Abstract
Nα acetylation is one of the most abundant protein modifications in eukaryotes and is catalyzed by N-terminal acetyltransferases (NATs). NatA, the major NAT in Saccharomyces cerevisiae, consists of the subunits Nat1p, Ard1p, and Nat5p and is necessary for the assembly of repressive chromatin structures. Here, we found that Orc1p, the large subunit of the origin recognition complex (ORC), required NatA acetylation for its role in telomeric silencing. NatA functioned genetically through the ORC binding site of the HMR-E silencer. Furthermore, tethering Orc1p directly to the silencer circumvented the requirement for NatA in silencing. Orc1p was Nα acetylated in vivo by NatA. Mutations that abrogated its ability to be acetylated caused strong telomeric derepression. Thus, Nα acetylation of Orc1p represents a protein modification that modulates chromatin function in S. cerevisiae. Genetic evidence further supported a functional link between NatA and ORC: (i) nat1Δ was synthetically lethal with orc2-1 and (ii) the synthetic lethality between nat1Δ and SUM1-1 required the Orc1 N terminus. We also found Sir3p to be acetylated by NatA. In summary, we propose a model by which Nα acetylation is required for the binding of silencing factors to the N terminus of Orc1p and Sir3p to recruit heterochromatic factors and establish repression.
N-terminal acetylation is one of the most common protein modifications in eukaryotes and is found on ca. 85% of all mammalian and 50% of all yeast proteins (35). It occurs cotranslationally at the α-N position of the initiator methionine of nascent polypeptides when 20 to 50 amino acids are protruding from the ribosome. Alternatively, the methionine is cleaved off by specific aminopeptidases, and the newly exposed residue is then acetylated by one of several N-terminal acetyltransferases (NATs). Nα acetylation is an irreversible process, unlike the reversible posttranslational Nɛ acetylation of internal lysine residues in histones and transcription factors (8) and thus is likely to be functionally distinct.
Despite the prevalence of this modification, little is known about its biological relevance. It was earlier suggested to protect proteins from degradation, but this hypothesis is no longer favored (35). Insight into the function of Nα acetylation comes from the analysis of NATs in Saccharomyces cerevisiae. Here, three NAT complexes exist that are known as NatA, NatB, and NatC. One function of NatB is that tropomyosin requires NatB-dependent acetylation to interact with actin (41). NatC acetylates the major coat protein gag of the L-A double-stranded RNA virus, which is a prerequisite for the assembly of virus particles (48). The three NAT complexes contain as their catalytic subunit the proteins Ard1p, Nat3p, and Mak3p, respectively, which are members of the GNAT (GCN5-related N-acetyltransferase) superfamily of acetyltransferases. Recently, the novel GNAT-homolog Nat4p was identified to specifically acetylate histones H2A and H4 (43).
The NAT complexes are distinguished by their substrate specificity. NatA accounts for the majority of N-terminally acetylated proteins and acetylates substrates with an alanine or serine and, less frequently, a threonine or glycine at the penultimate position. NatA acetylation occurs after removal of the initial methionine, whereas the other two NATs, NatB and NatC, acetylate the N-terminal methionine (35). Substrate specificity is determined predominantly, but not exclusively, by the penultimate amino acid, and other degenerate signals are proposed to lie within the first 50 amino acids of the nascent polypeptide. NATs also exist in higher eukaryotes, and sequence homologs have been identified in the genomes of all model organisms (35). Thus far, homologs of Nat1p and Ard1p have been implicated in development and cellular proliferation, and human NATH was found to be overexpressed in malignant tumors (11).
In addition to its catalytic subunit Ard1p, NatA consists of Nat1p and Nat5p (14, 31). Nat1p anchors NatA to the ribosome, can be cross-linked to nascent polypeptide chains, and is required for NatA's enzymatic activity. Nat5p is a GNAT-family acetyltransferase whose function within the complex remains to be characterized (14). The deletion of NAT1 or ARD1 results in the same pleiotropic phenotypes of slow growth, temperature sensitivity, chromosomal instability, inability to enter G0, and failure to sporulate as homozygous diploids (31, 32, 52). The overexpression of both Nat1p and Ard1p is required to increase NatA activity in vivo (34). Importantly, NatA also functions in transcriptional repression. nat1Δ and ard1Δ cause strong derepression of the HML silent mating-type locus and of subtelomeric reporter genes (1, 31). This suggests that one or several proteins require N-terminal acetylation by NatA in order to function in transcriptional silencing.
The term silencing describes the stable repression of genes within certain genomic regions through the formation of specialized heterochromatin-like structures. This type of gene regulation exists in all eukaryotes and has essential functions in cellular differentiation and developmental processes (18). In S. cerevisiae, heterochromatin-like domains include the silent mating-type loci HML and HMR, the telomeres and the rDNA locus. The HM loci are repressed by flanking silencer sequences, E and I, which consist of binding sites for Rap1p, Abf1p, and the origin recognition complex (ORC) (38). Besides its role in silencing, the six-subunit ORC complex is the eukaryotic replication initiator complex (3, 15) and is highly conserved between yeast and larger eukaryotes. However, the initiation function of ORC is not required for silencing. Rather, ORC at the silencers recruits the silent information regulator Sir1p via the N terminus of its large subunit, Orc1p (13). Consistent with this, the deletion of the first 235 amino acids of Orc1p results in impaired silencing, while having no effect on replication (4). Silencing at the HM loci is then established by the interaction of Sir1p and Rap1p with Sir2p, Sir3p, and Sir4p. Sir3p and Sir4p bind to the hypoacetylated N-terminal tails of histones H3 and H4, which are deacetylated by the NAD+-dependent histone deacetylase Sir2 (20, 24). Thus, the multimeric silencing complex spreads along the chromosome and generates a repressive chromatin structure in this region (21). Interestingly, the N-terminal 214 amino acids of Orc1p and Sir3p are very similar (50% identity, 63% similarity), both containing a BAH (for bromo-adjacent homology) domain of ca. 140 amino acids that is predicted to mediate protein-protein interactions (4, 6). However, despite their similarity, Sir3p cannot bind to Sir1p, and the binding of Orc1p to Sir1p occurs via a small nonconserved subdomain of Orc1p between amino acids 100 and 129 (54). Notably, point mutations within the BAH domain of Sir3p lead to defective HM and telomeric silencing, whereas the domains for interaction with Sir4p, Rap1p and histones are located in the C terminus of the protein (44).
Under normal conditions, Sir2p, Sir3p, and Sir4p are essential for silencing. Interestingly, a Sir-independent mode of silencing has been characterized that is mediated by SUM1-1, a dominant mutation in the SUM1 gene that encodes a mitotic repressor of meiotic genes. The Sum1-1 protein binds to ORC at the silencers and recruits the Sir2p homolog Hst1p to establish silencing that does not require the Sir proteins (47).
Similar to the HM loci, the telomeres also contain binding sites for ORC and Abf1p in the subtelomeric core X region and for Rap1p in the terminal TG1-3 repeats. Together, they recruit Sir2p, Sir3p, and Sir4p to form heterochromatin-like structures (19). In contrast to HM silencing, the repression of telomere proximal genes is semistable and independent of functional Sir1p (9). Further stabilization is proposed to be achieved by the formation of a loop structure at each chromosomal end and the clustering of several telomeres into foci (19).
The silencing mechanism at the nucleolar ribosomal DNA (rDNA) locus differs from HM and telomeric silencing in that it requires only Sir2p and not the other Sir proteins. Together with Net1p and Cdc14p, Sir2p is part of the RENT silencing complex, which locates to two distinct regions within the rDNA repeats (22).
The involvement of NatA in silencing suggests that the silencing function of one or several silencing proteins is regulated by Nα acetylation. However, the identity of these proteins is not known. Ribosomal proteins are frequent targets of NatA (2), but they are unlikely to play a role in silencing. Histone H2B is another known NatA substrate, but the deletion of its N terminus has no effect on silencing (27). Genetic interactions have been described between NAT1 or ARD1 and two silencing determinants, SIR1 and SIR3 (44, 45), thus rendering them possible candidates. However, neither has directly been implicated in NatA-dependent silencing.
