Abstract
Protein kinases form one of the largest protein families and are found in all species, from viruses to humans. They catalyze the reversible phosphorylation of proteins, often modifying their activity and localization. They are implicated in virtually all cellular processes and are one of the most intensively studied protein families. In recent years, they have become key therapeutic targets in drug development as natural mutations affecting kinase genes are the cause of many diseases. The vast amount of data contained in the primary literature and across a variety of biological data collections highlights the need for a repository where this information is stored in a concise and easily accessible manner. The UniProt Knowledgebase meets this need by providing the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information. Here, we describe the expert curation process for kinases, focusing on the Caenorhabditis elegans kinome. The C. elegans kinome is composed of 438 kinases and almost half of them have been functionally characterized, highlighting that C. elegans is a valuable and versatile model organism to understand the role of kinases in biological processes.
Keywords: curation, database, kinase, UniProtKB
Introduction
Kinase-mediated protein phosphorylation is one of the most abundant and conserved post-translational modifications (PTMs) in eukaryotic organisms. Analysis of the human phosphoproteome showed that probably one-third of proteins are phosphorylated [1]. This reversible protein modification controls several aspects of protein dynamics including enzymatic activity, cellular localization, protein–protein interaction and stability, thereby allowing tight control of cellular processes such as cell proliferation, differentiation, apoptosis, development and cell migration [2–4]. Kinases play an essential role in translating external cues into biological responses via the regulation of complex signaling cascades [5,6]. It is therefore not surprising that defects in kinases lead to a variety of diseases including diabetes, various types of cancer, and inflammatory and neurodegenerative diseases [7].
Protein kinases are one of the largest protein families, representing ∼2% of the human proteome [8]. They catalyze the transfer of the γ-phosphate of ATP onto the hydroxyl group of serine, threonine and tyrosine residues. In humans, 67% of kinases phosphorylate serine and threonine residues [8]. Kinases are divided into two main groups: eukaryotic kinases (ePKs), the most abundant group, share a conserved kinase domain that folds into two lobes. The pocket formed by the two lobes contains the ATP and substrate-binding sites. Atypical kinases (aPKs), on the other hand, lack significant sequence similarity to the ePK kinase domain although they have proven kinase activity [8,9].
Kinases have become key therapeutic targets in drug development by the pharmaceutical industry due to the disease implications [10]. For instance, Parkinson's disease, a severe neurodegenerative disease, is often caused by mutations in the leucine-rich repeat serine/threonine-protein kinase 2 (LRRK2) gene [UniProt Knowledgebase (UniProtKB) Q5S007], and a considerable amount of effort has been invested into designing LRRK2 inhibitors to treat this condition [11]. Owing to their essential role in regulating cell proliferation, it is not surprising that many cancers are often linked to mutations in kinase genes or are caused by uncontrolled regulation of kinase activity. For example, mutations affecting the receptor tyrosine protein kinase ERBB2 (UniProtKB P04626) have been implicated in the development of some cancers including lung, breast and gastric cancers [12]. Although several kinase inhibitors have been developed and are currently used to treat some cancers, for other diseases such as muscular dystrophy which can result from mutations in the titin gene (UniProtKB Q8WZ42), few treatments are available [13].
The vast amount of data contained in the primary literature and across a variety of biological data collections highlights the need for a repository where this information is stored in a concise and easily accessible manner. The UniProtKB meets this need by providing the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information.
An important step in the curation of kinases in UniProtKB was achieved in 2008 when the annotation of both the human and mouse kinomes was completed based on the available knowledge at the time [14]. As new data are published, this annotation is continuously reviewed and updated, ensuring that our users have access to up-to-date information. Model organisms other than the mouse have played a crucial role in elucidating kinase functions. This prompted us to extend curation efforts to the Caenorhabditis elegans kinome. C. elegans shares many basic cellular mechanisms with vertebrates and, consequently, it is commonly used to study organ development, stress responses, metabolism and aging. Therefore, its kinome constitutes an ideal target for expert curation. Here, we present the curation of the C. elegans kinome. Besides giving an overview of the C. elegans kinome, we also provide a detailed description of the curation process for kinases in UniProtKB.
UniProt Knowledgebase
The UniProtKB is produced by the UniProt Consortium, a collaboration between the European Bioinformatics Institute (EMBL-EBI), the SIB Swiss Institute of Bioinformatics (SIB) and the Protein Information Resource (PIR) [15]. The database consists of two parts [16]. In the reviewed UniProtKB/Swiss-Prot section, protein entries are manually curated by expert biocurators based on a critical review of experimental data provided by peer-reviewed literature and result from sequence analysis tools. The unreviewed UniProtKB/TrEMBL section consists of entries that are enriched with automatic annotation based on two rule-based systems: UniRule and Statistical Automated Annotation System (SAAS) [15,16], as well as predictions from a suite of sequence analysis methods to enrich the records with extra sequence-specific information. In addition, cross-references are provided to 157 specialized resources including nucleotide sequence resources, model organism databases and protein family databases.
UniProtKB curation
General curation
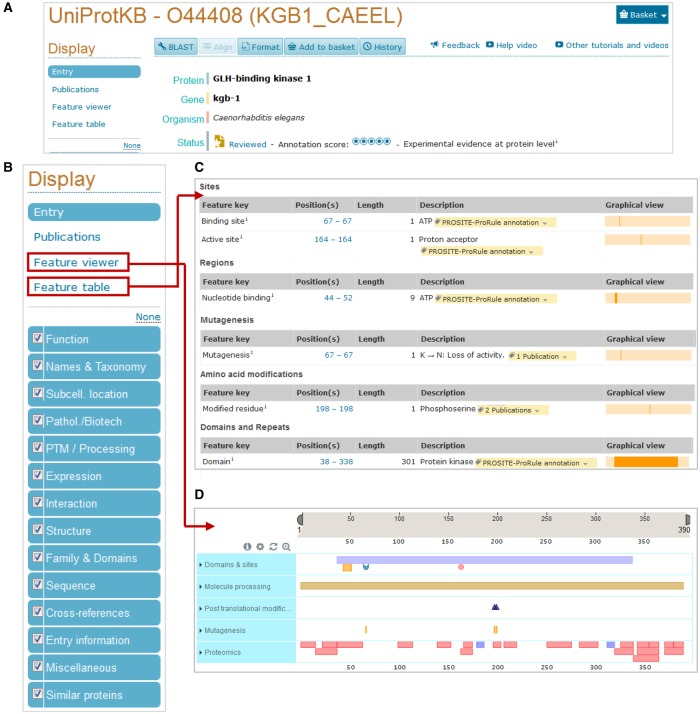
A UniProtKB entry provides a large amount of information in a structured and concise way (Figure 1). The expert curation process involves several steps [16,17]. (1) Entries corresponding to the same gene and same species are identified using BLAST against UniProtKB. (2) Sequences are merged and discrepancies are identified and reported. (3) Sequence analysis programs are run to predict sequence features that are then manually assessed, and the appropriate results are selected for annotation. (4) Literature databases are searched for relevant papers from which relevant information is extracted after being critically assessed by the curator. We prioritize publications with (i) a high impact in the scientific community that contains functional data for previously uncharacterized proteins, (ii) new 3D-structural information, (iii) enzymatic reactions that may complete the annotation of known metabolic pathways or networks, (iv) PTMs and their consequences, (v) novel splice variants and (vi) disease-causing variants and polymorphisms. We do not aim to curate all published papers and instead select a representative subset to provide a complete overview of the available information for a given protein [17]. Conflicting results are carefully assessed and, when appropriate, are reported in a ‘caution’ comment in the protein entry. (5) All information is attributed to its original source as described in more detail below. (6) Each completed entry undergoes both automated procedures and manual checks by another curator to ensure that it meets the required quality standards before integration into UniProtKB/Swiss-Prot.
Figure 1. UniProtKB website entry.
(A) View of the top of an entry containing the recommended protein name and links to various tools such as BLAST and sequence alignment. (B) The various sections found in a UniProtKB entry are shown on the left on the website page. Sequence features can be viewed in a table (C) or using the graphical feature viewer (D). The red squares indicate where they are in the website entry.
During the curation of an entry, a recommended protein name is added and any alternative names used in the literature are also included (Figure 1A). A diverse and detailed amount of information, related to the role of the protein such as its function, subcellular location, tissue distribution, interactions with other proteins and protein family membership, is included (Figure 1B). In addition, information is also provided about the presence and localization of functional domains, important residues, post-translationally modified residues, variants and mutagenesis sites (Figure 1C,D). Information regarding isoforms, sequence conflicts and 3D structure is also added when available. For enzymes such as kinases, specific information is recorded including enzyme commission (EC) number, catalytic activity, biophysicochemical properties, enzyme regulation, enzymatic pathways and active sites.
Data evidence
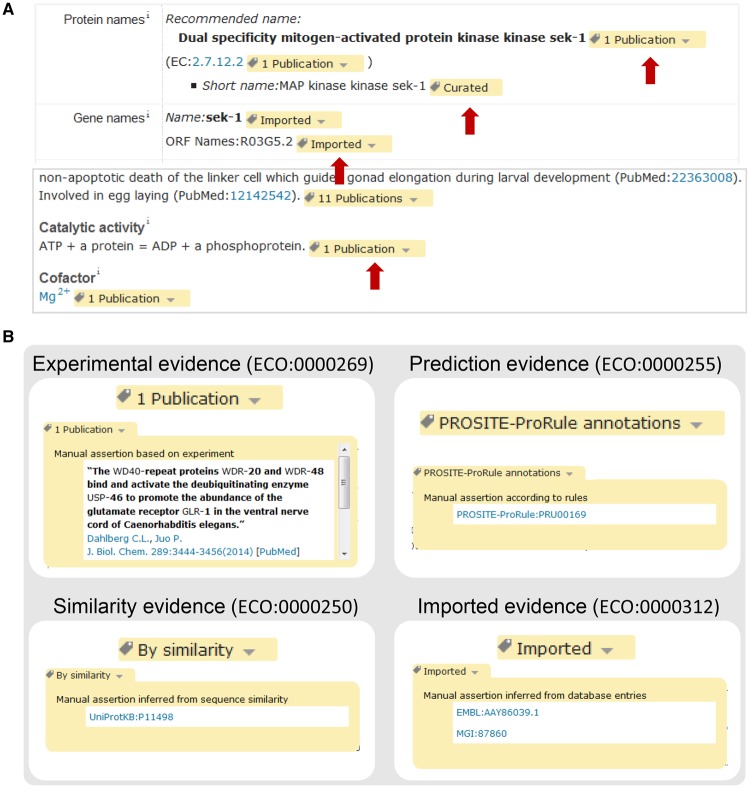
The information in a UniProtKB record comes from a range of different sources, so a crucial aspect and particular strength of UniProtKB annotation is that each piece of information in an entry is linked to its original source so that users can easily identify its origin and evaluate it. UniProtKB makes use of a subset of evidence codes from the Evidence and Conclusion Ontology (ECO) to indicate data origin (Table 1) [18]. These ECO codes are transformed into simple labels on the UniProt website (Figure 2A). For instance, for information inferred from experimental data, we provide a link to the original paper (Figure 2B). For information which has been transferred from a related experimentally characterized protein, the accession number of the characterized protein is indicated, providing a link to the entry with experimental evidence (Figure 2B). Information which has been predicted by the UniProt automatic annotation systems or by the sequence analysis programs that are used during the manual curation process is linked to its original source (Figure 2B). If information has been imported from another database, the database name and the identifier of the entry from which the information has been imported are provided (Figure 2B). This system allows users to tell where all of the data in a UniProtKB record have come from.
