an RNeasy kit (QIAGEN). For reverse transcriptase PCR (RT-PCR), 1 μg of total RNA from each sample was reverse transcribed using Moloney murine leukemia virus RT and random hexanucleotide primers (Roche) according to the supplier's protocol. cDNA (1 μl) and Taq polymerase (Bioline) were used in each subsequent PCR with primers to detect expression of GAPDH (ADS733 [5′-GCATTGCTGATGATCTTGAGG-3′] and ADS734 [5′-TCGGAGTCAACGGATTTG-3′]), c-fos (ADS735 [5′-CAGTGCCAACTTCATTCC-3′] and ADS736 [5′-GAACTCTAGTTTTTCCTTCTCC-3′]), egr-1 (ADS770 [5′-CCCTTCCCCAATTACTATTCC-3′] and ADS771 [5′-CCCCCAAATCATCACTCC-3′]), TR3 (ADS740 [5′-ATACACCCGTGACCTCAACC-3′] and ADS741 [5′-TACATCCCCAGCATCTTCC-3′]), and vinculin (ADS1029 [5′-CCAAGGTCAGAGAAGCCTTCC-3′] and ADS1030 [5′-CGTAGCTGTTCAAGGTCTGGT-3′]) (42). PCR products were detected using 2% agarose gels, and ethidium bromide-stained bands were visualized using a Bio-Rad Gel Doc system. For Northern analysis, 5 μg of RNA was subjected to formaldehyde gel electrophoresis and transferred to Hybond-XL membranes (Pharmacia Biotech) according to the manufacturer's instructions. The filters were prehybridized at 68°C for 3 h in 0.5 M sodium phosphate buffer (pH 7.2)-7% sodium dodecyl sulfate (SDS)-1 mM EDTA and hybridized in the same solution with a human Mcl-1 full-length cDNA fragment labeled by random priming (Roche). After the hybridization, nonspecifically bound radioactivity was removed by four washes in 40 mM phosphate buffer-1% SDS-1 mM EDTA at 68°C for 20 min.
Microarray experiments were performed using human focus and human U133A GeneChip oligonucleotide arrays (Affymetrix, Inc.) according to the manufacturer's instructions. Briefly, 15 μg of total RNA was used to prepare first-strand cDNA by use of an oligo(dT)-T7 primer. Following second-strand synthesis, biotinylated cRNA targets were generated using an Enzo BioArray HighYield RNA transcript-labeling kit (Affymetrix, Inc.) by in vitro transcription with biotinylated UTP and CTP. The fragmented cRNA targets together with labeled controls were hybridized to the arrays at 45°C with rotation at 60 rpm for 16 h according to the manufacturer's instruction. Following hybridization the arrays were processed using Affymetrix fluidics protocol Midi-euk2 for the Focus arrays and EukGE-WS2 for the U133A arrays and stained with R-phycoerythrin conjugated to streptavidin (Molecular Probes, Inc.). Images of the arrays were then acquired using Microarray Suite (MAS) version 5.0 software and an Affymetrix 2500 GeneChip scanner. Normalization and further analyses were carried out using RMAExpress software (http://stat-www.berkeley.edu/users/bolstad/RMAExpress/RMAExpress.html). Microarray data were generated using three independent samples from PA-stimulated and two independent samples from unstimulated EcR293(Elk-En)#1.3 cells.
Chromatin immunoprecipitation (IP).
Cells were plated on 10-cm-diameter dishes, serum starved for 24 h, and than stimulated with 10 nM phorbol miristate acetate (PMA) for 15 min. Proteins were cross-linked to DNA by incubation with 1% formaldehyde for 10 min at 37°C. The cells were washed twice with ice-cold 0.125 M glycine in 1× PBS and scraped into 1 ml of PBS containing protease inhibitor cocktail (Boehringer) and 1 mM PMA. Cells were pelleted and lysed with lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0). Lysates were sonicated to produce DNA sizes between 200 to 500 bp. Samples were diluted 1 in 10 with IP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.0], 16.7 mM NaCl, protease inhibitor cocktail) and immunoprecipitated with anti-Elk-1 (Santa Cruz) or nonspecific immunoglobulin G (IgG) (Upstate) overnight at 4°C. After 1 h of incubation with salmon sperm DNA, 30 μl of protein A dynal beads (Dynal Biotech) was added to the samples. After 1 h the beads were washed with the following buffers: TSE I (20 mM Tris [pH 8.1], 2 mM EDTA, 150 mM NaCl, 1% Triton, 0.1% SDS), TSE II (20 mM Tris [pH 8.1], 2 mM EDTA, 500 mM NaCl, 1% Triton, 0.1% SDS), buffer III (10 mM Tris [pH 8.1], 0.25 M LiCl, 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate), and TE (10 mM Tris [pH 8.0], 1 mM EDTA). DNA was eluted with elution buffer (0.1 M NaHCO3, 1% SDS). NaCl (0.2 M) was added to the eluted samples and to the input, and cross-linking was reversed by overnight incubation at 65°C. After digestion with proteinase K, DNA was cleaned using a QIAGEN PCR cleanup kit. Promoter-specific primers used to amplify the DNA by PCR were as follows: for human Egr-1 promoter, ADS1269 (TGCAGGATGGAGGTGCC) and ADS1270 (AGTTCTGCGCGCTGGGATCTCTC); for human Mcl-1 promoter, ADS1271 (ACTCAGAGCCTCCGAAGACC) and ADS1272 (CGCCAGCGAACTCTTTTT); and for SRF intronic sequence, ADS1273 (GCCACAGGGCAGTAGATGTT) and ADS1274 (TCAGGCCCAAGTATCCACTC).
Hoechst staining and fluorescence microscopy.
Cells were harvested and fixed on ice with 1% formaldehyde in PBS. Cells were then spun down and resuspended in a solution of 10 ng of Hoechst stain (Sigma)/ml in PBS. Visualization of staining was carried out using a pseudoconfocal fluorescence microscope. Pictures were generated using picture publisher software. EGFP was detected using a fluorescent microscope (oil immersion lens).
BrdU incorporation and fluorescence-activated cell sorting (FACS).
Cells were plated out onto 10-cm-diameter dishes, and at the required time points BrdU (Sigma) was added to the cells at a final concentration of 10 μM. The cells were then incubated at 37°C for 1 h (times stated in figures are times without the 1 h of BrdU incubation). For flow cytometry, cells were harvested and fixed by the dropwise addition of 5 ml of ice-cold 70% ethanol. Cellular DNA was denatured with 2 M HCl, and the cells were stained with a mouse anti-BrdU antibody and a secondary fluorescein isothiocyanate (FITC)-conjugated antibody (Dako). Cells were then incubated with 10 mg of propidium iodide/ml to allow examination of the DNA content of cells. Analyses were carried out on a Becton-Dickinson flow cytometer for both BrdU uptake and cell cycle profile.
RESULTS
Elk-En fusion proteins repress SRE-dependent gene regulation.
