Abstract
Multivesicular body morphology and size are controlled in part by PtdIns(3,5)P2, produced in mammalian cells by PIKfyve-directed phosphorylation of PtdIns(3)P. Here we identify human Vac14 (hVac14), an evolutionarily conserved protein, present in all eukaryotes but studied principally in yeast thus far, as a novel positive regulator of PIKfyve enzymatic activity. In mammalian cells and tissues, Vac14 is a low-abundance 82-kDa protein, but its endogenous levels could be up-regulated upon ectopic expression of hVac14. PIKfyve and hVac14 largely cofractionated, populated similar intracellular locales, and physically associated. A small-interfering RNA-directed gene-silencing approach to selectively eliminate endogenous hVac14 rendered HEK293 cells susceptible to morphological alterations similar to those observed upon expression of PIKfyve mutants deficient in PtdIns(3,5)P2 production. Largely decreased in vitro PIKfyve kinase activity and unaltered PIKfyve protein levels were detected under these conditions. Conversely, ectopic expression of hVac14 increased the intrinsic PIKfyve lipid kinase activity. Concordantly, intracellular PtdIns(3)P-to-PtdIns(3,5)P2 conversion was perturbed by hVac14 depletion and was elevated upon ectopic expression of hVac14. These data demonstrate a major role of the PIKfyve-associated hVac14 protein in activating PIKfyve and thereby regulating PtdIns(3,5)P2 synthesis and endomembrane homeostasis in mammalian cells.
PIKfyve synthesizes phosphatidylinositol (3,5)P2 [PtdIns(3,5)P2] and PtdIns(5)P in mammalian cells (7, 17). Although no direct information is presently available, the PtdIns(3,5)P2 and PtdIns(5)P pools within the cell periphery (i.e., outside the cell nucleus) are likely restricted to the membranes of the late endocytic structures where a subfraction of PIKfyve enzyme resides (23). While PtdIns(5)P's role at these locations is presently unknown, functional studies with PIKfyve point mutants deficient in the PtdIns(3,5)P2-generating activity (7, 9) support a critical role for PtdIns(3,5)P2 in these structures. Thus, ectopic expression of these mutants in different mammalian cell types induces a dramatic dominant-negative effect in the form of dilated PIKfyve-positive vesicles along with a progressive accumulation of large cytoplasmic vacuoles of endocytic origin, with all defects being restored upon cytoplasmic microinjection of PtdIns(3,5)P2 but not PtdIns(5)P (7, 9). Ultrastuctural studies identified the dilated endocytic compartments as multivesicular bodies (MVBs), which, in addition to a significant gain of limiting membranes, display a lower number of intralumenal vesicles and membrane whorls (10). These data indicate that in mammalian cells the PtdIns(3,5)P2 pool regulates the morphogenesis and function of MVBs. Upstream regulators of PIKfyve activity and phosphoinositide (PI) product generation in mammals are presently unknown.
PIKfyve belongs to an evolutionarily ancient gene family with structurally related members that are present as single-copy genes in all eukaryotes with sequenced genomes (20). Fab1, the yeast ortholog of PIKfyve, controls PtdIns(3,5)P2 synthesis in Saccharomyces cerevisiae (5, 14). The most prominent phenotype resulting from FAB1 inactivation in S. cerevisiae includes a grossly enlarged and poorly acidified vacuole concomitant with a depleted PtdIns(3,5)P2 pool (5, 14). Because identical phenotypes, along with a depleted PtdIns(3,5)P2 pool, are manifested by deletion of VAC7 and VAC14 genes in S. cerevisiae, Fab1, Vac7, and Vac14 likely function in the same pathway (1, 2, 4, 6). The role of Vac14 as an upstream activator of Fab1 is further supported by data demonstrating that overexpression of FAB1 suppresses vacuolar defects in vac14Δ and restores steady-state levels of PtdIns(3,5)P2 (2, 4). Likewise, expression of the fab1 mutant allele that bypasses the requirement for Vac7 suppresses the vacuolar morphology defects and restores PtdIns(3,5)P2 synthesis in vac7Δ cells (6). The comparable phenotypic changes in yeast and mammalian cells due to perturbed PIKfyve/Fab1-directed PtdIns(3,5)P2 production together with the structural similarity between these orthologs suggest a common regulation of these enzymes. Whereas Vac7 has no structural homologue in any database, homologues of yeast Vac14 have been found in the genome of all eukaryotes sequenced to date. With the premise that Vac14 could serve as a PIKfyve regulator, we have obtained the cDNA of human Vac14 (hVac14). Here we report characterization of mammalian Vac14 protein and its identification as a bona fide upstream activator of PIKfyve activity.
MATERIALS AND METHODS
hVac14 antibody production and other antibodies.
Clone MGC-984, carrying the C-terminal region of hVac14, was purchased from ATCC (GenBank accession number BC000536). The EcoRI-XhoI fragment (encompassing residues 523 to 782 of the hVac14 sequence; accession number AK056433) was ligated with a corresponding digestion of pGEX5X-3 in frame with glutathione S-transferase (GST). Rabbit polyclonal anti-hVac14 antiserum (Covance, Denver, Pa.) was directed against the purified GST-Vac14 (amino acids 523 to 782) peptide fragment. Unless otherwise stated, the crude antiserum (WS047) was used for immunoprecipitation and the affinity-purified antibody (purified on the GST-Vac14 [523 to 782] peptide as described previously [22]) was used for Western blotting. Polyclonal anti-PIKfyve (R7069 and R7096, directed against the N and C termini, respectively) and antihemagglutinin (anti-HA) antibodies (R4289; a gift of Mike Czech) were described elsewhere (16, 24). Polyclonal antibodies against Rab4, IRAP, and cation-independent mannose 6-phosphate receptor (CI-MPR) were gifts of I. Mellman, P. Pilch, and B. Hoflack, respectively. Anti-green fluorescent protein (GFP) and anti-myc monoclonal antibodies were from Clontech and ATCC, respectively.
Vac14 and other constructs.
The cDNA clone (AK056433) representing full-length hVac14 was purchased from DOB and NBRC of NITE, Kisarazu-shi, Chiba, Japan. The 5′ untranslated region was eliminated by PCR. A PCR fragment was generated from nucleotide 269 (the initial Met) to the internal SmaI site at nucleotide 641 (sense primer, 5′-GCCGTCTAGAAATGAACCCGGAGAAG, flanked by XbaI; antisense primer, 5′-GTATCCCGGGCCACCTTGACGATG, flanked by SmaI) with the AK056433 clone as a template. pEGFP-HA-hVac14 was generated in a triple ligation consisting of the following fragments: the XbaI/SmaI-digested PCR product; SmaI/SalI of a pCMV5 construct, in which the SmaI/XbaI fragment of the AK056433 clone was subcloned; and XbaI/SalI of pEGFP-C2, engineered with the HA tag sequence next to and in frame with GFP, as detailed elsewhere (23). pCMV5-HA-hVac14 was generated by ligating the XbaI fragment from pEGFP-HA-hVAc14 into the XbaI-digested pCMV5 engineered with the HA tag sequence in frame with hVac14. The PCR-amplified fragment and the correct orientation of all constructs were verified by sequencing and restriction endonuclease mapping. (His)6-hVac14 was generated in pRSETb (Invitrogen) by subcloning XhoI-KpnI (from pEGFP-HA-hVAc14) and KpnI-HindIII fragments (from pBluescript II SK+ hVAc14) into XhoI-HindIII-digested pRSETb. The protein was produced in an Escherichia coli strain (BL21DE3) and was purified as described previously (24).
Myc-tagged wild-type pCMV5-PIKfyve (pCMV5-PIKfyveWT) or pCMV5-PIKfyveK1831E was generated by ligating the XbaI-SalI fragments from pCMV5-HA-PIKfyveWT or pCMV5-PIKfyveK1831E, respectively (24), and a double-stranded oligonucleotide flanked with EcoRI and XbaI sites, encoding the 12-amino-acid epitope of human c-myc oncogene product (EQKLISEEDLLR), into EcoRI-SalI-digested pCMV5. Expression of Myc-tagged proteins was confirmed by Western blotting.
Cell cultures and tissues.