Table 1. ECO codes used during the UniProtKB manual curation process.
| ECO code | Term name | Usage |
|---|---|---|
| ECO:0000269 | Experimental evidence used in manual assertion | Information for which there is published experimental evidence |
| ECO:0000303 | Non-traceable author statement used in manual assertion | Information based on author statements in scientific articles for which there is no experimental support |
| ECO:0000250 | Sequence similarity evidence used in manual assertion | Information which has been propagated from a related experimentally characterized protein |
| ECO:0000312 | Imported information used in manual assertion | Information which has been imported from another database and manually verified |
| ECO:0000305 | Curator inference used in manual assertion | Information which has been inferred by a curator based on his/her scientific knowledge or on the scientific content of an article |
| ECO:0000255 | Match to sequence model evidence used in manual assertion | Information originating from the UniProt automatic annotation systems or any of the sequence analysis programs used during the manual curation process and which has been manually verified |
| ECO:0000244 | Combinatorial evidence used in manual assertion | Information which is manually curated based on a combination of experimental and computational evidence |
Figure 2. Evidences in a UniProtKB website entry.
(A) Each piece of information is associated with ECO codes that are replaced with easy-to-understand labels on the website (red arrows). (B) Examples of how ECO codes for experimental, similarity, predicted and imported data are displayed on the UniProt website. The hyperlinked source can be seen by clicking on the arrow on the right.
Gene Ontology annotation
The use of standardized vocabularies to describe various biological functions provides a powerful way to enrich biological databases. The Gene Ontology (GO) is an established standard to describe genes and gene products in terms of their associated biological processes, cellular components and molecular functions in a species-independent manner, allowing a consistent description of gene products across databases [19]. UniProtKB entries are enriched with GO terms that are assigned based on many electronic methods as well as through manual curation of experimental data from the literature [20]. In addition, GO terms are imported from other GO Consortium database members into the relevant UniProtKB entries along with details of the annotation source. UniProt provides selected GO terms on the UniProt website, but a link at the end of the GO term list provides access to the complete set of available GO terms via the QuickGO browser (https://www.ebi.ac.uk/QuickGO/) [20].
Kinase identification and curation
How are kinases identified and curated?
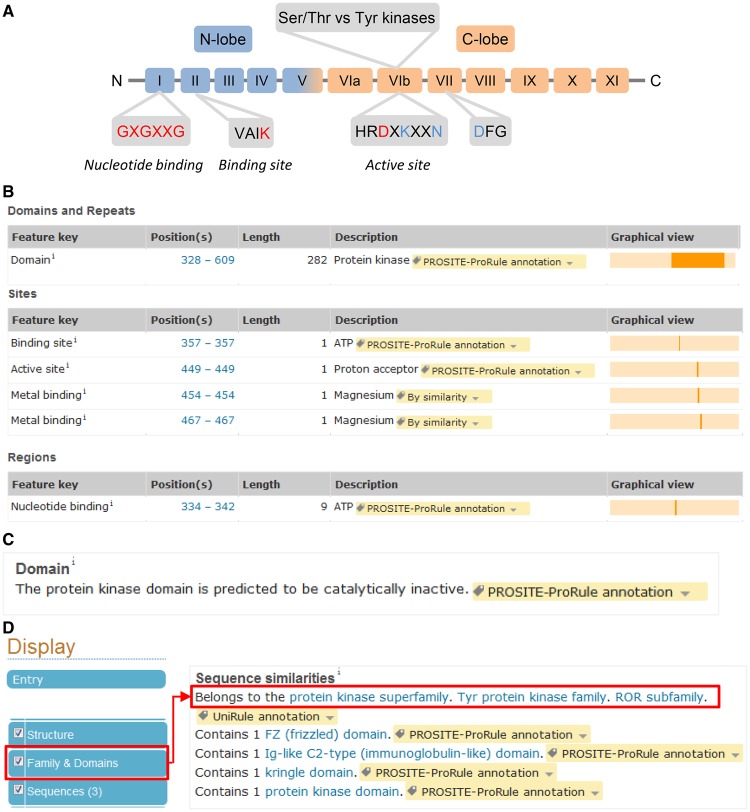
A sequence hallmark for ePKs is the presence of a so-called kinase domain [21]. Usually ∼250–300 amino acids in length, it contains 11 subdomains with conserved residues essential for catalytic activity, features that protein classification resources such as PROSITE [22] and Pfam [23] use to identify potential kinases (Figure 3A). The kinase domain usually contains four conserved motifs involved in the catalytic process [21]. (1) The GXGXXG motif, or the glycine-rich loop, mediates ATP binding by covering and anchoring the nontransferred phosphate groups and enclosing the adenine ring. (2) The VAIK motif contains an essential lysine residue which, by participating in the anchorage and orientation of the ATP, is required for maximal activity. (3) The HRDXKXXN motif, or the catalytic loop, contains an aspartic acid residue that acts as a proton acceptor of the substrate hydroxyl group during the transfer of the ATP phosphoryl group. The lysine residue helps to neutralize the negative charge of the phosphate group, whereas the asparagine residue stabilizes the loop and chelates the secondary Mg(2+) that bridges the α- and γ-phosphate groups. (4) The last motif, DFG, is also important for kinase activity as the aspartic acid residue chelates the primary Mg(2+) that bridges the β- and γ-phosphate groups and thereby helps to orient the γ-phosphate group for transfer. In addition, the residues next to the third motif are used to differentiate between tyrosine and serine/threonine kinases. The first three conserved motifs and the kinase domain are annotated in UniProtKB (Figure 3B).
Figure 3. Characteristics of the kinase domain.
(A) Structure of the kinase domain. The 11 subdomains are labeled I to XI. The amino acids in red are recorded in the feature table of a kinase entry. The residues in blue are important but are not recorded in UniProtKB. (B) Example of site and domain annotation (UniProtKB Q8I7M8). (C) Comment added in the ‘Family and Domains’ section for pseudokinases. (D) Protein kinase classification is found in the ‘Family and Domains’ section (UniProtKB G5EGK5).
The presence or absence of the aspartic acid in the active site is used to predict the catalytic activity status. In addition to predicted activity based on sequence analysis, we also search the literature for experimental proof of kinase activity. This step is important as the presence of an active site does not guarantee that a kinase will be active. For example, human RYK (UniProtKB P34925) has all the sites required for catalysis including the aspartic acid residue, but the kinase shows little activity in vitro [24]. Conversely, sometimes important residues are missing or are in unexpected positions. For instance, rat Wnk1 (UniProtKB Q9JIH7) has been experimentally shown to have catalytic activity although the conserved lysine involved in ATP binding is replaced by a cysteine. Another lysine residue at a different position appears to fulfill this role [25]. When the aspartic acid residue is missing from the predicted active site and there is no experimental evidence for activity, the following comment is added in the ‘Family & Domains’ section: ‘The protein kinase domain is predicted to be catalytically inactive’ (Figure 3C).
aPKs have proved to be more difficult to identify by sequence analysis due to the lack of sequence similarity to the conventional ePK kinase domain [8,9]. The catalytic residues are sometimes conserved and, often, aPK and ePK share a similar kinase domain 3D structure. Unfortunately, there are few rules to identify them and their annotation as kinases is often based on the literature and annotated orthologs.
In vitro kinase assays are the best way so far to determine if a protein has kinase activity or not. Usually, the assay is done in a test tube and contains the kinase and a substrate that can be a peptide or a full-length protein in a buffer containing ATP and a divalent cation [usually Mg(2+)] [26]. Several readouts are used to assess the phosphorylation of substrates such as incorporation of radioactive phosphate, Western blot with phospho-antibodies and mass spectrometry. Although kinase assays are the standard method of measuring kinase activity, they are not performed in a ‘physiological environment’ (i.e. in cells). In vivo, most kinases are kept inactive and require prior activation involving PTMs or binding to other proteins. The isolation procedures or their production in a different cell type or a cell from a different species can also affect the kinase activity. In some cases, experimental evidence shows very low kinase activity, suggesting that the kinase is likely to be inactive. Also, conflicting results are found in the literature or experimental results do not support sequence analysis prediction of kinase activity. Experimental data assessing kinase activity in vivo, although crucial, can be challenging to interpret. In vivo kinase activity is often demonstrated by looking at substrate phosphorylation in cells or whole organisms in which the kinase has been knocked down or which express an inactive kinase generated by mutating the lysine involved in ATP-binding to an arginine. Sometimes, low molecular mass inhibitors are used. Although they can be a useful tool when knockdown experiments are not possible, their lack of specificity may lead to incorrect conclusions. The recent development of fluorescent kinase sensors to measure kinase activity directly in cells will provide useful information to assess kinase activity in more physiological conditions [27]. Below is an example illustrating how curators interpret and report this kind of data.
C. elegans drl-1 (UniProtKB Q86ME2) is predicted to be inactive as residues involved in the catalytic activity are missing. However, dlr-1 appeared to be active in an in vitro kinase assay [28]. One possible explanation for the discrepancy is that immunoprecipitation of drl-1 from COS cells pulled down another kinase that is responsible for the activity detected. Moreover, they used a promiscuous substrate, myelin basic protein (MBP), which is known to be phosphorylated by multiple kinases [29]. As there is no other evidence to suggest that dlr-1 is an active kinase, we annotated the protein as an inactive kinase by adding a ‘Domain’ comment to describe that the protein kinase domain is predicted to be catalytically inactive and a ‘Caution’ comment mentioning that some activity has been detected although residues involved in the catalytic activity are absent.
Curators critically assess all the available evidence and record it in a way that conveys how reliable the evidence is. For example, C. elegans kgb-1 (UniProtKB O44408) has extensive in vitro and in vivo experimental evidence, pak-2 (UniProtKB G5EFU0) kinase activity has been inferred by similarity with pak-1 (UniProtKB Q17850), which has been experimentally shown to have kinase activity, whereas, for spe-6 (UniProtKB Q95PZ9), the only evidence for kinase activity is based on manual evaluation of results from sequence analysis tools.
Kinase families
Kinases are divided into three groups based on the residues they phosphorylate: serine/threonine kinases (Ser/Thr kinases) that phosphorylate serine and/or threonine residues, tyrosine kinases (Tyr kinases) that phosphorylate tyrosine residues and dual-specificity kinases that phosphorylate serine/threonine and tyrosine residues. To make sense of such a large family of proteins, Hunter and then Manning developed a hierarchical classification for kinases, taking into account not only sequence similarities in the kinase domain but also using additional information from domains outside the catalytic domain, from phylogeny and from known functions [8,21]. ePKs were divided into nine groups: eight of the groups, including AGC, CMGC, CK1, CAMK, receptor-type guanylate cyclase (RGC), STE, tyrosine kinase-like (TKL) and OTHER, cover Ser/Thr kinases, whereas the tyrosine kinase (TK) group comprises Tyr kinases. The CMGC and STE groups also include most of the dual-specificity kinases. The OTHER group contains Ser/Thr kinases that do not fit into any of the other groups. Kinases with no similarity to conventional ePKs were classified into one unique group aPKs. These 10 groups were further divided into families and subfamilies. In UniProtKB, this classification is used but in a slightly modified form. Families and subfamilies in UniProtKB correspond to groups and families, respectively, in Manning's and Hanks' classification [8,21]. Kinases of the ‘OTHER’ group are classified with the general Ser/Thr protein kinase family name. For kinases related to NEK1 (classified as OTHER in Manning et al. [8]), a family has been specifically created in UniProtKB. For the classification of aPKs, family names are based on Manning's subfamily classification [8]. This information is recorded in the ‘Family & Domains’ section (Figure 3D). Members from all these families have been identified across multiple metazoan organisms with the exception of yeast, which appears to lack Tyr kinases [30].
