Abstract
Dynamic changes in transcription factor function can be mediated by switching its interaction with coactivators and corepressors. Erythroid Krüppel-like factor (EKLF) is an erythroid cell-specific transcription factor that plays a critical role in β-globin gene activation via its interactions with CBP/p300 and SWI/SNF proteins. Unexpectedly, it also interacts with Sin3A and histone deacetylase 1 (HDAC1) corepressors via its zinc finger domain. We now find that selected point mutants can uncouple activation and repression and that an intact finger structure is not required for interactions with Sin3A/HDAC1 or for transrepression. Most intriguingly, EKLF repression exhibits stage specificity, with reversible EKLF-Sin3A interactions playing a key role in this process. Finally, we have located a key lysine residue that is both a substrate for CBP acetylation and required for Sin3A interaction. These data suggest a model whereby the stage of the erythroid cell alters the acetylation status of EKLF and plays a critical role in directing its coactivator-corepressor interactions and downstream transcriptional effects.
A pivotal control point for the regulation of gene expression is at the initial generation of the transcript (8, 46). The eukaryotic cell has developed a wide range of control within which a gene can be activated from a dormant state or repressed from an activated state (59). These states can be temporally fixed by the additional controls imposed by chromatin structure and epigenetic marks upon both histones and DNA (60). Proteins that impose such determinants have been generally segregated into transcriptional activators and repressors (37). However, a recent realization that adds an additional layer of complexity is that such a demarcation of function is oversimplified, as some factors can do double duty as activators and repressors (3, 20, 21). These activities are controlled by a variety of external stimuli and provide a means by which the surprisingly low number of genes in the mammalian genome can exert multiple effects on genomic targets (33).
Erythroid Krüppel-like factor (EKLF/KLF1) controls adult β-globin gene expression by the interaction of its three zinc fingers with the CAC element (5′CCACACCCT3′) located within the proximal promoter (4, 40, 48). Genetic ablation of EKLF leads to loss of the DNase-hypersensitive site at the β-promoter and absence of β-globin gene expression, resulting in embryonic lethality due to a profound β-thalassemia and toxic accumulation of α-globin chains (34, 44, 50, 63). Murine yolk sac (primitive) erythroid cells express embryonic β-like globins and appear normal, but the lethality arises at the time of the switch to adult β-globin expression, which in the mouse occurs in the definitive erythroid cells of the fetal liver (58). EKLF's ability to interact with p300/CBP (67) and with the SWI/SNF complex (2) suggests a means by which EKLF integrates these components at the β-promoter and induces transcription initiation. These interactions are interrelated, as p300/CBP acetylates EKLF and enables it to more efficiently interact with SWI/SNF proteins (68). Of particular importance are EKLF-BRG1 interactions (9, 27).
Given the molecular and genetic evidence for EKLF activation, it was unexpected to find that EKLF can also interact with corepressors (Sin3A and histone deacetylase 1 [HDAC1]) and transcriptionally repress promoters in vivo (13). These observations have coincided with other intimations of additional EKLF function. For example, EKLF is expressed in yolk sac erythroid cells and in early hematopoietic cells, neither of which express adult β-globin (25, 53, 57, 70). In addition, hemoglobin rescue of EKLF-null erythroid cells is not sufficient to yield morphologically normal cells (49). Finally, inducible expression of EKLF yields cells with a decreased proliferation rate (14). Although some of these may still relate to EKLF function as an activator, it is now also possible that EKLF repression plays a role in these phenotypes.
As a result, we have more fully investigated EKLF repression by performing a structure-function analysis of this effect. This has enabled us to uncouple activation and repression functions of EKLF, with respect to both functional tests and protein interactions. Of particular interest, we find that the cellular environment has a dramatic effect on these attributes.
MATERIALS AND METHODS
Cell lines, plasmids, and antibodies.
The cell lines, plasmids, and antibodies used in this study were previously described (13, 68), including baculovirus pFL/HDAC1 (23) and plasmid pHS2βLuc (10). EBHX11 cells grown in Iscove’s modified Dulbecco’s medium plus fetal bovine serum and monothioglycerol were supplemented with either leukemia inhibitory factor (LIF; Gibco) or erythropoietin (Epo; Amgen) as previously described (28).
Mutagenesis was performed with the Quik Change kit (Stratagene) and changed the first zinc-coordinating histidine within each EKLF finger to asparagine: H313N, H343N, and H371N. Such a change has been shown to disrupt the individual finger structure with no effect on adjacent fingers (15) and, unlike mutation of the second histidine (56), completely alters DNA binding.
Transfections, immunoprecipitations, assays, and analyses.
Transfections, luciferase and chloramphenicol transferase assays, immunoprecipitations, and Western bloting were performed as described before (13, 68), including normalization to pXGH5 growth hormone plasmid as an internal control. Electroporation was used to transfect DNA into EBHX cells as follows. Ten million EBHX11L cells were mixed with 40 μg of total DNA and electroporated (Bio-Rad Gene Pulser) at 0.4 kV and 150 μF; EBHX11E cells were mixed with 30 μg of total DNA and electroporated at 500 μF. Glutathione S-transferase (GST) pull-down assays were performed as described previously (52), using in vitro-translated or baculovirus-expressed FL-HDAC1 (23) that was purified with M2-agarose (Sigma).
RESULTS
Generation of selectively altered EKLF activation and repression derivatives.