HEK293, stable HEK293 (TetOn) inducibly expressing PIKfyveWT, COS-7, PC12, HIRcB, Jurkat, and CHO-T cells or 3T3-L1 fibroblasts were cultured under conditions described in previous studies (7-10, 17, 24). Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0] containing 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate) supplemented with 1× protease inhibitor cocktail (1 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml, 5 μg of aprotinin/ml, 1 μg of pepstatin/ml, and 1 mM benzamidine) and 1× phosphatase inhibitor cocktail (25 mM β-glycerophosphate, 10 mM sodium pyrophosphate, 50 mM NaF, and 2 mM NaVO3) or were homogenized for fractionation (see below). Tissues dissected from female or male mice were first homogenized in HES++ buffer (20 mM HEPES-NaOH [pH 7.5], 1 mM EDTA, and 255 mM sucrose, supplemented with 1× protease and 1× phosphatase inhibitor cocktails) and then lysed in RIPA buffer.
vac14 siRNA and cell transfection.
Smart pool human vac14 small interfering RNA (siRNA) and cyclophilin A siRNA duplexes were designed and synthesized by Dharmacon on a fee-for-service basis. HEK293 cells were transiently transfected with siRNA duplexes (100 nM) with Lipofectamine 2000 (Invitrogen) as a transfection reagent. Experiments were performed 72 h posttransfection. HEK293 or COS-7 cells were transfected with the indicated cDNAs by Lipofectamine 2000 (for biochemical assays) or Lipofectamine (for immunofluorescence) and were used 24 h posttransfection.
Confocal and light microscopy.
For confocal microscopy, COS-7 cells grown on coverslips were transfected with the constructs indicated in the respective figure legend. Twenty four hours following transfections, cells were washed, fixed in 4% formaldehyde, permeabilized, rewashed, and stained with monoclonal anti-myc or polyclonal anti-CI-MPR antibodies as described elsewhere (10, 23). Detection was achieved with Texas red-coupled goat anti-mouse immunoglobulin G (IgG) (Molecular Probes) or CY3-coupled goat anti-rabbit IgG (Kirkegaard & Perry Laboratories). Coverslips were mounted on slides by using a Slow Fade Antifade kit (Molecular Probes). Slides were observed and photographed with an LSM 510 confocal light microscope (Carl Zeiss, Thernwood, N.Y.) at the John D. Dingell Veterans Affairs Medical Center (Detroit, Mich.) with a 40× 1.2-numeric-aperture planapochromatic water immersion objective lens. The confocal microscope was operated with pinhole settings between 1.1 and 1.4 Airy units in each of the fluorescence channels. Images were recorded at 2,048 by 2,048 pixels at 12 bits/channel.
Light microscopy of live HEK293 cell lines was performed with a digital imaging fluorescence microscope (Nikon Eclipse TE200) using a 40× Hoffman Modulation Contrast objective. Images were captured by a SPOT RT Slider charge-coupled device camera (Diagnostic Instruments) and were processed by SPOT 3.2 software.
Subcellular fractionation and equilibrium centrifugation in iodixanol gradient.
Transiently transfected HEK293 cells or HEK293 stable line cells induced to express PIKfyveWT (clone 9) were homogenized in HES++ buffer at 4°C and were fractionated into total membranes and cytosols as described previously (8). For equilibrium centrifugation, total membrane fractions resuspended in HES++ buffer were mixed with iodixanol (OptiPrep; Sigma) in a Quick-Seal centrifuge tube to 30% iodixanol and 128 mM sucrose concentrations. A self-generating gradient was performed by centrifugation to equilibrium for 4 h as specified previously (8). Fractions (∼0.37 ml) collected from the bottom of the tube were analyzed for protein concentration and by immunoblotting.
Immunoblotting, immunoprecipitation, and PIKfyve kinase activity.
Immunoblotting with the indicated antibodies was performed subsequent to protein separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransfer onto nitrocellulose membranes as previously described (7-9). A chemiluminescence kit (Pierce) was used to detect horseradish peroxidase-bound secondary antibodies. Endogenous PIKfyve and Vac14 or their HA-tagged forms were immunoprecipitated from RIPA lysates (supplemented with 1× protease and 1× phosphatase inhibitor cocktails) of tissues and cells by using anti-PIKfyve, anti-Vac14, or anti-HA antibodies. Control immunoprecipitates with preimmune sera were run in parallel. Immunoprecipitations were carried out for 16 h at 4°C, with protein A-Sepharose CL-4B added in the final 1.5 h of incubation. Immunoprecipitates were washed with RIPA buffer plus 1× protease inhibitor cocktail and then were processed by Western blotting. For the lipid kinase assay, washed immunoprecipitates were subjected to 15 min of incubation at 30°C in an assay buffer (25 mM HEPES [pH 7.4], 120 mM NaCl, 2.5 mM MgCl2, 2.5 mM MnCl2, 5 mM β-glycerophosphate, and 1 mM dithiothreitol) supplemented with 50 μM ATP, [γ-32P]ATP (12.5 μCi), and 100 μM PtdIns (from soybean; Avanti Polar Lipids, Inc). Lipids were extracted and analyzed by thin-layer chromatography with the acidic solvent system (acetic acid-propanol), except for experiments presented in the supplemental figures, where a chloroform-methanol-water-ammonia solvent system was used as described previously (16). The presence of MnCl2 (2.5 mM) and relatively low concentrations of MgCl2 (2.5 mM) during the kinase reaction completely eliminates PtdIns 3-P synthesis by PI 3-kinase activity that may present in PIKfyve immunoprecipitates. This precludes the contribution of PI 3-K activity to PtdIns(3,5)P2 production, as has been shown elsewhere (18) and is further detailed here (compare Fig. S1 and S2 in the supplemental data). PIKfyve displays a preference to use natural instead of synthetic PtdIns or PtdIns(3)P, even if the latter presents at a minute quantity, as has been previously shown and is detailed here (16, 21) (see Fig. S1 and S2 in the supplemental data). This is most likely due to an inadequate quality of the synthetic substrate preparations, i.e., presence of catalysts, unnatural lipid side chain, etc., to optimally support PIKfyve activity. The natural PtdIns substrate used in the in vitro assay contains PtdIns(3)P as a contaminant, and therefore both PtdIns(5)P and PtdIns(3,5)P2 could be synthesized at high levels and monitored simultaneously. Generated lipid products were detected by autoradiography. Protein or lipid levels were quantified from the intensity of the bands or spots with a laser densitometer (Molecular Dynamics) by volume integration. Several films of different exposure times were quantified to assure that the signals were within the linear range of the film.
[32P]orthophosphate labeling, lipid extraction, and HPLC.
HEK293 cells (transfected with vac14 or control cyclophilin A siRNA duplexes) or the stable HEK293-PIKfyveWT cell line (transiently transfected with pEGFP-HA-hVac14) was labeled in phosphate- and serum-free Dulbecco's modified Eagle medium (DMEM) for 2.5 h at 37°C with [32P]orthophosphate as described previously (7, 17). Cells were washed in the presence of 1× phosphatase inhibitors and were scraped with CH3OH-1 M HCl (1:1) in the presence of 5 mM EDTA and 5 mM tetrabutylammonium hydrogen sulfate. Extracted radiolabeled lipids were deacylated and analyzed by high-performance liquid chromatography (HPLC) on a Whatman 5-μ Partisphere SAX (H2PO4−) column eluted with a shallow ammonium phosphate gradient as detailed elsewhere (7, 16, 17). Radioactivity was analyzed with an online Radiomatic 525TR flow scintillation analyzer (Packard). (Glycerophosphorylinositol ([3H]GroPIns) 4-P, [3H]GroPIns 4,5-P2, and [3H]GroPIns 3-P deacylated from [3H]PtdIns(4)P (NEN), [3H]PtdIns(4,5)P2 (NEN), and [3H]PtdIns(3)P, respectively, were coinjected as internal HPLC standards. [32P]GroPIns 3,5-P2 and [32P]GroPIns 3,4-P2 external standards were deacylated from [32P]PtdIns(3,5)P2 and [32P]PtdIns(3,4)P2, which were enzymatically synthesized with PIKfyve and PI 3-kinase as described previously (16, 17). FLO-ONE radiochromatography software (Packard) was used for data evaluation and documentation. Individual peak radioactivity was quantified by area integration and is presented as a percentage of the combined radioactivity from the 32P-labeled GroPIns 3-P, GroPIns 4-P, GroPIns 3,5-P2, GroPIns 3,4-P2, and GroPIns 4,5-P2 peaks (total radioactivity).
Quantitation of endomembrane vacuolation by neutral red.
Seventy-two hours posttransfection with the siRNA duplexes, HEK293 cells were treated with NH4Cl (10 mM) for 40 min at 37°C. Cells were then allowed to take up neutral red for 4 min as detailed previously (11). Following washings, neutral red was extracted from cells with acidified alcohol. The absorbance (540 nm) of the samples was measured with a spectrophotometer (Beckman DU-50). The net neutral red accumulation was calculated by subtracting the absorbance values for nontreated cells from those of NH4Cl-treated cells.
