Abstract
Cyclin-dependent kinase (CDK) is required for the initiation of chromosomal DNA replication in eukaryotes. In Saccharomyces cerevisiae, the Clb5 and Clb6 cyclins activate Cdk1 and drive replication origin firing. Deletion of CLB5 reduces initiation of DNA synthesis from late-firing origins. We have examined whether checkpoints are activated by loss of Clb5 function and whether checkpoints are responsible for the DNA replication defects associated with loss of Clb5 function. We present evidence for activation of Rad53 and Ddc2 functions with characteristics suggesting the presence of DNA damage. Deficient late origin firing in clb5Δ cells is not due to checkpoint regulation, but instead, directly reflects the decreased abundance of S-phase CDK, as Clb6 activates late origins when its dosage is increased. Moreover, the viability of clb5Δ cells depends on Rad53. Activation of Rad53 by either Mrc1 or Rad9 contributes to the survival of clb5Δ cells, suggesting that both DNA replication and damage pathways are responsive to the decreased origin usage. These results suggest that reduced origin usage leads to stress or DNA damage at replication forks, necessitating the function of Rad53 in fork stabilization. Consistent with the notion that decreased S-CDK function creates stress at replication forks, deletion of RRM3 helicase, which facilitates replisome progression, greatly diminished the growth of clb5Δ cells. Together, our findings indicate that deregulation of S-CDK function has the potential to exacerbate genomic instability by reducing replication origin usage.
Duplication of eukaryotic chromosomes involves the initiation of DNA synthesis from multiple origins of replication distributed along each chromosome. Although chromosomal DNA replication is restricted to the S-phase period of the cell cycle, individual replication origins initiate DNA synthesis (or “fire”) at different times during S phase in a regulated fashion such that each replication origin has a characteristic initiation time within S phase. The exact nature of this regulation is not fully understood but appears to be influenced by two apparently unrelated factors: the local chromatin environment of each origin, which establishes the relative order of origin firing prior to S phase (reviewed in reference 43); and checkpoint signaling pathways, which modulate the extent to which the initiation of certain origins is delayed, particularly in response to replication defects or DNA damage (21, 29, 31, 33).
The pre-replicative complex (pre-RC) assembles at and governs the function of each replication origin (reviewed in reference 4). Activation of the pre-RC results in its conversion into two, divergent, replication fork complexes (replisomes) through the recruitment of additional DNA synthesis factors, such as Cdc45 and DNA polymerases. Pre-RC activation requires the function of cyclin-dependent kinase (CDK), which consists of a catalytic subunit (Cdk1) controlled by a cyclin subunit whose expression and stability are cell cycle regulated. In Saccharomyces cerevisiae, two B-type cyclins, Clb5 and Clb6, accumulate at the beginning of each cell cycle and stimulate the origin activation function of Cdk1, commencing S phase (9, 15, 30).
Clb5-Cdk1 appears to account for the majority of S-phase CDK function as mutation of CLB5, but not CLB6, results in a lengthened S phase, suggesting that fewer pre-RCs are activated in CLB5-deficient cells (9, 30). Consistent with this, late origin activation is specifically deficient in the absence of CLB5, whereas deletion of CLB6 has no apparent effect on DNA synthesis or late origin firing (8). Thus, Clb5 appears capable of activating a full complement of origins. In the absence of both Clb5 and Clb6, S phase becomes dependent on later-expressed cyclins Clb1 to -4 and begins significantly later than in either single mutant (30). Together, these results suggest that Clb6 can drive pre-RC activation, but only early in S phase. Whether this reflects absence of Clb6 during late S phase, a specific ability of Clb5 to target late origins for activation (perhaps due to the local chromatin structure or chromatin-specific substrates), or a requirement for a higher threshold of Clb-Cdk1 activity for late origin firing remains unresolved.
An implied consequence of decreased late origin function in clb5Δ cells is an increase in average replicon size, particularly in later-replicating regions of the genome. This apparently has the effect of delaying completion of chromosomal DNA replication, which may elicit a checkpoint-mediated mitotic delay (S-M checkpoint). A second possible consequence of increased replicon size is an increased propensity for replisome defects, which may result in replication stress or DNA damage, either of which may elicit an intra-S checkpoint response (reviewed in reference 24). These cell cycle checkpoints in response to replication defects or DNA damage involve signaling pathways that sense the presence of a replication defect or damaged DNA and initiate cellular responses aimed at rectifying the problem. The Mec1 kinase plays a major role in sensing sites of DNA damage or replication stress, apparently through recruitment by Ddc2 to such sites and activating the effector kinases, Rad53 and Chk1 (reviewed in reference 19). The activation of these effector kinases appears to involve their phosphorylation by Mec1, which is mediated by one of two adapter proteins, Rad9 and Mrc1. Rad9 mediates the activation of Rad53 and Chk1 in response to DNA damage, whereas Mrc1 mediates the activation of Rad53 in response to replication inhibition. Activated Chk1 and Rad53 together delay mitotic progression, providing time for the cell to cope with the insult, while Rad53 also induces DNA repair genes, increases deoxyribonucleotide concentrations, inhibits late origin activation, and stabilizes replisomes. In combination, these responses are critical for the viability of cells subjected to replicative stress or DNA damage.
Recent studies argue that replisome stabilization by Rad53 is of paramount importance in replicating cells exposed to a replication inhibitor (e.g., hydroxyurea [HU]) or DNA-damaging agent (e.g., methyl methanesulfonate [MMS]) (5, 18, 34, 37). In the absence of MEC1 or RAD53, aberrant replication fork structures are detected at stalled replisomes; these cells rapidly lose viability and are unable to resume DNA synthesis, even after the inhibitory compound is removed. Mrc1 appears to function exclusively during S phase and is the primary mediator of Rad53 function in cells blocked in replication by HU, whereas Rad9 functions in DNA damage responses throughout the cell cycle. However, overlap between these two pathways exists, as deletion of both MRC1 and RAD9 creates cells that are much more sensitive to HU than either single mutant and have comparable sensitivity to cells lacking RAD53 (1). This indicates either that Rad9 can substitute for Mrc1 in relaying a replication fork stress signal from Mec1 or, more likely, that replication stress creates DNA damage detectable by Mec1 and Rad9, thereby activating Rad53's replisome stabilization function. Neither the mechanism(s) nor a target through which Rad53 acts to stabilize replication forks is known.
The similar effects on late origin firing caused by CLB5 deletion or by intra-S checkpoint activation suggested the possibility that loss of late origin firing in clb5Δ cells is the result of checkpoint activation. Decreased origin activation due to decreased S-CDK levels should increase the average length of DNA synthesized by each replisome, possibly creating a requirement for Rad53's stabilization function. Alternatively, lack of Clb5 could cause an inherent defect in replisome function (independent of replicon size) that stimulates checkpoint activation. To determine whether the loss of Clb5 function is associated with a checkpoint, we have examined the effects of CLB5 deletion on checkpoint signaling and the effects on origin function and cell viability of disrupting different checkpoint functions in clb5Δ mutant cells.
MATERIALS AND METHODS
Plasmid and strain constructions.
Strains are described in Table 1. Most gene deletions were constructed by PCR-based methods using kanMx, HIS5, and TRP1 selectable markers as described previously (17). Ddc2-GFP(S65T) was introduced by PCR-mediated gene modification as described previously (17). Rad53-HA3 was introduced with plasmid p404-RAD53-HA or p406-RAD53-HA. CLB5 and CLB6 deletions were constructed with plasmids ESD221 (clb5::URA3), ESD262 (clb5::hisG), and ESD291 (clb6::LEU2) (30). pRS405-mrc1AQ contains the 5-kb XhoI mrc1AQ fragment from pAO138 (23); this plasmid was digested with NdeI for integration at the mrc1Δ::kanMx locus. To construct pclb5-CLB6, the CLB6 open reading frame was amplified by PCR, sequenced to confirm that no mutations were introduced, and inserted into plasmid pC5C2-3NF (6). For replacement of CLB5 by CLB6 at the native CLB5 locus, pclb5-CLB6 was digested with StuI and KpnI and transformed into clb5Δ::URA3 cells; transformants were selected on 5-fluoroorotic acid and confirmed by PCR.
TABLE 1.