In the present study, we have identified a silencing-relevant target of NatA. We found that NatA was required for all forms of silencing in S. cerevisiae and was necessary for the structural and functional integrity of the telomeric foci. Genetically, nat1Δ functioned through the ORC binding site of the HMR-E silencer. The requirement for NatA in silencing could be bypassed by artificially tethering Orc1p, but not the other Orc proteins, to the silencer, thus suggesting that Orc1p was a silencing-relevant NatA target. We found Orc1p to be fully Nα acetylated in wild-type and completely unacetylated in nat1Δ strains. Mutations in the penultimate residue of Orc1p that inhibited its ability to be acetylated by NatA caused a severe loss of telomeric silencing, as does the deletion of NAT1. The lack of acetylation did not affect the interaction of Orc1p with Sir1p, since HM silencing was not impaired in the orc1 mutants and still depended on functional SIR1 in nat1Δ strains. Nα acetylation was specifically required for the silencing function of Orc1, since the unacetylated orc1 mutants showed no replication defect. Genetic interactions further supported a functional link between NatA and ORC: nat1Δ was synthetically lethal with the replication-defective orc2-1 mutation. Furthermore, nat1Δ displayed synthetic lethality with SUM1-1. Intriguingly, this lethality was suppressed by a deletion in the N terminus of Orc1p, thus suggesting that Nα acetylation regulated the interaction of Orc1p with Sum1-1p. We further identified Sir3p as a target of NatA. Since the mutation of the penultimate amino acid of Sir3p also caused silencing defects, we likewise propose that Sir3p's silencing function requires NatA-dependent Nα acetylation.
In summary, we have provided the first direct evidence for the functional dependence of the silencing factors Orc1p and Sir3p on NatA-mediated N-terminal acetylation. Thus, N-terminal protein acetylation joins other irreversible protein modifications, such as lysine or arginine methylation, as a novel mechanism for modulating chromatin function.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strains and plasmids used in the present study are listed in Tables 1 and 2, respectively. Yeast was grown and manipulated according to standard procedures. Endogenous ORC1 was disrupted in a diploid strain carrying pAE405 and the two HMR alleles HMR SS ΔI and HMR SS abf1−ΔI by using the PCR-mediated knockout technique. The complete open reading frame of ORC1 plus 200 bp of upstream sequence was replaced by a fragment containing SpHIS5 GFP. Haploid orc1Δ strains were obtained by sporulation and tetrad dissection. For orc1-A2V and orc1-A2P strains, point mutant orc1 alleles created by site-directed mutagenesis on integrative plasmids were introduced into the LEU2 locus of AEY2866 and AEY2867, followed by elimination of pAE405 on 5-fluoorotic acid (5-FOA) medium. sir3-A2P strains were constructed by integrative transformation of PstI-linearized pAE1027, which carries a KpnI/HindIII fragment of sir3-A2P (1 to 502 bp) and its promoter region. The A2P point mutation was introduced by site-directed mutagenesis. TAP-tagged versions of ORC1 and SIR3 controlled by their natural promoters were integrated into the URA3 locus of AEY1558 and AEY2706 by transforming the strains with the NcoI-linearized plasmids pAE877, pAE989, pAE990, pAE1007, pAE1023, and pAE1026. Integrants were selected by using standard genetic techniques and were verified by Western blotting. Telomeric URA3 was inserted into the appropriate strains by transforming SalI/EcoRI-linearized pVII-L URA3-TEL (17). Endogenous ORC1 was hemagglutinin (HA) tagged by duplicative integration by using XbaI-linearized pSB991 (gift from S. Bell). Gene knockouts with kanMX were performed as described previously (51). The suppression of the nat1Δ SUM1-1 synthetic lethality by orc1Δ1-235 was determined as follows. Strain JRY7176 (39) was transformed with a URA3-SIR2 plasmid in order to give the strain mating ability and to create diploids with AEY3134. The URA3-SIR2 plasmid was then lost from the diploid by counterselection on 5-FOA containing media. The diploid was sporulated, tetrads were dissected, and segregants were analyzed for their genotype. Segregants that were Trp+ and Leu+ genotypically were orc1Δ::TRP1 and also LEU2::orc1Δ1-235, because orc1Δ alone is lethal. To select segregants among these with nat1Δ::LEU2, the fact was exploited that SIR2 and NAT1 are neighboring genes within the yeast genome, making recombination between them highly unlikely. Thus, His− segregants from the cross by interference were also nat1Δ::LEU2. Ten such segregants were chosen; proteins were extracted and subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting with an α-myc antibody to determine their SUM1-1 status. Several segregants were identified that showed a strong signal, and they were presumed to have the genotype orc1Δ::TRP1 LEU2::orc1Δ1-235 nat1Δ::LEU2 7myc-SUM1-1.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Sourcea |
|---|---|---|
| AEY1 | MATα ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 (=W303-1B) | |
| AEY2 | MATaade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 (=W303-1A) | |
| AEY5 | MATα HMR SS ΔI | |
| AEY71 | MATα HMR-E Δ300-256 (ΔABF) | A. Brand |
| AEY80 | MATanat1-5::LEU2 | R. Sternglanz |
| AEY81 | MATα HMR-E Δ331-324 (ΔRAP) | A. Brand |
| AEY84 | MATα HMR-E Δ352-358 (ΔACS) | A. Brand |
| AEY574 | MATα HMR SS ΔI orc2-1 | J. Rine |
| AEY1017 | MATα TEL VII-L::URA3 | J. Berman |
| AEY1224 | MATaSUM1-1 | D. Shore |
| AEY1227 | MATα nat1-5::LEU2 | |
| AEY1273 | MATα HMR SS ΔI nat1Δ::LEU2 | |
| AEY1275 | MATα HMR SS ΔI 5xGal4-RAP-ABF | |
| AEY1276 | MATα HMR SS ΔI 5xGal4-RAP-ABF nat1Δ::LEU2 | |
| AEY2144 | MATα HMR-E Δ331-324 (ΔRAP) nat1Δ::LEU2 | |
| AEY2146 | MATα HMR-E Δ352-358 (ΔACS) nat1Δ::LEU2 | |
| AEY2148 | MATα HMR-E Δ300-256 (ΔABF) nat1Δ::LEU2 | |
| AEY2371 | MATα TEL VII-L::URA3 nat1Δ::LEU2 | |
| AEY2947 | AEY1276 sir1Δ::kanMX | |
| AEY3008 | MATα sum1Δ::URA3 nat1Δ::LEU2 | |
| AEY3068 | MATaORC1-HA-URA3 | |
| AEY3070 | MATanat1-5::LEU2 ORC1-HA-URA3 | |
| AEY3134 | MATaADE2 lys2Δ nat1Δ::LEU2 | |
| AEY3161 | MATaorc2-1 nat1Δ::LEU2 pRS316-ORC2 | |
| AEY2866 | MATα ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 (=W303-1B) HMR SS ΔI orc1Δ::HIS5-GFP pAE405 | |
| AEY2867 | MATaade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100 (=W303-1A) HMR SS ΔI orc1Δ::HIS5-GFP pAE405 | |
| AEY2903 | AEY2866 orc1-A2V::LEU2 without pAE405 | |
| AEY2912 | AEY2867 nat1Δ::kanMX | |
| AEY2913 | AEY2867 orc1-A2V::LEU2 without pAE405 | |
| AEY2916 | AEY2866 nat1Δ::kanMX | |
| AEY3038 | AEY2913 TEL VII-L::URA3 | |
| AEY3102 | AEY2867 orc1-A2P::LEU2 without pAE405 | |
| AEY3103 | AEY2866 orc1-A2P::LEU2 without pAE405 | |
| AEY3105 | AEY3102 TEL VII-L::URA3 | |
| AEY3373 | AEY2867 sir3-A2P | |
| AEY3378 | AEY2866 sir3-A2P | |
| AEY743 | MATaade2Δ::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 orc1Δ::TRP1 HIS3::HMR-URA3P-ADE2-E; pSPB162 (pURA3 ORC1) | S. Bell |
| AEY2721 | AEY743 orc1-A2V::LEU2 without pSPB162 | |
| AEY3101 | AEY743 orc1-A2P::LEU2 without pSPB162 | |
| AEY3109 | AEY743 nat1Δ::LEU2 | |
| AEY1558 | MATaleu2 trp1 ura3-52 prc1-407 pep4-3 prb1-112 | E. Jones |
| AEY2706 | AEY1558 nat1Δ::kanMX | |
| AEY2719 | AEY1558 ORC1(1-250)-TAP::URA3 | |
| AEY2758 | AEY1558 nat1Δ::kanMX ORC1(1-250)-TAP::URA3 | |
| AEY3107 | AEY1558 orc1-A2P (1-250)-TAP::URA3 | |
| AEY3110 | AEY1558 orc1-A2V (1-250)-TAP::URA3 | |
| AEY3171 | AEY1558 SIR3(1-235)-TAP::URA3 | |
| AEY3173 | AEY1558 nat1Δ::kanMX SIR3(1-235)-TAP::URA3 | |
| AEY3334 | AEY1558 sir3-A2T (1-235)-TAP::URA3 | |
| AEY3371 | AEY1558 sir3-A2P (1-235)-TAP::URA3 | |
| AEY160 | MATα his3Δ200 leu2Δ1 ura3-167 trp1Δ633 met15Δ1 RDN::Ty1::MET15 | J. Boeke |
| AEY2786 | AEY160 nat1Δ::kanMX |
Unless indicated otherwise, strains were constructed during the course of this study or were from the laboratory strain collection. Groups of strains between lines of space are isogenic.