Sites important for kinase activity
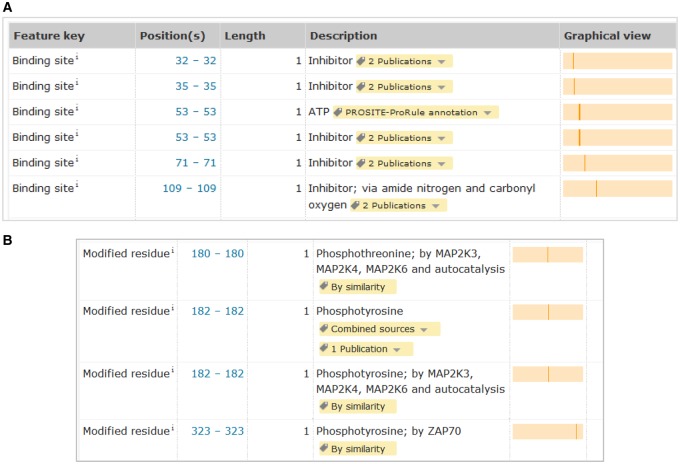
Residues important for catalytic activity are annotated together with information showing whether they have been experimentally determined or computationally predicted (Figure 3B). For pseudokinases that lack enzyme activity due to the absence of catalytic sites, nucleotide-binding sites are still included when they are conserved. Although pseudokinases lack the capacity to hydrolyze ATP, the binding to ATP can still play a role in maintaining the correct folding of the protein or in promoting binding to other proteins [31]. In addition, we annotate metal-binding sites, substrate-binding sites, post-translationally modified residues and inhibitor-binding sites (Figures 3B and 4).
Figure 4. Annotation of important residues.
Sites involved in inhibitor binding (A) and post-translationally modified residues (B) are annotated (UniProtKB P47811).
Substrates
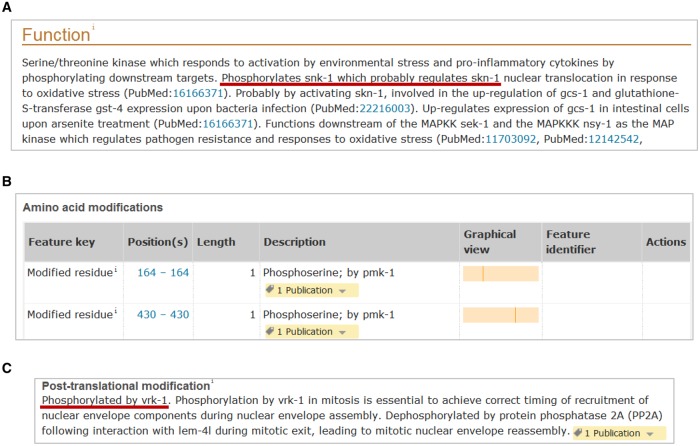
Kinase substrates are indicated in the ‘Function’ section of the kinase entry (Figure 5A). Only physiological substrates are generally reported. In vitro substrates are usually not reported unless there is convincing evidence that they could be genuine in vivo substrates. Substrates, such as MBP or histones, are often used in in vitro kinase assays, but rarely are bona fide in vivo substrates and therefore are not reported. The identification of genuine kinase substrates is challenging because kinases are capable of phosphorylating multiple substrates, especially in in vitro assays, and in vivo some substrates are context-specific [32]. Closely related kinases often share common substrates and are capable of substituting for each other. A substrate is considered genuine in UniProtKB when (1) there is experimental evidence that it is directly phosphorylated in vitro by the kinase and (2) the absence of the kinase or its inactivation in vivo abolishes phosphorylation of the substrate. Below are two examples illustrating the difficulties in assessing kinase substrates.
Figure 5. Kinase substrate annotation.
(A) Substrates are annotated in the ‘Function’ section. Example: pmk-1 (UniProtKB Q17446). (B) Phosphorylated residues and the phosphorylating kinase are recorded under ‘Amino acid modifications’. Example: snk-1 (UniProtKB P34707). (C) When the position of the phosphorylated site is unknown, the phosphorylation data are reported in the ‘Post-translational modification’ subsection. Example: baf-1 (UniProtKB Q03565).
CSNK1A1/CK1 (UniProtKB P48729) was named casein kinase (CK) for its capacity to phosphorylate casein in in vitro assays and other acidic proteins [33]. It turned out that the kinase responsible for casein phosphorylation in vivo is FAM20C (UniProtKB Q8IXL6), a Ser/Thr protein kinase localized in the Golgi, which phosphorylates secreted proteins [34]. Another example is the phosphorylation of the ribosomal subunit protein RPS6 (UniProtKB P62753). For a long time, the kinases responsible for its phosphorylation were thought to be RPS6KA1/RSK1 (UniProtKB Q15418) and RPS6KA3/RSK2 (UniProtKB P51812), but it turned out that RPS6KB1/S6K1 (UniProtKB P23443) and RPS6KB1/S6K2 (UniProtKB Q9UBS0) are the main physiological S6 kinases in cells, and Rsk kinases probably modulate RPS6 phosphorylation only under certain conditions [35–37].
In addition to curation of substrate data in kinase entries, entries are also curated to include information about the modified residues and the kinases responsible for these modifications (Figure 5B). When modified residues are known, this information is shown in the ‘Amino acid modification’ table but when the position is not known, the phosphorylation is recorded in the ‘Post-translational modifications’ subsection (Figure 5C).
EC number, catalytic activity and kinetics
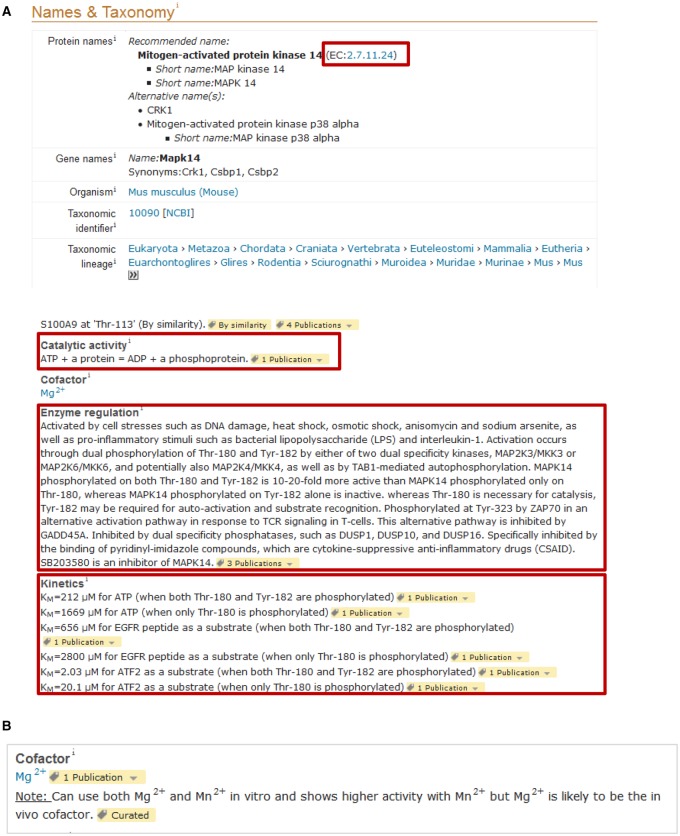
In UniProtKB, kinase activity is recorded in two separate sections (Figure 6A). An EC number corresponding to the enzymatic reaction is included in the ‘Names & Taxonomy’ section. Several EC numbers describing kinase enzymatic activity exist, based on the residues phosphorylated and substrate preferences. EC numbers with the 2.7.11.- root are specific for Ser/Thr kinases with 2.7.11.1 for kinases which have no substrate specificity or whose substrate is not known. EC 2.7.10.1 and 2.7.10.2 correspond to receptor and nonreceptor Tyr kinases (RTKs), respectively. EC 2.7.12.1 and 2.7.12.2 are for dual-specificity kinases. The complete reaction corresponding to the EC number is added in the ‘Function’ section under ‘Catalytic activity’ (Figure 6A). For pseudokinases, we do not add an EC number or a catalytic activity comment. When available, we also report kinetic parameters including the Michaelis–Menten constant (Km) and maximal velocity (Vmax), which can be found under ‘Kinetics’ in the ‘Function’ section (Figure 6A).
Figure 6. Specific annotation for kinases.
(A and B) Kinases and other enzymes contain specific sections related to their enzymatic activity including EC number, catalytic activity, cofactor and kinetics. We follow the recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (IUBMB) for the description of the enzymatic activity. Example: UniProtKB P47811.
Cofactors
The majority of kinases have an absolute requirement for a divalent cation as part of the phospho-transfer catalytic process [38]. Usually, magnesium [Mg(2+)] is the divalent cation used by most kinases. There are a few exceptions such as the peripheral plasma membrane protein CASK (UniProtKB O14936), which does not require divalent cations [39]. In in vitro kinase assays, manganese [Mn(2+)] is often present in the kinase buffer together with magnesium or alone. In some cases, such as for Tyr kinases, experimental evidence suggests that Mn(2+) works better than Mg(2+) in vitro [40,41]. Although Mn(2+) can be a cofactor in vitro, it is not clear whether it can also be used in vivo. Indeed, cells from organisms living in a normal environment contain millimolar levels of magnesium but micromolar levels of manganese, so magnesium is likely to be the in vivo cofactor [42,43]. Figure 6B shows an example of how this information is reported in an entry.
Enzyme regulation
Most kinases are usually inactive or have low basal activity in steady-state conditions. A few of them appear to be constitutively active; one example is the Ser/Thr kinase PDPK1 (UniProtKB O15530). Typically, their activation occurs in response to external/intracellular stimulus following changes in the cell environment and is tightly regulated. The importance of tight control of kinase activation is highlighted by the fact that uncontrolled activation of kinases is often the cause of disease. For example, a translocation of tyrosine kinase ABL1 (UniProtKB P00519) with the B-cell receptor gene results in deregulation of ABL1 activation and is found in patients with chronic myeloid leukemia [44,45]. UniProtKB provides information about enzyme regulatory mechanisms in the ‘Function’ section (Figure 6A). The most commonly used mechanism involves phosphorylation/dephosphorylation of residues by other kinases and phosphatases [46]. Besides phosphorylation, other regulatory mechanisms exist including ubiquitination, glycosylation, methylation and allosteric interactions. In addition to providing information about the role of PTMs in enzyme regulation, further details may be provided in the ‘PTM/Processing’ section where the PTMs may be described in more detail and where any known modified sites are indicated (UniProtKB P00533 and Figure 4B). In this section, we also record information about low-weight molecules that are known to directly activate or inhibit kinase activity.