Yeast strains used in this study
| Straina | Genotype | Source or reference |
|---|---|---|
| OAy470 | MATaade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1Δ::hisG | 3 |
| DGy159 | sml1Δ::HIS3 mec1Δ::TRP1 bar1Δ::LEU2 | 2 |
| FHy20 | rad53Δ::RAD53-HA3 (TRP1) | 2 |
| JAy30 | clb5Δ::kanMx | 2 |
| DGy138 | clb6Δ::kanMx rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy147 | clb5Δ::HIS5 rad53Δ::RAD53-HA3 (TRP1) clb6Δ::LEU2 | This study |
| DGy149 | sml1Δ::HIS3 clb5Δ::kanMx | This study |
| DGy155 | sml1Δ::HIS3 rad53Δ::kanMx BAR1 | This study |
| DGy160 | sml1Δ::HIS3 bar1Δ::LEU2 | This study |
| DGy162 | clb5Δ::kanMx sml1Δ::HIS3 mec1Δ::TRP1 bar1Δ::LEU2 | This study |
| DGy163 | sml1Δ::HIS3 mec1Δ::TRP1 bar1Δ::LEU2 clb5Δ::kanMx rad53Δ::RAD53-HA3 (URA3) | This study |
| DGy183 | rad9Δ::HIS5 | This study |
| DGy190 | rad9Δ::HIS5 clb5Δ::URA3 | This study |
| DGy207 | clb5Δ::kanMx sml1Δ::HIS3 bar1Δ::LEU2 | This study |
| DGy213 | sml1Δ::HIS3 clb5Δ::kanMx bar1Δ::URA3 rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy226 | clb5Δ::URA3 | This study |
| DGy400 | rad9Δ::HIS5 rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy410 | rad9Δ::HIS5 clb5Δ::URA3 rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy430 | DDC2-GFP (TRP1) | This study |
| DGy431 | clb5Δ::kanMx DDC2-GFP (TRP1) | This study |
| DGy438 | clb5::CLB6 | This study |
| DGy439 | clb5::CLB6 clb6Δ::kanMx | This study |
| DGy448 | clb5::CLB6 rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy449 | clb5::CLB6 clb6Δ::kanMx rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy511 | sml1Δ::HIS3 bar1Δ::LEU2 rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy516 | mrc1Δ::kanMx::mrc1AQ(LEU2) clb5Δ::HIS5 | This study |
| DGy529 | mrc1Δ::URA3::mrc1AQ(LEU2) bar1Δ::kanMx rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy530 | mrc1Δ::kanMx::mrc1AQ(LEU2) clb5Δ::HIS5 rad53Δ::RAD53-HA3 (URA3) | This study |
| DGy531 | sml1Δ::HIS3 mec1Δ::TRP1 bar1Δ::LEU2 rad53Δ::RAD53-HA3 (TRP1) | This study |
| DGy532 | sml1Δ::HIS3 mrc1Δ::TRP1::mrc1AQ (LEU2) BAR1 | This study |
| DGy534 | DGy532 × OAy980 diploid | This study |
| SSy60 | mrc1Δ::URA3::mrc1AQ (LEU2) bar1Δ::kanMx | This study |
| SSy68 | rrm3Δ::TRP1 BAR1 | This study |
| OAy850 | clb5Δ::HIS5 rad53Δ::RAD53-HA3 (TRP1) | This study |
| OAy874 | MATα sml1Δ::HIS3 mec1Δ::TRP1 | This study |
| OAy931 | OAy874 × DGy207 diploid | This study |
| OAy945 | MATα sml1Δ::HIS3 clb5Δ::URA3 | This study |
| OAy966 | MATα sml1Δ::HIS3 rad53-11 BAR1 | This study |
| OAy967 | DGy155 × OAy945 diploid | This study |
| OAy969 | OAy971 × OAy972 diploid | This study |
| OAy970 | DGy149 × OAy966 diploid | This study |
| OAy971 | MATα sml1Δ::HIS3 clb5Δ::URA3 mec1Δ::TRP1 | This study |
| OAy972 | sml1Δ::HIS3 tel1Δ::kanMx BAR1 | This study |
| OAy977 | sml1Δ::HIS3 mec1Δ::TRP1 BAR1 | This study |
| OAy978 | MATα sml1Δ::HIS3 clb5Δ::URA3 BAR1 | This study |
| OAy980 | MATα sml1Δ::HIS3 clb5Δ::URA3 rad9Δ::kanMx BAR1 | This study |
| OAy981 | OAy977 × OAy980 diploid | This study |
| OAy982 | tel1Δ::kanMx sml1Δ::HIS3 BAR1 | This study |
| OAy983 | MATα mec1Δ::TRP1 sml1Δ::HIS3 BAR1 | This study |
| OAy988 | OAy978 × OAy987 diploid | This study |
| OAy989 | OAy978 × SSy68 diploid | This study |
| OAy990 | sml1Δ::HIS3 mec1Δ::TRP1 tel1Δ::kanMx | This study |
| OAy991 | sml1Δ::HIS3 clb5Δ::URA3 mec1Δ::TRP1 tel1Δ::kanMx | This study |
Strains are like OAy470 unless otherwise noted.
Yeast methods.
Experiments were performed at 30°C except as noted. Yeast extract-peptone-dextrose (YEPD) medium was used for all experiments. For spore analysis, freshly grown diploid cells were sporulated for 5 days at 23°C, dissected with a Singer MSM200 micromanipulator, and germinated at 23 or 30°C as indicated. After 3 days at 30°C or 5 days at 23°C, plates were imaged and genotyped by replica plating and PCR analysis when necessary. DNA content analysis has been described previously (2).
Analysis of replication structures.
Two-dimensional (2D) gel analysis was performed as described at http://fangman-brewer.genetics.washington.edu, except that replication structures were enriched on benzoylated naphthoylated DEAE-cellulose (Sigma). Fifty micrograms of DNA was digested with NcoI and BamHI for analysis of ARS603 (3.6 kb), ARS603.5 (2.6 kb), and ARS1011 (2.4 kb) or with EcoRI for analysis of ARS305 (5.8 kb) and ARS1413 (5.3 kb). Samples being compared were run in a single large gel and transferred together to a single membrane for hybridizations. DNA probes were labeled with the MegaPrime DNA labeling kit (Amersham Pharmacia) and detected on a PhosphorImager (Storm 860; Molecular Dynamics).
Analysis of Rad53.
Protein extracts were prepared by trichloroacetic acid precipitation as described previously (10) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), using 10% polyacrylamide (77:1) gels. 16B12 (Covance-BabCo) anti-hemagglutinin (HA) antibody was used at 1:5,000 in conjunction with SuperSignal Elisa Femto (Pierce) for quantitative chemiluminescent detection of protein with a ChemiDoc XRS 170-8070 (Bio-Rad) and Quantity One Analysis software (Bio-Rad). Quantification of Rad53 phosphorylation appears to indicate a higher proportion of phosphorylated Rad53 than visual inspection of the images suggests; however, this is apparently due to the presence of various Rad53 species with reduced mobility that do not migrate as distinct bands because quantification of multiple subsaturated exposures of the blots yielded identical results.
Microscopy.
An Olympus IX71 microscope with a ×60, 1.4-NA PlanApo oil immersion objective was used. Green fluorescent protein (GFP) fluorescence was detected using a Chroma fluorescein isothiocyanate filter set (excitation, 485/20 nm; emission, 515/30 nm), and the images were captured with a Roper Scientific DV42059 camera. The images were visualized using SoftWoRx software (Applied Precision). Fields of cells were photographed in several focal planes through the nuclei, until the foci were visible. Exposure time, gain, and binning functions were held constant. The percentage of cells with Ddc2 foci and cellular morphologies were determined from analysis of at least 100 cells at each time point.
RESULTS
The failure of late origin firing in clb5Δ cells is not due to checkpoint inhibition.