RESULTS
Identification of endogenous Vac14 in mammalian cells and tissues.
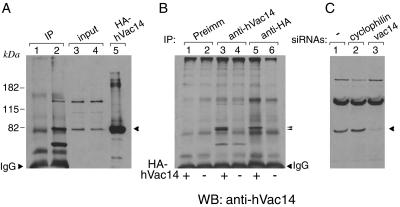
Although sequences homologous to that of yeast Vac14 have been identified in the genome of mammals, information about Vac14 protein distribution, abundance, and function in mammalian cells and tissues is unavailable. To address these questions we first generated anti-hVac14 antibodies. The electrophoretic mobility of immunoreactive endogenous Vac14 was assessed relative to the mobility of ectopically expressed HA-hVac14 in COS-7 cells. A single band that migrated in SDS-PAGE gels identically with the 82.5-kDa prestained protein standard (Invitrogen) was detected by anti-hVac14 antibodies in control or HA-hVac14-transfected COS-7 cells (Fig. 1A). Such a mobility is in the range of the predicted molecular weight of 87,972 for the deduced protein sequence of full-length hVac14 (782 amino acids). The authenticity of the 82-kDa band as endogenous mammalian Vac14 was confirmed by two additional experimental approaches. First, anti-hVac14 antiserum, but not the preimmune serum, immunoprecipitated the 82-kDa protein band from COS-7 cell lysates (Fig. 1A, lanes 1 and 2). A single band, slightly above the 82-kDa marker, was detected in anti-HA immunoprecipitates derived from lysates of HA-hVac14-transfected cells but not from nontransfected cells (Fig. 1B). As expected, the 82-kDa band appeared as a doublet if anti-hVac14 antibodies were used for immunoprecipitation, consistent with the subtle mobility differences between endogenous Vac14 and HA-hVac14 (Fig. 1B). Second, a selective and almost complete knockdown of the anti-hVac14-immunoreactive 82-kDa protein band was seen in lysates of HEK293 cells transfected with human vac14 siRNA duplexes but not with control siRNA duplexes derived from cyclophilin A (Fig. 1C). Thus, the 82-kDa protein band represents the authentic endogenous mammalian Vac14.
FIG. 1.
hVac14 electrophoretic mobility and protein knockdown by siRNA. (A) COS-7 cell lysates derived from two 100-mm confluent dishes/condition were immunoprecipitated with preimmune (lane 1) or anti-hVac14 sera (lane 2) as described in Materials and Methods. Immunoprecipitates were resolved by SDS-PAGE (6% acrylamide) along with the input (lanes 3 and 4) or lysates of HA-hVac14-expressing COS-7 cells as a molecular size marker (lane 5) and then were immunoblotted with anti-hVac14 antibodies. The arrowhead shows the endogenous Vac14 band determined by the position of HA-hVac14. (B) Lysates derived from COS-7 cells transiently transfected with pCMV5-HA-hVac14 (+) or pCMV5 alone (−) were immunoprecipitated with the indicated antibodies (equivalent to 1/3 of confluent 100-mm-diameter dish/condition). Immunoprecipitates were resolved by SDS-PAGE (6% acrylamide) and immunoblotted with anti-hVac14 antibodies. The two arrowheads point to a doublet in lane 3 consisting of immunoprecipitated HA-hVac14 and endogenous Vac14 that is increased under transfection. In nontransfected cells, only a faint band of endogenous Vac14 could be seen due to the small amount of protein subjected to immunoprecipitation (lane 4). (C) HEK293 cells were transfected with the indicated siRNA duplexes. Seventy-two hours posttransfection, cell lysates were resolved by SDS-PAGE (6% acrylamide) and immunoblotted with anti-hVac14 antibodies. The arrowhead depicts endogenous hVac14 and its selective elimination by vac14 siRNAs directed to the human sequence. In all three panels, shown are chemiluminescence detections of representative immunoblots out of four to five independent experiments with similar results. IP, immunoprecipitate; WB, Western blot; Preimm, preimmune.
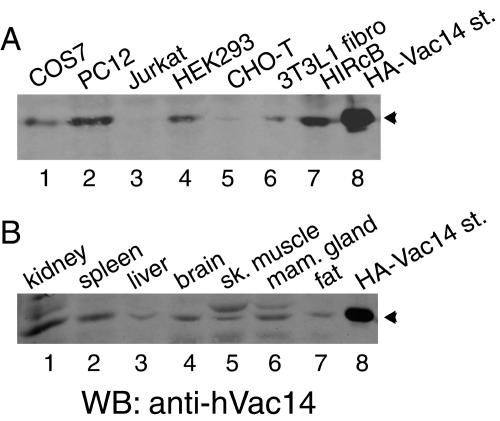
Consistent with Vac14 being an evolutionarily conserved protein present in all genome or expressed sequence tag databases from yeast to humans, we documented protein expression in several cultured mammalian cells, including COS-7, HEK293, PC12, HIRcB, and 3T3-L1 fibroblasts, but we did so only if substantial amounts of the proteins of the lysates were analyzed by Western blotting (Fig. 2A). In CHO-T and Jurkat cells, however, endogenous Vac14 was below the detectability of Western blotting (Fig. 2A). The protein was, however, detected in those cell types if it was concentrated by immunoprecipitation with anti-hVac14 antibodies prior to immunoblotting (data not shown). Quantitative Western blot analysis using increasing amounts of recombinantly produced (His)6-hVac14 estimated the hVac14 endogenous levels to be at 25 to 50 ng of the RIPA-soluble proteins from HEK293 cells per mg (data not shown). Vac14 was also present in all mouse tissues examined, with relatively higher levels in kidney, brain, mammary gland, and spleen and lower levels in liver, muscle, and fat (Fig. 2B). Thus, our data are consistent with the conclusion that mammalian Vac14 is a widespread but relatively low-abundance protein.
FIG. 2.
Vac14 is ubiquitously expressed in mammalian cells and tissues. Lysates derived from indicated cell lines (A, 200 μg of protein) or mouse tissues (B, 120 μg of protein) were analyzed by SDS-PAGE (6% acrylamide) and immunoblotting with anti-hVac14 antibodies. Shown are chemiluminescence detections of representative immunoblots out of two to three independent experiments for the indicated cell lines or tissues yielding similar results. st., standard; sk, skeletal; mam., mammary; WB, Western blot.
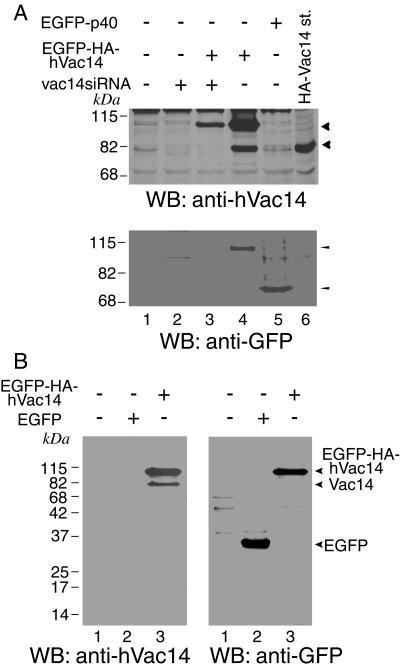
Very intriguing is our observation that, for reasons presently unknown, ectopic transfection with either pCMV5-HA-Vac14 (Fig. 1B, lane 3 versus lane 4) or pEGFP-HA-Vac14 (Fig. 3A, upper panel, lane 4 versus lane 1, and 3B, left panel, lane 3 versus lanes 1 and 2) in both COS-7 or HEK293 cells dramatically (more than eightfold) up-regulated endogenous Vac14 protein expression. The effect was specific, because expression of other enhanced green fluorescent protein (EGFP)-tagged (Fig. 3A, lane 5) and EGFP-HA-tagged proteins (data not shown) or EGFP protein alone (Fig. 3B, lane 2) had no effect on endogenous Vac14. The increment in the endogenous levels was completely eliminated if HEK293 cells were cotransfected with siRNA duplexes directed against human vac14 (Fig. 3A, lane 3). The possibility of a cleavage was ruled out by the lack of detectable peptide bands, other than EGFP-Vac14, specifically immunoreacting with the anti-GFP antibody (Fig. 3B, lane 3). These data suggest that ectopically expressed hVac14 positively feeds back on the endogenous protein expression under a mechanism that is presently unknown. Together, these results are consistent with the conclusion that in mammalian cells the steady-state expression of endogenous Vac14 protein is at relatively low levels but, under stimuli that remain to be identified and for reasons that we touch upon below, it could be dramatically up-regulated.