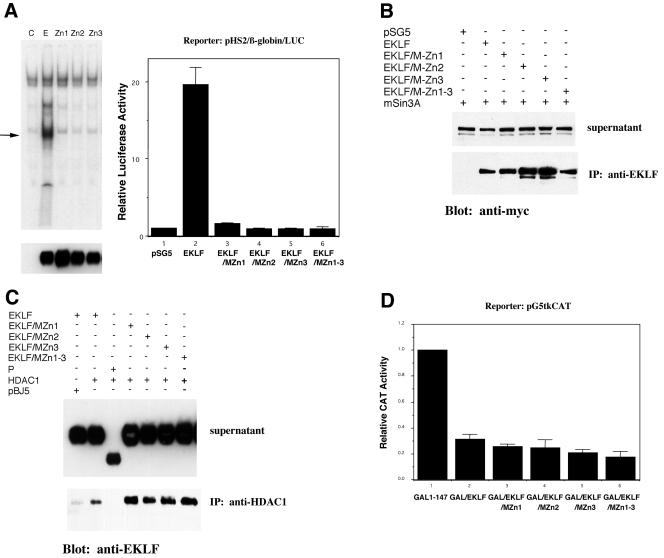
We attempted to uncouple EKLF repression and activation functions by the rational design of site-directed mutants. Given our previous data that EKLF cannot bind DNA and repress simultaneously (13), a straightforward prediction was that an EKLF mutant that no longer binds DNA would lose its activation function yet retain its repression function. To test this idea, we generated site-directed mutants that altered a critical histidine residue known to be important for coordinating zinc within each zinc finger. These full-length derivatives were tested for DNA binding after transfection in COS7 cells by making extracts and monitoring binding to the β-globin CACCC element. As seen in Fig. 1A, transfection of wild-type EKLF leads to a novel gel shift not present in mock-transfected COS7 cells. This binding is not seen when EKLF constructs that contain point mutations in either zinc finger 1, 2, or 3 are tested in a similar fashion, even though their expression levels are similar to wild type. It is not surprising then that all of these have lost their ability to transactivate the native β-globin promoter when tested in erythroleukemic K562 cells (Fig. 1A).
FIG. 1.
Characterization of EKLF derivatives that are deficient in activation yet retain repression. (A) DNA binding (left) and transactivation (right) by wild-type EKLF (lane E), single-zinc-finger mutants (MZn1, MZn2, and MZn3), or a triple mutant (MZn1-3) were tested in vitro by a gel shift assay on a β-globin promoter CACCC element oligonucleotide (left) and in vivo by cotransfection with the natural β-globin promoter-luciferase reporter into K562 cells (right). Extracts for gel shift analysis were prepared from COS7 cells after transfection with the indicated constructs. The arrow indicates the novel shift seen upon EKLF transfection, and the protein levels of the wild type and mutant constructs are shown by anti-EKLF Western blot analysis below each lane. pSG5 is the empty EKLF expression vector (lane C). Luciferase activity in extracts was normalized to cotransfected growth hormone. IP, immunoprecipitation. (B) The wild type or mutants with EKLF mutations in each (MZn1, MZn2, and MZn3) or all (MZn1-3) of the zinc fingers were cotransfected into K562 cells with myc-Sin3A. Extracts were immunoprecipitated with anti-EKLF and blotted and probed with anti-myc after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A portion of the extract was saved as supernatant to monitor myc-Sin3A expression levels. pSG5 is the empty EKLF expression vector. (C) The wild type or EKLF mutants with mutations in each (MZn1, MZn2, and MZn3) or all (MZn1-3) of the zinc fingers or with deletion of all the zinc fingers (P) were cotransfected into K562 cells with HDAC1. Extracts were immunoprecipitated with anti-HDAC1, blotted, and probed with anti-EKLF after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A portion of the extract was saved as supernatant to monitor EKLF expression levels. pBJ5 is the empty HDAC1 expression vector. (D) Transrepression by wild-type GAL/EKLF, single-zinc-finger mutants (MZn1, MZn2, and MZn3), or triple mutant (MZn1-3) derivatives was tested by cotransfection with the pG5tkCAT reporter in K562 cells. Cotransfection with GAL 1-147 provides the baseline (nonrepressed) reporter activity. Chloramphenicol acetyltransferase (CAT) activity in extracts was normalized to cotransfected growth hormone.
We next tested whether these mutants were still able to interact with Sin3A and HDAC1 by coimmunoprecipitation experiments after transfection into K562 cells. Figure 1B shows that each of the mutated EKLF derivatives—even one with mutations in all three zinc fingers—is able to interact with Sin3A to the same extent as the wild type. Similarly, EKLF's interaction with HDAC1 is retained with all of these constructs (Fig. 1C). However, as seen before (13), the zinc finger domain is absolutely required for this interaction. We conclude that the zinc finger structure of EKLF is not required for interaction with Sin3A and HDAC1.
The final test of this series was to monitor the repression activity of each EKLF mutant. These changes were introduced into the GAL/EKLF chimera previously demonstrated to repress transcription of the G5tkCAT reporter (13). This reporter retains a basal level of activity since it contains the thymidine kinase minimal promoter downstream of five GAL4 DNA binding sites. As seen previously, wild-type GAL/EKLF represses this promoter ∼70% in K562 cells (Fig. 1D). This repression level is not altered by using any of the zinc finger mutants; indeed, the three-finger mutant appears to be a slightly more efficient repressor (Fig. 1D). From these data, we conclude that neither the EKLF zinc finger structure nor EKLF DNA binding is required for repression and that we have successfully generated an EKLF derivative that represses but no longer activates transcription.