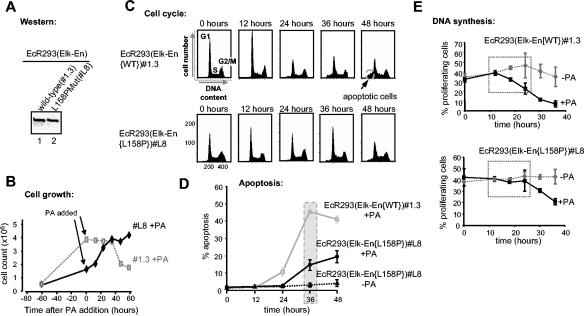
The TCFs have been implicated in regulating the expression of immediate-early genes such as c-fos in response to stress and mitogenic signaling (Fig. 1A). In particular, their association with the ERK MAPK cascade and transduction of mitogenic signals suggests a role for the TCFs in regulating cell cycle entry and controlling cellular proliferation. To investigate such a potential role and the phenotypic consequences of disrupting TCF activity, we created a dominant-negative form of Elk-1 that consists of full-length Elk-1 fused to the Drosophila En repression domain (Elk-En). The expression of this fusion protein, Elk-En, is predicted to repress the activity of TCF target genes and hence disrupt downstream cellular processes. Due to the conservation of the TCF structure, Elk-En is likely to inhibit the activity of SAP-1- and SAP-2-regulated genes in addition to Elk-1 targets. Thus, this approach should overcome problems associated with redundancy of function between the TCFs that is suggested by the phenotypes associated with knockout studies (2, 5, 8). Fusions of other transcription factors to the En repression domain have been successfully used to study their biological functions (3, 24). Stable cell lines were created in human EcR293 cells that inducibly express Elk-En in response to stimulation with PA (Fig. 1B). One cell line, EcR293(Elk-En)#1.3, that exhibits undetectable levels of Elk-En in the absence of PA but high levels following PA induction (Fig. 1C) was selected for further study.
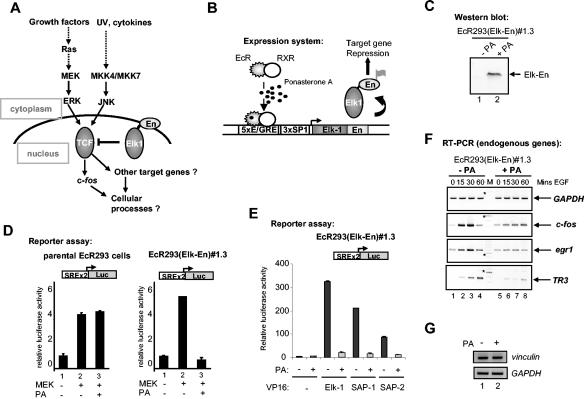
FIG. 1.
Induction of Elk-En represses SRE-dependent gene regulation. (A) Schematic indicating the role of the TCFs as modulators of transcriptional effects in response to signals from MAPK pathways. Elk-En fusion proteins block signaling through Elk-1 to downstream target genes. En represents the En repression domain. (B) The PA-inducible expression system contained in EcR293(Elk-En) cell lines. EcR and RXR represent the ecdysone and retinoid X receptors that, upon binding to PA, activate the expression of Elk-En. Elk-En contains a C-terminal FLAG tag. (C) Western blot of FLAG-tagged Elk-En in EcR293(Elk-En)#1.3 cells before and after PA treatment for 24 h. (D) Repression of SRE-Luc reporter genes in response to PA-induction of Elk-En in EcR293(Elk-En)#1.3 cells. Parental EcR293 or EcR293(Elk-En)#1.3 cells were transfected with SRE-luciferase reporter constructs (0.25 μg) in the presence or absence of a vector encoding constitutively active MEK (0.5 μg) where indicated. Cells were incubated for 24 h in 10% FBS in the presence or absence of 5 μM PA. Data are presented as means and standard deviations (n = 3) normalized for β-galactosidase activity from a cotransfected pCH110 reporter construct. (E) Reporter gene analysis using SRE-Luc reporter in EcR293(Elk-En)#1.3 cells. Where indicated, plasmids encoding 0.5 μg of Elk-1, SAP-1, or SAP-2-VP16 fusions were transiently transfected to activate the reporters. Elk-En expression was induced by adding 5 μM PA. (F) Repression of EGFP induction of endogenous TCF target genes by Elk-En following PA induction in EcR293(Elk-En)#1.3 cells. Cells were synchronized by incubation in 0.05% FBS for 48 h; where indicated, 5 μM PA was added for the final 12 h. Cells were then stimulated with 50 nM EGFP, and the expression of GAPDH, c-fos, egr-1, and TR3 was analyzed by RT-PCR at the indicated times. For clarity, the color on the original image has been inverted. The asterisks indicate the locations of the 600-bp bands in the marker (M) lane. (G) RT-PCR analysis of the SRF-regulated gene vinculin. EcR293(Elk-En)#1.3 cells were serum starved for 24 h and incubated with PA for 7 h (where indicated) before stimulation with 10% serum for 1 h. GAPDH is shown as a control.
To assess whether Elk-En can repress transcription through SREs, EcR293(Elk-En)#1.3 cells were transfected with a luciferase reporter construct driven by two tandem SREs. Cotransfection with MEK, a constitutively active form of the ERK activator, led to a large increase in reporter activity (Fig. 1D). However, when the cells were treated with PA to induce Elk-En expression, no reporter activation was observed (Fig. 1D, right panel). In contrast, PA treatment had no effect on SRE-luciferase activity in the parental EcR293 cells (Fig. 1D, left panel). Thus, induction of Elk-En expression leads to potent repression of SRE-driven reporters in EcR293(Elk-En)#1.3 cells. Due to the structural homology shared by the TCFs, it is predicted that Elk-En would repress all TCF-mediated gene regulation. To demonstrate that this is indeed the case, we investigated whether Elk-En could compete with constitutively active Elk-1, SAP-1, and SAP-2 fusions to the VP16 transcriptional activation domain in regulating SRE-driven reporter constructs (Fig. 1E). Upon induction with PA, Elk-En was able to efficiently inhibit transcriptional activation by each of the TCF-VP16 fusion proteins.
The TCFs are known to activate the transcription of the immediate-early genes such as c-fos through SREs in response to a variety of stimuli, including epidermal growth factor (EGF). To verify that Elk-En represses transcription of endogenous SRE-regulated genes, we analyzed the effect of Elk-En expression on the putative target genes c-fos, egr-1, and TR3. EcR293(Elk-En)#1.3 cells were treated with PA and then stimulated with EGF. In the absence of PA, EGF leads to rapid induction of both c-fos and egr-1 expression followed by rapid shutoff. The kinetics of TR3 induction are somewhat slower (Fig. 1F, lanes 1 to 4). However, in the presence of PA, gene induction is severely inhibited in all three cases (Fig. 1F, lanes 5 and 6). Elk-1 and SRF can also interact in solution in vitro in the absence of DNA (52). We therefore tested whether Elk-1-independent SRF target genes such as vinculin (18) were repressed by Elk-En through protein-protein interactions. However, vinculin was not repressed by Elk-En either in the presence (Fig. 1G) or the absence (data not shown) of serum. Thus, the induction of Elk-En in EcR293(Elk-En)#1.3 cells leads specifically to the repression of endogenous TCF target genes in addition to repressing TCF-responsive reporter constructs.
Induction of Elk-En causes growth arrest.