FIG. 3.
Ectopic expression of Vac14 triggers endogenous protein expression. (A) HEK293 cells first transfected with the indicated pEGFP constructs were cotransfected with the human vac14 siRNA duplexes 5 h later as indicated. Cell lysates, collected 72 h posttransfection, were resolved along with an HA-hVac14 molecular size marker by SDS-PAGE (6% acrylamide) and were immunoblotted with the indicated antibodies, with a stripping step in between. The arrowheads in the upper panel depict the mobility of expressed EGFP-hVac14 or endogenous hVac14 and their selective elimination by Vac14 siRNAs. The arrowheads in the lower panel depict equal expression of EGFP-hVac14 and control EGFP-p40, yet only the former increases endogenous hVac14 levels (upper panel). (B) COS-7 cells were transfected with the indicated pEGFP constructs. Cell lysates, collected 40 h posttransfection, were resolved by SDS-PAGE (12% acrylamide) and immunoblotted with the indicated antibodies, with a stripping step in between. WB, Western blot.
Vac14 cofractionates and colocalizes with PIKfyveWT.
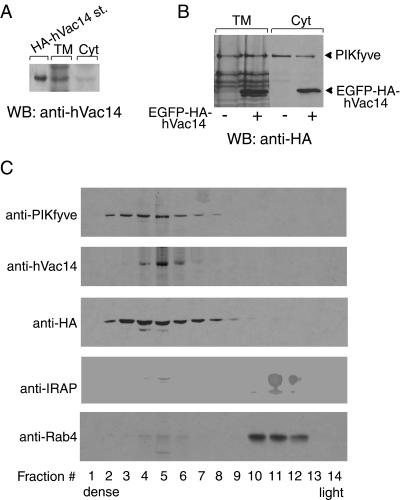
In contrast to yeast Vac14, no transmembrane domain is predicted for the mammalian orthologs by any available algorithm. Therefore, if it is membrane associated, mammalian Vac14 is expected to be peripherally bound. To gain insight into the intracellular distribution of endogenous hVac14, HEK293 cells were fractionated into cytosols and total membranes and were analyzed by immunoblotting. About one half of total hVac14 was detected in the membrane fraction (Fig. 4A). Ectopically expressed EGFP-HA-hVac14 in the HEK293-PIKfyveWT cell line paralleled the distribution of endogenous hVac14 and was equally partitioned between total membranes and cytosol, much like PIKfyveWT (Fig. 4B). To obtain further characteristics for the membrane-associated hVac14 structures in the context of PIKfyve membranes, we used a recently characterized equilibrium density gradient sedimentation of HEK293 cells stably expressing PIKfyveWT (8). Consistent with previous studies, we recovered immunoreactive PIKfyveWT in the denser bottom half of the 30% iodixanol gradient, whereas endosomal proteins IRAP and Rab4 were found essentially in the top, lighter fractions (Fig. 4C). Importantly, both endogenous and ectopically expressed hVac14 that was detected with anti-HA antibodies almost fully cofractionated with PIKfyveWT (Fig. 4C). Clearly, the results from the equilibrium sedimentation in density gradients are consistent with the notion that hVac14 and PIKfyve may reside at the same intracellular membranes.
FIG. 4.
Vac14 and PIKfyve partition similarly between cytosol and membranes and cofractionate in equilibrium gradient sedimentation. (A) HEK293 cells were fractionated into cytosol (Cyt) and total membranes (TM). Aliquots of the fractions along with an HA-hVac14 molecular size marker were resolved by SDS-PAGE and immunoblotted with anti-hVac14. The loading in TM versus Cyt is threefold more on the basis of cell number. Shown is a chemiluminescence detection of a representative blot out of three independent fractionations with similar results. (B and C) HEK293 stable cell line (clone 9) transiently transfected with pEGFP-HA-hVac14 and induced to express PIKfyveWT was fractionated into TM and Cyt. Both fractions were analyzed by immunoblotting with anti-HA to detect expressed EGFP-HA-hVAc14 or HA-PIKfyve as indicated (B). The loading in TM versus Cyt is twofold more on the basis of cell number (B). TM fractions derived from the stably induced and transiently transfected conditions were subjected to equilibrium sedimentation in 30% iodixanol, as described in Materials and Methods. (C) Fractions were collected from the bottom of the gradient. Aliquots were analyzed by SDS-PAGE (6% acrylamide for PIKfyve, hVac14, EGFP-HA-hVac14, and IRAP or 10.5% acrylamide for Rab4) and immunoblotting with the indicated antibodies, with a stripping step in between. The anti-HA blot depicts the sedimentation profile of expressed EGFP-HA-hVac14. Shown are chemiluminescence detections of blots from a representative experiment out of three independent fractionations yielding similar results. WB, Western blot.
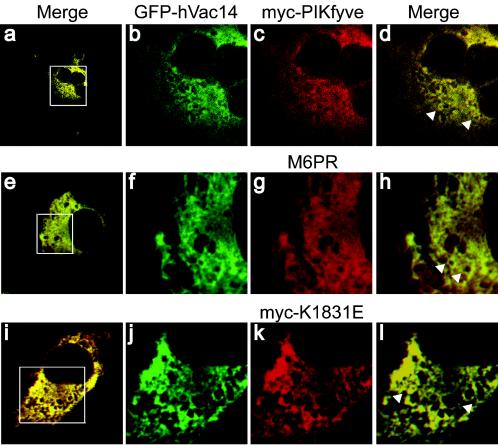
This important question was further addressed by confocal microscopy in COS-7 cells ectopically expressing pEGFP-HA-hVac14 and Myc-PIKfyveWT (or the dominant-negative mutant Myc-PIKfyveK1831E). It should be noted that the low levels of both endogenous PIKfyve and hVac14 in cells hamper the in situ detection of the authentic proteins. On the other hand, COS cells are convenient for fluorescence microscopy, first because of the large cell size and second because the localization of ectopically expressed PIKfyveWT most likely reflects that of the endogenous protein, as has been previously concluded based on data obtained in 3T3-L1 adipocytes (23). As shown in Fig. 5, colocalization of PIKfyveWT (or PIKfyveK1831E) and hVac14WT was markedly pronounced, particularly at the perinuclear region, as assessed on the merged images showing a substantial population of vesicles in yellow. However, distinct populations of single-positive green- or red-stained vesicles were also observed, indicating that although the two proteins largely overlap, a unique localization pattern is associated with each of them. Because PIKfyve partly resides on MVBs in transfected COS cells (23), we next examined possible localization of hVac14 to MVBs by staining for CI-MPR that localizes predominantly on membranes of late endosomes or MVBs in cells of kidney origin. As illustrated in Fig. 5e to h, the substantial overlap of the fluorescence staining for the CI-MPR marker and expressed GFP-hVac14 is consistent with the notion that a significant population of membrane-bound hVac14 localizes to late endosomes or MVBs. Because PIKfyve is one of the proteins that maintain MVB integrity, morphology, and size (7, 9), we anticipated that hVac14 may also be engaged in this function.
FIG. 5.
Confocal microscopy reveals substantial colocalization of ectopically expressed hVac14 with PIKfyve or MPR. Twenty-four hours posttransfection with pEGFP-HA-hVac14 alone or together with pCMV5-Myc-PIKfyveWT or pCMV5-Myc-PIKfyveK1831E, COS-7 cells were fixed in formaldehyde and permeabilized. Expressed Myc-PIKfyveWT or Myc-PIKfyveK1831E was detected with anti-myc monoclonal antibody and Texas red-conjugated secondary anti-mouse antibody, whereas CI-MPR was detected with the polyclonal anti-MPR and CY3-conjugated secondary anti-rabbit antibodies. Expression of EGFP-HA-hVac14 was visualized by GFP fluorescence. Cells were viewed in a Zeiss LSM 510 confocal microscope. Merged images of the green and red channel, presented in panels a, e, and i, illustrate a typical expressing and coexpressing cell per condition and depict a substantial area of colocalization of hVac14 with PIKfyveWT, CI-MPR, or PIKfyveK1831E (yellow). The boxed areas in panels a, e, and i are enlarged images, the green (b, f, and j) or red (c, g, and k) channels and their respective merge (d, h, and l). Note that the yellow signal appears on the limiting membrane of perinuclear vesicles (representative vesicles are pointed to by arrowheads in panels d, h, and l).
Vac14 protein knockdown renders cells prone to vacuolation.