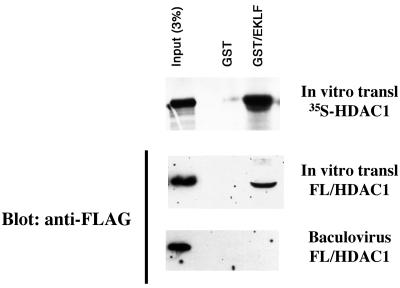
Our next task was to uncouple these two activities in the other direction: i.e., to see if an EKLF derivative could be found that activates but no longer represses transcription. To help us understand the protein interaction we should focus on, we first determined whether the EKLF interaction with HDAC1 was direct or not. HDAC1 is typically known to be recruited by corepressors, such as Sin3A, to promoters (32). We initially performed an in vitro pull-down experiment with GST-EKLF and in vitro-labeled and translated HDAC1 and found that these interact (Fig. 2). However, reticulocyte lysates have significant levels of Sin3A (16), so we obtained a baculovirus construct that expressed FL-HDAC1 and tested this the same way. As a control, we found that in vitro-translated FL-HDAC1 interacted with GST-EKLF, but when this protein was first purified from baculovirus, it no longer interacted with EKLF, implying that another protein (likely Sin3A) was responsible for recruiting HDAC1 to EKLF and thus that this interaction is not direct. As a result, we focused the rest of our analyses on the EKLF-Sin3A interaction.
FIG. 2.
Indirect interaction between EKLF and HDAC1. In vitro pull-down assays were performed with GST or GST/EKLF that was incubated with radiolabeled, in vitro-translated HDAC1 (top), in vitro-translated FL/HDAC1 (middle), or purified FL/HDAC1 (bottom). The top panel is the autoradiograph, and the other two panels are from Western blot analysis with anti-FLAG antibodies.
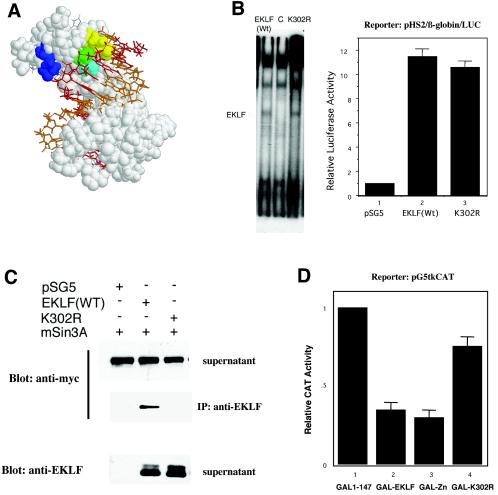
Our earlier studies had localized the Sin3A interaction domain of EKLF to its zinc finger region (13). As a result, our mutagenesis strategy required that we find a zinc finger mutant that still retains DNA binding yet has lost its ability for a specific protein interaction. We were aided immensely in this task by our earlier molecular modeling of EKLF (17). This structure had been based on the Zif268 cocrystal structure and had accurately predicted EKLF's inability to interact with β-thalassemia point mutations within the β-globin CACCC element. This model enabled us to focus our attention on EKLF zinc finger amino acids that were not directly involved in DNA binding, that were not rigorously conserved across other zinc finger proteins, and that faced away from the helix interface surface. Out of a limited number of possibilities, we focused our attention on lysine 302 (K302) as fulfilling these prerequisites (Fig. 3A). EKLF protein containing a replacement of this lysine with another basic residue (arginine; K302R) was tested for DNA binding to the CACCC element and transactivation of the β-globin promoter; both were normal (Fig. 3B). However, EKLF K302R was no longer able to bind Sin3A, as tested by coimmunoprecipitation experiments, even though expression levels were similar to wild type (Fig. 3C). The functional test of repression in vivo was performed by incorporating the K302R mutation in the GAL/EKLF chimera and testing this in the G5tkCAT repression assay. In this case, most of EKLF's repression activity was lost (Fig. 3D). These studies demonstrate that EKLF activation and repression functions are separable and suggest that EKLF protein interactions with Sin3A are critical for efficient repression to occur.
FIG. 3.
Selection and testing of an EKLF derivative that is deficient in repression yet retains activation. (A) A space-filling molecular model (17) of the three EKLF zinc fingers interacting with the double-strand CAC oligonucleotide (G-rich strand in red and C-rich strand in gold) is shown. The three XYZ amino acids (30) critical for binding within zinc finger 1 (K306, yellow; H309, green; A312, light blue) and the location of K302 (dark blue) are as indicated. (B) DNA binding (left) and transactivation (right) by wild-type (Wt) EKLF and K302R mutant EKLF were tested in vitro by a gel shift assay on a β-globin promoter CACCC element oligonucleotide (left) and in vivo by cotransfection with the natural β-globin promoter-luciferase (LUC) reporter into K562 cells (right). Extracts for gel shift analysis were prepared from COS7 cells after transfection with the indicated constructs. The novel shift seen upon EKLF transfection is indicated on the left. pSG5 is the empty EKLF expression vector. Luciferase activity in extracts was normalized to cotransfected growth hormone. (C) The wild type (WT) or K302R EKLF mutant was cotransfected into K562 cells with myc-Sin3A. Extracts were immunoprecipitated (IP) with anti-EKLF, blotted, and probed with anti-myc or anti-EKLF (as indicated) after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A portion of the extract was saved as supernatant to monitor myc-Sin3A and EKLF expression levels. pSG5 is the empty EKLF expression vector. (D) Transrepression by wild-type GAL/EKLF, GAL/ZnF, or the GAL/EKLF K302R mutant was tested by cotransfection with the pG5tkCAT reporter in K562 cells. Cotransfection with GAL 1-147 provides the baseline (nonrepressed) reporter activity. Chloramphenicol acetyltransferase (CAT) activity in extracts was normalized to cotransfected growth hormone.