To study the effect of Elk-En expression on cell growth properties, we first monitored cell growth following administration of PA to EcR293(Elk-En)#1.3 cells. In the absence of PA, the cells showed a continual increase in numbers over 5 days. However, upon PA treatment, growth rapidly stopped and the numbers of cells began to decline shortly afterwards (Fig. 2A). To probe whether the reduced growth was due to a decrease in cell division or an increase in cell death, we first investigated the cell cycle profiles of EcR293(Elk-En)#1.3 cells in the absence and presence of PA. Very little difference in the overall distribution of cells in different phases of the cell cycle could be observed following PA treatment (Fig. 2B; compare upper and lower panels). However, one key difference was the appearance of a sub-G1 peak in the PA-treated cells that is indicative of the presence of apoptotic cells. Although no arrest and accumulation in a particular phase of the cell cycle was apparent, we also probed whether the cells were still proliferating and undergoing DNA synthesis. In the absence of PA treatment, a large proportion of EcR293(Elk-En)#1.3 cells continued synthesizing DNA (Fig. 2C, upper panels—also see Fig. 5E). However, in the presence of PA a severe reduction of cells undergoing DNA synthesis is apparent (Fig. 2C, lower panels-also see Fig. 5E). Thus, the induction of Elk-En expression blocks cellular proliferation and initiates cell cycle arrest irrespective of the position in the cell cycle.
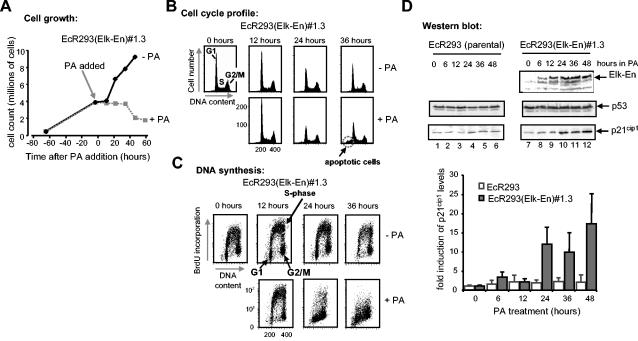
FIG. 2.
Induction of Elk-En causes growth arrest. (A) PA induction of Elk-En stops cell growth in EcR293(Elk-En) cells. EcR293(Elk-En)#1.3 cells were grown for 65 h to 30% confluency in 10% FBS and then treated with 5 μM PA for the indicated times. Cells remaining on the plate were trypsinized and were counted at the indicated times (averages of three counts are shown). (B and C) Cell cycle profile (B) and proliferation status (C) of EcR293(Elk-En)#1.3 cells in response to Elk-En induction. EcR293(Elk-En)#1.3 cells were grown for 24 h and then for the indicated times in the presence or absence of 5 μM PA. At the indicated time points, cells were harvested and the DNA content of the cells was determined by propidium iodide staining and FACS analysis (both axes represent linear scales) (B). Prior to harvesting, cells were incubated with BrdU for 1 h and the BrdU-dependent fluorescence was analyzed following FITC anti-BrdU staining. BrdU fluorescence was plotted on the y axis (logarithmic scale), with propidium iodide stain (DNA content) plotted on the x axis (linear scale) (C). Data are representative of two independent experiments. (D) Elk-En expression leads to induction of p21cip1 expression. EcR293 and EcR293(Elk-En)#1.3 cells were incubated in 10% FBS for 36 h, and 5 μM PA was added to the culture medium for a further 0 to 48 h. At the times indicated, extracts were assayed by Western blotting for the presence of Elk-En (top panel), p53 (middle panels), and p21cip1 (bottom panels). All blots are representative of at least two separate experiments. Data from the p21cip1 blots are shown quantitatively below each panel. Results plotted represent means plus standard deviations of severalfold enhancement compared to unstimulated-cell results for each cell line (data are averages of three separate experiments).
FIG. 5.
EcR393 cells expressing the Elk-En(L158P) mutant exhibit reduced growth arrest and apoptotic induction. (A) Western blot analysis of wild-type Elk-En and the Elk-En(L158P) following PA induction for 24 h in the EcR293-Elk-En cell lines #1.3 and L8, respectively. (B) PA induction of Elk-En does not stop cell growth in EcR293(Elk-En{L158P}) cells. EcR293(Elk-En{L158P})#L8 cells were grown and treated with PA as described for Fig. 2A. The data from the EcR293(Elk-En)#1.3 cell line results shown for Fig. 2A are shown in the background for comparison. (C) Cell cycle profile of EcR293(Elk-En{L158P})#L8 cells in response to Elk-En induction. EcR293(Elk-En{WT})#1.3 and EcR293(Elk-En{L158P})L8 cells were grown and treated with PA and analyzed by FACS as described for Fig. 2B. The apoptotic sub-G1 peak is highlighted. (D) Reduced levels of apoptotic cell death occur in EcR293-Elk-En(L158P)#L8 cells. Apoptotic cells were visualized by Hoechst staining following treatment of EcR293(Elk-En{WT})#1.3 and EcR293(Elk-En{L158P})L8 cells with PA as described for Fig. 3B. (E) The percentage of EcR293(Elk-En{WT})#1.3 and EcR293(Elk-En{L158P})L8 cells undergoing DNA synthesis following PA induction of Elk-En was analyzed by studying BrdU incorporation by FACS analysis (Fig. 2C). Results shown represent the averages for two dishes for each time point and are representative of at least two separate experiments. The dotted boxes highlight the differences during early time points between untreated and PA-treated cells.
Both p53 and p21cip1 are known to be involved in cell cycle inhibition (reviewed in references 13 and 32). We therefore probed whether either of these molecular markers was upregulated in response to Elk-En induction. p53 levels were invariant, irrespective of the presence or absence of PA. However, treatment of EcR293(Elk-En)#1.3 cells led to the accumulation of p21cip1 (Fig. 2D). The kinetics of p21cip1 induction were delayed in comparison to Elk-En expression, consistent with this being a downstream effect, and the onset of p21cip1 induction between 12 and 24 h correlated with the onset of growth arrest (Fig. 2C). Thus, Elk-En expression leads to cell cycle arrest, inhibition of proliferation, and the induction of the cell cycle inhibitor p21cip1.
Expression of Elk-En induces apoptosis.
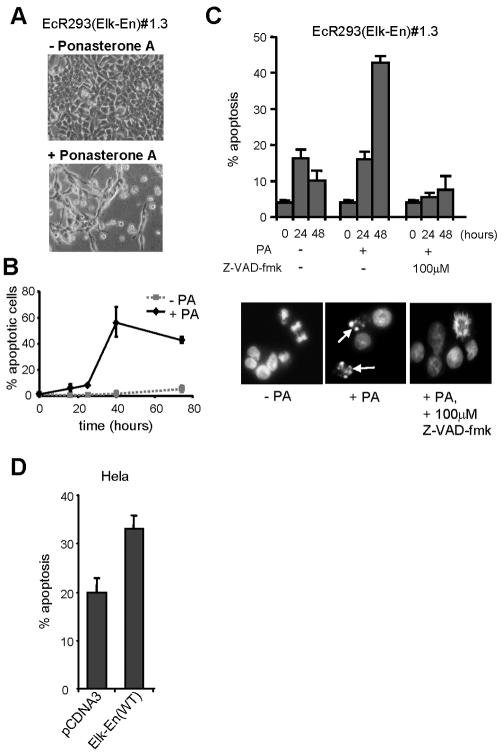
The reduction in cell numbers and the appearance of a sub-G1 peak following PA treatment of EcR293(Elk-En)#1.3 cells (Fig. 2) suggests that Elk-En expression induces apoptotic cell death in addition to causing growth arrest. Furthermore, following PA treatment, cultures of EcR293(Elk-En)#1.3 cells contain cellular debris and cells of altered morphology (Fig. 3A). To investigate whether cells were dying of apoptosis, we first used Hoechst staining to analyze cells to look for the altered nuclear morphology that is characteristic of apoptotic cells (Fig. 3B). In the absence of PA, a low basal level of apoptotic cells was observed. However, upon PA treatment, a huge increase in apoptosis was seen, consistent with a role for Elk-En in promoting apoptosis. Apoptotic cell death was also confirmed using a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assay (data not shown). To further characterize this phenomenon, we incubated cells with the Z-VAD-fmk caspase inhibitor and scored apoptotic cells following Hoechst staining. Treatment of EcR293(Elk-En)#1.3 cells with Z-VAD-fmk blocked the ability of PA to induce apoptosis (Fig. 3C). Thus, Elk-En expression leads to apoptotic cell death in a caspase-dependent manner.