TABLE 2.
Plasmids used in this studya
| Plasmid | Description | Sourceb |
|---|---|---|
| pAE100 | pRS316 ADH1P-GAL4(1-147)-SIR1 | J. Rine |
| pAE108 | pRH98-1 GPDP-GAL4(1-147)-ORC2 | J. Rine |
| pAE109 | pRH98-1 GPDP-GAL4(1-147)-ORC5 | J. Rine |
| pAE405 | pRS316 ORC1 | |
| pAE408 | pTT64 GAL4(1-147)-ORC1(5-267) | R. Sternglanz |
| pAE516 | pRH98-1 GPDP-GAL4(1-147)-ORC6 | |
| pAE580 | pRS316 SIR3-GFP | D. Shore |
| pAE595 | pRH98-1 GPDP-GAL4(1-147)-ORC3 | |
| pAE597 | pRH98-1 GPDP-GAL4(1-147)-ORC4 | |
| pAE877 | pRS306 ORC1(1-250)-TAP | |
| pAE989 | pRS306 orc1-A2P(1-250)-TAP | |
| pAE990 | pRS306 orc1-A2V(1-250)-TAP | |
| pAE1007 | pRS306 SIR3(1-235)-TAP | |
| pAE1023 | pRS306 sir3-A2T(1-235)-TAP | |
| pAE1026 | pRS306 sir3-A2P(1-235)-TAP | |
| pAE1027 | YIplac204 sir3-A2P(1-502) |
Cloning details are available from the authors upon request.
Unless indicated otherwise, plasmids were constructed during the course of this study or were from the laboratory plasmid collection.
Silencing and plasmid loss assays.
Qualitative and quantitative mating assays were performed as described previously (10) by using AEY264 (MATa his4) and AEY265 (MATα his4) as mating tester strains. Silencing of the TEL VII-L::URA3 and HMR-ADE2 reporter genes was analyzed by spotting serial dilutions on 5-FOA or adenine-deficient plates, respectively. Expression of the RDN::Ty1::MET15 rDNA reporter construct was assayed as described previously (42). In plasmid loss assays, strains AEY5, AEY3103, and AEY2903 transformed with plasmid pJR338 (ARS1 SUP11-1 URA3 [a gift from J. Rine]) were taken from selective medium and plated in various dilutions onto rich medium. After 3 days of growth at 30 or 23°C for the orc2-1 strain AEY574, the portion of fully and half-red colonies on the total number of colonies was determined.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were performed essentially as described previously (39). For fixation, cell cultures were incubated with 2% formaldehyde for 30 min at room temperature. For preclearing and isolation of the precipitated complexes, protein G-agarose beads (Sigma) were used. Immunoprecipitation was performed overnight with α-HA antibody (Covance). In the subsequent PCR, primers 5′-CCATGGCCCAGTCTCACTAAATCAGTACG-3′ and 5′-GTGAATAAAGAGTGTTACAATAGTGAGGTGC-3′ were used to amplify the CoreX sequence of chromosome XI-L. The primers described in reference 39 were used to simultaneously amplify SSC1.
IEF and immunoblotting.
Isoelectric focusing (IEF) gels (pH 3 to 10) were loaded with whole-cell extracts prepared by glass bead lysis as described earlier and run according to the supplier's instruction (Bio-Rad). SDS-polyacrylamide gel electrophoresis and Western blotting were performed according to standard protocols with PAP (Sigma), α-myc (Invitrogen), or α-HA antibodies (Covance) as appropriate. The theoretical pI was calculated by using online resources (http://us.expasy.org/tools/pi_tool.html).
In-gel digestion and peptide mass fingerprinting.
TAP-tagged Orc1p N-terminal peptides were purified from strains derived from the protease-deficient strain AEY1558 by using the standard TAP protocol (37). Purified Orc1p was separated on a 12% acrylamide gel and visualized by Coomassie G-250 staining. The protein band was excised and divided into two probes. The probes were cleaved in situ as described previously (40) by using either AspN (37°C) or GluC (25°C) protease (both from Roche) at a final concentration of 11.7 ng/μl or 25 ng/μl, respectively. The reduction and carbamidomethylation step was omitted. The digest supernatant (0.5 μl) was applied on a fast-evaporation nitrocellulose-α-cyano-4-hydroxycinnamic acid layer (50) and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry with a Bruker Reflex mass spectrometer (Bruker Daltonics) in the reflector mode equipped with pulsed-ion extraction and a nitrogen laser (337 nm). For selected peptides, the amino acid sequence was determined by analysis of fragment ions generated by post source decay (7) by using the FAST method (Bruker).
RESULTS
NatA was required for repression of the HM loci, telomeres, and the rDNA locus.
nat1Δ was previously described to cause pronounced derepression at the natural HML locus and at marker genes inserted in subtelomeric regions (Fig. 1A and B) (1, 31). In contrast, due to functional redundancy within the HMR-E silencer, wild-type HMR is not affected by nat1Δ unless it is weakened by a deletion of the Rap1p binding site (45). To further evaluate the role of NatA in silencing, we tested its effect on the synthetic HMR-E silencer (HMR SSΔI). This silencer variant is a minimal silencer that lacks much of the functional redundancy of natural HMR (30). Significantly, nat1Δ caused complete derepression at HMR SSΔI, as monitored by the loss of mating ability due to the coexpression of a information from HMR in the MATα strain (Fig. 1A). This supported the notion that NatA had a function at HMR that was masked by the functional redundancy of natural HMR.
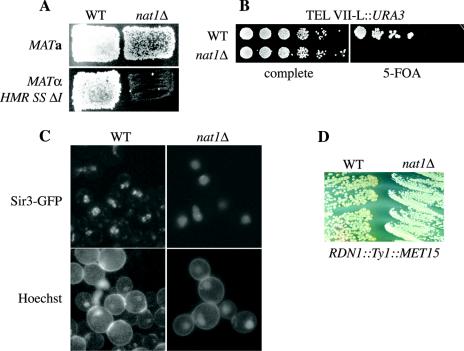
FIG. 1.
NatA activity was required for HM, telomeric, and rDNA silencing and for the integrity of telomeric foci. (A) The deletion of NAT1 resulted in derepression of HML and HMR SS ΔI, as measured by the reduced mating ability of MATa and MATα strains. Patch-mating assays were performed with MATa strains AEY2 (WT) and AEY80 (nat1Δ) and MATα HMR SS ΔI strains AEY5 (WT) and AEY1273 (nat1Δ). (B) Silencing of URA3 inserted near the left telomere of chromosome VII depended on functional NatA. Serial dilutions of strains AEY1017 (WT) and AEY2371 (nat1Δ) were assayed on 5-FOA medium counterselecting for URA3 expressing cells. (C) The association of GFP-tagged Sir3p with telomeric foci was inhibited in nat1Δ cells. Strains AEY160 (WT) and AEY2786 (nat1Δ) were transformed with pAE580. Bar, 2 μm. (D) Silencing of MET15 inserted into the rDNA locus was impaired by nat1Δ, as indicated by the brighter colony color of strain AEY 2786 (nat1Δ) compared to AEY160 (WT) on lead indicator medium.
NatA is also required for silencing of subtelomeric reporter genes (Fig. 1B) (1). Since the insertion of these genes generates truncated versions of the telomeres, we sought to determine whether the structure of native chromosomal ends also depended on NatA. To this end, we investigated the localization of green fluorescent protein (GFP)-tagged Sir3p, which naturally colocalizes with Rap1p and Sir4p in perinuclear foci (19), in wild-type and nat1Δ strains. Interestingly, Sir3-GFP dissociated from the perinuclear foci in the absence of NAT1 and became distributed throughout the nucleus (Fig. 1C). This distinct pattern was unlikely to be caused by a lower concentration of Sir3-GFP because the deletion of NAT1 had no effect on the steady-state level of TAP-tagged Sir3 (see Fig. 6A). This result suggested that NatA was required for the structural integrity of telomeric clusters at the nuclear periphery.