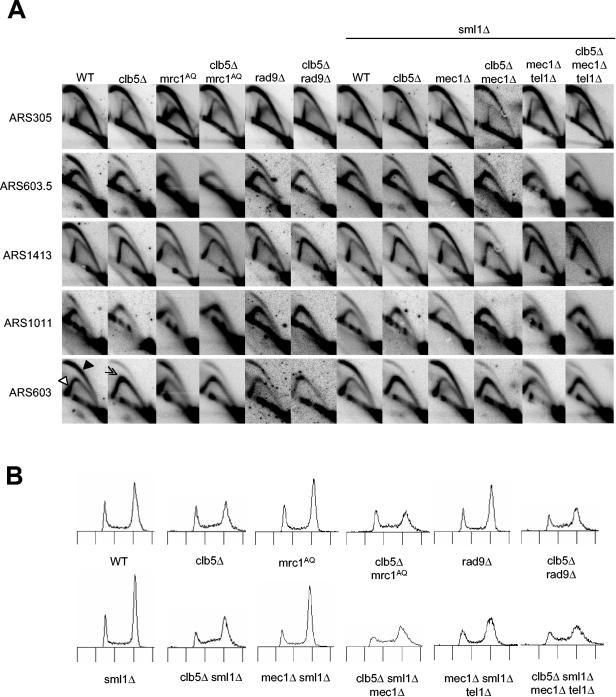
In a previous study of replication origin function in clb5Δ cells, initiation of late origins was greatly diminished, whereas initiation of early origins was essentially normal (8). We hypothesized that the failure of late origin activation in clb5Δ cells results from checkpoint inhibition of late origins, which is dependent on Mec1, Rad53, and Mrc1 (1, 29, 33). To test this idea, we analyzed initiation of three late (ARS603, ARS1011, and ARS1413) and two early (ARS305 and ARS603.5) replication origins of wild-type and clb5Δ strains and checkpoint-defective derivatives of these strains. We analyzed origin firing by examining replication initiation structures using 2D agarose gel (2D gel) electrophoresis of unsynchronized, cycling cells. This allows a determination of the overall frequency (efficiency) of origin usage because all replication structures occurring at the origin are represented as a composite (11).
Analysis of wild-type cells showed efficient activation of all of these origins based on the strong signals for “bubble” and “large Y” structures (Fig. 1A, indicated in the first ARS603 panel by filled and empty arrowheads, respectively). In clb5Δ mutants, initiations of ARS1011 and ARS603 were impaired severely as shown by the reduction in the “bubble” and “large Y” structures and the corresponding increase in the “small Y” structures (indicative of passive replication of the origin by a replication fork emanating from a flanking origin), particularly apparent at the apex of the “Y” arc (Fig. 1A, double arrowhead in the second ARS603 panel). In contrast to ARS603 and ARS1011, we observed that initiation from late origin ARS1413 (and ARS501; data not shown) was reduced less in clb5Δ cells. Unexpectedly, we also observed reduced initiation of the early origin ARS603.5 in clb5Δ cells (Fig. 1A). However, as expected, initiation of the very-early-firing origin, ARS305 (and ARS607; data not shown), was unaffected by CLB5 deletion. Thus, in agreement with the results of the previous study, late origins initiate replication inefficiently in the absence of CLB5. However, in contrast to the previous study, we observed that loss of CLB5 has a more modest effect on some late origins and reduces initiation of at least one relatively early origin (Fig. 1A).
FIG. 1.
Late origin initiation in clb5Δ cells is not blocked by checkpoints. (A) Unsynchronized, mid-log-phase cultures of wild-type (WT; OAy470), clb5Δ (JAy30), mrc1AQ (SSy60), clb5Δ mrc1AQ (DGy516), rad9Δ (DGy183), clb5Δ rad9Δ (DGy190), sml1Δ (DGy160), clb5Δ sml1Δ (DGy207), mec1Δ sml1Δ (DGy159), clb5Δ mec1Δ sml1Δ (DGy162), mec1Δ tel1Δ sml1Δ (OAy990), and clb5Δ mec1Δ tel1Δ sml1Δ (OAy991) strains were subjected to 2D gel analysis. One set of blots was probed sequentially for ARS603, ARS603.5, and ARS1011, and a second set of blots was probed sequentially for ARS305 and ARS1413. (B) DNA content of the cultures in panel A.
To determine the effect of specifically eliminating the intra-S replication checkpoint, we introduced the mrc1AQ allele into clb5Δ cells. This allele, which contains mutations in all of its potential Mec1 phosphoacceptor sites, significantly reduces or eliminates Mrc1-mediated phosphorylation of Rad53, but does not have the replication defect associated with a null mutation of MRC1 (24). Elimination of the replication checkpoint did not increase activation efficiency of late origins in clb5Δ cells (Fig. 1A). The presence of Mrc1AQ did not appear to alter initiation of either early or late origins in clb5Δ or wild-type cells. This strongly suggests that the failure of late origin firing in clb5Δ cells is not due to inhibition by the intra-S replication checkpoint.
DNA-damaging agents (e.g., MMS) also can lead to inhibition of late origin firing (33). Thus, if loss of Clb5 function were creating DNA damage, Rad9 might be involved in relaying DNA damage signals from Mec1, leading to late origin inhibition. However, deletion of RAD9 did not increase late origin firing in clb5Δ cells (Fig. 1A). To eliminate the possible redundancy of Mrc1 and Rad9 in relaying signals from Mec1, we tested the effect of deleting MEC1. Deletion of SML1 suppresses the lethality of cells lacking MEC1 or RAD53 (45) and was used to enable analysis of these strains; its deletion did not otherwise affect the results. Analysis of late origin firing in clb5Δ mec1Δ sml1Δ cells showed that Mec1 does not block late origin firing in clb5Δ cells (Fig. 1A). All the origins initiated replication with normal frequencies in sml1Δ and mec1Δ sml1Δ cells (Fig. 1A). We also tested the effect of deleting TEL1 in addition to MEC1, because Tel1 can substitute, at least partially, for Mec1 in the activation of Rad53 (28, 39). As discussed in detail below, elimination of Mec1 and Tel1 severely compromised the proliferation of clb5Δ cells. Nevertheless, we were able to grow cultures of these cells and analyze replication structures (Fig. 1A). The results show that Tel1 does not inhibit late origin firing in clb5Δ mec1Δ cells. Thus, neither the DNA replication nor DNA damage checkpoints are involved in suppressing the activity of late origins in clb5Δ cells.
The DNA content of unsynchronized cells shows the distribution of the population of cells in different periods of the cell cycle, providing an indication of the relative durations spent in each cell cycle phase. Comparison of clb5Δ with wild-type cells shows an increase in the length of S phase in clb5Δ cells, as more cells have a DNA content between 1C and 2C (Fig. 1B). The proportions of S phase cells in cultures of mrc1AQ, rad9Δ, mec1Δ, and mec1Δ tel1Δ cells were similar to those in wild-type cells, suggesting no increase in the length of S phase in these checkpoint mutant cells (Fig. 1B). In contrast, the proportions of S-phase cells in cultures of clb5Δ mrc1AQ, clb5Δ rad9Δ, clb5Δ mec1Δ, and clb5Δ mec1Δ tel1Δ cells were similar to those in clb5Δ cells, indicating that the lengthened S phase of clb5Δ cells is not due to replication inhibition by these checkpoint functions (Fig. 1B). Together with the analyses of late origin function, these data demonstrate that neither replication nor damage checkpoints are responsible for the observed replication defects of clb5Δ cells.
Decreased S-CDK activity triggers Rad9-mediated activation of Rad53.
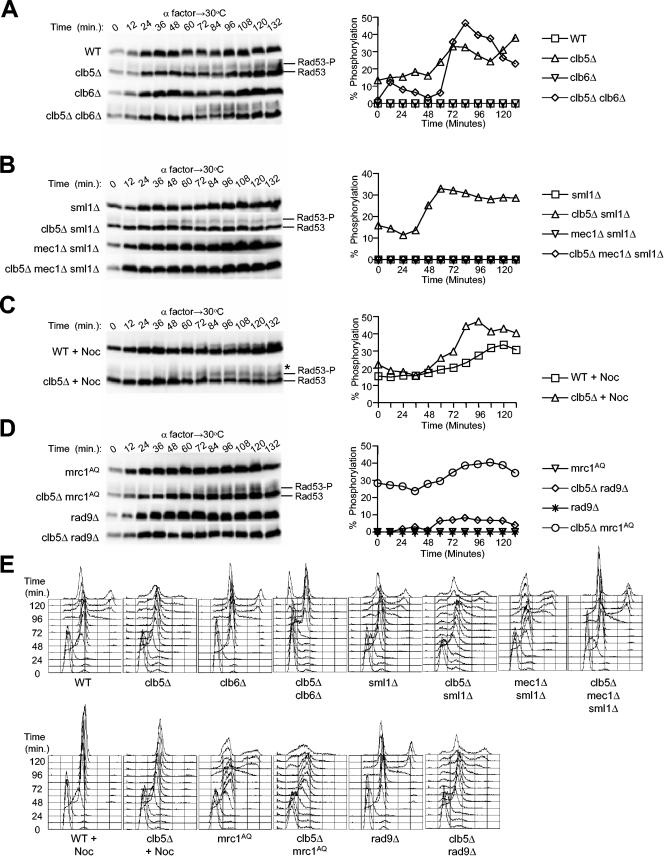
We had wished to directly address the role of Rad53 in origin regulation in clb5Δ cells; however, we found that the combined deletion of CLB5 and RAD53 was lethal (see below). This suggested that Rad53 was active in clb5Δ cells, possibly contributing to replisome stabilization, which is critical when cells experience replicative stress. One hallmark of Rad53 kinase activation is its phosphorylation in a Mec1-dependent manner (25, 28, 35). To determine whether Rad53 was activated in clb5Δ cells, we monitored Rad53 phosphorylation, which decreases its mobility in SDS-PAGE. Cells were synchronized in G1 with α-factor and released into S phase. Every 12 min, we analyzed Rad53 mobility by SDS-PAGE and measured the proportion of all slower-migrating forms of Rad53 using quantitative luminescence detection, while monitoring S-phase progression by DNA content analysis.