The C. elegans kinome
Introduction
The nematode C. elegans has been used in the past 50 years as a multicellular model organism to study a wide variety of biological processes including development, neurogenesis, metabolism, aging, stress and behavioral responses [47]. Although C. elegans is a relatively simple organism, it shares many genes and molecular signaling pathways with more complex organisms including humans. A study showed that ∼38% of its genes have counterparts in humans, making it an extremely useful model to understand human biology and the development of diseases [48]. In 2002, Manning et al. published a comprehensive list of the human kinome together with the kinome of model organisms including C. elegans whose genome sequence was completed in 1998 [8,49]. When the C. elegans and human kinomes were compared, it was estimated that 55% of C. elegans kinases have human orthologs and 81% have close homologs, supporting the use of C. elegans in understanding kinase functions in human biology [48,50]. For instance, genetic studies of the C. elegans Ser/Thr kinase lrk-1 (UniProtKB Q9TZM3), a homolog of human LRRK2 (UniProtKB Q5S007) involved in Parkinson's disease, have helped to shed light on its role in the development of the nervous system and thus provided some clues to understand the progressive neurodegeneration caused by the mutation of the LRRK2 gene [51].
Identification of C. elegans kinome members in UniProtKB
The first step in the curation of the C. elegans kinome was to create a list of kinases and their corresponding UniProtKB accession numbers. To identify ePKs, we took advantage of the InterPro entry IPR000719 cross-reference, which defines the protein kinase domain, to query the database. InterPro cross-references are automatically added to UniProtKB entries and provide a classification of proteins into families as well as predicting domains and important sites [52]. The search identified 573 entries corresponding to 413 unique genes (release 2015_12). The remaining 169 entries are either isoforms or redundant sequences.
In 2005, Manning et al. published an updated list of the C. elegans kinome [50]. Before comparing it with our list, we checked the status of the genes in this list to ensure that they were still considered to be protein-coding (Supplementary Tables S1–S3). The revised list contains 393 ePKs. Comparing the two lists showed that 388 kinases are common to both lists. Twenty-five kinases were only present in the UniProtKB list (Supplementary Table S4) and five were only present in the Manning list. These five ePKs do not have a ‘typical’ kinase domain recognized by IPR000719 cross-reference, but have a ‘Protein kinase-like domain’ (InterPro entry IPR011009).
aPKs are usually difficult to identify as their kinase domain is not as well conserved as in ePKs. The Manning list contains 19 aPKs. None have a cross-reference to IPR000719 although 12 have a cross-reference to ‘Protein kinase-like domain’ InterPro entry IPR011009 (Supplementary Tables S5 and S6). There are probably more aPKs but, without experimental evidence, we decided not to add other candidates identified with IPR011009 as this InterPro entry is also found in other enzymes such as choline kinases. The difficulty in identifying potential aPKs is illustrated by the aPK, H03A11.1 (UniProtKB Q9XTW2), an ortholog of human FAM20C, which does not contain a typical protein kinase or kinase-like domain and is not found using either IPR000719 or IPR011009, but which has been shown to have kinase activity [53]. In summary, combining a UniProtKB search and the published Manning list, we generated a new list of the C. elegans kinome containing 438 unique kinases (418 ePKs and 20 aPKs) (Supplementary Table S10).
Curation of the C. elegans kinome
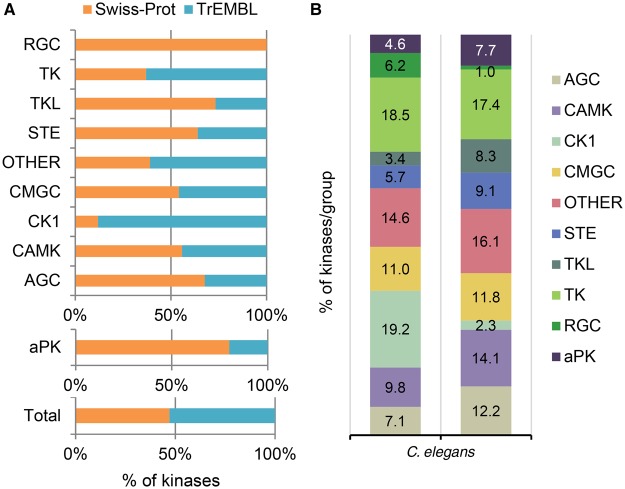
The next step was to assess the extent of the curation already available in UniProtKB. Of the 438 kinases contained in the new list, 25% (112 UniProtKB/Swiss-Prot entries) were annotated, with 73% of them having experimental evidence. We analyzed the remaining 326 UniProtKB/TrEMBL entries for associated experimental evidence available in the literature. This resulted in the functional annotation of 94 UniProtKB/TrEMBL entries and the update of 37 UniProtKB/Swiss-Prot entries with experimental data extracted from 235 references. The extent of the annotation varied; for some entries, only limited experimental data were available (gcy-20, UniProtKB O62179), whereas for others, a more comprehensive picture was obtained (cam-1, UniProtKB G5EGK5). As part of the curation process, 503 GO terms were added, including 465 related to the ‘biological process’ category, 20 related to the ‘cellular component’ category and 18 related to the ‘molecular function’ category. This extensive curatorial effort shows that almost half of the C. elegans kinome (206 kinases) has been characterized so far (Figure 7A) and new data will continue to be added as they become available.
Figure 7. Curation status overview.
(A) Curation status of C. elegans kinases by group. The percentage of kinases for each group that have been manually curated (Swiss-Prot) is shown. (B) Comparison of kinase distribution into the various groups between C. elegans and human (adapted from Manning et al. for human [8]).
Kinome characteristics
Here, we will describe some characteristics of the C. elegans kinome that have been highlighted during the expert curation process and provide some comparisons with the human kinome.
The C. elegans kinome contains members from all of the 10 kinase groups (Figure 7B). Curation affected kinases in all of these groups with the most substantial updates involving the RGC, receptor tyrosine and STE-like kinases (Figure 7A).
Although phosphorylation is the main function of kinases, it was surprising how very few studies provide experimental evidence for catalytic activity with kinase activity usually being inferred based on activity in mammalian counterparts or on the effect of mutating the predicted active site. Indeed, only 45 (21%) of the manually curated kinases have had their catalytic activity confirmed experimentally.
While the majority of kinases have only one catalytic domain, some contain two kinase domains. In humans, there are 14 ePKs with two kinase domains, of which eight have both domains functional. Eight C. elegans kinases have two predicted kinase domains. Based on similarity with human homologs, rskn-1 (UniProtKB Q21734) and rskn-2 (UniProtKB Q18846) have two functional domains, the N-terminal domain belonging to the AGC family and the C-terminal domain to the CAMK family. gcn-2 (UniProtKB D0Z5N4) and unc-89 (UniProtKB O01761) have two kinase domains, one of which is predicted to be inactive. Besides these four kinases, C. elegans contains four other entries lacking functional characterization that have two predicted kinase domains. kin-33 (UniProtKB H2KZK1), a putative member of the CAMK family, has two predicted functional kinase domains, whereas three putative members of the CK1 family, H05L14.1 (UniProtKB G5EEA3), F59A6.4 (UniProtKB Q21026) and T05A7.6 (UniProtKB Q22203), have the first kinase domain predicted to be inactive.
Identification of kinase substrates is an essential aspect of understanding kinase biological functions. During the curation of the C. elegans kinome, 19 substrates were also updated. A large-scale study of the C. elegans phosphoproteome showed that up to 2400 proteins are phosphorylated (9% of the proteome) [54], and work will continue to identify and curate these substrates.
Although 3D structures have been instrumental in understanding protein kinase domain architecture and the catalytic mechanism, only seven C. elegans kinases have their 3D structure solved, whereas >50% of human or mouse kinase 3D structures have been determined.
Alternative protein sequences
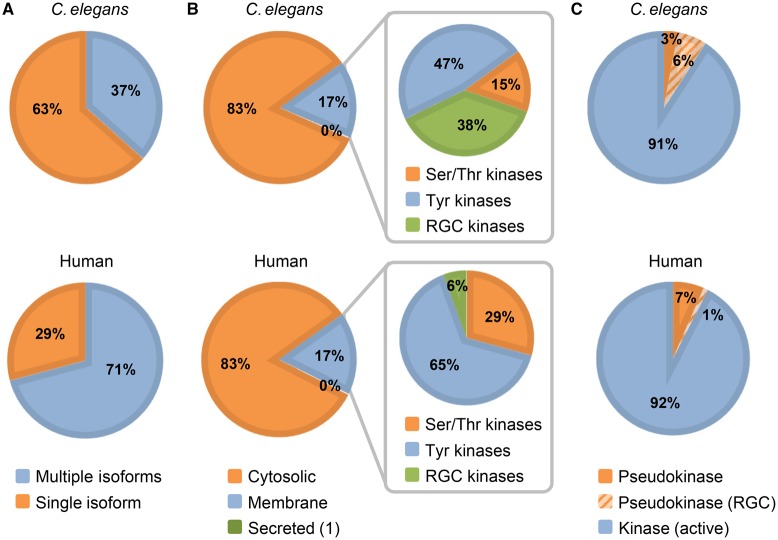
In human, splice variants or isoforms have been identified for more than two-thirds of the kinome (Figure 8A) [14]. They provide an additional level of control over biological processes, either through expression in different tissues, such as the insulin receptor (UniProtKB P06213), or different subcellular compartments, such as fibroblast growth factor receptor FGFR3 (UniProtKB P22607), or by lacking important domains or residues, such as isoform 2 of tyrosine protein kinase receptor FLT1 (UniProtKB P17948), which lacks both transmembrane and cytoplasmic domains. The UniProtKB record for each alternatively spliced protein provides the alternative protein sequences in the ‘Sequences’ section and describes how the isoform sequences differ from the displayed canonical sequence. For example, C. elegans ttn-1 (UniProtKB G4SLH0) has nine isoforms, seven generated by alternative splicing and two by alternative promoter usage. Where available, we record isoform-specific data such as function, tissue distribution and subcellular location. In C. elegans, 36% of kinases have at least one isoform in addition to the main canonical sequence, with the majority having between one and three isoforms (Figure 8A). Although several kinases have confirmed or predicted isoforms, few of them have a demonstrated role in vivo. There are only five kinases where a functional role has been shown for a splice variant: cam-1 (UniProtKB G5EGK5-1), gcy-28 (UniProtKB Q86GV3-1), egl-15 (UniProtKB Q10656-2), pct-1 (UniProtKB Q8I7M8-1) and unc-89 (UniProtKB O01761).
Figure 8. Overview of the C. elegans and human kinomes.
(A) Pie chart shows the % of kinases having one or multiple isoforms. (B) Kinase subcellular localization. The expanded section shows to which group the membrane kinases belong. (C) Distribution of active versus inactive kinases.