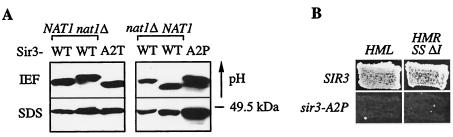
FIG. 6.
Sir3p was N terminally acetylated by NatA. (A) The isoelectric point of the Sir3p N terminus was shifted toward a more basic pH upon the deletion of NAT1 and when the penultimate amino acid of Sir3 was changed to proline but not to threonine. Whole-cell extracts of strains expressing TAP-tagged N-terminal peptides (amino acids 1 to 235) of Sir3p (AEY3171 [WT]) and AEY3173 [nat1Δ]), Sir3-A2T (AEY3334), and Sir3-A2P (AEY3371) were applied to IEF and SDS gels, which were subsequently analyzed in immunoblots with the PAP antibody. The double band of the Sir3-A2P sample in the IEF gel suggested the coexistence of Nα-acetylated and unacetylated protein. (B) The sir3-A2P mutation caused derepression at HML and HMR controlled by the synthetic HMR-E silencer (HMR SS ΔI), as measured in patch mating assays of MATa strains AEY2867 (SIR3) and AEY3373 (sir3-A2P) and MATα strains AEY2866 (WT) and AEY3378 (sir3-A2P), respectively.
We next sought to determine whether NatA also functioned in rDNA silencing. To this end, we tested the effect of nat1Δ on the expression of a MET15 reporter gene integrated at the rDNA locus, whose expression can be monitored on lead indicator medium (42). nat1Δ strains showed a brighter colony color than wild-type strains on this medium, indicating that MET15 was derepressed by nat1Δ (Fig. 1D). Thus, NatA functioned in all forms of silencing in S. cerevisiae. Since they share mechanistic similarities, one possibility is that a silencing factor(s) common to all three silenced regions is the target of NatA.
Tethering Orc1p or Sir1p to the silencer bypassed the requirement for NatA in silencing.
The involvement of NatA in silencing indicated that one or more silencing factors in yeast depended upon Nα acetylation for proper function. In order to narrow down the number of potential candidates, we sought to genetically characterize the precise role of nat1Δ in silencing. We first sought to determine whether NatA functions through silencing components that act upstream or downstream of Sir1p. For these experiments, we took advantage of the fact that silencing at HMR can be achieved by replacing the ORC binding site (ACS) of the synthetic HMR-E silencer by unrelated Gal4 binding sites and expressing fusions of the ORC subunits or of Sir1p to the Gal4-DNA-binding domain (12). Tethering of Gal4-Sir1p bypasses the requirement for ORC in silencing (12), which supports the notion that ORC recruits Sir1p to the silencer. Importantly, Gal4-Sir1p-mediated silencing was independent of NAT1 (Fig. 2A), indicating that NatA functioned upstream of Sir1p, and hence through ORC, in silencing.
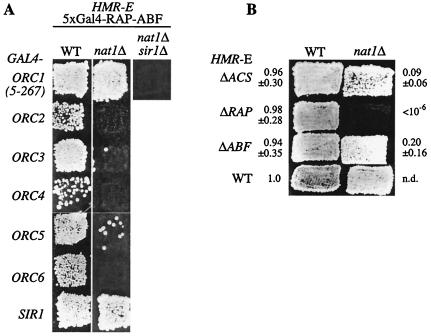
FIG. 2.
The silencing function of NatA was genetically linked to ORC1. (A) Tethered silencing by Orc1p, but not the other ORC subunits, was independent of NAT1 and required SIR1. In MATα strains AEY1275 (WT), AEY1276 (nat1Δ), and AEY 2947 (nat1Δ sir1Δ), the ORC binding site of the synthetic HMR-E silencer was replaced by five Gal4-binding sites (HMR SS ΔI and 5xGal4-RAP-ABF). The strains carried plasmids encoding the Gal4 DNA-binding domain fused N terminally to Orc1p (5 to 267 amino acids) (pAE408), Orc2p (pAE108), Orc3p (pAE595), Orc4p (pAE597), Orc5p (pAE109), Orc6p (pAE516), and Sir1p (pAE100) and were tested for HMR silencing in patch-mating assays. (B) The deletion of the binding site for Rap1p, but not for ORC or Abf1p, from HMR-E disrupted HMR silencing in nat1Δ mutants. HMR silencing was tested by the α-mating ability of wild-type and nat1Δ strains with wild-type HMR-E (AEY1 and AEY1227) and HMR-E lacking the binding site for ORC (AEY84 and AEY2146), Rap1p (AEY81 and AEY2144), and Abf1p (AEY71 and AEY2148). The results from quantitative mating assays are given relative to a value of 1.0 for AEY1 (WT).
We next tested whether the tethering of individual ORC subunits required NAT1 to establish silencing. The rationale of these experiments was that if N-terminal acetylation were required for the association of an ORC subunit to the silencer, direct tethering of this subunit by an N-terminal fusion to Gal4p would relieve its requirement for NatA. In the case of Orc1, the N-terminal part (encoding amino acids 5 to 267) was used, because it is sufficient to establish tethered silencing (49). Significantly, we found that tethered silencing of the subunits Orc2, Orc3, Orc4, and Orc6 disrupted in nat1Δ strains (Fig. 2A). The Orc5 fusion also showed strong derepression in the nat1Δ background, although we consistently observed a low amount of residual silencing. In strong contrast to these subunits, tethered Orc1p (amino acids 5 to 267) was able to provide robust silencing in the absence of NAT1. Interestingly, this silencing still depended upon Sir1p, since the NatA independent Gal4-Orc1p-mediated silencing was inhibited in a sir1Δ strain (Fig. 2A). These observations suggested that Orc1p required the N-terminal acetylation by NatA in order to be recruited sufficiently to the silencer and that the acetylation did not affect Orc1p's ability to interact with Sir1p. Consistent with this, Orc1p carries an alanine at the penultimate position, making it a likely candidate for Nα acetylation by NatA.
To determine whether NatA acts via ORC in the silencing context of natural HMR, we tested through which of the HMR-E elements nat1Δ functioned. To this aim, we exploited the fact that this form of silencing shows some functional redundancy in that the mutation of the ORC or Rap1p binding site alone has little effect but that the deletion of both causes strong derepression. The Abf1p binding site plays a minor role in this context, because the double mutation of the ORC and Abf1p binding sites only causes mild derepression (5, 26). Importantly, the loss of a silencer element can be achieved either by deleting a binding site in cis or by mutating the respective protein in trans. Thus, we reasoned that if NatA acted through ORC in silencing, nat1Δ should impair silencing in combination with the loss of the Rap1 binding site but not with a deletion of the binding site for ORC. Notably, silencing was completely abolished in nat1Δ strains with HMR-E lacking the Rap1p binding site, thereby suggesting ORC, but not Rap1p, as the NatA target (Fig. 2B). In contrast, nat1Δ did not cause significant derepression when the ORC binding site was deleted, suggesting that NatA functioned via this element (Fig. 2B). Not surprisingly, nat1Δ caused no further derepression in combination with the deletion of the Abf1 binding site, which was in line with the fact that double ORC-Abf1 binding site mutants showed a mild silencing defect (26). Altogether, these findings suggested that nat1Δ acted on HMR-E through the ORC binding site, and we therefore reasoned that ORC was the target of NatA in silencing.
Orc1p was N terminally acetylated by NatA.
Since the above genetic experiments suggested Orc1p as a silencing-relevant substrate of NatA, we directly tested whether Orc1p was N terminally acetylated in a NatA-dependent fashion. For this purpose, an additional copy of ORC1 encoding the first 250 amino acids fused to the TAP (for tandem affinity purification) tag (Orc1-TAP) under control of the natural ORC1 promoter was introduced into wild-type and nat1Δ strains. We reasoned that this size of the Orc1p N-terminal peptide was sufficient for Nα acetylation by NatA, since Nat1p contacts nascent polypeptide chains of ca. 80 amino acids (14). The TAP tag allows the fast and simple purification of large amounts of the tagged protein by two successive affinity purification steps (see below) (37). Since Nα acetylation shifts the isoelectric point (pI) of a given protein toward a more acidic pH (28), we used IEF gels to determine whether nat1Δ altered the pI of Orc1-TAP. Significantly, Orc1-TAP focused at a more basic pH when isolated from a nat1Δ strain compared to a wild-type strain (Fig. 3A), suggesting that Orc1p was acetylated by NatA.