In wild-type cells, Rad53 migrated as the unmodified form throughout the cell cycle, indicating no checkpoint signaling in these cells (Fig. 2A). However, we observed slower-migrating forms of Rad53 in cells lacking CLB5, indicating that checkpoint activation of Rad53 had occurred (Fig. 2A). Phosphatase treatment eliminated the slower-migrating forms, demonstrating they were phosphorylated forms of Rad53, and the slower-migrating forms were associated with increased autokinase activity (data not shown). Also as expected, the slower-migrating forms of Rad53 were dependent on Mec1, whereas deletion of SML1 did not affect the level of Rad53 phosphorylation in wild-type or clb5Δ cells (Fig. 2B). The proportion of Rad53 that was modified peaked during late S/G2 phase. For example, in clb5Δ cells, Rad53 phosphorylation peaked at 72 min (Fig. 2A), which was after the bulk of chromosomal DNA had been replicated (Fig. 2E). In clb5Δ sml1Δ cells, the pattern was similar: Rad53 phosphorylation peaked at 60 min (Fig. 2B), after the bulk of DNA synthesis had been completed (Fig. 2E). Although the absolute level of Rad53 phosphorylation in cells lacking Clb5 is significantly below that caused by treatment with high doses of a replication inhibitor or DNA-damaging compound typically used, their effects on chromosomal replication are also much more profound.
FIG.2.
Rad53 phosphorylation indicates checkpoint activation in clb5Δ cells. Strains with the following genotypes, which all express Rad53-HA3, were blocked in G1 phase with α-factor at 23°C and released synchronously into S phase at 30°C: wild type (WT; FHy20), clb5Δ (OAy850), clb6Δ (DGy138), clb5Δ clb6Δ (DGy147), sml1Δ (DGy511), clb5Δ sml1Δ (DGy213), mec1Δ sml1Δ (DGy531), clb5Δ mec1Δ sml1Δ (DGy163), rad9Δ (DGy400), clb5Δ rad9Δ (DGy410), mrc1AQ (DGy529), and clb5Δ mrc1AQ (DGy530). (Note that 10-μg/ml nocodazole was included where indicated [+Noc].) At the indicated intervals, proteins were extracted and analyzed by immunoblotting with anti-HA antibody (A to D). Chemiluminescence was used for quantitative detection of Rad53. Percent phosphorylation was calculated as (Rad53-P)/(Rad53-P + Rad53), where Rad53-P included all slower-migrating forms of Rad53. (E) DNA content of cells in panels A to D.
Analysis of cells lacking CLB6 supports the concurrence between accumulation of activated Rad53 and later stages of DNA replication. Deletion of CLB6 alone did not result in checkpoint activation, consistent with the lack of any apparent effect on DNA replication (Fig. 2A). However, deletion of both CLB5 and CLB6 resulted in delayed entry into S phase and a corresponding delay in the accumulation of activated Rad53. In clb5Δ clb6Δ cells, Rad53 phosphorylation peaked at 84 min (Fig. 2A), by which time DNA replication was complete or nearly so (Fig. 2E). Rad53 activation declined as these cells divided and entered G1 (Fig. 2A and E, 108 to 132 min). Together, these results suggest that the activation of Rad53 is a direct result of undergoing DNA replication in the absence of Clb5 function.
Because the completion of DNA synthesis is delayed in cells lacking CLB5, we considered the possibility that checkpoint activation might have resulted from attempted anaphase entry with incompletely replicated DNA, causing DNA damage. To determine whether Rad53 activation resulted from mitosis, we monitored Rad53 phosphorylation in the presence of nocodazole, which blocks spindle assembly and mitosis. Nocodazole arrest results in a modification of Rad53 that reduces its gel mobility, even in wild-type cells (Fig. 2C). Nevertheless, additional slower-migrating forms of Rad53 were observed in nocodazole-arrested clb5Δ cells (Fig. 2C, asterisk). Thus, Rad53 phosphorylation was observed in the presence of nocodazole, indicating that checkpoint activation did not result from anaphase entry. On the contrary, checkpoint activation appeared to delay cell division briefly in clb5Δ cells as large-budded cells persisted in the population at 96 and 108 min when most wild-type cells had divided (Fig. 3C); the emergence of unbudded and small-budded cells in the second cell cycle also was delayed relative to the wild type (Fig. 3C). Careful inspection of the DNA content profiles also points to a delay in cell division of clb5Δ cells (Fig. 2E). Whereas most wild-type cells had completed the second S phase by 120 min (note the 4C peak, in addition to the 2C peak and the lack of newly emerging cells with 1C DNA content), a significant number of clb5Δ cells continued to undergo cytokinesis at 120 to 132 min (note the populations of cells with <2C DNA content). Furthermore, this delay in cell division of clb5Δ cells was abolished by deletion of MEC1 (Fig. 2E).
FIG. 3.
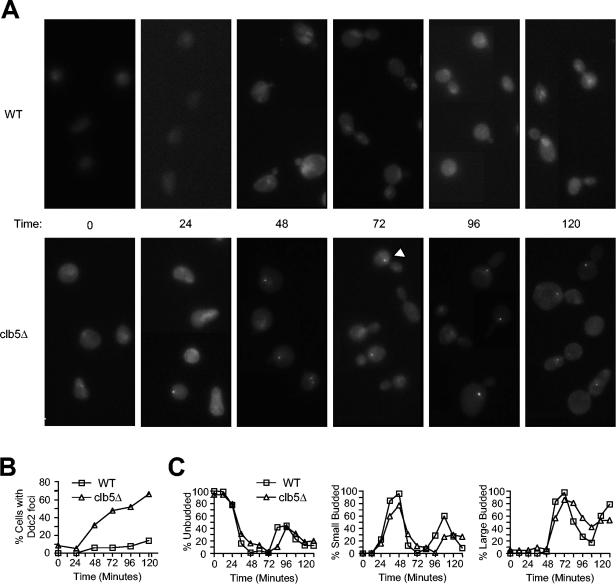
Ddc2 localization indicates presence of DNA damage in clb5Δ cells. Wild-type (DGy430) and clb5Δ (DGy431) strains, which express Ddc2-GFP, were blocked in G1 phase with α-factor at 23°C and released synchronously into S phase at 30°C. At the indicated intervals, cells were fixed and analyzed for Ddc2-GFP fluorescence localization (A and B) and cellular morphology. The arrowhead shows an example of a Ddc2-GFP focus. (B) The percentage of cells with at least one Ddc2-GFP focus was determined and plotted. (C) Cell morphologies were determined and plotted as unbudded (left), small budded (middle), and large budded (right).
The increase in Rad53 phosphorylation in late S/G2 phase suggested that incompletely replicated or damaged DNA accumulated during S phase in the absence of Clb5 function. To explore in more depth the cause of Rad53 activation, we analyzed its dependence on Mrc1 and Rad9. Rad53 phosphorylation did not depend on Mrc1, as the peak level of Rad53 phosphorylation in clb5Δ mrc1AQ cells was not reduced compared to that in clb5Δ cells (Fig. 2D). Instead, Rad9 is responsible for the majority of Rad53 activation in clb5Δ cells, because the proportion of phosphorylated Rad53 was diminished greatly in clb5Δ rad9Δ cells (Fig. 2D). These data suggest that the replication defect of clb5Δ cells produces aberrant DNA, which is detected by Mec1 and Rad9, leading to Rad53 activation.