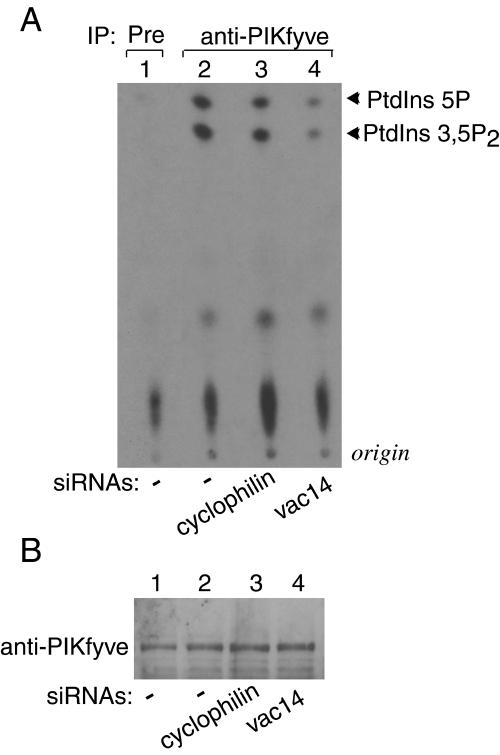
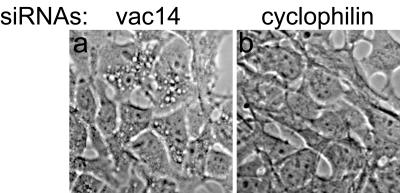
Based on data for yeast documenting similar phenotypes in vac14Δ and fab1Δ mutants (2, 4) and on the above demonstrated colocalization, we expected that hVac14 protein elimination would dilate MVB-related membrane structures and induce cytoplasmic vacuolation in cells, much like the dominant-negative PIKfyve mutants that are deficient in PtdIns(3,5)P2 production. Therefore, we tested whether hVac14 protein knockdown will induce the characteristic abnormal phenotype in HEK293 cells (7, 9). As demonstrated above (Fig. 1C and 3A), HEK293 cell transfection with human vac14 siRNA Smart pool duplexes arrested efficiently (more than 85%) and selectively both endogenous and ectopic hVac14 protein expression. Expression of other proteins (Fig. 1 and 3A, see unchanged nonspecific bands) or PIKfyve (see Fig. 7B) was practically unaltered by the vac14 siRNA duplexes. Under conditions of endogenous hVac14 protein knockdown, we observed no obvious changes of normal HEK293 cell morphology 24 to 72 h posttransfection. However, these cells were highly susceptible to developing cytoplasmic vacuoles upon short treatment with low concentrations of weak bases (NH4Cl, 10 mM, 40 min) (Fig. 6). Importantly and consistent with previous results (7), these mild conditions of NH4Cl treatment did not produce any phenotypic changes in control HEK293 cells transfected with cyclophilin A siRNAs (Fig. 6). Quantitation of the extent of the endomembrane vacuolation by neutral red uptake assay (11) demonstrated a 2- ± 0.3-fold increase of the optical density in vac14 siRNA- versus cyclophilin A siRNA-transfected cells in response to NH4Cl treatment. These data indicate that the elimination of hVac14 protein renders cells prone to developing an abnormal endomembrane phenotype seen upon expression of kinase-deficient dominant-negative PIKfyve mutants (7, 9).
FIG. 7.
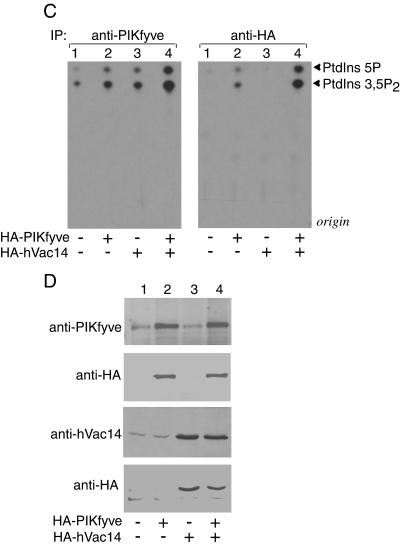
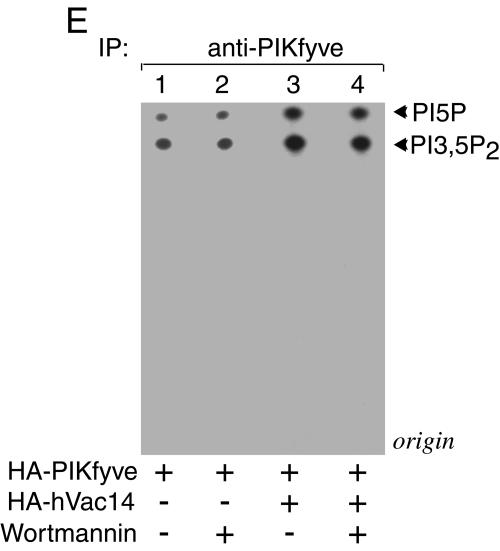
Modulations in hVac14 protein expression levels affect PIKfyve lipid kinase activity. (A and B) HEK293 cells were transfected with the indicated siRNAs. PIKfyve lipid kinase activity (A) or protein expression levels (B) were assayed 72 h posttransfection as described below. (C to E) HEK293 cell line (clone 9) was transfected or not transfected with pCMV5-HA-hVac14 and then was treated with or without doxycycline to induce HA-PIKfyveWT protein expression as indicated. PIKfyve lipid kinase activity (C and E) or protein expression levels (D) were assayed 40 h posttransfection or induction. (A, C, and E) Immunoprecipitates of cell lysates prepared with the indicated antibodies or preimmune sera (Pre) were immobilized on protein A-Sepharose beads and, after washings, were subjected to lipid kinase assay as described in Materials and Methods. (E) Samples were first preincubated with or without wortmannin (20 nM, 15 min) and then were subjected to lipid kinase assay. Shown are autoradiograms of the thin-layer chromatography-resolved lipids from representative experiments out of three (A) and six (C) experiments with similar results. (B and D) Lysates derived from cells in panels A and C were analyzed by SDS-PAGE (6% acrylamide) and immunoblotting with the indicated antibodies. Shown are chemiluminescence detections of representative immunoblots out of three to five experiments for each condition. IP, immunoprecipitate.
FIG. 6.
hVac14 protein knockdown renders cells susceptible to vacuolation. HEK293 cells were transfected with siRNA Smart pool directed to human vac14 or cyclophilin A as indicated. Seventy-two hours posttransfection, cells were treated with NH4Cl (10 mM) for 40 min at 37°C and then observed live by a light microscope (TE200; Nikon) with a 40× Hoffman modulation contrast objective. Shown are images of live cells captured by a SPOT RT Slider camera. NH4+ treatment induced multiple cytoplasmic vacuoles in cells transfected with vac14 siRNAs (seen in 85% ± 10%, mean ± standard errors of the mean; n = 3) but was completely ineffective in cells transfected with cyclophilin A siRNAs.
Vac14 regulates PIKfyve activity in vitro.
Although we did not observe cytoplasmic vacuoles upon attenuation of hVac14 protein expression, the cell susceptibility to such changes implies that the hVac14 protein knockdown may have affected PIKfyve kinase activity. Consistent with this prediction, we found that hVac14 protein elimination (see Fig. 1C) resulted in a marked decrease of the PIKfyve lipid kinase activity in vitro without affecting PIKfyve protein levels (Fig. 7A and B). Parallel transfection with control cyclophilin A siRNAs did not result in significant changes in PIKfyve lipid kinase activity or protein levels (Fig. 7A and B). Quantitation of three separate experiments revealed a decrease of 3.2- ± 0.3-fold in both PtdIns(3,5)P2 and PtdIns(5)P production due to siRNA-directed vac14 gene silencing, as calculated from the intensity of the radioactive PI spots on thin-layer chromatography plates. This result is consistent with the notion that, in a cellular context, hVac14 is upstream of PIKfyve and regulates the PIKfyve enzymatic activity. The observed ∼30% residual PtdIns(3,5)P2-producing activity of PIKfyve remaining despite the hVac14 protein knockdown may explain why the abnormal cell phenotype was not manifested under these conditions. Indeed, studies with Fab1 as well as our data with PIKfyve mutants selectively defective in PtdIns(3,5)P2 production (5, 9) revealed the necessity of a drastic loss in PtdIns(3,5)P2 production (88% or more) to render cells morphologically abnormal.