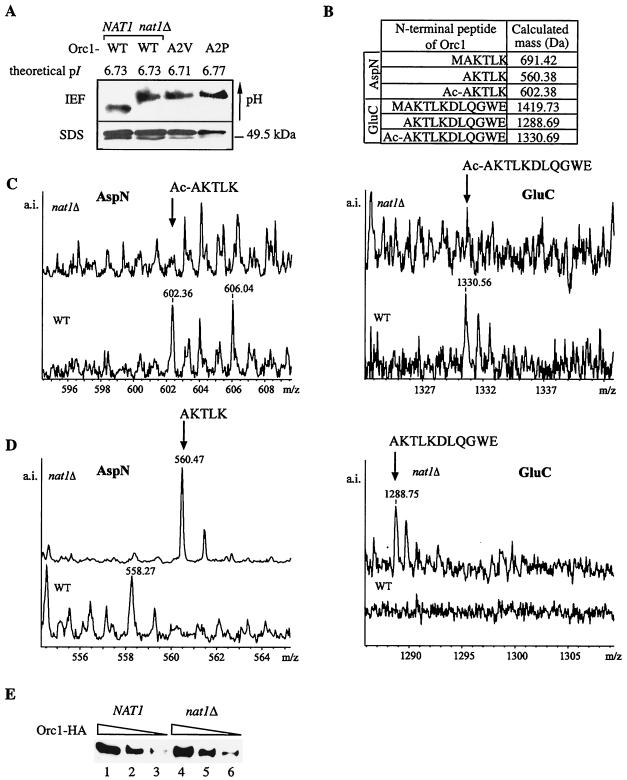
FIG. 3.
Orc1p was N terminally acetylated by NatA. (A) The isoelectric point (pI) of the Orc1p N terminus shifted to a more basic pH either by the deletion of NAT1 or by the mutation of the penultimate alanine to valine or proline. Whole-cell protein extracts of strains AEY2719 (WT), AEY2758 (nat1Δ), AEY3107 (orc1-A2P), and AEY3110 (orc1-A2V) were applied to IEF and SDS gels. TAP-tagged Orc1p (amino acids 1 to 250) was detected in subsequent immunoblots with a PAP-anitbody. The faster-migrating band in the SDS gel was identified as Orc1p by MALDI-TOF analysis and probably is a proteolytic fragment. (B) Theoretical molecular mass of N-terminal peptides of Orc1p generated by proteolysis with AspN or GluC endopeptidase. The molecular mass as calculated by using http://us.expasy.org/tools/peptide-mass.html increases by 42 Da due to Nα acetylation. (C) MALDI-TOF spectra of Orc1-TAP derived from a wild-type, but not from a nat1Δ strain, identified the mass of an acetylated N-terminal peptide of Orc1p. Orc1-TAP was purified for MALDI-TOF analysis from AEY2719 (WT) and AEY2758 (nat1Δ). The data obtainedfrom the AspN and GluC cleaved samples were consistent for each strain with minimal differences to the theoretical value due to the precision of measurements. (D) The MALDI-TOF spectrum of Orc1-TAP from the nat1Δ strain, but not from wild-type strain, contained the mass of an unacetylated N-terminal Orc1 peptide. Analysis was performed as in Fig. 3C. (E) Orc1p was not destabilized by nat1Δ. Whole-cell protein extracts of wild-type (NAT1) (AEY3068) and nat1Δ (AEY 3070) strains expressing HA-tagged Orc1p were analyzed by Western blotting with an α-HA antibody. Equal protein concentrations were loaded on lanes 1 and 4, 2 and 5, and 3 and 6, respectively.
It has previously been proposed that NATs can also provide Nɛ acetylation (35). Therefore, to test whether the pI shift corresponded to Nα acetylation of Orc1p, we used mass spectrometry to measure differences in acetylation in N-terminal peptides derived from Orc1-TAP that was isolated from wild-type or nat1Δ strains. Acetylation extends the mass of NAT substrates by 42 Da, which is the size of the bound acetyl group (36). Orc1-TAP samples purified with the TAP protocol from wild-type or nat1Δ backgrounds were digested individually with the AspN and GluC endopeptidases in order to obtain N-terminal peptides of a suitable size. We obtained a set of two different protein solutions of the wild-type and the nat1Δ derived samples, which were examined in independent experiments. In the subsequent analysis, the measured mass of the N-terminal peptide from the wild-type and the nat1Δ probe was compared to the calculated value on the basis of the amino acid sequence (Fig. 3B). In the AspN as well as in the GluC cleaved samples, neither the wild-type nor the nat1Δ strain-derived N-terminal fragments matched the calculated mass of a peptide containing the initial methionine (data not shown). This supported the notion that the initiator methionine was removed from proteins with alanine at the penultimate position. Furthermore, the mass of the N-terminal peptide of the wild-type sample was larger by 42 Da than the calculated value in the AspN, as well as the GluC, cleaved sample (Fig. 3C). However, in both cases this size increase was not found in the nat1Δ strain (Fig. 3D). The identity of the N-terminal peptide from the AspN cleaved protein of the nat1Δ strain was further verified by its fragment spectrum. The fragment pattern further indicated that the acetyl group was attached to the N-α position of the N-terminal residue and not to N-ɛ of one of the two internal lysine residues present in the peptide (data not shown). In summary, the mass spectrometric data demonstrated that Orc1p was N-terminally acetylated in the presence of Nat1p and not acetylated in its absence, strongly suggesting that it was a direct target of NatA.
The hypothesis has been put forward that Nα acetylation improves protein stability. In order to test whether this was also true for Orc1p, we compared the abundance of HA-tagged Orc1p in wild-type and nat1Δ strains (Fig. 3E). The amount of Orc1p was not decreased by nat1Δ, indicating that Nα acetylation was not required for the stability of the protein.
Unacetylated orc1 mutants caused telomeric derepression.
We next sought to determine whether the observed N-terminal acetylation of Orc1p was of significance for its silencing function. To do this, we generated orc1 alleles in which the penultimate amino acid was changed from alanine to valine or proline and tested their effect on silencing. Proline, as well as valine, promotes the cleavage of the initiator methionine but prevents N-terminal acetylation (23). In order to test whether the respective mutants were acetylated or not, we tested the isoelectric properties of the TAP variants Orc1-A2P and Orc1-A2V that were constructed analogous to wild-type Orc1-TAP. Significantly, the isoelectric point of Orc1-A2P-TAP and Orc1-A2V-TAP was at a more basic pH than wild-type Orc1p, although the calculated pI was roughly the same for all Orc1p versions (Fig. 3A). The shift was comparable to that of wild-type Orc1-TAP in the nat1Δ background, showing that the mutations of the penultimate amino acid to valine or proline had inhibited the ability of Orc1p to be acetylated by NatA.
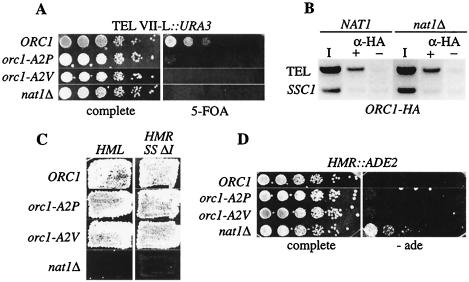
We then sought to determine whether these mutations had an impact on telomeric silencing, since the deletion of NAT1 strongly affects silencing of subtelomeric genes (Fig. 1B). For this purpose, we monitored the repression of a URA3-reporter gene inserted in the subtelomeric region of chromosome VII-L (17). Comparable to nat1Δ, orc1-A2P and orc1-A2V caused a strong derepression of the subtelomeric URA3 reporter, as indicated by diminished growth on URA3 counterselective medium (5-FOA; Fig. 4A). This showed that the loss of N-terminal acetylation of Orc1p compromised its function in telomeric silencing.
FIG. 4.
Nα acetylation of Orc1p was essential for telomeric silencing. (A) A URA3 gene inserted near the left telomere of chromosome VII was derepressed in unacetylated orc1-A2P and orc1-A2V mutants. URA3 expression was tested in serial dilution assays of strains AEY1017 (ORC1), AEY3038 (orc1-A2V), AEY3105 (orc1-A2P), and AEY2371 (nat1Δ) on 5-FOA-containing medium. (B) Telomeric association of Orc1 was not affected by nat1Δ. Anti-HA ChIPs from ORC1-HA strains AEY3068 (NAT1) and AEY3070 (nat1Δ) were analyzed by PCR with primers specific to the telomeric CoreX element of chromosome XI-L (TEL) and as a negative control to the SSC1 gene. The inverse image of an ethidium bromide-stained gel shows the products of 1:100-diluted input samples (I) and of 1:4 diluted samples that were either precipitated with α-HA antibody (+) or mock treated (−). (C) Loss of Nα acetylation of Orc1p did not impair silencing of HML and HMR SSΔI. Patch-mating assays were performed to test HML silencing by using MATa strains AEY2867 (ORC1), AEY3102 (orc1-A2P), AEY2913 (orc1-A2V), and AEY2912 (nat1Δ) and to test HMR SS ΔI silencing by using the MATα strains AEY2866 (ORC1), AEY3103 (orc1-A2P), AEY2903 (orc1-A2V), and AEY2916 (nat1Δ). (D) nat1Δ, but not unacetylated orc1 mutants caused slight derepression of ADE2 inserted at the HMR locus. Serial dilutions of strains AEY743 (WT), AEY3101 (orc1-A2P), AEY2721 (orc1-A2V), and AEY3109 (nat1Δ) were grown on medium lacking adenine.