Cellular influences on EKLF function.
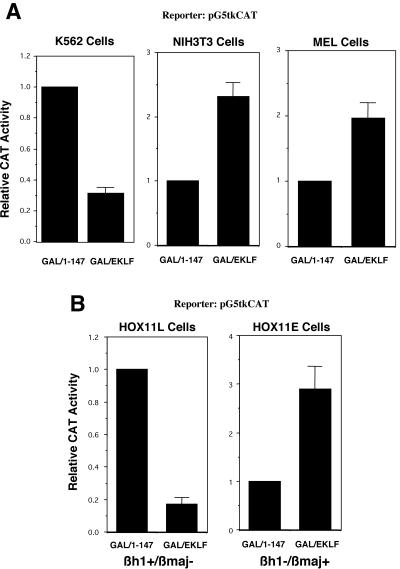
We next wished to address the potential physiological relevance of EKLF-corepressor interactions by determining whether the cellular milieu may play a part in altering EKLF function in vivo. Such a consideration arises from the observation that EKLF is expressed in primitive erythroid cells and in hematopoietic cells where adult β-globin is not expressed (25, 53, 57, 70). As a simple first approach, we tested for in vivo repression, using the G5tkCAT reporter, in NIH 3T3 cells as a non-erythroid cell type. The results show that, unlike the erythroid K562 line, GAL/EKLF does not repress this reporter but rather yields a mild (∼2.5-fold) activation (Fig. 4A).
FIG. 4.
Cell and stage specificity of EKLF transrepression. (A) Transrepression by wild-type GAL/EKLF was tested in K562 (human fetal-type erythroid), NIH 3T3 (murine nonerythroid), and MEL (murine adult-type erythroid) cells and compared to basal levels of reporter activity after cotransfection with GAL 1-147. Chloramphenicol acetyltransferase (CAT) activity in extracts was normalized to cotransfected growth hormone. (B) HOX11-immortalized erythroid cells (EBHX11) growing in LIF (11L; left panel) or Epo (llE; right panel) were transfected with pG5tkCAT and GAL 1-147 or GAL/EKLF. The statuses of embryonic (βh1) and adult (βmaj) globins in these two cell lines are indicated below each panel. Chloramphenicol acetyltransferase activity in extracts was normalized to cotransfected growth hormone.
K562 cells are considered a human “fetal” erythroid line in that they express the fetal γ-globin gene and not the adult β-globin gene. On the other hand, MEL cells are considered an “adult” erythroid line, as they express only the adult β-globin gene. In vivo tests with these cells demonstrate that EKLF does not repress the reporter in MEL cells but shows a mild (∼2-fold) activation (Fig. 4A). These results suggest that the EKLF repression or activation function is not only cell specific but also stage specific.
However, there are some limitations to these experiments. First, these are two different immortalized cell lines with considerably different provenances. Second, K562 cells are human and MEL cells are murine. To remove these issues from consideration, we investigated the HOX11-immortalized cell lines, particularly EBHX11 (28). This particular line is erythroid but can alter its gene expression pattern between primitive (expresses embryonic βh1) and definitive (expresses adult βmaj), depending on whether it is grown in the presence of LIF (EBHX11L) or Epo (EBHX11E), respectively. We first established transient transfection conditions with each of these sublines and then tested EKLF-repressive abilities on the G5tkCAT reporter. EBHX11L cells, which express embryonic βh1 globin and thus exhibit a primitive erythroid phenotype, behaved as K562 cells: EKLF acted as a strong repressor, decreasing reporter activity by ∼80% (Fig. 4B). However, EKLF repression was not observed when the same transfection analysis was performed in EBHX11E cells, which express adult βmaj globin and exhibit a definitive phenotype (Fig. 4B). Indeed, as seen with MEL cells, a mild activation was observed. These studies, using the same cell line simply grown under different conditions, demonstrate that EKLF function can be altered by what are likely subtle differences in cellular milieu, exhibiting strong stage specificity.
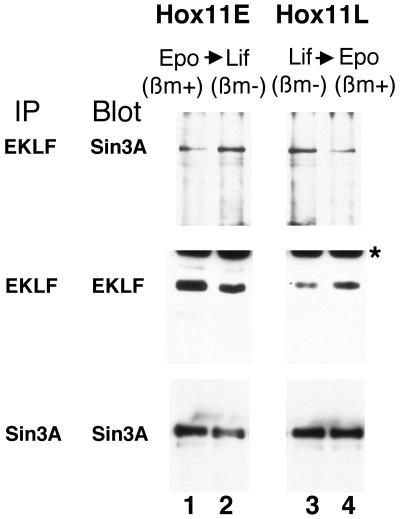
Based on our protein interaction analyses, we postulated that EKLF interactions with Sin3A might have also changed depending on the growth condition of the EBHX11 cells. We therefore performed coimmunoprecipitation experiments on the endogenous EKLF and Sin3A proteins, under two conditions: (i) interactions were monitored in EBHX11L cells before and after switching from LIF to Epo for 48 h, and (ii) interactions were monitored in EBHX11E cells before and after switching from Epo to LIF for 48 h. The results shown in Fig. 5 demonstrate a number of points. First, levels of Sin3A are not significantly altered under any condition (compare the Sin3A signals across all four lanes in the bottom panel). Second, EKLF levels are higher in EBHX11E cells than those in EBHX11L cells (compare EKLF signals in lanes 1 and 3 in the middle panel). Third, switching EBHX11E cells to growth in LIF decreases EKLF levels (compare EKLF signals in lanes 1 and 2 in the middle panel); conversely, switching EBHX11L cells to growth in Epo increases EKLF levels (compare EKLF signals in lanes 3 and 4 in the middle panel). This part of the results demonstrates that EKLF levels fluctuate slightly depending on the phenotype of the erythroid cell.