Having established that low levels of hVac14 protein negatively regulate PIKfyve activity in vitro, we next examined whether increased protein expression of hVac14 could have the opposite effect. This was tested in anti-PIKfyve or anti-HA immunoprecipitates derived from lysates of an HA-hVac14-transfected HEK293 stable cell line in the presence or absence of induced HA-PIKfyveWT expression (Fig. 7C and D). Intriguingly, we observed 2.5- ± 0.5-fold induction of the in vitro synthesis of PIKfyve lipid products in anti-PIKfyve immunoprecipitates due to HA-hVac14 expression, seen in four independent experiments. The possibility that this increase is due to Vac14-dependent activation of PI 3-kinase, found associated with PIKfyve under certain conditions (16, 18), was ruled out by the inability of wortmannin added to the in vitro assay to eliminate the increment in PtdIns(3,5)P2 (Fig. 7E). In anti-HA immunoprecipitates, HA-hVac14 expression also increased lipid production, but it did so only under induced HA-PIKfyveWT expression, indicating that this effect is strictly PIKfyve related. Because PIKfyve protein levels, either induced or noninduced, remained unchanged under HA-hVac14 ectopic expression (Fig. 7D), the likely explanation is that overexpressed hVac14 increases the intrinsic activity of PIKfyve.
Vac14 controls intracellular PtdIns(3)P consumption and PtdIns(3,5)P2 accumulation.
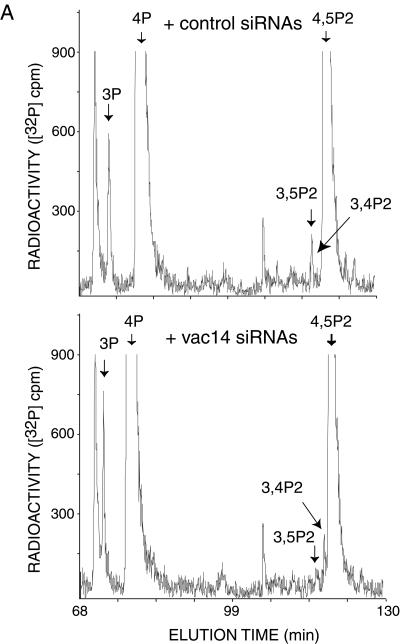
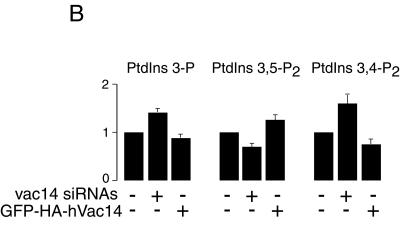
To understand whether Vac14-dependent regulation of PIKfyve activity controls PtdIns(3,5)P2 intracellular production, we examined the PI levels in vivo by HPLC-inositol head group analysis. As illustrated in Fig. 8A and B, elimination of hVac14 protein expression achieved by vac14 siRNA-directed silencing in HEK293 cells resulted in a small but significant decrease in [32P]PtdIns(3,5)P2 accumulation. Intriguingly, [32P]PtdIns(3)P was found elevated under hVac14 protein depletion, consistent with its accumulation due to down-regulated conversion into PtdIns(3,5)P2. PtdIns(3,4)P2 was also significantly elevated under these conditions, most likely due to PtdIns(3)P mass action (Fig. 8). The opposite changes were seen in the PIKfyveWT-HEK293 cell line transfected with pEGFP-HA-hVac14. A small but significant increase in PtdIns(3,5)P2 was documented in both induced and noninduced coexpression of PIKfyveWT. [32P]PtdIns(3)P and [32P]PtdIns(3,4)P2 accumulation were slightly reduced due to EGFP-HA-hVac14 expression (Fig. 8B). Together these results indicate that modulations in hVac14 protein expression affect intracellular PtdIns(3,5)P2 levels, consistent with Vac14 functioning as a regulator of the PtdIns(3,5)P2-synthesizing enzyme PIKfyve in a mammalian cell context.
FIG. 8.
Modulations in hVac14 protein expression levels alter intracellular PtdIns(3,5)P2 in 32P-labeled HEK293 cells. (A) HEK293 cells were transfected with the indicated siRNAs derived from the corresponding sequences of human vac14 or cyclophilin A. Seventy-two hours posttransfection, cells were labeled with [32P]orthophosphate as described in Materials and Methods. Cell lipids were extracted, deacylated, and coinjected with [3H]GroPIns 4-P, [3H]GroPIns 3-P, and [3H]GroPIns 4,5-P2 as internal HPLC standards (elution times are indicated by arrows). The elution times of [32P]GroPIns 3,5-P2 and [32P]GroPIns 3,4-P2 standards (arrows) were determined from parallel HPLC runs. Fractions were monitored for 3H and 32P radioactivity by an online flow-scintillation analyzer. Shown are HPLC elution profiles of a typical labeling experiment out of five experiments with similar results. (B) Quantitation of results shown in panel A (siRNAs). Also presented is the quantitation of three independent labelings in the HEK293 cell line (clone 9) expressing PIKfyveWT, cotransfected with a control pEGFP vector or pEGFP-HA-hVac14, and labeled 24 h posttransfection with [32P]orthophosphate as shown in panel A. For quantitation, the radioactive peaks were first calculated as a percentage of total PI radioactivity and then were expressed as a percentage of the PtdIns(3)P, PtdIns(3,5)P2, or PtdIns(3,4)P2 relative level in the corresponding control condition. Data are presented as means ± standard errors of the means.
Physical interaction of PIKfyve and hVac14.
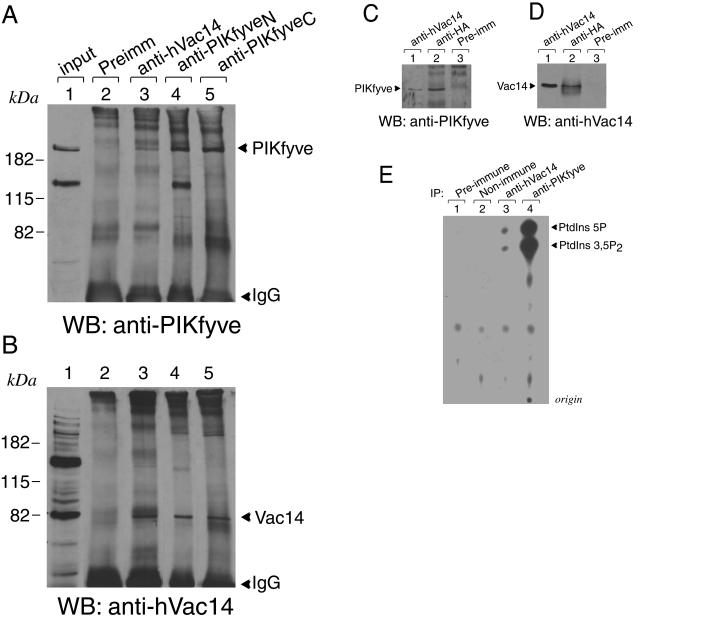
PIKfyve and hVac14 colocalization observed by biochemistry and morphology, together with the alterations of PIKfyve enzymatic activity in vitro by modulating hVac14 protein expression demonstrated above, suggest physical association between the two proteins. However, no physical interaction has been found for the two yeast orthologs by coimmunoprecipitation (2, 4). To test whether this holds true for the mammalian counterparts, we performed coimmunoprecipitations for both ectopically expressed proteins in the HEK293 stable cell line and endogenous forms in PC12 cells, a cell type found to contain authentic PIKfyve and Vac14 (see Fig. 2) in substantial amounts. To our surprise, we observed unequivocal coimmunoprecipitation of PIKfyve with anti-hVac14 and, vice versa, of mammalian Vac14 with anti-PIKfyve in both native cells and transfected cell systems expressing either one of the proteins. As illustrated in Fig. 9A and B, immunoreactive endogenous Vac14 was detected in PIKfyve immunoprecipitates derived from lysates of PC12 cells. Importantly, the endogenous Vac14 protein band was apparent in immunoprecipitates of two unrelated anti-PIKfyve antibodies directed to the C or N terminus of the molecule (Fig. 9A and B), eliminating any possibility for cross-reactivity. Likewise, endogenous hVac14 immunoreactive with anti-hVac14 antibodies on Western blots was found in anti-HA immunoprecipitates derived from the HEK293 cell line stably expressing HA-PIKfyveWT (Fig. 9C and D). Concordantly, in reciprocal immunoprecipitations with anti-hVac14 in PC12 cells and the HEK293 cell line stably expressing HA-PIKfyveWT, we have detected immunoreactive PIKfyve (Fig. 9A and C). Control coimmunoprecipitations with corresponding preimmune sera coupled with Western blotting with the reciprocal antibodies were negative (Fig. 9A to D). Together these results are consistent with the notion that in a cellular context, PIKfyve and Vac14 are assembled in a complex.