Ddc2 localization suggests the presence of DNA damage in clb5Δ cells.
Cellular recognition of damaged DNA involves binding of Ddc2 to sites of DNA damage (20, 27). In cells expressing Ddc2 fused to GFP, DNA damage causes the accumulation of Ddc2-GFP into one or more brightly fluorescent foci (20). To address directly whether DNA damage results from loss of Clb5 function, we examined the cellular localization of Ddc2 in wild-type and clb5Δ cells released synchronously from G1. In wild-type cells, Ddc2-GFP was present throughout the nucleus and did not accumulate into distinct foci, regardless of cell cycle stage (Fig. 3A); the percentage of cells with at least one Ddc2-GFP focus was quantified (Fig. 3B). In clb5Δ cells, very few brightly fluorescent foci were observed prior to S phase or during early S (Fig. 3A and B, 0 and 24 min). As cells progressed farther into S, one or more foci of Ddc2-GFP were observed, suggesting that Ddc2-GFP recognized and associated with sites of DNA damage (Fig. 3A and B, 48 and 72; e.g., arrowhead). At 72 min, when the majority of cells were large budded, about half of the cells showed one or more Ddc2-GFP foci (Fig. 3B and C); fewer than 10% of large-budded wild-type cells showed a focus (Fig. 3B and C), and no such foci were observed in cells lacking Ddc2-GFP (data not shown). Ddc2-GFP foci persisted in most clb5Δ cells at 96 and 120 min, when DNA content analysis suggested that DNA replication was complete (Fig. 3A and B, and see Fig. 2E for DNA content). Ddc2-GFP foci were observed very infrequently in unbudded cells, indicating that Ddc2-GFP disperses prior to cell division. This is consistent with repair of DNA damage prior to mitosis and cell division, as expected if a checkpoint were delaying mitotic progression of clb5Δ cells. Analysis of unsynchronized cells confirmed the correspondence between cell cycle stage and Ddc2-GFP focus formation (data not shown). Together with the analysis of Rad53 phosphorylation, these results provide strong evidence that DNA damage occurs during S phase and peaks during late S and G2 in clb5Δ cells.
Viability of clb5Δ mutants depends on Rad53 activation.
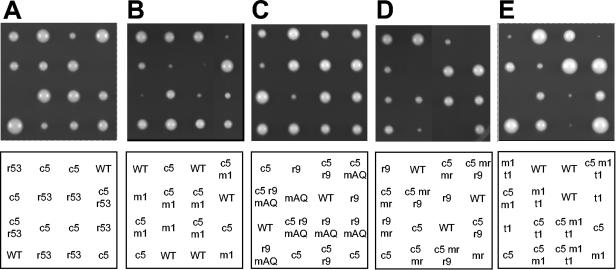
As mentioned previously, deletion of Rad53 in clb5Δ cells is lethal, which contrasted sharply with the viability of clb5Δ mec1Δ cells. In our initial analysis at 23°C, dissection of a diploid heterozygous for clb5Δ and rad53Δ (homozygous for sml1Δ) yielded no viable clb5Δ rad53Δ spores, whereas each of the single mutants was obtained at normal frequencies (Table 2). In contrast, dissection of a diploid heterozygous for clb5Δ and mec1Δ (homozygous for sml1Δ) yielded similar numbers of clb5Δ mec1Δ segregants to those of either single mutant (Table 2). This result suggested that the function of Rad53 in sustaining the viability of clb5Δ cells could function independently of the Mec1-dependent signaling pathways, which normally lead to stimulation of Rad53's kinase activity. In subsequent analyses of spore viabilities at 30°C, we again found lethality of combined CLB5 and RAD53 deletion (Table 2 and Fig. 4A). However, we found a significant requirement for Mec1 in the viability of clb5Δ cells at 30°C; spore viability of clb5Δ mec1Δ cells was about 40% that of either single mutant or the wild type (Table 2). Furthermore, the very small colony size of clb5Δ mec1Δ cells indicated a growth defect at 30°C (Fig. 4B). Together, these results suggest a greater requirement for Mec1 in sustaining the viability of clb5Δ cells at the higher temperature.
TABLE 2.
Rad53 activation is required for viability of CLB5 mutant cells
| Diploid strain | Temp (°C) | No. of viable, haploid spores | |||
|---|---|---|---|---|---|
| OAy967 (CLB5/clb5Δ RAD53/rad53Δ sml1Δ/sml1Δ) | CLB5 RAD53 | clb5Δ RAD53 | CLB5 rad53Δ | clb5Δ rad53Δ | |
| 23 | 21 | 16 | 12 | 0 | |
| 30 | 36 | 41 | 38 | 0 | |
| OAy931 (CLB5/clb5Δ MEC1/mec1Δ sml1Δ/sml1Δ) | CLB5 MEC1 | clb5Δ MEC1 | CLB5 mec1Δ | clb5Δ mec1Δ | |
| 23 | 26 | 28 | 27 | 27 | |
| 30 | 32 | 28 | 36 | 14 | |
| OAy970 (CLB5/clb5Δ RAD53/rad53-11 sml1Δ/sml1Δ) | CLB5 RAD53 | clb5Δ RAD53 | CLB5 rad53-11 | clb5Δ rad53-11 | |
| 23 | 66 | 63 | 62 | 15 | |
| 30 | 67 | 62 | 62 | 0 | |
| DGy534 (CLB5/clb5Δ MRC1/mrc1AQ RAD9/rad9Δ sml1Δ/sml1Δ) | CLB5 MRC1 RAD9 | clb5Δ MRC1 RAD9 | CLB5 mrc1AQ RAD9 | CLB5 MRC1 rad9Δ | |
| 30 | 33 | 26 | 26 | 24 | |
| clb5Δ mrc1AQ RAD9 | clb5Δ MRC1 rad9Δ | CLB5 mrc1AQ rad9Δ | clb5Δ mrc1AQ rad9Δ | ||
| 26 | 24 | 37 | 23 | ||
| OAy981 (CLB5/clb5Δ MRC1/mrc1Δ RAD9/rad9Δ sml1Δ/sml1Δ) | CLB5 MRC1 RAD9 | clb5Δ MRC1 RAD9 | CLB5 mrc1Δ RAD9 | CLB5 MRC1 rad9Δ | |
| 23 | 17 | 11 | 18 | 27 | |
| 30 | 27 | 22 | 23 | 29 | |
| clb5Δ mrc1Δ RAD9 | clb5Δ MRC1 rad9Δ | CLB5 mrc1Δ rad9Δ | clb5Δ mrc1Δ rad9Δ | ||
| 23 | 16 | 18 | 14 | 9 | |
| 30 | 24 | 22 | 20 | 0 | |
| OAy969 (CLB5/clb5Δ MEC1/mec1Δ TEL1/tel1Δ sml1Δ/sml1Δ) | CLB5 MEC1 TEL1 | clb5Δ MEC1 TEL1 | CLB5 mec1Δ TEL1 | CLB5 MEC1 tel1Δ | |
| 23 | 31 | 26 | 33 | 34 | |
| clb5Δ mec1Δ TEL1 | clb5Δ MEC1 tel1Δ | CLB5 mec1Δ tel1Δ | clb5Δ mec1Δ tel1Δ | ||
| 29 | 28 | 29 | 17 | ||
| OAy988 (CLB5/clb5Δ RAD53/rad53Δ HHT-F2/hht-f2Δasml1Δ/sml1Δ) | CLB5 RAD53 HHT-F2 | clb5Δ RAD53 HHT-F2 | CLB5 rad53Δ HHT-F2 | CLB5 RAD53 hht-f2Δ | |
| 30 | 27 | 28 | 22 | 34 | |
| clb5Δ rad53Δ HHT-F2 | clb5Δ RAD53 hht-f2Δ | CLB5 rad53Δ hht-f2Δ | clb5Δ rad53Δ hht-f2Δ | ||
| 0 | 18 | 34 | 9 | ||
HHT-F2 is HHT2-HHF2.
FIG. 4.