FIG. 5.
Endogenous EKLF-Sin3A protein interactions in EBHX11 cells grown under different conditions. Extracts were generated from EBHX11 cells grown in Epo (Hox11E) or LIF (Hox11L) or after cytokine replacement (2 days) as indicated. Extracts were immunoprecipitated (IP) and probed after Western blotting with anti-EKLF or -Sin3A antibodies as indicated. The statuses of adult βmaj (βm) globins in these two cell lines under the different conditions are indicated above each panel. The asterisk indicates a nonspecific signal on the Western blot.
However, the critical top panel of Fig. 5 demonstrates in two ways that EKLF-Sin3A interactions change dramatically, depending on the growth conditions. First, even though EKLF levels are lower in EBHX11L, the EKLF-Sin3A interaction is higher in EBHX11L than EBHX11E cells (lane 3 versus 1). Second, altering growth conditions for 48 h is sufficient to increase this interaction when EBHX11E cells are switched to LIF (lane 1 versus 2) and to decrease this interaction when EBHX11L cells are switched to Epo (lane 3 versus 4). These results are consistent with the functional tests seen with EKLF in EBHX11 and suggest that EKLF repression of the reporter in EBHX11L cells is due to its higher level of interaction with Sin3A. These studies demonstrate that EKLF association with Sin3A is both stage specific and reversible.
DISCUSSION
EKLF has a defined role as a critical activator of adult β-globin transcription during definitive erythropoiesis (4, 48). However, our present studies identify EKLF repression as a separable attribute from activation and also suggest that this switch in activity is dramatically affected by the stage specificity (i.e., primitive versus definitive) of the erythroid environment.
Structural aspects of EKLF-corepressor interactions.
The ability of transcription factors to form alternate complexes with coactivators and corepressors has been observed in a number of systems (3, 20, 21). A particularly well-characterized paradigm is that of the nuclear hormone receptor (NR) family, which interacts with Sin3A and HDAC at its repressed, target promoter until an external stimulus (ligand) alters its configuration (39). This results in a dramatic change of NR complex component proteins that replaces corepressors with CBP, leading to transcriptional activation of its target promoter. By analogy, EKLF might undergo such a conformational change dependent on a cellular milieu (rather than on a direct external stimulus). It might also be reasonable to assume that the three highly structured zinc fingers would be a less likely target for such conformational changes (18, 31) and that the large proline-rich domain may provide the flexibility required for such alterations (64). Indeed, the proline-rich domain is critical for transcriptional activation in vivo (6, 12) and in vitro (2).
And yet of particular relevance is the importance of K302 in this process, as it is required for Sin3A interaction and is also acetylated by CBP (68). This amino acid is located within the amino-proximal β-sheet of EKLF zinc finger 1, directly adjacent to a highly basic stretch in the proline-rich region. This interaction is therefore quite different from that seen between Sin3A and the Sin3-interacting domain of KLF11 (47), which is not present in EKLF. Based on structural modeling studies, K302 likely plays little role in EKLF-DNA interactions (17). However, its role in two very different sets of interactions identifies it as a nodal point that may play a critical role in EKLF function (Fig. 6) (see below). In contrast to activation, repression does not require the proline-rich region (13), and as a result our studies suggest that the zinc finger domain forms the core EKLF repression module. Our results carry this idea further, as neither ability to bind DNA nor an intact finger structure is required for repression. This suggests the unprecedented idea that zinc finger structure may also be conformationally modulated as needed in vivo.
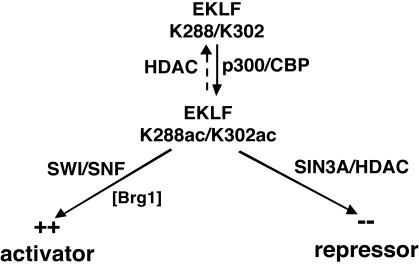
FIG. 6.
Model for EKLF activation or repression. EKLF is modified by p300/CBP at two sites: K288 and K302. K288ac may play a role in optimal interaction with SWI/SNF via BRG1. At the same time, K302 is critical for EKLF-Sin3A interaction, raising questions about potential cross-regulation between these two pathways and the downstream targets that may be affected. The possibility that acetylated EKLF (K302ac) binds with higher affinity to HDACs and is deacetylated as a result of this interaction has not been tested but is also implied by this scheme.
Our previous coimmunoprecipitation and inhibitor studies (13) also lead us to postulate that the EKLF-Sin3A interaction results in recruitment of HDAC1. These components are likely part of a much larger in vivo complex that acts to repress target genes in the cell (24, 43, 69). As a result, the molecular details of EKLF-protein interactions during repression are not likely to be simple.
Biological implications of stage-specific repression by EKLF.