We next sought to determine whether the telomeric silencing phenotype of nat1Δ was caused by reduced binding of ORC to the telomeres. To this end, we tested the association of HA-tagged Orc1 to the ORC binding site in the CoreX region of chromosome XI-L in wild-type and nat1Δ strains by using ChIP. We could readily detect Orc1-HA at the CoreX region in a wild-type strain (Fig. 4B). Interestingly, it was also bound to the CoreX in a nat1Δ strain, suggesting that ORC binding to the telomere was not inhibited by the absence of Nα acetylation. Thus, one alternative explanation for NatA's effect on ORC is that it alters the recruitment of other silencing factors to the telomere (see Discussion).
HM silencing was not impaired in unacetylated orc1 mutants.
We next tested whether HM silencing was also impaired by the lack of N-terminal acetylation of Orc1p. No silencing defect was detectable at HML and the synthetic HMR SSΔI in the orc1-A2P and orc1-A2V mutants, which was in contrast to the strong defect caused by nat1Δ (Fig. 4C). The mutants also caused no silencing defect of the sensitive ADE2 reporter inserted at HMR, whereas nat1Δ caused a slight derepression of ADE2 in this context (Fig. 4D). One explanation is that HM silencing is more robust than telomeric silencing and thus is less sensitive to the orc1 mutations. Furthermore, this suggested that more NatA silencing targets exist in HM silencing. In light of the tethered silencing experiment shown above, other ORC subunits may be among these NatA targets, which might result in a reduced stability of the ORC complex in the nat1Δ mutants (see Discussion).
The lack of N-terminal acetylation of Orc1p did not affect ORC's replication function.
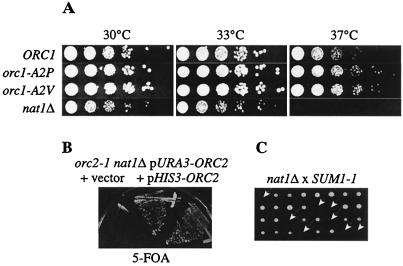
Since ORC functions as the replication initiator complex besides its role in silencing, we also sought to determine whether the lack of Orc1p acetylation affected its replication function. We therefore tested orc1-A2P and orc1-A2V strains for temperature sensitivity, a phenotype that is associated with replication defects in orc2-1 and orc5-1 mutants (29). Both unacetylated orc1 mutants grew as well as wild-type strains and were not temperature sensitive, suggesting that replication was not affected (Fig. 5A). In contrast, the growth of nat1Δ strains was considerably diminished at 37°C, suggesting that the temperature sensitivity of nat1Δ was due to defects other than Nα acetylation of Orc1p.
FIG. 5.
nat1Δ was synthetically lethal with orc2-1 and SUM1-1. (A) Unacetylated orc1-A2V and orc1-A2P mutants were not temperature sensitive. Serial dilutions of strains AEY2866 (ORC1), AEY 3103 (orc1-A2P), AEY2903 (orc1-A2V), and AEY2916 (nat1Δ) were grown for 2 days on complete medium at the indicated temperatures. (B) Viability of the orc2-1 nat1Δ double mutants was rescued by plasmid-borne ORC2. AEY3161 (orc2-1 nat1Δ pURA3-ORC2) transformed either with pJR1818 (pHIS3-ORC2) (12) or with pRS313 (vector) was tested for ORC2 dependence by counter selection for pURA3-ORC2 on 5-FOA medium. (C) SUM1-1 nat1Δ double mutants were inviable. SUM1-1 nat1Δ segregants of tetrads dissected from a cross between SUM1-1 (AEY1224) and nat1Δ (AEY3008) are marked by arrows.
We furthermore used a plasmid stability assay as a second measurement of replication function in the mutants. In this assay, the strains were transformed with a plasmid that carries ARS1 and the suppressor tRNA SUP11-1, which converts the colony color of the ade2 mutants from red to white. The stability of this plasmid was measured as the frequency of red sectoring colonies on nonselective medium. orc1-A2P and orc1-A2V mutants displayed frequencies of ca. 13 and 11%, respectively, which resembled the wild-type frequency of 12%. In contrast, 71% of the colonies of the replication-defective orc2-1 strain were fully or half red. Taken together, these results showed that the Orc1 N-terminal mutations did not cause a replication defect.
nat1Δ was synthetically lethal with orc2-1.
We further evaluated the functional link between NatA and ORC by investigating genetic interactions between nat1Δ and orc2-1. Interestingly, we found that nat1Δ orc2-1 double mutants were unable to survive. In crosses between nat1Δ and orc2-1 strains, double mutant segregants did not grow up except for a few cases, where pinprick colonies appeared after prolonged incubation, but which were unable to form colonies when restreaked (data not shown). In addition, the viability of orc2-1 nat1Δ double mutants was dependent on the presence of an Orc2p encoding plasmid (Fig. 5B). Since orc2-1 causes a replication defect, this observation suggested that nat1Δ compromised replication even further such that the double mutants were unable to replicate.
Synthetic lethality between nat1Δ and SUM1-1 was suppressed by orc1Δ1-235.
We next sought to determine the dependence of Sir-independent, SUM1-1-dependent silencing on NatA. However, in a set of genetic crosses in which nat1Δ and SUM1-1 segregated, we observed synthetic lethality between nat1Δ and SUM1-1 (Fig. 5C). The segregation of the unmarked SUM1-1 mutation was determined by monitoring sum1Δ::URA3 in the segregants from sum1Δ::URA3/SUM1-1 heterozygous diploids. Interestingly, nat1Δ was not synthetically lethal with sum1Δ (data not shown), suggesting that the lethality was due to novel properties of the mutant Sum1-1 protein.
Since Sum1-1p has been shown to interact with the N terminus of Orc1p and because NatA acetylates this very N terminus, we hypothesized that the lethality may be connected to the lack of Orc1p acetylation. The ability of Sum1-1p to function in silencing is inhibited by the deletion of amino acids 1 to 235 of Orc1p (39). Hence, we tested whether this deletion also inhibited the synthetic lethality of SUM1-1 with nat1Δ. Significantly, strains with orc1Δ1-235 as the sole source of Orc1p that were both nat1Δ and SUM1-1 were readily recovered from a cross (see details in Materials and Methods) and showed normal growth characteristics (data not shown). This showed that the synthetic lethality of nat1Δ with SUM1-1 was inhibited by deletion of the N terminus of Orc1p.
Sir3p was acetylated by NatA.
In a previous study, Stone et al. (44) observed decreased telomeric silencing and an enhanced sir1Δ mating defect when the penultimate alanine of Sir3p was exchanged for a threonine. This sir3-A2T mutation was epistatic to nat1Δ, suggesting that the phenotype was caused by the lack of Nα acetylation of Sir3. We therefore tested the isoelectric properties of the TAP-tagged N-terminal peptide of the mutant protein. Surprisingly, Sir3(1-235)-A2T focused like wild-type Sir3p in the IEF gel, indicating that Nα acetylation was not affected in the sir3-A2T mutant (Fig. 6A). In contrast, the pI of Sir3(1-235)-TAP shifted to a more basic pH upon the deletion of NAT1, suggesting that Sir3p was Nα acetylated by NatA.
In order to test whether this acetylation plays a role in silencing, we tested the mating ability of a sir3-A2P mutant. This mutation caused the loss of Nα acetylation of Sir3p, as judged by the shift of the pI to a more basic pH, although a portion of Sir3-A2P apparently remained acetylated (Fig. 6A). Since silencing was disrupted at HML and HMR controlled by the synthetic HMR SS ΔI silencer in the sir3-A2P mutant (Fig. 6B), we concluded that Sir3p was another silencing factor whose Nα acetylation by NatA was required for silencing.