FIG. 3.
Induction of Elk-En induces apoptosis. (A) EcR293(Elk-En)#1.3 cell results following incubation in 10% FBS for 36 h in the presence or absence of 5 μM PA. (B) EcR293(Elk-En)#1.3 cells were incubated in 10% FBS in the presence or absence of 5 μM PA, and samples were taken for Hoechst staining at the indicated times. Cells (3 × 100) were examined microscopically for healthy or apoptotic nuclei, and the percentage of apoptotic cells was calculated. Means ± standard deviations are plotted. (C) EcR293(Elk-En)#1.3 cells were treated and analyzed as described for panel B except that in one case, the caspase inhibitor Z-VAD-fmk (100 μM) was added at the same time as PA. Data shown are the averages of three cell counts taken from each of two separate slides and are representative of two individual experiments. Representative pictures of cells left untreated, those treated for 48 h with PA, and those treated with Z-VAD-fmk are shown. Apoptotic nuclei are indicated with arrows. (D) Elk-En stimulates apoptosis in HeLa cells. HeLa cells were cotransfected with GFP and Elk-En expression plasmids, and the percentage of apoptotic cells was counted following Hoechst staining. HeLa cells transfected with empty pCDNA3 vector are shown as a control. Data are representative of four independent experiments.
To investigate whether the apoptotic effect of Elk-En expression was specific to 293 cells or was a more general phenomenon, we also investigated the consequences of transiently expressing Elk-En fusions in HeLa cells. As observed with 293 cells, Elk-En expression resulted in apoptotic induction in HeLa cells (Fig. 3D). Similar results were obtained with other cell lines (IAK and ADS; data not shown). Thus, inhibition of TCF activity by Elk-En induction leads to apoptotic cell death in a number of different cell types.
An Elk-En(L158P) mutant is defective in SRE-mediated repression.
Elk-1 is thought to act primarily through the formation of ternary complexes with SRF on SREs (reviewed in references 46 and 50). However, it is possible that Elk-1 could act in combination with other transcription factors (e.g., Pax-5) (16, 40) or in an autonomous binding mode. In addition, it is possible that overexpression of Elk-En fusions could result in the repression of genes usually regulated by different ETS-domain transcription factors by virtue of their overlapping DNA binding specificities. We therefore wished to create a control system whereby we could differentiate between these possibilities.
Protein-protein interactions between the B-box regions of Elk-1 and SRF are essential for efficient ternary complex formation (23, 55), and the introduction of point mutations into this region inhibits Elk-1 recruitment to SREs in vitro and in vivo (Fig. 4B) (28). In contrast, binding to high-affinity ets motifs such as the E74 site is unaffected by mutations in the B-box. Thus, by the creation of mutant Elk-En fusion proteins that contain a defective B-box, it should be possible to differentiate between SRF-dependent (requiring intact B-box) and SRF-independent (no B-box requirement) effects of Elk-En.
A mutant version of Elk-En was created that contains a single mutation, L158P, in the B-box. This was previously shown to disrupt recruitment of truncated Elk-1 proteins via SRF (28) and probably does so by altering the trajectory of the B-box along the hydrophobic binding groove on SRF (Fig. 4A) (19). Gel retardation analysis demonstrates that the L158P mutation also disrupts ternary complex formation in the context of Elk-En fusion proteins. Only residual binding activity can be detected (Fig. 4C). In contrast, autonomous binding of Elk-En to the E74 site is unaffected by this mutation (Fig. 4D).
Stable cell lines that inducibly express Elk-En(L158P) (Fig. 1B) were created using an ecdysone-inducible system. One such line, EcR293(Elk-En{L158P}L8, was selected for further analysis, as it expressed amounts of Elk-En equivalent to those seen with EcR293(Elk-En)#1.3 cells (Fig. 5A). EcR293(Elk-En{L158P})L8 cells were initially functionally verified by comparison with EcR293(Elk-En)#1.3 cells in reporter gene assays (Fig. 4E to H). Reporter genes were activated by transfection with the constitutively active Elk-1 construct, Elk-VP16. While PA induction of wild-type Elk-En caused efficient repression of a SRE-Luc reporter (containing both SRF and ets binding sites), induction of Elk-En(L158P) promoted little repression. In contrast, both wild-type and mutant versions of Elk-En efficiently repressed an E74-Luc reporter (containing just ets motifs). Thus, as predicted by the DNA binding studies, Elk-En(L158P) is defective in SRE-mediated repression but retains its ability to repress transcription from autonomous binding sites. Therefore, the EcR293(Elk-En{L158P})L8 cells are an appropriate control system to differentiate between downstream effects of Elk-En that occur in an SRF-dependent and -independent manner.
Elk-En promotes growth arrest and apoptotic induction in a ternary complex-dependent manner.
We then wished to determine whether the growth arrest and enhanced apoptosis observed upon induction of Elk-En expression were due to the inhibition of ternary Elk-SRF-SRE complex-dependent functions of Elk-1. To do this, we compared the responses of EcR293(Elk-En)#1.3 cells (expressing wild-type Elk-En) and EcR293(Elk-En{L158P})L8 cells (expressing the L158P mutant form of Elk-En) to induction of Elk-En by PA. In contrast to the growth arrest and reduction in cell numbers observed with EcR293(Elk-En)#1.3 cells, PA treatment does not appear to affect the growth of EcR293(Elk-En{L158P})L8 cells, which continue to divide after addition of PA (Fig. 5B). Similarly, while PA treatment leads to the production of a large sub-G1 population in cells expressing wild-type Elk-En, a much smaller peak is observed with EcR293(Elk-En{L158P})L8 cells, suggesting reduced levels of apoptosis (Fig. 5C). This observation was confirmed by counting apoptotic cells after Hoechst staining. Treatment of EcR293(Elk-En)#1.3 cells with PA leads to substantial levels of apoptosis, whereas much reduced levels are observed with the EcR293(Elk-En{L158P})L8 cell line. This is most apparent at the 36-h time point, when 45% of the cells are apoptotic upon Elk-En(WT) induction but only 14% are apoptotic upon Elk-En(L158P) induction (Fig. 5D). Finally, we compared the numbers of proliferating cells in the two different cell lines. While a large decrease in proliferating cells is observed between 10 and 20 h of induction of wild-type Elk-En expression, no decreases in proliferative rates are observed in the EcR293(Elk-En{L158P})L8 cells until after 20 h. Thus, in comparison to the results seen with wild-type Elk-En, the induction of Elk-En(L158P) causes reduced growth arrest and proliferation and much-reduced levels of apoptosis.