A particularly intriguing observation is that, on a repressible promoter, EKLF function is stage specific. This immediately raises the issue of alternate EKLF function, as it is expressed in the yolk sac (57) and in early hematopoietic cells (25, 53, 70), neither of which express adult β-globin and likely provide a considerably different cellular environment in which EKLF can act. Although primitive and definitive cells can arise from a common precursor (29), they are divergently affected by genetic ablation (35, 42, 45, 61, 65). Similarly, distinct gene expression patterns, in addition to different renewal and multipotential capabilities of early versus late hematopoietic cells, segregate these two related cell types, giving rise to unequal cellular environments that differentially respond to extracellular cytokines (1, 41, 62). The end result is that transcriptional control mechanisms are likely to be significantly dissimilar even among nominally related erythroid or hematopoietic cells. The EKLF genetic ablation studies do not illuminate the role of the cellular environment directly, as yolk sac-derived primitive erythropoiesis and early hematopoiesis are normal in the absence of EKLF and as disruption of adult β-globin onset is the most dramatic result (44, 50). However, EKLF-null cells are more easily immortalized than wild-type cells (14), and rescue of hemoglobinization does not lead to healthy erythroid cells (49). This leaves open the possibility that other targets, less drastically affected by EKLF absence than adult β-globin and yet compensated for by redundancy with other KLFs (5), are repression targets. Microarray studies of another erythroid transcription factor, GATA1, reveal a significant number of down- as well as up-regulated targets that appear after its induction (22, 55).
In addition to the absence of β-globin, ablation of EKLF results in loss of the hypersensitive site (HS) at the β-globin promoter, diminution of HS3 within the far upstream locus control region (LCR), and disruption of the active chromatin hub (19, 63; F. Grosveld, personal communication). The chromatin structure of the β-like globin cluster is in an open configuration well before the onset of any transcription (26). This suggests that EKLF plays a transcription-independent role in chromatin structure and may be poised at the β-globin promoter and at the LCR in a repressive conformation until a cellular signal switches it to an activator, resulting in high-level β-globin transcription only at the correct developmental time. Such a hypothesis must remain speculative until an EKLF antibody that is suitable for chromatin immunoprecipitation analyses becomes available. Recent studies demonstrate that MafK-associated repressive transcription factor complexes at the LCR are exchanged for an activation complex during erythroid differentiation, resulting in a derepression of the β-globin gene (7). In this context, it is intriguing that the EKLF zinc finger domain (which interacts with Sin3A/HDAC1) alone is sufficient to form a hypersensitive site at the β-globin promoter in vitro (27).
Unlike the case with activation, where the adult β-globin promoter was identified early in the analysis of EKLF function (40), repression targets of EKLF are not known. This is significantly complicated by our observation that EKLF DNA binding is not required for its repressive effects. As a result, monitoring whether a CACCC element is present within a putative target is irrelevant to any such search. DNA binding by SCL (51) and the glucocorticoid receptor (54) is not required for all of their transcriptional effects in vivo, and EKLF repression likely falls into this surprising paradigm.
The present study, in addition to our previous analyses of EKLF transactivation function, points to its acetylation status as being a critical nodal point for its ultimate function in the cell (Fig. 6). EKLF transactivation of the native β-globin promoter is superactivated by inclusion of p300/CBP in transient assays, and an intact acetyltransferase is required for this to occur (68). EKLF is also acetylated by p300/CBP at two sites: K288 and K302. K288 is required for optimal transactivation of the β-globin promoter, likely by enabling more efficient formation of a complex with SWI/SNF (68). This interaction results in an open chromatin structure at the β-globin promoter, as assessed by in vitro chromatin assembly (2).
However, unlike K288, mutation of K302 had little effect on transactivation, superactivation, or SWI-SNF interaction (68). Our present studies shed the first light on K302 function, and unexpectedly it interfaces with repression, as K302 is required for EKLF interaction with Sin3A. As a result, a model that incorporates these observations (Fig. 6) would postulate that the acetylation status of K288 and K302 plays a critical role in whether EKLF behaves as an activator or repressor. This status can be directly altered by the cellular milieu in at least two ways: (i) whether the erythroid cell is primitive or definitive and (ii) whether the appropriate time after hematopoiesis has arrived for transcription to be induced after chromatin structure has been poised, yet held inactive, possibly by EKLF itself. This could be analogous to MyoD, where its association with HDAC complexes in undifferentiated muscle cells prevents it from being active until the cells differentiate (36). A direct prediction is that the acetylation statuses of K288 and K302 will not be equivalent in EKLF proteins that are isolated from differently staged cellular sources. Acetylation-specific antibodies would be most useful to address this issue; unfortunately, we have not been successful in generating anti-acetyl-K288 or -K302 antibodies that recognize these residues in the context of the full-length protein.
Three questions immediately arise in light of this model. One is whether the acetylation status of K302 affects the affinity of interaction between EKLF and a Sin3A complex. For example, acetylation of YY1 increases its binding affinity for HDAC1 (66). Second is whether EKLF is deacetylated by HDACs at K288 and/or K302. If so, then this might provide a powerful control point for activation or repression by the simple change of acetylation status at either or both of these sites. A number of transcription factors have been shown to be deacetylation substrates, including MyoD (36), RelA (11), YY1 (66), and E2F1 (38). Third is the status of chromatin at the β-like globin cluster after rescue of EKLF-null cells with single- or double-acetylation-mutant EKLF proteins or with EKLF zinc finger mutants that cannot bind DNA. Future studies based on answers to these questions will further refine this model and demonstrate whether EKLF acetylation status plays a directive role in regulating its activation and repression targets.
Acknowledgments
We thank Gordon Keller, Christina Grozinger, and Stuart Schreiber for cells and constructs and the Bieker lab for constructive comments.
This work was supported by NIH grant DK46865 to J.J.B.
REFERENCES
- 1.Akashi, K., X. He, J. Chen, H. Iwasaki, C. Niu, B. Steenhard, J. Zhang, J. Haug, and L. Li. 2003. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 101:383-389. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. J. Bieker, and B. M. Emerson. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93-104. [DOI] [PubMed] [Google Scholar]
- 3.Barolo, S., and J. W. Posakony. 2002. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16:1167-1181. [DOI] [PubMed] [Google Scholar]
- 4.Bieker, J. J. 2000. EKLF and the development of the erythroid lineage, p. 71-84. In K. Ravid and J. D. Licht (ed.), Transcription factors: normal and malignant development of blood cells. Wiley-Liss, New York, N.Y.