RAD53 and genes required for its activation are required for viability of clb5Δ cells. Diploid strains OAy967, OAy931, DGy534, OAy981, and OAy969 (panels A, B, C, D, and E, respectively) were induced to sporulate at 23°C. Germination was carried out at 30°C, except for panel E, which was carried out at 23°C. The relevant genotypes are indicated in the panels below as follows: WT, wild type; r53, rad53Δ; c5, clb5Δ; m1, mec1Δ; t1, tel1Δ; r9, rad9Δ; mAQ, mrc1AQ; and mr, mrc1Δ. The genotypes of inviable spores were inferred by assuming 2:2 segregation of all genetic loci within each tetrad.
The greater requirement of Mec1 at higher temperature might reflect a greater requirement for Rad53 function at the higher temperature because of Mec1's role in Rad53 activation. Consistent with this idea, we found that we could isolate viable clb5Δ rad53-11 (mec2-1) cells from tetrad dissections performed at 23°C, although at only about 25% of the frequency of either single mutant or wild-type cells (Table 2). We recovered no viable clb5Δ rad53-11 cells at 30°C (Table 2). Rad53-11 appears to be defective in undergoing Mec1-dependent phosphorylation (35). Thus, the viability (albeit poor) of clb5Δ rad53-11 cells at 23°C and the much better viability of clb5Δ mec1Δ cells at 23°C than at 30°C together support the idea that Mec1-mediated activation of Rad53 is required at 30°C, but less critical for viability at 23°C.
To address further the requirement for Rad53 activation by upstream kinase signaling, we analyzed the contributions of Rad9 and Mrc1 to the viability of clb5Δ cells. Individually, neither Rad9 nor Mrc1 was required for maintaining the viability of clb5Δ cells. Dissection at 30°C of a diploid heterozygous for clb5Δ, rad9Δ, and mrc1AQ (homozygous for sml1Δ) yielded similar numbers of clb5Δ mrc1AQ and clb5Δ rad9Δ haploid segregants as any of the single mutant strains (Table 2). And the growth rate of the clb5Δ mrc1AQ and clb5Δ rad9Δ double mutants was similar to that of clb5Δ cells (Fig. 4C). However, elimination of both Rad9 and Mrc1 checkpoint functions significantly reduced the growth rate of clb5Δ cells compared with any of the single or double mutant strains, although spore viability was not decreased (Table 2 and Fig. 4C). These results suggest that Mrc1- and/or Rad9-dependent Rad53 activation pathways function in response to loss of Clb5 function.
In view of the lethality of clb5Δ rad53Δ cells and poor viability of clb5Δ mec1Δ cells at 30°C, we found the spore viability of clb5Δ rad9Δ mrc1AQ surprising. However, it appears that Mrc1AQ retains some ability to activate Rad53 (because rad9Δ mrc1AQ SML1 cells are viable) (23), so we tested the effect of MRC1 deletion. At 30°C, deletion of MRC1 reduced the growth rate of clb5Δ cells, suggesting that checkpoint signaling through Mrc1 occurs in clb5Δ cells (Fig. 4D). Although the spore viability of clb5Δ mrc1Δ cells was not reduced, we did not recover viable clb5Δ rad9Δ mrc1Δ spores, suggesting that residual function of mrc1AQ maintained the viability of clb5Δ rad9Δ mrc1AQ cells (Table 2 and Fig. 4D). At 23°C, we recovered viable clb5Δ rad9Δ mrc1Δ spores; however, their viability was reduced significantly as only about half as many were recovered as the double mutant strains (Table 2). Together, these results strongly suggest that the function of Rad53 in sustaining the viability of clb5Δ cells requires activation through a checkpoint-signaling pathway that involves Rad9 and/or Mrc1. Cells lacking Mrc1 have a replication defect, which is manifested by a slower progression through S phase (1), so we cannot rule out the possibility that the lethality of clb5Δ rad9Δ mrc1Δ cells derives at least in part from an enhancement of the clb5Δ replication defect by the mrc1Δ replication defect. Nevertheless, the poor growth of clb5Δ rad9Δ mrc1AQ cells and lethality of clb5Δ rad9Δ mrc1Δ cells are consistent with the effect of MEC1 deletion, in that the viability of clb5Δ cells at 30°C depends on an intact Rad53 activation pathway.
Tel1 contributes to the viability of clb5Δ cells in the absence of Mec1.
The reduced dependence on Mec1 of clb5Δ cells at 23°C might indicate a less-severe replication defect due to loss of CLB5 at 23°C and as a consequence a diminished checkpoint signal. However, Mec1-mediated phosphorylation of Rad53 occurred in clb5Δ cells to a similar degree at 23 and 30°C (data not shown), indicating that the replication defect(s) of clb5Δ cells elicited a checkpoint response under both conditions. As deletion of RAD53 but not MEC1 was lethal at 23°C, the results argue that Rad53 can sustain the viability of clb5Δ cells even without stimulation by Mec1. However, the decreased viability of clb5Δ rad9Δ mrc1Δ cells suggests that stimulation of Rad53 kinase activity is important. Although Mec1 plays a predominant role in the activation of Rad53 in DNA damage and replication checkpoints in S. cerevisiae, yeast cells contain the closely related protein Tel1, which can partially substitute for Mec1 in its absence, suggesting that these two proteins share some overlapping functions in checkpoint responses (19). Thus, it was possible that in the absence of Mec1, Tel1 was responsible for stimulating the critical function of Rad53 in maintaining the viability of clb5Δ cells. If correct, elimination of TEL1 should reduce the viability of clb5Δ mec1Δ cells at 23°C.
Dissection at 23°C of a diploid heterozygous for clb5Δ, mec1Δ, and tel1Δ (homozygous for sml1Δ) yielded similar numbers of clb5Δ tel1Δ haploid cells to those by either single mutant (Table 2). Thus, like Mec1, Tel1 alone is not required for the viability of clb5Δ cells. As predicted, the clb5Δ mec1Δ tel1Δ triple mutant strain showed significantly reduced viability; only about 60% as many clb5Δ mec1Δ tel1Δ spores were recovered as compared with mec1Δ tel1Δ spores (Table 2 and Fig. 4E). Also, the viable clb5Δ mec1Δ tel1Δ spores formed much smaller colonies than either the clb5Δ mec1Δ or clb5Δ tel1Δ strains. The mec1Δ tel1Δ cells formed very small colonies even in the presence of CLB5, so it is not possible to unambiguously ascribe the reduced growth rate of clb5Δ mec1Δ tel1Δ cells to loss of checkpoint function. Nevertheless, the results are consistent with Tel1 contributing to the viability and robust growth of clb5Δ mec1Δ cells at 23°C. The reduced viability at 30°C of clb5Δ mec1Δ cells suggests that Tel1 can substitute only partially for Mec1 at 30°C. Tel1 likely contributes to the viability of clb5Δ mec1Δ cells by stimulating the activity of Rad53 through phosphorylation, as overproduction of Tel1 partially restores Rad53 phosphorylation (in HU- or MMS-treated cells) in the absence of Mec1 (28, 39). Although we did not detect phosphorylation of Rad53 (based on a mobility shift) in clb5Δ mec1Δ cells (Fig. 2B), it is possible that Tel1-mediated activation of Rad53 can occur without affecting the mobility of Rad53 or that the amount of phosphorylated Rad53 in clb5Δ mec1Δ cells is below the level of detection in our assay, but sufficient for viability.
In summary, the analyses of clb5Δ cells lacking both Rad53 activation pathways (Mec1 and Tel1 or Mrc1 and Rad9) support the idea that Rad53 activation contributes to the viability of clb5Δ cells. However, none of these mutant combinations completely phenocopies the effect of RAD53 deletion, suggesting that a Mec1- and Tel1-independent function of Rad53 also is important for the viability of clb5Δ cells. A Mec1- and Tel1-independent role of Rad53 in the regulation of histone levels has been described recently (13). As deletion of one of the two copies of histone H3 and H4 genes partially suppressed the slow growth, DNA damage sensitivity, and elevated chromosome loss phenotypes of rad53Δ strains in the previous study, we tested whether the more severe phenotype of clb5Δ rad53Δ cells (compared with clb5Δ cells completely lacking factors that activate Rad53) derived from defective regulation of histone levels. We dissected spores of a diploid strain heterozygous for clb5Δ, rad53Δ, and hht2-hhf2Δ (one copy of the histone H3 and H4 genes). As before, we recovered no clb5Δ rad53Δ cells; however, we did recover viable clb5Δ rad53Δ hht2-hhf2Δ cells, although only at about one-third the frequency of wild-type or most single mutant cells (Table 2). In addition, the clb5Δ rad53Δ hht2-hhf2Δ cells grew significantly more slowly than wild-type cells (data not shown). These data are consistent with the conclusion that the lethality of clb5Δ rad53Δ cells is due in significant part to loss of Rad53 checkpoint function (as in the clb5Δ cells lacking the Rad53 activation pathways). Furthermore, the more severe phenotype of clb5Δ rad53Δ cells appears to be related to defective regulation of histone levels associated with loss of Rad53 function.