Additional domains
Besides their kinase domain, kinases often contain other domains, many of which provide additional ways to regulate kinase activity by promoting dynamic interactions with other proteins. For instance, the Src homology 2 (SH2) domain mediates docking to phosphorylated tyrosine residues and the Src homology 3 (SH2) domain binds to proline-rich regions on other proteins [55]. The pleckstrin homology (PH) domain that binds to membrane phosphoinositols allows changes in the kinase subcellular localization [56,57]. About 10% of C. elegans entries have predicted SH2, SH3 or PH domains. SH2 is by far the most commonly found and is mainly present, like in other species, in tyrosine kinases (Supplementary Table S7). These domains have been shown to play a crucial role in regulating kinase activity and function in mammals, but little is known about their role in C. elegans. dkf-1 (UniProtKB Q9XUJ7) is one of the few kinases where the PH domain has been shown to negatively regulate kinase activity in the absence of diacylglycerol, either by direct steric occlusion or distortion of the catalytic cleft [58]. Other domains, such as the Cdc42/Rac interactive binding motif [59], the Death domain [60] and the sterile α motif [61], have also been predicted for some C. elegans kinases (Supplementary Table S7).
Subcellular localization of kinases
As in humans, C. elegans kinase domains are not only found in ‘cytoplasmic’ proteins but also in transmembrane proteins (Figure 8B and Supplementary Table S8). ‘Cytosolic’ kinases localize mainly to the cytosol, but they are also present in the nucleus and the Golgi. So far, only one C. elegans kinase, H03A11.1 (UniProtKB Q9XTW2), is thought to be secreted based on its orthology with human FAM20C (UniProtKB Q8IXL6), which has been shown experimentally to be secreted. The majority of C. elegans kinases are single-pass membrane proteins that belong to either the RGC family (discussed below) or the TK family (Figure 8B).
Receptor-type guanylate cyclases
In C. elegans, this family consists of 27 members (6% of the kinome) whereas, in humans, only five members have been identified (<1% of the kinome) (Figure 7B). In both species, these receptors are involved in the production of the second messenger cyclic guanosine monophosphate. In humans, they are implicated in vision, cardiovascular function and skeletal growth in response to peptide hormones [62].
In C. elegans, they are involved in olfactory, light, thermal and pheromone-sensing pathways and thereby chemotaxis. They are mainly expressed in sensory neurons, often asymmetrically, allowing the integration of different environmental clues [63]. daf-11 (UniProtKB Q8I4N4) and odr-1 (UniProtKB B1Q257) are two of the best characterized RGC kinases. The kinase domain, which is always proximal to the membrane, is predicted to be inactive due to the lack of the active site aspartic acid residue although some of the receptors still contain the ATP- and metal-binding sites. To our knowledge, there is no experimental evidence of these receptors having any kinase activity. It is not clear what the role of the kinase domain is. It may act as a scaffold or, by binding to ATP, may result in conformational changes important for activation of the guanylate cyclase domain [64]. For example, deleting the kinase domain of the atrial natriuretic peptide receptor, one of the human RGCs, activates its guanylate activity independently of ligand binding [65]. In C. elegans, the function of the extracellular domain is still poorly understood, especially with regard to how it senses the variety of chemicals and environmental changes in pH and temperature. A search in UniProtKB with PROSITE signatures for both kinase (PS50011) and guanylate cyclase (PS50125) domains provides an overview of the distribution of RGCs across the taxonomic range. While they are found in almost all metazoans, the family has expanded into only a few nematode species including the genus Caenorhabditis and the parasitic roundworm Ancylostoma ceylanicum, raising interesting questions about the evolution of this particular kinase family.
Uncharacterized entries: CK1 and TK family members
The CK1 and TK families represent 19 and 18%, respectively, of the C. elegans kinome (Figure 7B). Despite being the two most abundant families, these are the least well characterized, probably because they contain kinases specific to C. elegans. Compared with humans, where the CK1 family has only 12 members (2% of the kinome), C. elegans has 84 members (Figure 7B). There is little experimental characterization available for these kinases, with experimental data so far available for only five of them. Interestingly, this expansion of the CK1 family in C. elegans suggests that they are likely to be involved in specific C. elegans functions. Although they may not be relevant for human biology, characterization of these kinases could provide information on nematode biology and shed some light on their function in nematode parasites. Similarly, the cytosolic TKs are also a poorly characterized family. Only 25% have been experimentally characterized. The majority of C. elegans cytosolic kinases (34) belong to the Fer family, which in humans has only two members, FER (UniProtKB P16591) and FES (UniProtKB P07332).
Pseudokinases
Pseudokinases represent 9% of the C. elegans kinome (Figure 8C and Supplementary Table S9). Besides the 27 RGC kinases, the C. elegans kinome contains 16 additional pseudokinases that are spread across all the groups. Half of them are transmembrane proteins and two have a second active kinase domain. For comparison, the human kinome has 39 inactive kinases (∼7% of the kinome) (Figure 8C). Assessing experimentally the constitutive lack of catalytic activity is challenging not only for kinases but also for enzymes in general. In the majority of cases, the identification of pseudokinases is based on the absence of the aspartic acid, an essential residue for the active site. Indeed, the evidence for the lack of kinase activity for 93% of the C. elegans pseudokinases is based on sequence analysis tools (Supplementary Table S9). Although they are the most common source to predict a pseudokinase, their results can be misleading. For instance, lin-18 (UniProtKB G5EGT9), a RYK human homolog (UniProtKB P34925), has the active site aspartic acid, but in vitro experiments performed with the human ortholog RYK (UniProtKB P34925) have failed to detect any kinase activity [24]. So far, only two C. elegans pseudokinases, strd-1 (UniProtKB G5ECN5) and kin-32 (UniProtKB Q95YD4), have experimental evidence supporting their lack of kinase activity (Supplementary Table S9). While pseudokinases lack the conserved catalytic residue, about half of them have the ATP-binding site conserved. Whether these sites are still functional or not will need further experimental evidence [66].
Studies in human and also in C. elegans suggest that pseudokinases act as scaffolds and therefore may participate in the assembly of multiprotein complexes. For instance, ksr-2 (UniProtKB G5EDA5) may act as a scaffold for the Ras/MAP kinase signaling cascade based on similarity with human homologs [67,68]. Similarly, strd-1 interacts with two active kinases, par-4 (UniProtKB Q9GN62) and sad-1 (UniProtKB Q19469), during the establishment of neuronal polarity and synaptic organization [69,70]. Interestingly, pat-4 (UniProtKB Q9TZC4), a protein involved in adhesion, uses its kinase domain to recruit other proteins [71,72].
Role of kinases in C. elegans biology
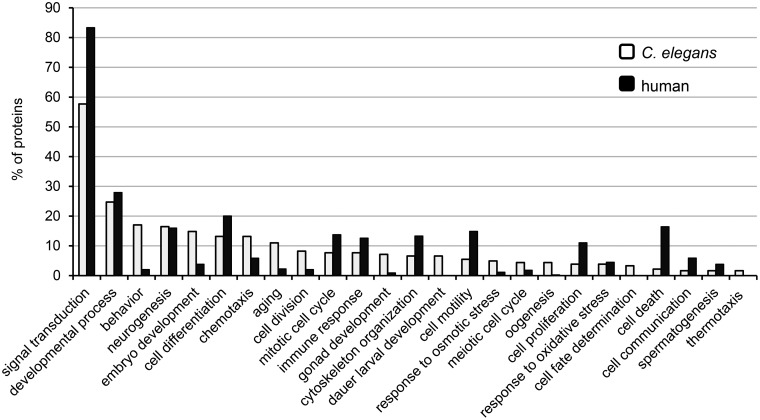
In humans, kinases control virtually all biological processes and previous analyses of the C. elegans kinome predicted, based on the high percentage of identity with human kinase genes, that its kinases were likely to have similar widespread functions [50,73]. To gain a general overview of C. elegans kinase functions, we took advantage of the GO terms associated with each of the 206 kinases that have been manually curated. These entries contain 2049 GO terms including 1379 biological process terms. To obtain a general view of which biological processes kinases are involved in, we used the GO slim tool in QuickGO (https://www.ebi.ac.uk/QuickGO/GMultiTerm#tab=introduction). A GO slim is a reduced list of GO terms that have been selected from the full set of terms available from the GO. It gives a broad overview of the ontology content for a given set of proteins. We created a customized list including 25 GO terms related to a broad range of biological processes covering the main biological functions and used it to analyze the 206 UniProtKB/Swiss-Prot kinase entries (Figure 9). Analysis of the manually annotated GO terms belonging to the ‘biological process’ category confirmed the widespread involvement of kinases in most biological processes (Figure 9). The GO term ‘signal transduction’ represents a third of the manual annotations made and is found in more than half of the UniProtKB/Swiss-Prot kinase entries, reflecting the crucial role of kinases in transducing extracellular cues into appropriate cellular responses. This aspect is discussed more in detail in the next section. Two other biological process-related GO terms heavily represented are neurogenesis and aging. This reflects the increasing attention in research that the development of the nervous system and the aging process have received in the last two decades. C. elegans has been widely used as a model to understand neurogenesis, mostly because the lineage, number and morphology of each neuron type is known and neuronal fate markers exist for all neurons [74]. For example, regulation of axonal/dendrite polarity in neurons by sad-1 (UniProtKB Q19469) was first elucidated in C. elegans before a similar role was demonstrated for the mammalian homolog Brsk1 (UniProtKB Q5RJI5) [75,76]. The aging process has also been well investigated in C. elegans, in particular the role played by the daf-2/insulin-like receptor-mediated signaling cascade (UniProtKB Q968Y9) in regulating longevity [77]. A similar role for this pathway in mammals and perhaps in humans has been partially confirmed [78]. Cell cycle is another biological process-related GO term that is present in several C. elegans kinase entries. This process, which is well conserved across all metazoans, is regulated by several kinases including the cyclin-dependent kinases (CDKs) whose main role is to promote progression through the various stages of the cell cycle. While these functions are conserved in orthologs, some of the C. elegans CDKs have additional functions, suggesting that the orthologs probably have other roles too [79]. For example, cdk-4 (UniProtKB Q9XTR1), in addition to its role in progression through the G1 phase, is also involved in sex determination during gonadogenesis by regulating the asymmetric division of the somatic gonadal precursor cell [80].
Figure 9. GO term analysis.
Percentage of proteins associated with a given GO terms. A GO slim analysis to assess the GO annotation associated with the 206 C. elegans UniProtKB/Swiss-Prot kinases and the 518 human UniProtKB/Swiss-Prot kinases was performed using the indicated general GO terms (https://www.ebi.ac.uk/QuickGO/GMultiTerm#tab=introduction).
For comparison, the same GO slim was used to analyze the 518 UniProtKB/Swiss-Prot kinase entries of the human kinome (Figure 9). Compared with the C. elegans kinome, ∼80% of kinases are associated with the GO term ‘signal transduction’ and most of them are involved in similar processes. The extent of the number of kinases involved in each process is likely to reflect constraints posed by how the kinase function can be assessed in each organism. Indeed, most experiments are mainly performed at the cellular level in humans, whereas C. elegans allows to assess kinase function at the level of the whole organism. For instance, in C. elegans, several kinases are involved in behavior, embryo development or aging, while in humans only a few appear to be involved.