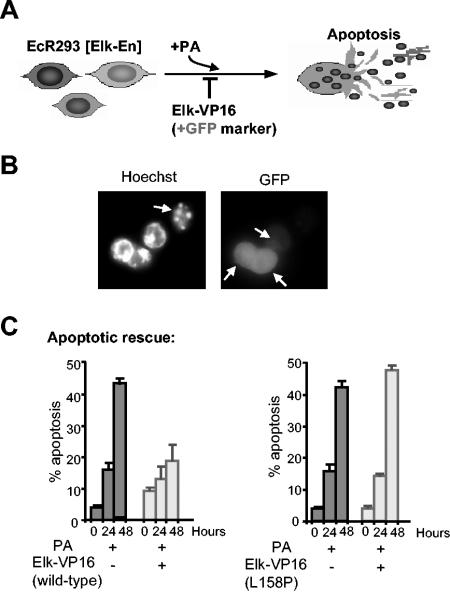
To further probe the ternary complex dependence of the Elk-En-mediated effects that we have observed and the sufficiency of Elk-1 to mediate these effects, we carried out rescue experiments with wild-type and L158P mutant versions of Elk-VP16 fusion proteins. These proteins function as constitutively active transcriptional activator proteins due to the presence of the potent VP16 activation domain and are predicted to oppose the repressive function of Elk-En fusion proteins. If TCF activity is specifically being altered by Elk-En expression, then Elk-VP16 should block apoptotic induction by Elk-En (Fig. 6A). In addition, if Elk-En is working through a ternary-complex-dependent manner, then Elk-VP16(L158P) should be unable to block apoptotic induction. EcR293(Elk-En)#1.3 cells were transfected with vectors encoding either wild-type Elk-VP16 or Elk-VP16(L158P) along with an EGFP marker and then stimulated with PA to induce Elk-En expression (Fig. 6A). The apoptotic cells were then scored by Hoechst staining, and transfected cells were observed by EGFP fluorescence. Cells transfected with Elk-VP16 appeared to be rescued from apoptotic death caused by Elk-En expression (Fig. 6B). Quantification of the data demonstrated that transfection of EcR293(Elk-En)#1.3 cells with Elk-VP16 resulted in almost complete rescue of cells from apoptosis (Fig. 6C). In contrast, no apoptotic rescue was observed upon transfection with Elk-VP16(L158P).
FIG. 6.
Elk-VP16 rescues cells from death induced by Elk-En. (A) Schematic of the experimental protocol. +GFP, EGFP. (B and C) Rescue of apoptosis by transfection of Elk-VP16 following induction of Elk-En with PA. EcR293(Elk-En)#1.3 cells were transfected with plasmids encoding EGFP (200 ng) and, where indicated, Elk-VP16 or Elk-VP16(L158P) (50 ng). At 24 h posttransfection (time 0), 5 μM PA was added where specified. Cells were analyzed by Hoechst staining at the indicated time points. Slides were examined for transfection efficiency by observing green fluorescing cells and for percentages of apoptosis by observing nuclear morphology. (B) Representative pictures are shown for cells transfected with 50 ng of Elk-VP16 and incubated with 5 μM PA for 48 h. The left panel shows Hoechst staining; the right panel shows GFP fluorescence. Arrows indicate apoptotic cells (Hoechst staining) and GFP-expressing cells (GFP fluorescence). (C) Data shown are the averages of three cell counts (percentages of total cell population) taken from each of two separate slides.
Collectively, these data demonstrate that Elk-En induces growth arrest and apoptosis in a ternary complex-dependent manner.
The antiapoptotic gene Mcl-1 is a key TCF-SRF target.
The data presented above indicate that the activity of genes regulated through ternary TCF-SRF complexes is required to promote proliferation and to block apoptotic cell death. To identify these target genes, we used Affymetrix microarray analysis to compare the mRNA expression profiles of unstimulated EcR293(Elk-En)#1.3 cells with those of the same cells stimulated with PA for 6 h to induce the expression of Elk-En. One of the most downregulated genes was Mcl-1 (ranked seventh; expression reduced to 56%). Northern analysis confirmed the microarray data, demonstrating a clear reduction in Mcl-1 mRNA levels following PA induction (Fig. 7A). Furthermore, in addition to inhibiting the basal levels of Mcl-1 expression, the induction of Elk-En blocks the increase in Mcl-1 expression caused by EGF stimulation (Fig. 7B). Mcl-1 has recently been shown to be a key antiapoptotic protein that acts at the top of the apoptotic cascade (36) and a key cellular target in adenoviral-mediated apoptosis (10), making it a likely target for Elk-En-mediated apoptosis. Moreover, a previous study demonstrated the functional importance of an SRE-like sequence in the Mcl-1 promoter (53).
To demonstrate a direct effect of Elk-En on Mcl-1 expression, we investigated the effect of Elk-En on the activity of a Mcl-1 promoter-driven luciferase reporter construct. Wild-type Elk-En caused efficient repression of the Mcl-1 promoter in a dose-dependent manner (Fig. 7C). However, in comparison, the mutant Elk-En(L158P) protein was much less efficient in repressing this promoter (Fig. 7C). Conversely, an Elk-VP16 fusion protein efficiently activated the Mcl-1 promoter (data not shown). To investigate whether SRF and Elk-1 could form ternary complexes on the Mcl-1 promoter in vitro, we carried out gel retardation assays on fragments containing this SRE. Both SRF and Elk-1 can bind this promoter independently (Fig. 7D, lanes 2 and 3). However, in the presence of SRF, Elk-1 is efficiently recruited into a ternary complex (Fig. 7D, lane 4). This is consistent with previous observations that Elk-1 can be found in complexes formed by nuclear extracts on this site (53). To verify these findings in vivo, we investigated whether endogenous Elk-1 could be found on the Mcl-1 promoter in vivo by chromatin IP analysis. Elk-1 was found to be associated with the promoters of both Mcl-1 and the known target gene Egr-1 (Fig. 7E, top two panels). No association with an SRF intronic fragment could be seen, demonstrating the specificity of binding (Fig. 7E, top two panels). Finally, we investigated whether Elk-En-mediated apoptosis could be rescued by transiently overexpressing Mcl-1. In comparison to cells transfected with empty expression vector, Mcl-1 expression was able to inhibit Elk-En-mediated apoptosis (Fig. 7F), thereby demonstrating the importance of this TCF target gene in protecting cells from apoptosis.
Together, these data therefore identify Mcl-1 as an important TCF target gene that protects cells from apoptosis in EcR293 cells.
DISCUSSION
The TCFs can be phosphorylated and activated in response to activation of all three of the major MAPK cascades in mammalian cells (reviewed in references 46 and 54). As such, they represent an important point of convergence for cell signaling pathways. Due to their association with target genes such as c-fos, it has been presumed that TCFs act to promote cell cycle entry and proliferation by activating the expression of this and other immediate-early genes. Here we demonstrate that the transcriptional activity of genes regulated by the ternary TCF-SRF-SRE complex is essential for cell growth and survival (Fig. 7G).
Immediate-early gene induction by TCFs.