- 5.Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355-34358. [DOI] [PubMed] [Google Scholar]
- 6.Bieker, J. J., and C. M. Southwood. 1995. The erythroid Krüppel-like factor transactivation domain is a critical component for cell-specific inducibility of a β-globin promoter. Mol. Cell. Biol. 15:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, M., J. A. Ranish, N. T. Kummer, J. Hamilton, K. Igarashi, C. Francastel, T. H. Chi, G. R. Crabtree, R. Aebersold, and M. Groudine. 2004. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol. 11:73-80. [DOI] [PubMed] [Google Scholar]
- 8.Brivanlou, A. H., and J. E. Darnell, Jr. 2002. Signal transduction and the control of gene expression. Science 295:813-818. [DOI] [PubMed] [Google Scholar]
- 9.Brown, R. C., S. Pattison, J. van Ree, E. Coghill, A. Perkins, S. M. Jane, and J. M. Cunningham. 2002. Distinct domains of erythroid Krüppel-like factor modulate chromatin remodeling and transactivation at the endogenous β-globin gene promoter. Mol. Cell. Biol. 22:161-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caterina, J. J., D. J. Ciavatta, D. Donze, R. R. Behringer, and T. M. Townes. 1994. Multiple elements in human beta-globin locus control region 5′ HS 2 are involved in enhancer activity and position-independent, transgene expression. Nucleic Acids Res. 22:1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L., W. Fischle, E. Verdin, and W. C. Greene. 2001. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293:1653-1657. [DOI] [PubMed] [Google Scholar]
- 12.Chen, X., and J. J. Bieker. 1996. Erythroid Krüppel-like factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. EMBO J. 15:5888-5896. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, X., and J. J. Bieker. 2001. Unanticipated repression function linked to erythroid Krüppel-like factor. Mol. Cell. Biol. 21:3118-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coghill, E., S. Eccleston, V. Fox, L. Cerruti, C. Brown, J. Cunningham, S. Jane, and A. Perkins. 2001. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood 97:1861-1868. [DOI] [PubMed] [Google Scholar]
- 15.DelRio, S., S. R. Menezes, and D. R. Setzer. 1993. The function of individual zinc fingers in sequence-specific DNA recognition by transcription factor IIIA. J. Mol. Biol. 233:567-579. [DOI] [PubMed] [Google Scholar]
- 16.Eilers, A. L., A. N. Billin, J. Liu, and D. E. Ayer. 1999. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J. Biol. Chem. 274:32750-32756. [DOI] [PubMed] [Google Scholar]
- 17.Feng, W. C., C. M. Southwood, and J. J. Bieker. 1994. Analyses of β-thalassemia mutant DNA interactions with erythroid Krüppel-like factor (EKLF), an erythroid cell-specific transcription factor. J. Biol. Chem. 269:1493-1500. [PubMed] [Google Scholar]
- 18.Frankel, A. D., J. M. Berg, and C. O. Pabo. 1987. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc. Natl. Acad. Sci. USA 84:4841-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillemans, N., R. Tewari, F. Lindeboom, R. Rottier, T. de Wit, M. Wijgerde, F. Grosveld, and S. Philipsen. 1998. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the beta-globin locus control region in vivo. Genes Dev. 12:2863-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 21.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 22.Grass, J. A., M. E. Boyer, S. Pal, J. Wu, M. J. Weiss, and E. H. Bresnick. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA 100:8811-8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 24.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 25.Hu, M., D. Krause, M. Greaves, S. Sharkis, M. Dexter, C. Heyworth, and T. Enver. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774-785. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez, G., S. D. Griffiths, A. M. Ford, M. F. Greaves, and T. Enver. 1992. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc. Natl. Acad. Sci. USA 89:10618-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller, G., C. Wall, A. Z. Fong, T. S. Hawley, and R. G. Hawley. 1998. Overexpression of HOX11 leads to the immortalization of embryonic precursors with both primitive and definitive hematopoietic potential. Blood 92:877-887. [PubMed] [Google Scholar]
- 29.Kennedy, M., M. Firpo, K. Choi, C. Wall, S. Robertson, N. Kabrun, and G. Keller. 1997. A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature 386:488-493. [DOI] [PubMed] [Google Scholar]
- 30.Klevit, R. E. 1991. Recognition of DNA by Cys2, His2 zinc fingers. Science 253:1367, 1395. [DOI] [PubMed] [Google Scholar]
- 31.Klug, A., and J. W. Schwabe. 1995. Zinc fingers. FASEB J. 9:597-604. [PubMed] [Google Scholar]
- 32.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 33.Levine, M., and R. Tjian. 2003. Transcription regulation and animal diversity. Nature 424:147-151. [DOI] [PubMed] [Google Scholar]
- 34.Lim, S. K., J. J. Bieker, C. S. Lin, and F. Costantini. 1997. A shortened life span of EKLF −/− adult erythrocytes, due to a deficiency of β-globin chains, is ameliorated by human γ-globin chains. Blood 90:1291-1299. [PubMed] [Google Scholar]
- 35.Lin, C. S., S. K. Lim, V. D'Agati, and F. Costantini. 1996. Differential effects of an erythropoietin receptor gene disruption on primitive and definitive erythropoiesis. Genes Dev. 10:154-164. [DOI] [PubMed] [Google Scholar]