Signaling cascades
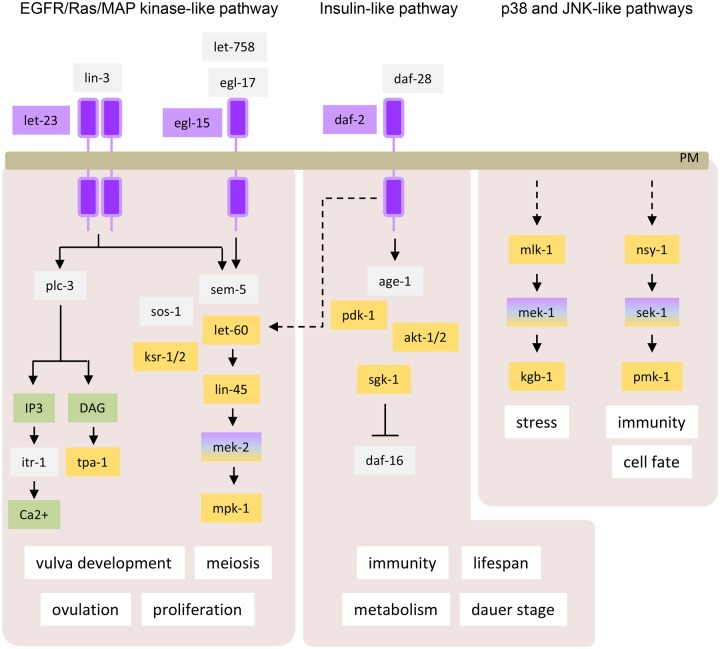
Based on the GO slim analysis described in the previous section, more than half of the C. elegans UniProtKB/Swiss-Prot kinase entries are associated with the GO term ‘signal transduction’. Indeed, protein kinases are often organized within phosphorylation signaling cascades where kinases regulate the activity of other downstream kinases and other proteins. Several of these signaling cascades are well conserved between species, and studies in C. elegans have been instrumental in identifying their components and understanding how they are sequentially organized. Among these pathways, the Ras/MAP kinase pathway, which regulates many cellular processes such as proliferation, is one of the best characterized. It consists of Ras, a small GTPase, which sequentially activates the kinases raf, MEK and ERK, which in C. elegans correspond to lin-45 (UniProtKB Q07292), mek-2 (UniProtKB Q10664) and mpk-1 (UniProtKB P39745) [81]. Although more rudimentary compared with mammals, C. elegans has developed several defense mechanisms against pathogen infections among which a p38-like pathway composed of nsy-1 (UniProtKB Q21029), sek-1 (UniProtKB G5EDF7) and pmk-1 (UniProtKB Q17446) plays an essential role [82]. As shown in Figure 10, other signaling cascades are also conserved in C. elegans, including the PI3K, p38-like and JNK-like, and phospholipase C/Ca2+ pathways. Several of these signaling cascades are activated downstream from receptors such as the RTKs, to relay extracellular cues into appropriate cellular responses. The C. elegans genome contains 33 RTKs of which 18 have been characterized including the EGFR-like egl-15 (UniProtKB Q10656) and let-23 (UniProtKB P24348), insulin-like daf-2 (UniProtKB Q968Y9), and Wnt-like cam-1 (UniProtKB G5EGK5) and lin-18 (UniProtKB G5EGT9) receptors. Similar to their functions in mammals, they play important roles in development, growth, metabolism and aging [81,83,84]. Although most of the kinases implicated in these pathways are conserved across species, the Janus tyrosine kinases, which are a component of the JAK/STAT pathway, appear to have no ortholog in C. elegans [85]. In mammals, JAK kinases act primarily downstream of cytokine receptors and play an essential role in immunity [86]. In C. elegans, no type I cytokine or type II cytokine receptor genes and only a distantly related STAT protein gene, sta-1 (UniProtKB Q9NAD6), have been identified [85]. In addition to RTKs, TGF-β receptors [daf-1 (UniProtKB P20792), daf-4 (UniProtKB P50488) and sma-5 (UniProtKB G5EBT1)] are also conserved in C. elegans where they control similar biological processes as in other species such as development and also worm-specific processes such as Dauer development (as used previously in the article), an alternative larval stage specialized for survival under harsh environmental conditions [87]. One advantage in studying the various pathways described above in C. elegans is the ease with which epistatic genetic studies can be performed, providing a unique insight not only into how signaling components are ordered but also how signaling pathways interact and modulate each other. For example, the specification of vulval cell precursor fate during vulvar development involves the co-operation between the Ras/MAP kinase and the Wnt pathways [81].
Figure 10. Three of the mammalian signaling pathways conserved in C. elegans.
Some of the C. elegans biological processes that they control are also shown. Tyr kinases are in purple, Ser/Thr kinases are in yellow, non-kinase proteins are in gray and metabolites in green. IP3, inositol 1,4,5-trisphosphate; DAG, diacylglycerol; PM, plasma membrane.
How to access and search UniProtKB data
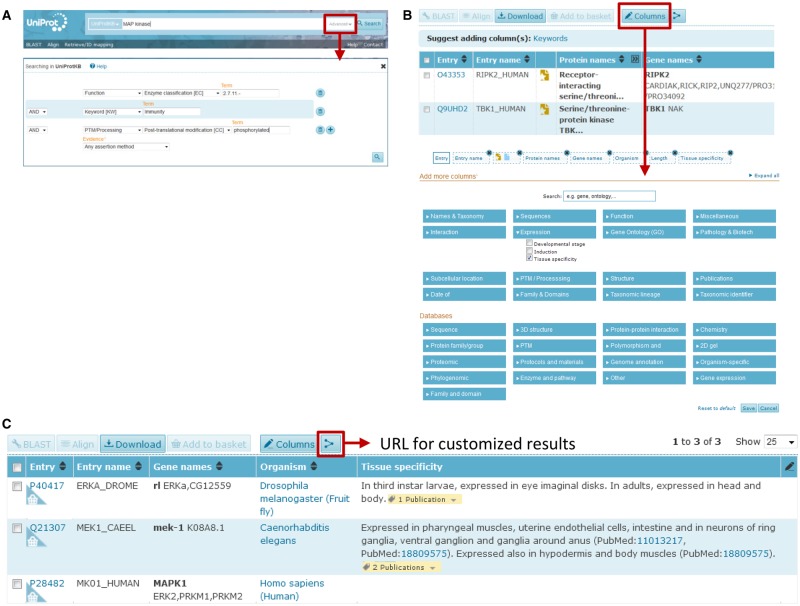
UniProtKB can be queried using the search box on the top of the website page either by typing terms directly into the box or by using the advanced search options (Figure 11A) [88]. The advanced search allows the user to restrict search terms to specific fields in a UniProtKB entry in advance or to combine multiple terms using Boolean logic (Figure 11A). For example, a user who is interested in finding all the Ser/Thr kinases that are involved in immune responses and are also phosphorylated can combine the field ‘Function’ and then ‘Enzyme classification [EC]’ in the drop-down menu where she/he enters the EC number corresponding to Ser/Thr kinases 2.7.11.-, the ‘Keyword [KW]’ field by entering ‘Immunity’ and ‘phosphorylated’ in the ‘Post-translational modification [CC]’ chosen from the ‘PTM/Processing’ drop-down menu (Figure 11A). The search can be further restricted to humans using the ‘Organism [OS]’ field, which will return nine protein entries. In this way, all the various sections of a UniProtKB entry can be queried and combined. The results can be stored in a basket and downloaded in a range of formats. The results view can be customized to include information of interest and hide unwanted data using the ‘Columns’ button (Figure 11B). Figure 10C shows how to use the ‘Columns’ button to compare the tissue localization of MEK1 homologs in three different species. In addition, it is possible to generate a unique URL for a customized results view, which can then be shared with others to aid in sharing data with collaborators. Programmatic access to the data is also provided as described at http://www.uniprot.org/help/programmatic_access.
Figure 11. The search bar and the column display tools in UniProtKB.
(A) The search bar with the advanced search button. An example of a search for Ser/Thr kinases using the advanced search (red square). (B) The ‘Columns’ tab is shown in red at the top of the result page. Clicking on this allows the customization of the sections displayed in the results page. (C) Comparison of mek-1 tissue expression in human, Drosophila and C. elegans using the ‘Columns’ tab.
Conclusion
Reversible protein phosphorylation is one of the most widespread PTMs used to regulate protein function and, unsurprisingly, kinases control virtually all cellular processes across the whole taxonomic range. Due to their importance, there is a huge wealth of information in the scientific literature not only for humans, but also for model organisms that have been instrumental in understanding kinase regulation and their biological functions. To facilitate the study of this family and also of other proteins, UniProtKB provides a searchable repository of data for a wide variety of species.
The C. elegans kinome represents ∼2% of the proteome and, similarly to other species, its kinases control virtually all aspects of nematode biology. Here, we provide an updated list of the C. elegans kinome consisting of 438 kinases including 418 ePKs and 20 aPKs. We also expertly curated >90 kinase entries, which brings the total number of kinases for which functional characterization is available to 206 kinases, representing almost 50% of the kinome. We will continue to curate members of the C. elegans kinome as more information becomes available. In the meantime, for the remaining UniProtKB/TrEMBL entries, for which no experimental data are available yet, automatic annotation already provides some predicted information based on sequence analysis and family rule-based annotation (for example, 17% have a predicted catalytic activity; mpk-2, UniProtKB H2KYF8). This information will contribute to a better understanding of kinase biological roles, not only in C. elegans, but also in more complex organisms, and will provide some insights into how this important protein family has evolved.
The curation process has highlighted some challenges in the study and the curation of kinases and their substrates. For instance, assessing kinase catalytic activity in living cells is still not straightforward, but the recent development of new techniques, such as fluorescently tagged substrate peptides or FRET-based assays, will provide valuable information on kinase activation in a physiological context [27,89]. In the last two decades, an important effort has been made to identify kinase substrates. The phosphoproteomes of several organisms either at the cell, organ or whole-body level in various conditions and developmental stages have been published [54,90,91]. In addition, several studies have also tried to identify consensus sites on substrates specific for a particular kinase [92]. The integration, annotation and display of this complex and vast amount of data are the challenges faced by UniProtKB, and work in this area is ongoing. The growing body of biomedical literature with over one million papers published every year constitutes another challenging aspect of curation, not only of kinases but also of proteins in general. Therefore, it is not surprising that concerns have been raised that literature curation is not sustainable. However, a recent study by UniProtKB has demonstrated that this process is actually sustainable and these results will be published soon (Poux et al. manuscript submitted).
By constantly updating and adapting our curation process to tailor it to the requirements of the scientific community, UniProtKB provides users with an essential tool for finding and comparing concise and accurate information, not only for kinases as shown here for the C. elegans kinases, but also for other proteins and organisms.
Database Depositions
We welcome feedback from researchers and actively encourage our user community to contact us using help@uniprot.org regarding updates or corrections to UniProtKB data. All UniProtKB data are freely available from the UniProt website at http://www.uniprot.org/.