To block TCF-mediated transcription, we created cell lines that contain an integrated inducible expression cassette that contains a gene encoding full-length Elk-1 fused to the En repression domain. Full-length Elk-1 was used to ensure that the correct interaction surfaces were present to permit promoter recruitment. The Drosophila En repression domain has been previously shown to convert other mammalian transcription factors into repressors (3). In reporter gene assays, the Elk-En fusion protein could inhibit SRE-dependent transcription and thereby block ERK MAPK signaling through these elements (Fig. 1D). This effect was SRE specific, as no effect was observed on other promoters that lack TCF binding motifs, including the simian virus 40 promoter contained in the pCH110 reporter vector (data not shown). Known TCF-dependent genes such as c-fos and egr-1 were shown to be repressed, and genes previously shown to be capable of forming ternary complexes in vitro such as TR3 were also identified as targets for Elk-1 in vivo (Fig. 1E). In contrast, the expression of GAPDH and the TCF-independent SRF target gene vinculin was unaffected. This is consistent with the observation that TCF targeting to the vinculin promoter is incapable of conferring TCF-specific signaling properties to SRF bound at this promoter (35). Microarray analysis identified a number of additional genes that were downregulated by Elk-En, including Mcl-1 (see below). Thus, the Elk-En fusion protein acts as a specific inhibitor of SRE-mediated transcription and can be used to probe the biological functions of Elk-1 and to identify novel target genes.
Inhibition of TCF-regulated gene activity results in growth arrest and apoptosis.
The expression of Elk-En in EcR293(Elk-En)#1.3 cells results in growth arrest and the induction of apoptosis. This is not a clonal effect, as five independent cell lines died of apoptosis upon induction of Elk-En expression (data not shown). Furthermore, this effect is not cell type specific, as apoptosis is also induced in both HeLa cells (Fig. 3D) and PC12 cells (data not shown). The onset of the block in cellular proliferation is between 12 and 24 h post-Elk-En induction (Fig. 2C). This correlates with the induction of the cell cycle inhibitor p21cip1 (Fig. 2D) and indicates that induction of p21cip1 expression is one of the likely molecular causes of growth arrest. However, p21cip1 is unlikely to be a direct target of the TCFs, due to the fact that Elk-En is a potent repressor protein and due to the delayed kinetics of p21cip1 expression. Examination of the cell cycle profiles of PA-induced EcR293(Elk-En)#1.3 cells does not indicate blockage at a particular phase of the cell cycle, although slight decreases in the G1 peak are apparent (Fig. 2B). This suggests that TCF-regulated gene activity potentially regulates both cell cycle entry and other later phases of the cell cycle.
p21cip1 has been proposed to be an inhibitor (reviewed in reference 13), and inducer (25, 31, 51) of apoptosis. In this study, p21cip1 might have contributed to apoptosis, as high levels of apoptosis are only observed at around 24 h post-Elk-En induction, when p21cip1 levels also increase. It is likely, however, that other molecular events contribute to apoptotic induction. Indeed, the expression of the key antiapoptotic Mcl-1 gene is downregulated in response to Elk-En induction (Fig. 7). Mcl-1 has recently been shown to be a key apoptotic regulator and to occupy a pivotal position at the top of the apoptotic pathway (36). Overexpression of Mcl-1 is able to rescue cells from Elk-En-induced apoptosis (Fig. 7F), demonstrating its importance in this process. Chromatin IP and reporter analysis demonstrate that this is a direct target gene (Fig. 7), as suggested by the results of promoter analysis carried out with K-562 cells in a previous study (53). Thus, the activity of the Mcl-1 gene appears to be a critical event for the blocking of cellular apoptosis in several different cell types. Indeed, our data are also consistent with the observation that Mcl-1 plays an essential role in the survival of U937 cells during PMA-induced differentiation (34).
Our results apparently differ from those of previous studies in which Rat1 and MCF7 cells that were stably transfected with Elk-1 or a splice variant, ΔElk-1, became more sensitive to apoptotic induction by calcium ionophore A23187 (45). However, it is not clear how direct the effect of Elk-1 is in these cell lines, as constitutively active promoters were used to drive Elk-1 expression, and the cells may have adapted to cope with this. Alternatively, the high levels of Elk-1 expressed in these lines might cause squelching of essential coregulators away from the endogenous proteins and hence loss of Elk-1 activity. Indeed, from our studies, such squelching would be predicted to enhance the apoptotic susceptibility.
TCFs regulate growth arrest and apoptosis in a ternary complex-dependent manner.
TCFs are thought to function mainly through SREs via forming ternary complexes with SRF. To investigate whether the biological effects we have observed are due to TCFs functioning in this manner, we used cell lines that express a mutant allele of Elk-En [Elk-En(L158P)] that cannot be recruited to SREs (Fig. 4). In contrast to the results seen with EcR293(Elk-En)#1.3 cells, EcR293(Elk-En{L158P})L8 cells exhibit reduced levels of apoptosis and growth arrest is not as apparent (Fig. 5). Again, this is not a clonal effect, as none of the independent cell lines that inducibly express Elk-En(L158P) underwent apoptosis at the levels observed with lines expressing wild-type Elk-En (data not shown). Furthermore, Elk-VP16(L158P) was incapable of rescuing cells from apoptosis whereas wild-type Elk-VP16 efficiently promoted cell survival. Interestingly, in addition to Elk-VP16, the reintroduction of wild-type Elk-1 can rescue cells from apoptosis, demonstrating that the presence of Elk-1 is sufficient for rescue (data not shown). Thus, activation of Elk-1 either by fusion of a potent activation domain or by endogenous signaling pathways is sufficient to overcome the block induced by repressive Elk-1 constructs, indicating a delicate balance in regulating cell death. Finally, a further advantage of using the mutant Elk-1 allele is that it controls for nonspecific effects such as squelching by the En repression domain. Thus, the use of the inhibitory Elk-En protein reveals a role for TCF-regulated gene activity in promoting proliferation and cell survival in a ternary complex-specific manner (Fig. 7G).
Despite the clear differences compared to the results seen with the EcR293(Elk-En)#1.3 cells, enhanced levels of apoptotic cell death and proliferative blockage are observed in the EcR293(Elk-En{L158P})L8 cells at later time points (Fig. 5). This could reflect the fact that residual levels of ternary complex formation can be observed with the Elk-1(L158P) mutant (Fig. 4C). At high expression levels, this would result in low-level inhibition of target genes; consequently, inhibitory thresholds would not be reached till a later time. Indeed, we observe that Elk-En(L158P) can also inhibit the expression of the Mcl-1 promoter, albeit to a lesser extent than that seen with the wild-type Elk-En (Fig. 7C), thereby providing a molecular explanation for these phenotypic observations. Alternatively, several different TCF-regulated pathways might be operative that guard against apoptosis. It is possible that an SRF-independent pathway becomes repressed at higher concentrations of Elk-En or with reduced kinetics. Again, this would lead to the toxicity that is revealed at the later time points. Such a possibility is suggested by the identification of tumor necrosis factor alpha (56), 9E3/cCAF (27), and α(1,3)-fucosyltransferase IV (59) as putative SRF-independent TCF target genes. Other SRF-independent target genes might exist that contribute to the biological properties of the TCFs. However, whatever the explanation, it is clear that we have identified a major ternary complex-dependent role for TCF-dependent gene regulation in promoting both proliferation and cell survival.
SRF was recently shown to play a critical role in protecting cells from apoptosis (4, 14). SRF is cleaved by caspases during apoptosis, and the reintroduction of a noncleavable SRF protein protects cells from apoptotic cell death. This indicates that SRF activity (and, by implication, ternary complex activity) is essential to block apoptosis and is consistent with our demonstration that inhibition of the activity of the TCFs, the partner proteins for SRF in the ternary complex, causes apoptotic cell death. A recent study has revealed that bcl-2, a gene related to Mcl-1 encoding an antiapoptotic protein, is an important target for SRF (43). However, this appears not to be regulated in a TCF-dependent manner, which is confirmed by the lack of significant changes in bcl-2 expression observed in our microarray experiments (data not shown). Thus, both TCF-dependent and -independent modes of apoptotic regulation are controlled through SRF.