FIG. 9.
hVac14 physically associates with PIKfyve. (A and B) PC12 cells, abundant in endogenous PIKfyve and Vac14 (lane 1 in panels A and B), were lysed in RIPA buffer and were immunoprecipitated with the preimmune serum from the Vac14 antibody production, anti-hVac14, and two anti-PIKfyve antibodies against the N terminus and C terminus (anti-PIKfyveN and anti-PIKfyveC, respectively) as indicated. Immunoprecipitates were resolved by SDS-PAGE (6% acrylamide) and were immunoblotted with anti-PIKfyve (A) and anti-hVac14 antibodies (B), with a stripping step in between. Shown are chemiluminescence detections of a blot from a representative experiment out of five independent coimmunoprecipitation experiments in this cell type yielding similar results. (C and D) Lysates of an HEK293 cell line (clone 9) induced to express HA-PIKfyveWT that were immunoprecipitated with anti-hVac14 (affinity purified), anti-HA antiserum, or preimmune serum from the Vac14 antibody production, as indicated, were resolved by SDS-PAGE (6% acrylamide) and were immunoblotted as described for panels A and B. (E) Immunoprecipitates of PC12 cell lysates prepared with anti-hVac14, anti-PIKfyve (against the N terminus), or irrelevant antisera (Non-imm) as well as with the preimmune serum of anti-hVac14 production were immobilized on protein A-Sepharose beads and, after washings, were subjected to lipid kinase assay as described in Materials and Methods. Extracted lipids were resolved by thin-layer chromatography. Shown is an autoradiogram from a representative experiment out of three with similar results. WB, Western blot; Preimm, preimmune; IP, immunoprecipitate.
If PIKfyve associates with Vac14, one would expect that Vac14 immunoprecipitates will display the ability to generate PIKfyve lipid products when subjected to a lipid kinase assay. We examined this important question with the endogenous proteins in PC12 cells to avoid any adverse interaction due to large amounts of proteins under ectopically expressed conditions. As illustrated in Fig. 9E, like anti-PIKfyve, anti-hVac14 immunoprecipitates derived from PC12 cell lysates readily synthesized both PtdIns(5)P and PtdIns(3,5)P2. In contrast, parallel immunoprecipitates of preimmune serum or irrelevant antiserum did not generate any lipid product (Fig. 9E). These data indicate that PIKfyve-Vac14 association could also be demonstrated by the presence of PIKfyve enzymatic activity coimmunoprecipitated with Vac14. These results also imply that the PIKfyve-Vac14 complex is rather stable, sustaining multiple washes at high-salt concentration involved in the lipid kinase assay.
DISCUSSION
Here we have identified and characterized mammalian Vac14, a structural homologue of yeast Vac14, as a positive regulator of the PtdIns(3,5)P2/PtdIns(5)P-producing enzyme PIKfyve. The protein is evolutionarily conserved, found in all eukaryotes with a sequenced genome, and expressed as an 82-kDa protein at various levels in numerous mammalian cells and tissues (Fig. 1 and 2). Based on quantitative estimation of hVac14 concentration in HEK293 cells, the protein could be categorized as being low in abundance. Mammalian Vac14 colocalized and physically associated with PIKfyve to up-regulate PIKfyve activity in vitro and PtdIns(3,5)P2 production in a cellular context (Fig. 5 and 7 to 9). hVac14 elimination rendered cells susceptible to morphological alterations (Fig. 1C, 3A, and 6) similar to those observed upon expression of PIKfyve dominant-negative mutants deficient in PtdIns(3,5)P2-producing activity (7, 9). Thus, hVac14 represents the first known associated regulator of PIKfyve enzymatic activity.
Besides structural homology, the hVac14/PIKfyve/PtdIns(3,5)P2 pathway in mammalian cells shares many functional similarities with the Vac14/Fab1/PtdIns(3,5)P2 pathway in yeast (1, 2, 4-10, 14). Thus, they both control MVB (mammals) or vacuole (yeast) size and morphology, as well as sorting and trafficking to lysosomes of a specific subset of proteins (Fig. 10). Furthermore, both yeast and mammalian cells display related phenotypes upon protein interference and gene inactivation of PIKfyve and FAB1 or VAC14 in the form of dilated MVBs (mammalian cells) or swollen vacuoles (yeast). However, a principal divergence of the two pathways is the sensitivity to osmoregulation. Unlike yeast cells, where steady-state levels of PtdIns(3,5)P2 rise 18- to 28-fold (2), many mammalian cells, including COS cells, CTLL2 cells (3, 12), 3T3-L1 fibroblasts, HEK293 cells (D. Sbrissa and A. Shisheva, unpublished observations), and likely others (13), do not increase PtdIns(3,5)P2 in response to hyperosmotic stress. Thus, the yeast pathway appears to be a mechanism for adaptation to sudden changes in the osmolarity of the environment, whereas the mammalian hVac14/PIKfyve/PtdIns(3,5)P2 pathway likely evolved in the relatively constant environment to serve specific roles related to MVB function, biogenesis, or other yet-to-be-identified cellular processes. Consistent with this notion, PIKfyve expressed in a fab1Δ strain from a yeast promoter restores basal levels of PtdIns(3,5)P2 but does not respond to hyperosmotic shock, whereas Fab1 expression complements this osmotic defect (13). Moreover, at least in vitro, hVac14 also increases PtdIns(5)P production by PIKfyve (Fig. 7 and 10), which adds another level of divergence within the pathways in yeast and mammalian cells, the latter but not the former showing PtdIns(5)P natural occurrence (13, 17). Furthermore, expression of Fab1 in yeast does not increase steady-state levels of PtdIns(3,5)P2, whereas PIKfyveWT expression elevates basal PtdIns(3,5)P2 production in several mammalian cell types (5, 7). Finally, yeast Vac14 and hVac14 are both positive upstream regulators of Fab1 and PIKfyve, yet the mechanism of this activation appears to be different. Thus, whereas yeast Vac14-Fab1 physical association has not been detected by various techniques, such as immunoprecipitation, cross-linking, and two-hybrid systems (2, 4), hVac14 readily associates with PIKfyve, as was unequivocally seen here, by coimmunoprecipitation with a number of different antibodies against the endogenous proteins or the HA epitope of ectopically expressed HA forms as well as by detecting PIKfyve lipid products in immunopurified Vac14 (Fig. 9). The challenge remains to identify physiological regulators of the hVac14/PIKfyve/PtdIns(3,5)P2 pathway.
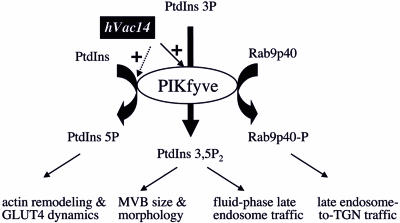
FIG. 10.
Model for PIKfyve enzymatic activities, their regulation by hVac14, and intracellular roles of phosphorylated products. PIKfyve is a dual-specificity enzyme that synthesizes PtdIns(3,5)P2 or PtdIns(5)P in a cellular context (7, 17) and phosphorylates protein substrates in vitro, including itself (8, 20). PIKfyve-phosphorylated products likely control distinct intracellular functions. PIKfyve-catalyzed synthesis of PtdIns(3,5)P2, positively regulated by physically associated hVac14, controls MVB size, MVB morphology, and traffic of fluid-phase markers at later stages of the endocytic pathway (9 and this study). hVac14's regulatory role in PIKfyve-catalyzed synthesis of PtdIns(5)P is confirmed only in vitro (this study). A possible role for PIKfyve-directed PtdIns(5)P synthesis in actin remodeling and GLUT4 dynamics has been suggested (19). PIKfyve-directed phosphorylation of the kelch β-propeller transport factor p40, a Rab9 effector that controls late endosome-to-TGN transport, is documented in vitro (8), and no direct evidence is presently available for the role of PIKfyve activity in this transport step. Possible activation of PIKfyve protein kinase by hVac14 has not been examined.