DISCUSSION
Posttranslational modifications play a critical role in the regulation of chromatin structure and function. One prominent example is given by the impact of histone acetylation on transcription initiation, elongation, and heterochromatin formation. In the present study, we identified a new target for the function of Nα acetylation in chromatin regulation. We found that Nα acetylation of Orc1p, the large subunit of the ORC, was essential for its function in telomeric silencing in yeast. Thus, this modification may constitute a novel chromatin regulatory mechanism comparable to ɛ-N lysine acetylation or methylation. In contrast to Nɛ acetylation, which readily can be removed by deacetylases, Nα acetylation is irreversible and thus may provide a stable mark similar to histone ɛ-N lysine methylation. This raises the question of how the modification can be removed in order to alter protein function upon demand. One possibility is that amino-terminal proteolysis may remove Nα acetylation from Orc1p, as is proposed for histone methylation. Alternatively, removal or turnover of Orc1p at ORC binding sites may be required in order to modulate its function in chromatin silencing, much like methylated histones are postulated to be exchanged on chromatin during replication and transcription (25).
How does Nα acetylation affect Orc1p's silencing function? Thus far, the recruitment of Sir1p to the HM silencers has been considered the exclusive task of Orc1p in silencing. However, genetic data indicate that ORC has additional silencing functions. Foremost, telomeric silencing completely depends upon ORC while being unaffected by the deletion of SIR1 (12), showing that ORC has a Sir1p-independent function in silencing the telomeres. Interestingly, we found that Nα acetylation did not affect Orc1p's ability to interact with Sir1p, a finding in agreement with the previous observation that SIR1 overexpression suppressed the nat1Δ silencing defect at HML (45). Thus, we propose that the Orc1p amino terminus interacts with an as-yet-unidentified silencing protein that functions primarily in telomeric silencing and that this interaction requires Nα acetylation of Orc1p by NatA. This view is supported by our finding that ORC binding to the telomeres was not inhibited by the absence of Nα acetylation (but see below). Crystallographic data show that the extreme N terminus of Orc1p is exposed on the surface of the protein in a structure distinct from the Sir1p interaction domain (54), thus rendering it a potential interaction module for another protein. In light of the high similarity between the Orc1p and Sir3p N termini, it is interesting that we also found Sir3p to be Nα acetylated by NatA. One possibility is that acetylated Orc1p and Sir3p both interact with the same hypothesized silencing factor. This novel interaction partner may specifically recognize the N terminus of Orc1p and/or Sir3p in its acetylated form. Precedence for modification-dependent protein interactions comes from bromodomain and chromodomain proteins that preferentially bind specific acetylated and methylated histone residues, respectively (25).
Interestingly, Nα acetylation affects Orc1p's function in silencing but not its function in replication initiation. Thus, Nα acetylation provides a possibility to regulate Orc1p's function in the two processes. Notably, in contrast to Orc1p acetylation, NatA does influence ORC's replication function, because nat1Δ orc2-1 double mutants were inviable. This suggested that other ORC subunits may be Nα acetylated by NatA and that this acetylation may impinge upon their ability to initiate replication, perhaps by affecting their ability to interact with other replication factors or by destabilizing the complex itself.
Further evidence for the hypothesis that Nα acetylation regulates interactions between Orc1p and other proteins comes from our observation of a synthetic lethal interaction between nat1Δ and SUM1-1 but not sum1Δ. Interestingly, the SUM1-1 mutation confers to the Sum1-1 protein the ability to interact with ORC but retains the ability to interact with the histone deacetylase Hst1p (39). Thus, Sum1-1p binds to the silencers via ORC, recruits Hst1p to the HM silencers, and establishes Sir2p-independent silencing at the HM loci. The binding of Sum1-1p to ORC is inhibited by deletion of the amino-terminal 235 amino acids of Orc1p, suggesting that Sum1-1p interacts with the Orc1p N terminus. Therefore, one interpretation of the inviability of nat1Δ SUM1-1 strains is that Sum1-1p interacts better (i.e., stronger and at more genomic locations) with Orc1p in its unacetylated form and that this inhibits replication initiation, which is ORC's essential function. This hypothesis is supported by the observation that the SUM1-1 nat1Δ synthetic lethality was inhibited by the deletion of the amino-terminal 235 amino acids of Orc1p. Thus, we postulate that Nα acetylation of Orc1p controls its ability to interact with Sum1-1p, as well as with other proteins.
N-terminal protein acetylation has long been hypothesized to protect proteins from degradation. However, several observations indicate that this does not hold true for the influence of NatA on silencing. First, we found that the level of Orc1p protein was indistinguishable between wild-type and nat1Δ strains. Second, the effect of nat1Δ on HML silencing and temperature sensitivity was suppressed by overexpression of the ribosome-bound chaperone Ssb1p (14), suggesting that nat1Δ caused a defect in protein folding rather than stability. However, Ssb1p overexpression did not suppress the telomeric silencing defect of nat1Δ or of the orc1 N-terminal mutants (data not shown), thus supporting the notion that acetylated Orc1p specifically recruits a novel protein to establish silencing rather than affecting Orc1p folding.
How does NatA acetylation influence silencing? Clearly, NatA has many cellular targets, which is reflected in the pleiotropic phenotypes of nat1Δ or ard1Δ mutants, but only few substrates are relevant for silencing. The unacetylated orc1 mutants show the same amount of telomeric derepression as nat1Δ, arguing that Orc1p acetylation by NatA plays a crucial role in telomeric silencing. At the HM loci, nat1Δ shows stronger derepression than the unacetylated orc1 mutants. One explanation is that telomeric silencing is less robust than HM silencing and thus can be perturbed more easily by the lack of Nα acetylation of Orc1p. In line with this, another orc mutation, orc5-1, also affects telomeric silencing more severely than HM silencing (12, 29). Another possibility is that the Nα acetylation of Orc1p plays a subordinate role in HM silencing, and thus NatA acts there primarily via other silencing factors. In agreement with this, the unacetylated sir3 mutant displayed a strong HM silencing phenotype. It is also possible that NatA has several more targets among the silencing components. One scenario is that additional ORC subunits are NatA targets. This notion is supported by an alternative interpretation of our tethered silencing results. Given that ORC's silencing function is to recruit Orc1p, and hence Sir1p, to the silencer, direct tethering of Orc1p may circumvent the requirement of proper ORC assembly in order to mediate silencing. The fact that no individual ORC subunit (with the exception of Orc1p) was able to bypass the requirement for NatA may mean that more than one subunit requires NatA acetylation for the recruitment of Orc1p and thus for silencing. In this model, NatA may affect the integrity of the ORC complex as a whole via Nα acetylation of several subunits and without affecting the stability of the individual subunits. Consistent with this, nat1Δ functioned genetically through the ORC binding site of the natural HMR. Also, the synthetic lethality of nat1Δ with orc2-1 suggests that the replication function of ORC was also impaired by nat1Δ. Intriguingly, ORC binding to telomeres was not reduced by nat1Δ. Perhaps the formaldehyde cross-linking used in the ChIP experiments prevents the detection of differences in ORC complex stability.
In summary, we propose that the effect of nat1Δ at the HM loci is the sum of several proteins lacking acetylation, among them Orc1p and Sir3p. The lack of acetylation results in a partial loss of function of the individual proteins, the cumulative effect of which causes derepression by delocalization of Sir3p and other Sir proteins.
Notably, both NatA and ORC are conserved between yeast and higher eukaryotes, posing the question of whether Nα acetylation is also required for Orc1p function in these organisms. Significantly, not only the replication initiation function but also the silencing function of ORC is conserved, since ORC is associated with heterochromatin in Drosophila and interacts physically with HP1, a central component of heterochromatin (33). Presently, it is not known whether Nα acetylation of DmORC1 plays a role in this interaction. However, DmORC1 also carries an alanine at the penultimate position, making it a likely target for NatA acetylation.
In higher organisms, homologs of the NatA subunits have been linked to developmental and differentiation processes. Mouse mNAT1 is expressed in the developing brain and is regulated by physiological levels of functional N-methyl-d-aspartate (NMDA) receptor in developing neurons (46). The mNAT1 homolog tubedown-1 is expressed highly in developing tissues and downregulated upon differentiation (16). Furthermore, a tubedown-1 variant, Tbdn100, was isolated in a transcription regulatory complex, suggesting that it may be a transcriptional coregulator (53). Interestingly, the human homolog NATH also is highly expressed in parts of the human brain and is overexpressed in malignant cells, for instance, in papillary thyroid carcinomas and several leukemia and carcinoma cell lines (11). It therefore has been hypothesized that NatA overexpression might simply correlate with high transcriptional activity. In light of our findings, it is tempting to speculate that NatA acetylation regulates cell proliferation by modifying ORC function in replication or in the control of gene expression. Thus, it will be interesting to identify chromatin factors in higher eukaryotes whose function depends on Nα acetylation by NatA.