In summary, our results suggest how mitogenic and survival pathways might trigger cell survival by utilizing the well-defined MAPK signaling route to the nucleus via the TCF component of the ternary complex (reviewed in reference 54). As the Ras-ERK MAPK pathway acts through the TCFs and has been implicated in regulating both proliferation and cell survival, it appears that the TCF-SRF complex is a key nuclear mediator of these processes. This has important implications for how Ras-ERK pathway signaling might lead to tumorigenesis, as blocking apoptosis is one of the prerequisites for tumor formation.
Acknowledgments
We thank Linda Shore and Anne Clancy for excellent technical support and Mike Jackson for help with the FACS analysis. We also thank Richard Treisman, Steve Edwards, and Xiaodong Wang for reagents, Caroline Dive for helpful advice, and Shen-Hsi Yang, Paul Shore, and Alan Whitmarsh for comments on the manuscript.
This work was funded by grants from the Wellcome Trust (WT), the AICR, and Cancer Research UK. E.R.V. was supported by a William Ross CRC studentship. A.K. was supported by the WT and Foundation for Polish Science Fellowship. A.D.S. was supported by a Research Fellowship from the Lister Institute of Preventive Medicine.
REFERENCES
- 1.Alexandropoulos, K., S. A. Qureshi, M. Rim, V. P. Sukhatme, and D. A. Foster. 1992. v-Fps-responsiveness in the Egr-1 promoter is mediated by serum response elements. Nucleic Acids Res. 20:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayadi, A., H. Zheng, P. Sobieszczuk, G. Buchwalter, P. Moerman, K. Alitalo, and B. Wasylyk. 2001. Net-targeted mutant mice develop a vascular phenotype and up-regulate egr-1. EMBO J. 20:5139-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badiani, P., P. Corbella, D. Kioussis, J. Marvel, and K. Weston. 1994. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 8:770-782. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotto, C., J. E. Ricci, F. Luciano, B. Mari, J. C. Chambard, and P. Auberger. 2000. Cleavage of the serum response factor during death receptor-induced apoptosis results in an inhibition of the c-FOS promoter transcriptional activity. J. Biol. Chem. 275:12941-12947. [DOI] [PubMed] [Google Scholar]
- 5.Cesari, F., S. Brecht, K. Vintersten, L. G. Vuong, M. Hofmann, K. Klingel, J. J. Schnorr, S. Arsenian, H. Schild, T. Herdegen, F. F. Wiebel, and A. Nordheim. 2004. Mice deficient for the Ets transcription factor Elk-1 show normal immune responses and mildly impaired neuronal gene activation. Mol. Cell. Biol. 24:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christy, B., and D. Nathans. 1989. Functional serum response elements upstream of the growth factor-inducible gene zif268. Mol. Cell. Biol. 9:4889-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarkson, R. W., C. A. Shang, L. K. Levitt, T. Howard, and M. J. Waters. 1999. Ternary complex factors Elk-1 and Sap-1a mediate growth hormone-induced transcription of egr-1 (early growth response factor-1) in 3T3-F442A preadipocytes. Mol. Endocrinol. 13:619-631. [DOI] [PubMed] [Google Scholar]
- 8.Costello, P. S., R. H. Nicolas, Y. Watanabe, I. Rosewell, and R. Treisman. 2004. Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nat. Immunol. 5:289-298. [DOI] [PubMed] [Google Scholar]
- 9.Criqui-Filipe, P., C. Ducret, S. M. Maira, and B. Wasylyk. 1999. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 18:3392-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuconati, A., C. Mukherjee, D. Perez, and E. White. 2003. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 17:2922-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton, S., and R. Treisman. 1992. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell 68:597-612. [DOI] [PubMed] [Google Scholar]
- 12.Datta, R., E. Rubin, V. Sukhatme, S. Qureshi, D. Hallahan, R. R. Weichselbaum, and D. W. Kufe. 1992. Ionizing radiation activates transcription of the EGR1 gene via CArG elements. Proc. Natl. Acad. Sci. USA 89:10149-10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dotto, G. P. 2000. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim. Biophys. Acta 1471:M43-M56. [DOI] [PubMed] [Google Scholar]
- 14.Drewett, V., A. Devitt, J. Saxton, N. Portman, P. Greaney, N. E. Cheong, T. F. Alnemri, E. Alnemri, and P. E. Shaw. 2001. Serum response factor cleavage by caspases 3 and 7 linked to apoptosis in human BJAB cells. J. Biol. Chem. 276:33444-33451. [DOI] [PubMed] [Google Scholar]
- 15.Ducret, C., S. M. Maira, Y. Lutz, and B. Wasylyk. 2000. The ternary complex factor Net contains two distinct elements that mediate different responses to MAP kinase signalling cascades. Oncogene 19:5063-5072. [DOI] [PubMed] [Google Scholar]
- 16.Fitzsimmons, D., W. Hodsdon, W. Wheat, S. M. Maira, B. Wasylyk, and J. Hagman. 1996. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 10:2198-2211. [DOI] [PubMed] [Google Scholar]
- 17.Gille, H., Sharrocks, A. D., and Shaw, P. E. 1992. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature 358:414-417. [DOI] [PubMed] [Google Scholar]
- 18.Gineitis, D., and Treisman, R. 2001. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276:24531-24539. [DOI] [PubMed] [Google Scholar]
- 19.Hassler, M., and T. J. Richmond. 2001. The B-box dominates SAP-1-SRF interactions in the structure of the ternary complex. EMBO J. 20:3018-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera, R. E., P. E. Shaw, and A. Nordheim. 1989. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature 340:68-70. [DOI] [PubMed] [Google Scholar]
- 21.Hill, C. S., R. Marais, S. John, J. Wynne, S. Dalton, and R. Treisman. 1993. Functional analysis of a growth factor-responsive transcription factor complex. Cell 73:395-406. [DOI] [PubMed] [Google Scholar]
- 22.Hipskind, R. A., V. N. Rao, C. G. Mueller, E. S. Reddy, and A. Nordheim. 1991. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature 354:531-534. [DOI] [PubMed] [Google Scholar]
- 23.Janknecht, R., and A. Nordheim, A. 1992. Elk-1 protein domains required for direct and SRF-assisted DNA-binding. Nucleic Acids Res. 20:3317-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaynes, J. B., and P. H. O'Farrell. 1991. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 10:1427-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang, K. H., W. H. Kim, and K. H. Choi. 1999. p21 promotes ceramide-induced apoptosis and antagonizes the antideath effect of Bcl-2 in human hepatocarcinoma cells. Exp. Cell Res. 253:403-412. [DOI] [PubMed] [Google Scholar]
- 26.Latinkic, B. V., M. Zeremski, and L. F. Lau. 1996. Elk-1 can recruit SRF to form a ternary complex upon the serum response element. Nucleic Acids Res. 24:1345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Q. J., S. Vaingankar, F. M. Sladek, and M. Martins-Green. 2000. Novel nuclear target for thrombin: activation of the Elk1 transcription factor leads to chemokine gene expression. Blood 96:3696-3706. [PubMed] [Google Scholar]