One intriguing, although not unexpected, result in our study is that the dramatic decrease of PIKfyve activity in vitro (>3.5-fold) under conditions of almost complete hVac14 protein elimination was not paralleled by a similar reduction in intracellular PtdIns(3,5)P2 levels (Fig. 7 and 8). Instead, we observed a 30% decrease in basal PtdIns(3,5)P2 under these conditions (Fig. 8B). Consistent with this modest reduction of intracellular PtdIns(3,5)P2 accumulation upon hVac14 protein knockdown, we observed the characteristic phenotype of massive cytoplasmic vacuolation only after a short cell pretreatment with weak bases at low doses (Fig. 6). A likely explanation of this phenomenon is a possible decrease in PtdIns(3,5)P2 turnover due to mistargeting PtdIns(3,5)P2-specific phosphatases. This is supported by studies both of yeast and mammalian cells (15, 25). Thus, in COS cells, PtdIns(3,5)P2 directs the localization of a PtdIns(3,5)P2-specific phosphatase, myotubularin, through the PtdIns(3,5)P2-binding GRAM domain of the enzyme (25). In yeast, Vac14 relays a complex regulation where it not only controls the steady-state and hyperosmotically increased levels of PtdIns(3,5)P2 through stimulating Fab1 activity but also activates and directs targeting of Fig4, a Sac domain-containing Mg2+-dependent PtdIns(3,5)P2-specific phosphatase (15). Biochemically uncharacterized mammalian orthologs of yeast Fig4 are found in databases. Future studies will reveal whether mammalian Fig4 antagonizes PIKfyve action and whether this is dependent on hVac14.
In summary, we have identified and functionally characterized mammalian Vac14, an 82-kDa protein. Yeast Vac14 and hVac14 are structural homologues that have both common and distinct functional features. By physically associating with and activating PIKfyve, hVac14 controls intracellular PtdIns(3,5)P2 production. Elimination of hVac14 protein expression is associated with cell susceptibility to phenotypic changes, seen upon expression of PIKfyve mutants deficient in PtdIns(3,5)P2 production.
Supplementary Material
Acknowledgments
We thank Linda McCraw for excellent secretarial assistance. We thank Mike Czech, Paul Pilch, I. Mellman, and B. Hoflack for antibodies and Chris Tierney, Queta Boese, and Zhen Jiang for helpful discussions of the siRNA approach.
This project was supported by National Institutes of Health (DK58058) and American Diabetes Association Research grants (to A.S.).
Footnotes
Supplemental material for this article may be found at http://mcb.asmusa.org.
REFERENCES
- 1.Bonangelino, C. J., N. L. Catlett, and L. S. Weisman. 1997. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 17:6847-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonangelino, C. J., J. J. Nau, J. E. Duex, M. Brinkman, A. E. Wurmser, J. D. Gary, S. D. Emr, and L. S. Weisman. 2002. Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 156:1015-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dove, S. K., F. T. Cooke, M. R. Douglas, L. G. Sayers, P. J. Parker, and R. H. Michell. 1997. Osmotic stress activates phosphatidylinositol-3,5-bisphospahte synthesis. Nature 390:187-192. [DOI] [PubMed] [Google Scholar]
- 4.Dove, S. K., R. K. McEwen, A. Mayes, D. C. Hughes, J. D. Beggs, and R. H. Michell. 2002. Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 12:885-893. [DOI] [PubMed] [Google Scholar]
- 5.Gary, J. D., A. E. Wurmser, C. J. Bonangelino, L. S. Weisman, and S. D. Emr. 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143:65-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gary, J. D., T. K. Sato, C. J. Stefan, C. J. Bonangelino, L. S. Weisman, and S. D. Emr. 2002. Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol. Biol. Cell 13:1238-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikonomov, O. C., D. Sbrissa, and A. Shisheva. 2001. Mammalian cell morphology and endocytic membrane homeostasis require PIKfyve enzymatic activity. J. Biol. Chem. 276:26141-26147. [DOI] [PubMed] [Google Scholar]
- 8.Ikonomov, O. C., D. Sbrissa, K. Mlak, R. Deeb, J. Fligger, A. Soans, R. L. Finley, Jr., and A. Shisheva. 2003. Active PIKfyve associates with and promotes the membrane attachment of the late endosome-to-TGN transport factor Rab9 effector p40. J. Biol. Chem. 278:50863-50871. [DOI] [PubMed] [Google Scholar]
- 9.Ikonomov, O. C., D. Sbrissa, K. Mlak, M. Kanzaki, J. Pessin, and A. Shisheva. 2002. Functional dissection of lipid and protein kinase signals of PIKfyve reveals the role of PtdIns(3,5)P2 production for endomembrane integrity. J. Biol. Chem. 277:9206-9211. [DOI] [PubMed] [Google Scholar]
- 10.Ikonomov, O. C., D. Sbrissa, M. Foti, J.-L. Carpentier, and A. Shisheva. 2003. PIKfyve controls fluid-phase endocytosis but not recycling/degradation of endocytosed receptors or sorting of procathepsin D by regulating multivesicular body morphogenesis. Mol. Biol. Cell 14:4581-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikonomov, O. C., D. Sbrissa, T. Yoshimori, T. L. Cover, and A. Shisheva. 2002. PIKfyve kinase and SKD1 AAA ATPase define distinct endocytic compartments. J. Biol. Chem. 277:46785-46790. [DOI] [PubMed] [Google Scholar]
- 12.Jones, D. R., A. González-García, E. Díez, C. Martinez, A. C. Carrera, and I. Mérida. 1999. The identification of phosphatidylinositol 3,5-bisphosphate in T-lymphocytes and its regulation by interleukin-2. J. Biol. Chem. 274:18407-18413. [DOI] [PubMed] [Google Scholar]
- 13.McEwen, R. K., S. K. Dove, K. T. Cooke, G. F. Painter, A. B. Holmes, A. Shisheva, Y. Ohya, P. J. Parker, and R. H. Michell. 1999. Complementation analysis in PtdInsP kinase-deficient yeast mutants demonstrates that Schizosaccharomyces pombe and murine Fab1p homologues are phosphatidylinositol 3-phosphate 5-kinases. J. Biol. Chem. 274:33905-33912. [DOI] [PubMed] [Google Scholar]
- 14.Odorizzi, G., M. Babst, and S. D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95:847-858. [DOI] [PubMed] [Google Scholar]
- 15.Rudge, S. A., D. M. Anderson, and S. D. Emr. 2004. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell 15:24-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sbrissa, D., O. C. Ikonomov, and A. Shisheva. 1999. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5′-phosphoinositides: effect of insulin. J. Biol. Chem. 274:21589-21597. [DOI] [PubMed] [Google Scholar]
- 17.Sbrissa, D., O. C. Ikonomov, R. Deeb, and A. Shisheva. 2002. Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway. J. Biol. Chem. 277:47276-47284. [DOI] [PubMed] [Google Scholar]
- 18.Sbrissa, D., O. C. Ikonomov, and A. Shisheva. 2001. Selective insulin-induced activation of class IA phosphoinositide 3-kinase in PIKfyve immune complexes from 3T3-L1 adipocytes. Mol. Cell. Endocrinol. 181:35-46. [DOI] [PubMed] [Google Scholar]
- 19.Sbrissa, D., O. C. Ikonomov, J. Strakova, and A. Shisheva. 2004. Role for a novel signaling intermediate, phosphatidylinositol 5-phosphate in insulin-regulated F-actin stress fiber breakdown and GLUT4 translocation. Endocrinology 145:4853-4865. [DOI] [PubMed]
- 20.Shisheva, A. 2001. PIKfyve: the road to PtdIns 5-P and PtdIns 3,5-P2. Cell Biol. Intern. 25:1201-1206. [DOI] [PubMed] [Google Scholar]
- 21.Shisheva, A., C. Demarco, O. Ikonomov, and D. Sbrissa. 2002. Insulin signaling: from cultured cells to animal models, p. 189-205. In G. Grunberger and Y. Zick (ed.), PIKfyve and acute insulin actions. Taylor & Francis, London, England.
- 22.Shisheva, A., S. J. Doxsey, J. M. Buxton, and M. P. Czech. 1995. Pericentriolar targeting of GDP-dissociation inhibitor isoform 2. Eur. J. Cell Biol. 68:143-158. [PubMed] [Google Scholar]
- 23.Shisheva, A., B. Rusin, O. C. Ikonomov, C. DeMarco, and D. Sbrissa. 2001. Localization and insulin-regulated relocation of 5′-phosphoinositide kinase PIKfyve in 3T3-L1 adipocytes. J. Biol. Chem. 276:11859-11869. [DOI] [PubMed] [Google Scholar]
- 24.Shisheva, A., D. Sbrissa, and O. Ikonomov. 1999. Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells. Mol. Cell. Biol. 19:623-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsujita, K., T. Itoh, T. Ijuin, A. Yamamoto, A. Shisheva, J. Laporte, and T. Takenawa. 2004. Myotubularin regulates the function of the late endosome through the GRAM domain-phosphatidylinositol 3,5-bisphosphate interaction. J. Biol. Chem. 279:13817-13824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.