Acknowledgements
We thank M. Courtot for the GO slim analysis. UniProt has been prepared by Alex Bateman, Maria Jesus Martin, Claire O'Donovan, Michele Magrane, Emanuele Alpi, Ricardo Antunes, Benoit Bely, Mark Bingley, Carlos Bonilla, Ramona Britto, Borisas Bursteinas, Hema Bye-A-Jee, Andrew Cowley, Alan Da Silva, Maurizio De Giorgi, Tunca Dogan, Francesco Fazzini, Leyla Garcia Castro, Luis Figueira, Penelope Garmiri, George Georghiou, Daniel Gonzalez, Emma Hatton-Ellis, Weizhong Li, Wudong Liu, Rodrigo Lopez, Jie Luo, Yvonne Lussi, Alistair MacDougall, Andrew Nightingale, Barbara Palka, Klemens Pichler, Diego Poggioli, Sangya Pundir, Luis Pureza, Guoying Qi, Steven Rosanoff, Rabie Saidi, Tony Sawford, Aleksandra Shypitsyna, Elena Speretta, Edward Turner, Nidhi Tyagi, Vladimir Volynkin, Tony Wardell, Kate Warner, Xavier Watkins, Rossana Zaru and Hermann Zellner at the European Bioinformatics Institute; Ioannis Xenarios, Lydie Bougueleret, Alan Bridge, Sylvain Poux, Nicole Redaschi, Lucila Aimo, Ghislaine Argoud-Puy, Andrea Auchincloss, Kristian Axelsen, Parit Bansal, Delphine Baratin, Marie-Claude Blatter, Brigitte Boeckmann, Jerven Bolleman, Emmanuel Boutet, Lionel Breuza, Cristina Casal-Casas, Edouard de Castro, Elisabeth Coudert, Beatrice Cuche, Mikael Doche, Dolnide Dornevil, Severine Duvaud, Anne Estreicher, Livia Famiglietti, Marc Feuermann, Elisabeth Gasteiger, Sebastien Gehant, Vivienne Gerritsen, Arnaud Gos, Nadine Gruaz-Gumowski, Ursula Hinz, Chantal Hulo, Florence Jungo, Guillaume Keller, Vicente Lara, Philippe Lemercier, Damien Lieberherr, Thierry Lombardot, Xavier Martin, Patrick Masson, Anne Morgat, Teresa Neto, Nevila Nouspikel, Salvo Paesano, Ivo Pedruzzi, Sandrine Pilbout, Monica Pozzato, Manuela Pruess, Catherine Rivoire, Bernd Roechert, Michel Schneider, Christian Sigrist, Karin Sonesson, Sylvie Staehli, Andre Stutz, Shyamala Sundaram, Michael Tognolli, Laure Verbregue and Anne-Lise Veuthey at the SIB Swiss Institute of Bioinformatics; Cathy H. Wu, Cecilia N. Arighi, Leslie Arminski, Chuming Chen, Yongxing Chen, John S. Garavelli, Hongzhan Huang, Kati Laiho, Peter McGarvey, Darren A. Natale, Karen Ross, C.R. Vinayaka, Qinghua Wang, Yuqi Wang, Lai-Su Yeh and Jian Zhang at the Protein Information Resource.
Abbreviations
- aPKs
atypical kinases
- CDKs
cyclin-dependent kinases
- EC number
Enzyme Commission number
- ECO
Evidence and Conclusion Ontology
- ePKs
eukaryotic kinases
- GO
Gene Ontology
- LRRK2
leucine-rich repeat serine/threonine-protein kinase 2
- MBP
myelin basic protein
- PH
pleckstrin homology
- PTMs
post-translational modifications
- RGC
receptor-type guanylate cyclase
- RTK
receptor Tyr kinase
- Ser/Thr kinases
serine/threonine kinases
- SH2
Src homology 2
- TK
tyrosine kinase
- TKL
tyrosine kinase-like
- Tyr kinases
tyrosine kinases
- UniProtKB
UniProt Knowledgebase.
Funding
UniProt is supported by the National Institutes of Health [U41HG007822, U41HG002273, R01GM080646, P20GM103446 and U01GM120953]; British Heart Foundation [RG/13/5/30112]; Parkinson's Disease United Kingdom [G-1307] and Swiss Federal Government through the State Secretariat for Education, Research and Innovation and European Molecular Biology Laboratory core funds. Funding for open access charge: National Institutes of Health [U41HG007822].
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P. et al. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 doi: 10.1016/j.cell.2006.09.026 [DOI] [PubMed] [Google Scholar]
- 2.Leto D. and Saltiel A.R. (2012) Regulation of glucose transport by insulin: traffic control of GLUT4. Nat. Rev. Mol. Cell. Biol. 13, 383–396 doi: 10.1038/nrm3351 [DOI] [PubMed] [Google Scholar]
- 3.Lim P.S., Sutton C.R. and Rao S. (2015) Protein kinase C in the immune system: from signalling to chromatin regulation. Immunology 146, 508–522 doi: 10.1111/imm.12510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hur E.-M. and Zhou F.-Q. (2010) GSK3 signalling in neural development. Nat. Rev. Neurosci. 11, 539–551 doi: 10.1038/nrn2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cargnello M. and Roux P.P. (2011) Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83. doi: 10.1128/MMBR.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendoza M.C., Er E.E. and Blenis J. (2011) The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 36, 320–328 doi: 10.1016/j.tibs.2011.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen P. (2014) Immune diseases caused by mutations in kinases and components of the ubiquitin system. Nat. Immunol. 15, 521–529 doi: 10.1038/ni.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manning G., Whyte D.B., Martinez R., Hunter T. and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 doi: 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 9.LaRonde-LeBlanc N. and Wlodawer A. (2005) The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim. Biophys. Acta 1754, 14–24 doi: 10.1016/j.bbapap.2005.07.037 [DOI] [PubMed] [Google Scholar]
- 10.Rask-Andersen M., Zhang J., Fabbro D. and Schiöth H.B. (2014) Advances in kinase targeting: current clinical use and clinical trials. Trends Pharmacol. Sci. 35, 604–620 doi: 10.1016/j.tips.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 11.Kachergus J., Mata I.F., Hulihan M., Taylor J.P., Lincoln S., Aasly J. et al. (2005) Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am. J. Hum. Genet. 76, 672–680 doi: 10.1086/429256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appert-Collin A., Hubert P., Crémel G. and Bennasroune A. (1015) Role of ErbB receptors in cancer cell migration and invasion. Front. Pharmacol. 6, 283 doi: 10.3389/fphar.2015.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmignac V., Salih M.A.M., Quijano-Roy S., Marchand S., Al Rayess M.M., Mukhtar M.M. et al. (2007) C-terminal titin deletions cause a novel early-onset myopathy with fatal cardiomyopathy. Ann. Neurol. 61, 340–351 doi: 10.1002/ana.21089 [DOI] [PubMed] [Google Scholar]
- 14.Quintaje S.B. and Orchard S. (2008) The annotation of both human and mouse kinomes in UniProtKB/Swiss-Prot: one small step in manual annotation, one giant leap for full comprehension of genomes. Mol. Cell. Proteomics 7, 1409–1419 doi: 10.1074/mcp.R700001-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43(D1), D204–D212 doi: 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magrane M. and Consortium U. (2011) UniProt knowledgebase: a hub of integrated protein data. Database 2011, bar009doi: 10.1093/database/bar009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poux S., Magrane M., Arighi C.N., Bridge A., O'Donovan C. and Laiho K. (2014) Expert curation in UniProtKB: a case study on dealing with conflicting and erroneous data. Database 2014, bau016 doi: 10.1093/database/bau016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chibucos M.C., Mungall C.J., Balakrishnan R., Christie K.R., Huntley R.P., White O. et al. (2014) Standardized description of scientific evidence using the evidence ontology (ECO). Database 2014, bau075 doi: 10.1093/database/bau075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gene Ontology Consortium (2015) Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 doi: 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntley R.P., Sawford T., Mutowo-Meullenet P., Shypitsyna A., Bonilla C., Martin M.J. et al. (2015) The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 43(D1), D1057–D1063 doi: 10.1093/nar/gku1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanks S.K., Quinn A.M. and Hunter T. (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52 doi: 10.1126/science.3291115 [DOI] [PubMed] [Google Scholar]
- 22.Sigrist C.J.A., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A. et al. (2013) New and continuing developments at PROSITE. Nucleic Acids Res. 41(D1), D344–D347 doi: 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L. et al. (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1), D279–D285 doi: 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katso R.M., Russell R.B. and Ganesan T.S. (1999) Functional analysis of H-Ryk, an atypical member of the receptor tyrosine kinase family. Mol. Cell. Biol. 19, 6427–6440 doi: 10.1128/MCB.19.9.6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu B.-e., English J.M., Wilsbacher J.L., Stippec S., Goldsmith E.J. and Cobb M.H. (2000) WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J. Biol. Chem. 275, 16795–16801 doi: 10.1074/jbc.275.22.16795 [DOI] [PubMed] [Google Scholar]
- 26.Hastie C.J., McLauchlan H.J. and Cohen P. (2006) Assay of protein kinases using radiolabeled ATP: a protocol. Nat. Protoc. 1, 968–971 doi: 10.1038/nprot.2006.149 [DOI] [PubMed] [Google Scholar]
- 27.Morris M.C. (2013) Fluorescent biosensors — probing protein kinase function in cancer and drug discovery. Biochim. Biophys. Acta 1834, 1387–1395 doi: 10.1016/j.bbapap.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 28.Chamoli M., Singh A., Malik Y. and Mukhopadhyay A. (2014) A novel kinase regulates dietary restriction-mediated longevity in Caenorhabditis elegans. Aging Cell 13, 641–655 doi: 10.1111/acel.12218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haubrich B.A. and Swinney D.C. (2016) Enzyme activity assays for protein kinases: strategies to identify active substrates. Curr. Drug Discov. Technol. 13, 2–15 doi: 10.2174/1570163813666160115125930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter T. and Plowman G.D. (1997) The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22, 18–22 doi: 10.1016/S0968-0004(96)10068-2 [DOI] [PubMed] [Google Scholar]
- 31.Zeqiraj E., Filippi B.M., Goldie S., Navratilova I., Boudeau J., Deak M. et al. (2009) ATP and MO25α regulate the conformational state of the STRADα pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 7, e1000126 doi: 10.1371/journal.pbio.1000126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue L. and Tao W.A. (2013) Current technologies to identify protein kinase substrates in high throughput. Front. Biol. 8, 216–227 doi: 10.1007/s11515-013-1257-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinna L.A., Baggio B., Moret V. and Siliprandi N. (1969) Isolation and properties of a protein kinase from rat liver microsomes. Biochim. Biophys. Acta 178, 199–201 doi: 10.1016/0005-2744(69)90152-1 [DOI] [PubMed] [Google Scholar]
- 34.Tagliabracci V.S., Pinna L.A. and Dixon J.E. (2013) Secreted protein kinases. Trends Biochem. Sci. 38, 121–130 doi: 10.1016/j.tibs.2012.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pende M., Um S.H., Mieulet V., Sticker M., Goss V.L., Mestan J. et al. (2004) S6k1−/−/S6K2−/− Mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24, 3112–3124 doi: 10.1128/MCB.24.8.3112-3124.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erikson E. and Maller J.L. (1986) Purification and characterization of a protein kinase from Xenopus eggs highly specific for ribosomal protein S6. J. Biol. Chem. 261, 350–355 PMID: [PubMed] [Google Scholar]