- 36.Mal, A., M. Sturniolo, R. L. Schiltz, M. K. Ghosh, and M. L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mannervik, M., Y. Nibu, H. Zhang, and M. Levine. 1999. Transcriptional coregulators in development. Science 284:606-609. [DOI] [PubMed] [Google Scholar]
- 38.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 40.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13:2776-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto, T., H. Iwasaki, B. Reizis, M. Ye, T. Graf, I. L. Weissman, and K. Akashi. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3:137-147. [DOI] [PubMed] [Google Scholar]
- 42.Mucenski, M. L., K. McLain, A. B. Kier, S. H. Swerdlow, C. M. Schreiner, T. A. Miller, D. W. Pietryga, W. J. Scott, and S. S. Potter. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell 65:677-689. [DOI] [PubMed] [Google Scholar]
- 43.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 44.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316-318. [DOI] [PubMed] [Google Scholar]
- 45.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 46.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 47.Pang, Y. P., G. A. Kumar, J. S. Zhang, and R. Urrutia. 2003. Differential binding of Sin3 interacting repressor domains to the PAH2 domain of Sin3A. FEBS Lett. 548:108-112. [DOI] [PubMed] [Google Scholar]
- 48.Perkins, A. 1999. Erythroid Kruppel like factor: from fishing expedition to gourmet meal. Int. J. Biochem. Cell Biol. 31:1175-1192. [DOI] [PubMed] [Google Scholar]
- 49.Perkins, A. C., K. R. Peterson, G. Stamatoyannopoulos, H. E. Witkowska, and S. H. Orkin. 2000. Fetal expression of a human agamma globin transgene rescues globin chain imbalance but not hemolysis in EKLF null mouse embryos. Blood 95:1827-1833. [PubMed] [Google Scholar]
- 50.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal β-thalassemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318-322. [DOI] [PubMed] [Google Scholar]
- 51.Porcher, C., E. C. Liao, Y. Fujiwara, L. I. Zon, and S. H. Orkin. 1999. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development 126:4603-4615. [DOI] [PubMed] [Google Scholar]
- 52.Quadrini, K. J., and J. J. Bieker. 2002. Kruppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of EKLF. J. Biol. Chem. 277:32242-32252. [DOI] [PubMed] [Google Scholar]
- 53.Reese, T. T., R. C. Gregory, E. R. Sharlow, R. E. Pacifici, J. A. Crouse, K. Todokoro, and D. M. Wojchowski. 1997. Epo-induced hemoglobinization of SKT6 cells is mediated by minimal cytoplasmic domains of the Epo or prolactin receptors without modulation of GATA-1 or EKLF. Growth Factors 14:161-176. [DOI] [PubMed] [Google Scholar]
- 54.Reichardt, H. M., K. H. Kaestner, J. Tuckermann, O. Kretz, O. Wessely, R. Bock, P. Gass, W. Schmid, P. Herrlich, P. Angel, and G. Schutz. 1998. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93:531-541. [DOI] [PubMed] [Google Scholar]
- 55.Rylski, M., J. J. Welch, Y.-Y. Chen, D. L. Letting, J. A. Diehl, L. A. Chodosh, G. A. Blobel, and M. J. Weiss. 2003. GATA-1-mediated proliferation arrest during erythroid maturation. Mol. Cell. Biol. 23:5031-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simpson, R. J., E. D. Cram, R. Czolij, J. M. Matthews, M. Crossley, and J. P. Mackay. 2003. CCHX zinc finger derivatives retain the ability to bind Zn(II) and mediate protein-DNA interactions. J. Biol. Chem. 278:28011-28018. [DOI] [PubMed] [Google Scholar]
- 57.Southwood, C. M., K. M. Downs, and J. J. Bieker. 1996. Erythroid Kruppel-like factor (EKLF) exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev. Dyn. 206:248-259. [DOI] [PubMed] [Google Scholar]
- 58.Stamatoyannopoulos, G., and F. Grosveld. 2001. Hemoglobin switching, p. 135-182. In G. Stamatoyannopoulos, P. W. Majerus, R. M. Perlmutter, and H. Varmus (ed.), The molecular bases of blood diseases. W. B. Saunders Co., Philadelphia, Pa.
- 59.Struhl, K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98:1-4. [DOI] [PubMed] [Google Scholar]
- 60.Vermaak, D., K. Ahmad, and S. Henikoff. 2003. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 15:266-274. [DOI] [PubMed] [Google Scholar]
- 61.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weissman, I. L., D. J. Anderson, and F. Gage. 2001. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol. 17:387-403. [DOI] [PubMed] [Google Scholar]
- 63.Wijgerde, M., J. Gribnau, T. Trimborn, B. Nuez, S. Philipsen, F. Grosveld, and P. Fraser. 1996. The role of EKLF in human β-globin gene competition. Genes Dev. 10:2894-2902. [DOI] [PubMed] [Google Scholar]
- 64.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, H., X. Liu, R. Jaenisch, and H. F. Lodish. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83:59-67. [DOI] [PubMed] [Google Scholar]
- 66.Yao, Y.-L., W.-M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, W., and J. J. Bieker. 1998. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. USA 95:9855-9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Krüppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 70.Ziegler, B. L., R. Muller, M. Valtieri, C. P. Lamping, C. A. Thomas, M. Gabbianelli, C. Giesert, H. J. Buhring, L. Kanz, and C. Peschle. 1999. Unicellular-unilineage erythropoietic cultures: molecular analysis of regulatory gene expression at sibling cell level. Blood 93:3355-3368. [PubMed] [Google Scholar]