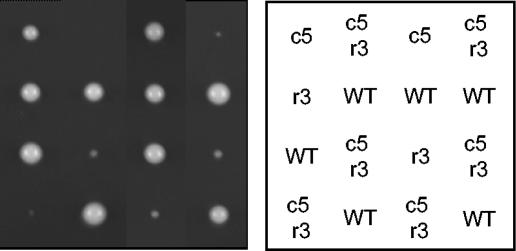
Rrm3 helicase is required for robust growth of clb5Δ cells.
Replication forks pause at numerous sites throughout the genome, such as tRNA genes, ribosomal DNA (rDNA), and centromeres (7, 14). Replication pause sites appear to be susceptible to DNA damage. Although its mechanism is not fully understood, Rrm3 helicase appears to facilitate the progression of replisomes through potential pause sites, as deletion of RRM3 increases fork pausing at many sites throughout the genome (14). Apparently as a consequence of increased fork stalling, DNA damage occurs and generates a checkpoint response that is required for the viability of rrm3Δ cells (14, 38). The increased replicon size of clb5Δ cells may enhance replicative stress by increasing the number of pause sites encountered by the average replisome because of the reduced number of converging replication forks. If true, clb5Δ cells might be expected to show a critical dependence on Rrm3 to facilitate progression of these replisomes and minimize DNA damage. To address whether Rrm3 contributes to the stability of replication forks in cells lacking Clb5 function, we determined the viability and growth characteristics of clb5Δ cells lacking RRM3 by dissection of a diploid heterozygous for clb5Δ and rrm3Δ. Cells lacking Clb5 and Rrm3 grew very poorly, forming only microcolonies after 3 days of growth at 30°C, whereas each single mutant formed robust colonies only slightly smaller than wild type (Fig. 5). The very poor growth of clb5Δ rrm3Δ cells is consistent with the idea that replication fork stress results from the loss of Clb5 activity and suggests that Rrm3 plays an important role in stabilizing replisomes in clb5Δ cells.
FIG. 5.
Rrm3 is required for robust growth of clb5Δ cells. Diploid strain OAy989 was induced to sporulate at 23°C and germinated at 30°C. The relevant genotypes are indicated in the panel to the right as follows: WT, wild type; r3, rrm3Δ; c5, clb5Δ; and c5r3, clb5Δ rrm3Δ. The genotypes of inviable spores were inferred by assuming 2:2 segregation within each tetrad of all genetic loci.
Increased CLB6 dosage suppresses the replication defects and checkpoint activation of clb5Δ mutant cells.
The replication defect(s) and checkpoint activation resulting from loss of Clb5 function may be due to a decrease of total Clb-Cdk activity present during S phase or due to loss of a Clb5-specific function. To determine whether low abundance of Clb6-Cdk limits its ability to drive late origin activation, we introduced an extra copy of CLB6 under control of the CLB5 promoter into clb5Δ cells (clb5::CLB6); the CLB5 and CLB6 genes are normally expressed with indistinguishable cell cycle timing (30). In the clb5Δ cells, expression of the extra copy of CLB6 significantly increased late origin firing (Fig. 6A). Interestingly, cells expressing only the single copy of CLB6 from the CLB5 promoter (clb5::CLB6 clb6Δ cells) exhibited more efficient late origin activation than clb5Δ cells, which have CLB6 under its own promoter (Fig. 6A). This result suggests that expression from the CLB5 promoter produces more Clb6 protein. The increased late origin activation resulting from increased CLB6 dosage argues that a low abundance of Clb6, rather than a Clb5-specific function, explains the defective late origin firing of clb5Δ cells.
FIG. 6.
Increased CLB6 dosage restores late origin function and suppresses Rad53 activation. (A) Cultures of clb5Δ (DGy226), clb5::CLB6 (DGy438), and clb5::CLB6 clb6Δ (DGy439) strains were analyzed as described in the legend to Fig. 1, except that ARS603.5 and ARS1413 are not shown. (B) DNA content of the cultures in panel A. (C to E) clb5::CLB6 (DGy448) and clb5::CLB6 clb6Δ (DGy449) cells, which both express Rad53-HA3, were blocked in G1 phase with α-factor at 23°C and released synchronously into S phase at 30°C. At the indicated times, cells were harvested for DNA content analysis (C) and immunoblot analysis of Rad53 (D and E) as described in the Fig. 2 legend. The data for clb5Δ cells from the experiment in Fig. 2A are shown in panel E for comparison.
DNA content analysis of cells expressing an extra copy of CLB6 also indicates that replication origin function is restored. In unsynchronized cells, the proportion of clb5::CLB6 and even clb5::CLB6 clb6Δ cells in S phase was reduced significantly compared with clb5Δ cells and was similar to that of wild-type cells (Fig. 6B). Increased Clb6 dosage also reduced the level of Rad53 activation. Particularly striking is the lack of increase in Rad53 phosphorylation that occurs during late S phase in clb5Δ cells (Fig. 6D and E). In clb5Δ cells expressing an extra copy of CLB6, there is a low basal level of Rad53 phosphorylation throughout the cell cycle, but no increase during S phase. Expression of only the CLB5 promoter-driven copy of CLB6 also reduced Rad53 activation, though less so (Fig. 6E). Together, these data are consistent with the notion that loss of CLB5 results in decreased S-CDK function during mid- to late S phase, impairing the activation of late-firing replication origins and timely genome duplication; as a result, the DNA damage checkpoint is activated. The DNA damage response is a direct result of decreased origin activation as increased S-CDK levels are accompanied by increased late origin firing and decreased Rad53 activation.
DISCUSSION
In this study, we investigated checkpoint regulation of S. cerevisiae cells lacking the S-phase cyclin Clb5. We were interested in the notion that previously characterized phenotypes of these cells might be a reflection of unsuspected checkpoint regulation due to diminished CDK activity. We have presented evidence for activation of the DNA damage and replication checkpoint pathways in clb5Δ cells. Rad9-dependent Rad53 phosphorylation and the localization of Ddc2 into foci occur as a result of undergoing S phase in the absence of Clb5 (Fig. 2 and 3). Furthermore, robust viability of clb5Δ cells requires Rad53 and at least one functional Rad53 activation pathway (Fig. 4 and Table 2). In spite of this essential checkpoint response, the lack of late origin firing in clb5Δ cells is not caused by either a DNA replication or DNA damage checkpoint (Fig. 1). Instead, checkpoint activation appears to result from the lack of late origin firing, because Rad53 and Ddc2 activation increases during late S and G2 after most genome replication is complete (Fig. 2 and 3). In addition, increasing the dosage of Clb6 suppressed the replication defects of clb5Δ cells, as well as the checkpoint activation, demonstrating that both are the direct result of decreased S-CDK function (Fig. 6).
Clb5 and Clb6 have similar replication origin specificities.
The original observation that clb5Δ and clb6Δ mutants exhibit different origin firing characteristics gave rise to at least two models postulating differences in the functions of Clb5 and Clb6 (8). In the first, Clb5-Cdk1 and Clb6-Cdk1 have equivalent substrate specificities, but different levels of associated kinase activity, either due to lower abundance of Clb6 during late S or to a weaker ability of Clb6 to stimulate Cdk1, in which case late origins are postulated to require a higher threshold of Cdk activity due to late-determining factors. In the second model, Clb5-Cdk1 and Clb6-Cdk1 have distinct substrate specificities such that Clb5-Cdk1, but not Clb6-Cdk1, is able to phosphorylate and stimulate the replication function of a late-origin-specific substrate. We proposed a third alternative in which intra-S checkpoint activation caused inhibition of late origin firing.