Acknowledgments
We thank A. Brand, S. Bell, J. Berman, J. Boeke, D. Gottschling, E. Jones, J. Rine, S. Rospert, L. Pillus, S. Schaper, D. Shore, and R. Sternglanz for strains and plasmids. We are grateful to U. Marchfelder and A. Barduhn for excellent technical assistance and P. Franke for help with recording the mass spectra. We also thank F. Hucho for his support of this work. We thank M. Shevak for help in preparing the figures; S. Rospert, J. Franke, and S. Schaper for critical reading of the manuscript; and the members of our laboratory for many helpful discussions.
This study was supported by the Deutsche Forschungsgemeinschaft (DFG grant EH 194/1-1 and 1-2) and the Max Planck Society. C.W. also acknowledges support by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in Saccharomyces cerevisiae. Cell 66:1279-1287. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, R. J., B. Polevoda, J. P. Reilly, and F. Sherman. 1999. The action of N-terminal acetyltransferases on yeast ribosomal proteins. J. Biol. Chem. 274:37035-37040. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., R. Kobayashi, and B. Stillman. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844-1849. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. P., J. Mitchell, J. Leber, R. Kobayashi, and B. Stillman. 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83:563-568. [DOI] [PubMed] [Google Scholar]
- 5.Brand, A. H., G. Micklem, and K. Nasmyth. 1987. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell 51:709-719. [DOI] [PubMed] [Google Scholar]
- 6.Callebaut, I., J. C. Courvalin, and J. P. Mornon. 1999. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446:189-193. [DOI] [PubMed] [Google Scholar]
- 7.Chaurand, P., F. Luetzenkirchen, and B. Spengler. 1999. Peptide and protein identification by matrix-assisted laser desorption ionization (MALDI) and MALDI-post-source decay time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 10:91-103. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, W. L., S. D. Briggs, and C. D. Allis. 2000. Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 12:326-333. [DOI] [PubMed] [Google Scholar]
- 9.Chien, C. T., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75:531-541. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenhofer-Murray, A. E., D. H. Rivier, and J. Rine. 1997. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145:923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fluge, O., O. Bruland, L. A. Akslen, J. E. Varhaug, and J. R. Lillehaug. 2002. NATH, a novel gene overexpressed in papillary thyroid carcinomas. Oncogene 21:5056-5068. [DOI] [PubMed] [Google Scholar]
- 12.Fox, C. A., A. E. Ehrenhofer-Murray, S. Loo, and J. Rine. 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276:1547-1551. [DOI] [PubMed] [Google Scholar]
- 13.Gardner, K. A., J. Rine, and C. A. Fox. 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautschi, M., S. Just, A. Mun, S. Ross, P. Rucknagel, Y. Dubaquie, A. Ehrenhofer-Murray, and S. Rospert. 2003. The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23:7403-7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavin, K. A., M. Hidaka, and B. Stillman. 1995. Conserved initiator proteins in eukaryotes. Science 270:1667-1671. [DOI] [PubMed] [Google Scholar]
- 16.Gendron, R. L., L. C. Adams, and H. Paradis. 2000. Tubedown-1, a novel acetyltransferase associated with blood vessel development. Dev. Dyn. 218:300-315. [DOI] [PubMed] [Google Scholar]
- 17.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at Saccharomyces cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751-762. [DOI] [PubMed] [Google Scholar]
- 18.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301:798-802. [DOI] [PubMed] [Google Scholar]
- 19.Grunstein, M. 1998. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell 93:325-328. [DOI] [PubMed] [Google Scholar]
- 20.Hecht, A., T. Laroche, S. Strahl-Bolsinger, S. M. Gasser, and M. Grunstein. 1995. Histone H3 and H4 N termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80:583-592. [DOI] [PubMed] [Google Scholar]
- 21.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, J., and D. Moazed. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17:2162-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, S., R. C. Elliott, P. S. Liu, R. K. Koduri, J. L. Weickmann, J. H. Lee, L. C. Blair, P. Ghosh-Dastidar, R. A. Bradshaw, K. M. Bryan, et al. 1987. Specificity of cotranslational amino-terminal processing of proteins in yeast. Biochemistry 26:8242-8246. [DOI] [PubMed] [Google Scholar]
- 24.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 26.Kasulke, D., S. Seitz, and A. E. Ehrenhofer-Murray. 2002. A role for the Saccharomyces cerevisiae RENT complex protein Net1 in HMR silencing. Genetics 161:1411-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kayne, P. S., U. J. Kim, M. Han, J. R. Mullen, F. Yoshizaki, and M. Grunstein. 1988. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55:27-39. [DOI] [PubMed] [Google Scholar]
- 28.Kimura, Y., M. Takaoka, S. Tanaka, H. Sassa, K. Tanaka, B. Polevoda, F. Sherman, and H. Hirano. 2000. Nα-acetylation and proteolytic activity of the yeast 20S proteasome. J. Biol. Chem. 275:4635-4639. [DOI] [PubMed] [Google Scholar]
- 29.Loo, S., C. A. Fox, J. Rine, R. Kobayashi, B. Stillman, and S. Bell. 1995. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol. Biol. Cell 6:741-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNally, F. J., and J. Rine. 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5648-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullen, J. R., P. S. Kayne, R. P. Moerschell, S. Tsunasawa, M. Gribskov, M. Colavito-Shepanski, M. Grunstein, F. Sherman, and R. Sternglanz. 1989. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouspenski, I. I., S. J. Elledge, and B. R. Brinkley. 1999. New yeast genes important for chromosome integrity and segregation identified by dosage effects on genome stability. Nucleic Acids Res. 27:3001-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pak, D. T., M. Pflumm, I. Chesnokov, D. W. Huang, R. Kellum, J. Marr, P. Romanowski, and M. R. Botchan. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311-323. [DOI] [PubMed] [Google Scholar]
- 34.Park, E. C., and J. W. Szostak. 1992. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 11:2087-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polevoda, B., and F. Sherman. 2003. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem. Biophys. Res. Commun. 308:1-11. [DOI] [PubMed] [Google Scholar]
- 36.Polevoda, B., and F. Sherman. 2001. NatC Nα-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J. Biol. Chem. 276:20154-20159. [DOI] [PubMed] [Google Scholar]
- 37.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 38.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 39.Rusche, L. N., and J. Rine. 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 41.Singer, J. M., and J. M. Shaw. 2003. Mdm20 protein functions with Nat3 protein to acetylate Tpm1 protein and regulate tropomyosin-actin interactions in budding yeast. Proc. Natl. Acad. Sci. USA 100:7644-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 43.Song, O. K., X. Wang, J. H. Waterborg, and R. Sternglanz. 2003. A Nα-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. J. Biol. Chem. 278:38109-38112. [DOI] [PubMed] [Google Scholar]
- 44.Stone, E. M., C. Reifsnyder, M. McVey, B. Gazo, and L. Pillus. 2000. Two classes of sir3 mutants enhance the sir1 mutant mating defect and abolish telomeric silencing in Saccharomyces cerevisiae. Genetics 155:509-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks, and R. Sternglanz. 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 11:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugiura, N., R. G. Patel, and R. A. Corriveau. 2001. N-Methyl-d-aspartate receptors regulate a group of transiently expressed genes in the developing brain. J. Biol. Chem. 276:14257-14263. [DOI] [PubMed] [Google Scholar]
- 47.Sutton, A., R. C. Heller, J. Landry, J. S. Choy, A. Sirko, and R. Sternglanz. 2001. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tercero, J. C., and R. B. Wickner. 1992. MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 267:20277-20281. [PubMed] [Google Scholar]
- 49.Triolo, T., and R. Sternglanz. 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381:251-253. [DOI] [PubMed] [Google Scholar]
- 50.Vorm, O., P. Roepstorff, and M. Mann. 1994. Improved resolution and very high sensitivity in MALDI TOF of matrix surfaces made by fast evaporation. Anal. Chem. 66:3281-3287. [Google Scholar]
- 51.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 52.Whiteway, M., and J. W. Szostak. 1985. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell 43:483-492. [DOI] [PubMed] [Google Scholar]
- 53.Willis, D. M., A. P. Loewy, N. Charlton-Kachigian, J. S. Shao, D. M. Ornitz, and D. A. Towler. 2002. Regulation of osteocalcin gene expression by a novel Ku antigen transcription factor complex. J. Biol. Chem. 277:37280-37291. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Z., M. K. Hayashi, O. Merkel, B. Stillman, and R. M. Xu. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21:4600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]