- 37.Roux P.P., Shahbazian D., Vu H., Holz M.K., Cohen M.S., Taunton J. et al. (2007) RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282, 14056–14064 doi: 10.1074/jbc.M700906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams J.A. (2001) Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 101, 2271–2290 doi: 10.1021/cr000230w [DOI] [PubMed] [Google Scholar]
- 39.Mukherjee K., Sharma M., Urlaub H., Bourenkov G.P., Jahn R., Südhof T.C. et al. (2008) CASK functions as a Mg2+-independent neurexin kinase. Cell 133, 328–339 doi: 10.1016/j.cell.2008.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper J.A. and King C.S. (1986) Dephosphorylation or antibody binding to the carboxy terminus stimulates pp60c-src. Mol. Cell. Biol. 6, 4467–4477 doi: 10.1128/MCB.6.12.4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grace M.R., Walsh C.T. and Cole P.A. (1997) Divalent ion effects and insights into the catalytic mechanism of protein tyrosine kinase Csk. Biochemistry 36, 1874–1881 doi: 10.1021/bi962138t [DOI] [PubMed] [Google Scholar]
- 42.Romani A. and Scarpa A. (1992) Regulation of cell magnesium. Arch. Biochem. Biophys. 298, 1–12 doi: 10.1016/0003-9861(92)90086-C [DOI] [PubMed] [Google Scholar]
- 43.Milne D.B., Sims R.L. and Ralston N.V. (1990) Manganese content of the cellular components of blood. Clin. Chem. 36, 450–452 PMID: [PubMed] [Google Scholar]
- 44.Lugo T.G., Pendergast A.M., Muller A.J. and Witte O.N. (1990) Tyrosine kinase activity and transformation potency of bcr-abl oncogene products. Science 247, 1079–1082 doi: 10.1126/science.2408149 [DOI] [PubMed] [Google Scholar]
- 45.Faderl S., Talpaz M., Estrov Z., O'Brien S., Kurzrock R. and Kantarjian H.M. (1999) The biology of chronic myeloid leukemia. N. Engl. J. Med. 341, 164–172 doi: 10.1056/NEJM199907153410306 [DOI] [PubMed] [Google Scholar]
- 46.Deribe Y.L., Pawson T. and Dikic I. (2010) Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 17, 666–672 doi: 10.1038/nsmb.1842 [DOI] [PubMed] [Google Scholar]
- 47.Corsi A.K., Wightman B. and Chalfie M.A. (2015) Transparent Window Into Biology: A Primer on Caenorhabditis elegans, pp. 1–31, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaye D.D. and Greenwald I. (2011) Ortholist: a compendium of C. elegans genes with human orthologs . PLoS ONE 6, e20085 doi: 10.1371/journal.pone.0020085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 doi: 10.1126/science.282.5396.2012 [DOI] [PubMed] [Google Scholar]
- 50.Manning G. (2005) Genomic Overview of Protein Kinases, pp. 1–19, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao C., El Khoury R., Wang W., Byrd T.A., Pehek E.A., Thacker C. et al. (2010) LRRK2-mediated neurodegeneration and dysfunction of dopaminergic neurons in a Caenorhabditis elegans model of Parkinson's disease. Neurobiol. Dis. 40, 73–81 doi: 10.1016/j.nbd.2010.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell A., Chang H.-Y., Daugherty L., Fraser M., Hunter S., Lopez R. et al. (2015) The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43(D1), D213–D221 doi: 10.1093/nar/gku1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao J., Tagliabracci V.S., Wen J., Kim S.-A. and Dixon J.E. (2013) Crystal structure of the Golgi casein kinase. Proc. Natl Acad. Sci. U.S.A. 110, 10574–10579 doi: 10.1073/pnas.1309211110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zielinska D.F., Gnad F., Jedrusik-Bode M., Wis´niewski J.R. and Mann M. (2009) Caenorhabditis elegans has a phosphoproteome atypical for metazoans that is enriched in developmental and sex determination proteins. J. Proteome Res. 8, 4039–4049 doi: 10.1021/pr900384k [DOI] [PubMed] [Google Scholar]
- 55.Pawson T. and Schlessingert J. (1993) SH2 and SH3 domains. Curr. Biol. 3, 434–442 doi: 10.1016/0960-9822(93)90350-W [DOI] [PubMed] [Google Scholar]
- 56.Haslam R.J., Koide H.B. and Hemmings B.A. (1993) Pleckstrin domain homology. Nature 363, 309–310 doi: 10.1038/363309b0 [DOI] [PubMed] [Google Scholar]
- 57.Saraste M. and Hyvönen M. (1995) Pleckstrin homology domains: a fact file. Curr. Opin. Struct. Biol. 5, 403–408 doi: 10.1016/0959-440X(95)80104-9 [DOI] [PubMed] [Google Scholar]
- 58.Feng H., Ren M. and Rubin C.S. (2006) Conserved domains subserve novel mechanisms and functions in DKF-1, a Caenorhabditis elegans protein kinase D. J. Biol. Chem. 281, 17815–17826 doi: 10.1074/jbc.M511898200 [DOI] [PubMed] [Google Scholar]
- 59.Burbelo P.D., Drechsel D. and Hall A. (1995) A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J. Biol. Chem. 270, 29071–29074 doi: 10.1074/jbc.270.49.29071 [DOI] [PubMed] [Google Scholar]
- 60.Feinstein E., Kimchi A., Wallach D., Boldin M. and Varfolomeev E. (1995) The death domain: a module shared by proteins with diverse cellular functions. Trends Biochem. Sci. 20, 342–344 doi: 10.1016/S0968-0004(00)89070-2 [DOI] [PubMed] [Google Scholar]
- 61.Stapleton D., Balan I., Pawson T. and Sicheri F. (1999) The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6, 44–49 doi: 10.1038/4917 [DOI] [PubMed] [Google Scholar]
- 62.Sharma R.K. and Duda T. (2014) Membrane guanylate cyclase, a multimodal transduction machine: history, present, and future directions. Front. Mol. Neurosci. 7, 56 doi: 10.3389/fnmol.2014.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ortiz C.O., Etchberger J.F., Posy S.L., Frøkjaer-Jensen C., Lockery S., Honig B. et al. (2006) Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173, 131–149 doi: 10.1534/genetics.106.055749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaleel M., Saha S., Shenoy A.R. and Visweswariah S.S. (2006) The kinase homology domain of receptor guanylyl cyclase C: ATP binding and identification of an adenine nucleotide sensitive site. Biochemistry 45, 1888–1898 doi: 10.1021/bi052089x [DOI] [PubMed] [Google Scholar]
- 65.Chinkers M. and Garbers D.L. (1989) The protein kinase domain of the ANP receptor is required for signaling. Science 245, 1392–1394 doi: 10.1126/science.2571188 [DOI] [PubMed] [Google Scholar]
- 66.Murphy J.M., Zhang Q., Young S.N., Reese M.L., Bailey F.P., Eyers P.A. et al. (2014) A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457, 323–334 doi: 10.1042/BJ20131174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang H., Koo C.Y., Stebbing J. and Giamas G. (2013) The dual function of KSR1: a pseudokinase and beyond. Biochem. Soc. Trans. 41, 1078–1082 doi: 10.1042/BST20130042 [DOI] [PubMed] [Google Scholar]
- 68.Clapéron A. and Therrien M. (2007) KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene 26, 3143–3158 doi: 10.1038/sj.onc.1210408 [DOI] [PubMed] [Google Scholar]
- 69.Narbonne P., Hyenne V., Li S., Labbe J.-C. and Roy R. (2010) Differential requirements for STRAD in LKB1-dependent functions in C. elegans. Development 137, 661–670 doi: 10.1242/dev.042044 [DOI] [PubMed] [Google Scholar]
- 70.Kim J.S.M., Hung W., Narbonne P., Roy R. and Zhen M. (2010) C. elegans STRADα and SAD cooperatively regulate neuronal polarity and synaptic organization. Development 137, 93–102 doi: 10.1242/dev.041459 [DOI] [PubMed] [Google Scholar]
- 71.Mackinnon A.C., Qadota H., Norman K.R., Moerman D.G. and Williams B.D. (2002) C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 12, 787–797 doi: 10.1016/S0960-9822(02)00810-2 [DOI] [PubMed] [Google Scholar]
- 72.Lin X., Qadota H., Moerman D.G. and Williams B.D. (2003) C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr. Biol. 13, 922–932 doi: 10.1016/S0960-9822(03)00372-5 [DOI] [PubMed] [Google Scholar]
- 73.Plowman G.D., Sudarsanam S., Bingham J., Whyte D. and Hunter T. (1999) The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc. Natl Acad. Sci. U.S.A. 96, 13603–13610 doi: 10.1073/pnas.96.24.13603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hobert O. (2010) Neurogenesis in the Nematode Caenorhabditis elegans, pp. 1–24, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kishi M., Pan Y.A., Crump J.G. and Sanes J.R. (2005) Mammalian SAD kinases are required for neuronal polarization. Science 307, 929–932 doi: 10.1126/science.1107403 [DOI] [PubMed] [Google Scholar]
- 76.Crump J.G., Zhen M., Jin Y. and Bargmann C.I. (2001) The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron 29, 115–129 doi: 10.1016/S0896-6273(01)00184-2 [DOI] [PubMed] [Google Scholar]
- 77.Lapierre L.R. and Hansen M. (2012) Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol. Metab. 23, 637–644 doi: 10.1016/j.tem.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bitto A., Wang A.M., Bennett C.F. and Kaeberlein M. (2015) Biochemical genetic pathways that modulate aging in multiple species. Cold Spring Harb. Perspect. Med. 5 doi: 10.1101/cshperspect.a025114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lim S. and Kaldis P. (2013) Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 140, 3079–3093 doi: 10.1242/dev.091744 [DOI] [PubMed] [Google Scholar]
- 80.Tilmann C. and Kimble J. (2005) Cyclin D regulation of a sexually dimorphic asymmetric cell division. Dev. Cell 9, 489–499 doi: 10.1016/j.devcel.2005.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sundaram M.V. (2013) Canonical RTK-Ras-ERK Signaling and Related Alternative Pathways, pp. 1–38, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim D.H. and Ewbank J.J. (2015) Signaling in the Innate Immune Response, pp. 1–51, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawa H. and Korswagen H.C. (2013) Wnt Signaling in C. elegans, pp. 1–30, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murphy C.T. and Hu P.J. (2013) Insulin/Insulin-Like Growth Factor Signaling in C. elegans, pp. 1–43, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boulay J.-L., O'Shea J.J. and Paul W.E. (2003) Molecular phylogeny within type I cytokines and their cognate receptors. Immunity 19, 159–163 doi: 10.1016/S1074-7613(03)00211-5 [DOI] [PubMed] [Google Scholar]
- 86.O'Shea J.J. and Plenge R. (2012) JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 36, 542–550 doi: 10.1016/j.immuni.2012.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gumienny T.L. and Savage-Dunn C. (2013) TGF-β Signaling in C. elegans, pp. 1–34, WormBook; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pundir S., Magrane M., Martin M.J., O'Donovan C. and UniProt Consortium (2015) Searching and navigating uniProt databases. Curr. Protoc. Bioinformatics 50, 1.27.1–1.27.10 doi: 10.1002/0471250953.bi0127s50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnson S.A. and Hunter T. (2005) Kinomics: methods for deciphering the kinome. Nat. Methods 2, 17–25 doi: 10.1038/nmeth731 [DOI] [PubMed] [Google Scholar]
- 90.Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J. et al. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 doi: 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- 91.Bridon G., Bonneil E., Muratore-Schroeder T., Caron-Lizotte O. and Thibault P. (2012) Improvement of phosphoproteome analyses using FAIMS and decision tree fragmentation. Application to the insulin signaling pathway in Drosophila melanogaster S2 cells. J. Proteome Res. 11, 927–940 doi: 10.1021/pr200722s [DOI] [PubMed] [Google Scholar]
- 92.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V. and Skrzypek E. (2015) Phosphositeplus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43(D1), D512–D520 doi: 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Reference deleted.