- 28.Ling, Y., J. H. Lakey, E. C. Roberts, and A. D. Sharrocks. 1997. Molecular characterization of the B-box protein-protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J. 16:2431-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maira, S. M., J. M. Wurtz, and B. Wasylyk. 1996. Net (ERP/SAP2), one of the Ras-inducible TCFs, has a novel inhibitory domain with resemblance to the helix-loop-helix motif. EMBO J. 15:5849-5865. [PMC free article] [PubMed] [Google Scholar]
- 30.Mansour, S. J., Matten, W. T., Hermann, A. S. Candia, J. M. Rong, S. Fukasawa, K. Vande, G. F. Woude, and N. G. Ahn. 1994. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265:966-970. [DOI] [PubMed] [Google Scholar]
- 31.Matsushita, H., R. Morishita, I. Kida, M. Aoki, S. Hayashi, N. Tomita, K. Yamamoto, A. Moriguchi, A. Noda, Y. Kaneda, J. Higaki, and T. Ogihara. 1998. Inhibition of growth of human vascular smooth muscle cells by overexpression of p21 gene through induction of apoptosis. Hypertension 31:493-498. [DOI] [PubMed] [Google Scholar]
- 32.May, P., and E. May. 1999. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 18:7621-7636. [DOI] [PubMed] [Google Scholar]
- 33.Mora-Garcia, P., and K. M. Sakamoto. 2000. Granulocyte colony-stimulating factor induces Egr-1 up-regulation through interaction of serum response element-binding proteins. J. Biol. Chem. 275:22418-22426. [DOI] [PubMed] [Google Scholar]
- 34.Moulding, D. A., R. V. Giles, D. G. Spiller, M. R. White, D. M. Tidd, and S. W. Edwards. 2000. Apoptosis is rapidly triggered by antisense depletion of MCL-1 in differentiating U937 cells. Blood 96:1756-1763. [PubMed] [Google Scholar]
- 35.Murai, K., and R. Treisman. 2002. Interaction of serum response factor (SRF) with the Elk-1 B box inhibits RhoA-actin signaling to SRF and potentiates transcriptional activation by Elk-1. Mol. Cell. Biol. 22:7083-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nijhawan, D., M. Fang, E. Traer, Q. Zhong, W. Gao, F. Du, and X. Wang. 2003. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 17:1475-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, M. A., A. E. Rogers, and R. Treisman. 1995. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET). EMBO J. 14:2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qureshi, S. A., X. M. Cao, V. P. Sukhatme, and D. A. Foster. 1991. v-Src activates mitogen-responsive transcription factor Egr-1 via serum response elements. J. Biol. Chem. 266:10802-10806. [PubMed] [Google Scholar]
- 39.Rim, M., S. A. Qureshi, D. Gius, J. Nho, V. P. Sukhatme, and D. A. Foster. 1992. Evidence that activation of the Egr-1 promoter by v-Raf involves serum response elements. Oncogene 7:2065-2068. [PubMed] [Google Scholar]
- 40.Roberts, E. C., R. W. Deed, J. D. Norton, and A. D. Sharrocks. 2001. Id helix-loop-helix proteins antagonize the activity of Pax transcription factors by inhibiting DNA binding. Mol. Cell. Biol. 21:524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 42.Schratt, G., B. Weinhold, A. S. Lundberg, S. Schuck, J. Berger, H. Schwarz, R. Weinberg, U. Rüther, and A. Nordheim. 2001. Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Mol. Cell. Biol. 21:2933-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schratt, G., U. Philippar, D. Hockemeyer, H. Schwarz, S. Alberti, and A. Nordheim. 2004. SRF regulates Bcl-2 expression and promotes cell survival during murine embryonic development. EMBO J. 23:1834-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seth, A., F. A. Gonzalez, S. Gupta, D. L. Raden, and R. J. Davis. 1992. Signal transduction within the nucleus by mitogen-activated protein kinase. J. Biol. Chem. 267:24796-24804. [PubMed] [Google Scholar]
- 45.Shao, N., T. Chai, J. Q. Cui, N. Wang, K. Aysola, E. S. Reddy, and V. N. Rao. 1998. Induction of apoptosis by Elk-1 and deltaElk-1 proteins. Oncogene 17:527-532. [DOI] [PubMed] [Google Scholar]
- 46.Sharrocks, A. D. 2002. Complexities in ETS-domain transcription factor function and regulation; lessons from the TCF subfamily. Biochem. Soc. Trans. 30:1-9. [DOI] [PubMed] [Google Scholar]
- 47.Sharrocks, A. D., F. von Hesler, and P. E. Shaw. 1993. The identification of elements determining the different DNA binding specificities of the MADS box proteins p67SRF and RSRFC4. Nucleic Acids Res. 21:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharrocks, A. D., H. Gille, and P. E. Shaw. 1993. Identification of amino acids essential for DNA binding and dimerization in p67SRF: implications for a novel DNA-binding motif. Mol. Cell. Biol. 13:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharrocks, A. D., A. Galanis, and S.-H. Yang. 2000. Docking domains and substrate specificity determination for the MAP kinases. Trends Biochem. Sci. 24:448-453. [DOI] [PubMed] [Google Scholar]
- 50.Shaw, P. E., and J. Saxton. 2003. Ternary complex factors: prime nuclear targets for mitogen-activated protein kinases. Int. J. Biochem. Cell Biol. 35:1210-1226. [DOI] [PubMed] [Google Scholar]
- 51.Sheikh, M. S., M. Garcia, Q. Zhan, Y. Liu and A.J. Fornace, Jr. 1996. Cell cycle-independent regulation of p21Waf1/Cip1 and retinoblastoma protein during okadaic acid-induced apoptosis is coupled with induction of Bax protein in human breast carcinoma cells. Cell Growth Differ. 7:1599-1607. [PubMed] [Google Scholar]
- 52.Shore, P., and A. D. Sharrocks. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14:3283-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Townsend, K. J., P. Zhou, L. Qian, C. K. Bieszczad, C. H. Lowrey, A. Yen, and R. W. Craig. 1999. Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J. Biol. Chem. 274:1801-1813. [DOI] [PubMed] [Google Scholar]
- 54.Treisman, R. 1996. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 8:205-215. [DOI] [PubMed] [Google Scholar]
- 55.Treisman, R., R. Marais, and J. Wynne. 1992. Spatial flexibility in complexes between SRF and its accessory proteins. EMBO J. 11:4631-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai, E., J. V. Falvo, A. V. Tsytsykova, A. K. Barczak, A. M. Reimold, L. H. Glimcher, M. J. Fenton, D. C. Gordon, I. F. Dunn, and A. E. Goldfeld. 2000. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 20:6084-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vickers, E. R., and A. D. Sharrocks. 2002. The use of inducible engrailed fusion proteins to study the cellular functions of eukaryotic transcription factors. Methods 26:270-280. [DOI] [PubMed] [Google Scholar]
- 58.Wasylyk, B., J. Hagman, and A. Gutierrez-Hartmann. 1998. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signalling pathway. Trends Biochem. Sci. 23:213-216. [DOI] [PubMed] [Google Scholar]
- 59.Withers, D. A., and S. Hakomori. 2000. Human alpha (1,3)-fucosyltransferase IV (FUTIV) gene expression is regulated by elk-1 in the U937 cell line. J. Biol. Chem. 275:40588-40593. [DOI] [PubMed] [Google Scholar]
- 60.Yang, S.-H., P. R. Yates, A. J. Whitmarsh, R. J. Davis, and A. D. Sharrocks. 1998. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol. Cell. Biol. 18:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, S. H., P. Shore, N. Willingham, J. H. Lakey, and A. D. Sharrocks. 1999. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 18:5666-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]