Our results clearly eliminated the hypothesis that checkpoint regulation blocks late origin firing in clb5Δ cells. Mutation of factors required to block late origin firing in response to replication defects or DNA damage did not enable late origin firing in clb5Δ cells (Fig. 1A). However, increased dosage of CLB6 did enhance late origin firing in clb5Δ cells, demonstrating that Clb6 is capable of activating late origins (Fig. 6A). Importantly, increased CLB6 dosage also restored the wild-type replication rate (Fig. 6B and C) and significantly reduced Rad53 activation (Fig. 6D and E). Together, these results argue that the replication defect(s) and resulting checkpoint response of clb5Δ cells are due to a general decrease in S-CDK activity, rather than loss of a Clb5-Cdk1-specific function. We note, however, that increased CLB6 dosage did not fully restore late origin firing (compare Fig. 6A with 1A) or eliminate Rad53 activation (Fig. 6D and E). Although this may reflect still insufficient CLB6 dosage, we cannot exclude the possibility that one or more Clb5-specific substrates exist, whose lack of phosphorylation in clb5Δ cells is at least partly responsible for these residual defects.
Reduced S-CDK function causes DNA damage.
In the absence of CLB5, late origins initiate replication inefficiently, increasing average replicon size. This increase is likely to be particularly dramatic in large, late-replicating regions of the genome, some of which are hundreds of kilobases in length (26). Unusually large replicons may have an increased propensity for defects in replication fork progression due to normal fork impediments or limited processivity, leading to DNA damage. Hundreds of sites in the genome present obstacles or potential obstacles to fork progression, including tRNA genes, rDNA repeats, and unfired replication origins (7, 14, 41). A reduction in replication origin usage not only creates more potential pause sites at each unfired origin but also reduces the number of replication forks relative to the number of pause sites. Hence, the amount of time a fork spends at pause sites before a converging fork arrives may increase, which in turn may increase the likelihood of replication fork collapse and associated DNA damage. The effect of RRM3 deletion is consistent with the notion of increased replisome stress in the absence of Clb5; deletion of RRM3 seriously impaired the growth of clb5Δ cells (Fig. 5). Because loss of Rrm3 significantly increases replisome pausing and causes DNA damage, its loss probably exacerbates replication stress and/or resulting DNA damage of clb5Δ cells.
The proposition that decreased S-CDK function creates replication stress at forks is also supported by the critical importance of Rad53-activating pathways in the proliferation of clb5Δ cells (Table 2 and Fig. 4). As Rad53 stabilizes stressed replication forks, the loss of viability resulting from RAD53 disruption in clb5Δ cells is consistent with replisome stress or instability. Because Rad53 activation primarily occurs late in S phase and is dependent on Rad9, which mediates DNA damage signals, DNA damage appears to be the primary signal for Rad53 activation in clb5Δ cells (Fig. 2). The formation of Ddc2-containing foci in late S and G2 also directly points to the presence of DNA damage in cells that undergo S phase in the absence of Clb5 function (Fig. 3). We do observe a low level of Rad53 phosphorylation in G1-synchronized cells lacking Clb5 that might reflect a low level of Rad53 phosphorylation in all clb5Δ cells in a population. However, we think it is more likely to reflect a persistent DNA damage signal in a fraction of cells, because Ddc2 foci were observed only in a small fraction of unbudded cells.
In addition to stabilizing replication forks through stimulation of Rad53, DNA replication and damage checkpoints delay cell cycle progression through Mec1- and Rad9-dependent activation of Chk1. Hence, the viability of clb5Δ cells might depend on Chk1-dependent delay of mitotic entry preventing the lethal segregation of incompletely replicated DNA. There appears to be a delay in mitotic progression in clb5Δ cells, as cell division is delayed (Fig. 2E and 3C). This mitotic delay may allow the accumulated mitotic cyclins to activate remaining unfired origins and hasten completion of genome replication, while also permitting time for repair of any DNA damage. Nevertheless, mitotic delay does not appear to constitute the vital checkpoint function in clb5Δ cells, because deletion of RAD9 or RAD9 and CHK1 (Table 2 and Fig. 4C and D; data not shown for CHK1) did not affect the growth or viability of clb5Δ cells to the same extent as RAD53 deletion, which does not eliminate the Chk1-mediated mitotic delay (18). Thus, the vital function of checkpoint signaling in clb5Δ cells appears to be fork stabilization by Rad53, although a supplemental role by Chk1 and Rad53 in mitotic delay seems likely.
Indeed, a previous study analyzing the replication of a yeast artificial chromosome (YAC) containing a relatively large (170 kbp) region devoid of replication origins found evidence of a Rad9-dependent delay in cell division (40). Replication of the origin-free region caused an extension in the length of S phase, and loss of Rad9 led to mitotic instability of the YAC and DNA deletions within the origin-free region of the YAC. The apparently more critical role of Rad9 in replication of the YAC, which could be replicated only by a single replisome, compared to chromosomal replication in clb5Δ cells may be due to the ability of mitotic cyclins to enable completion of S phase by activating any unfired origins prior to mitosis, reducing the length of mitotic delay. The reduced requirement of Rad9 in clb5Δ cells may also reflect the involvement of Mrc1 because more replication forks likely are under stress in clb5Δ cells than in wild-type cells carrying the YAC.
Although the failure of late origin firing in clb5Δ cells cannot be attributed to checkpoint regulation, there appears to be a replication checkpoint response in clb5Δ cells, which probably occurs too late to inhibit origin function. Evidence for a replication checkpoint is based on the decreased growth rate of clb5Δ mrc1Δ cells and the strong effect on cell growth and viability of a combined MRC1 and RAD9 mutation compared with the effect of deleting only RAD9 (Fig. 4D and Table 2). Interestingly, mrc1AQ (and mrc1Δ) appeared to increase the level of Rad53 phosphorylation in clb5Δ cells (Fig. 2D; data not shown for mrc1Δ). Perhaps loss of Mrc1 checkpoint signaling at stressed forks allows for more DNA damage accumulation, resulting in a stronger Rad9-dependent Rad53 activation. Also, deletion of RAD9 did not entirely eliminate Rad53 phosphorylation, indicating that Mrc1 mediated Rad53 activation in clb5Δ rad9Δ cells (Fig. 2D). Indeed, the idea of replication fork stress due to increased replicon size might be expected to generate both a replication stress response through Mrc1, followed by a Rad9-mediated damage response, as DNA damage might result from replication stress or attempts to repair destabilized replication fork complexes. The occurrence of replication fork stress likely to elicit a replication checkpoint is supported by the critical role of Rrm3 in maintaining robust proliferation of clb5Δ cells (Fig. 5), as Rrm3 facilitates the progression of replication forks through sites of potential fork pausing.
Overlapping functions of Mec1 and Tel1 in response to replication stress.
A dual contribution by Mec1 and Tel1 may be involved in the checkpoint response to loss of Clb5 function. Deletion of MEC1 or TEL1 in clb5Δ cells had no effect on spore viability at 23°C; however, deletion of both significantly reduced viability and growth rate (Table 2 and Fig. 4E). Because telomeres are late replicating, loss of Clb5 function may have particularly strong effects on telomere replication. Considering the critical function of Tel1 in telomere maintenance, a role for Tel1 in sensing defective telomere replication in clb5Δ cells and activating Rad53 is plausible.
A related explanation for the dual involvement of Mec1 and Tel1 lies in the types of DNA damage that they may primarily sense and respond to. Association of Mec1 with chromatin involves interactions with the single-stranded DNA binding protein RPA, suggesting that single-stranded DNA at sites of DNA damage and/or excess unwound DNA at stressed replication forks is sensed by Mec1 (46). In contrast, Tel1 association with chromatin appears to occur at sites of double-strand DNA breaks (22, 39). The replication defects associated with decreased S-CDK activity may give rise to both types of DNA damage, explaining the involvement of both proteins. Nevertheless, Mec1 and Tel1 are able to effectively substitute for each other, as the deletion of both reduces the viability and growth rate of clb5Δ cells compared with clb5Δ mec1Δ or clb5Δ tel1Δ cells (Table 2 and Fig. 4E). The ability to substitute may occur because either pathway can lead to Rad53 activation, which is critical for viability, rather than an ability of Mec1 to sense DNA damage normally sensed by Tel1, and vice versa. For example, Rad53 activated at Tel1-sensed sites of damage may act to stabilize stressed replication forks or sites of DNA damage that may go undetected in the absence of Mec1. The function of Tel1 in clb5Δ cells probably involves Mrc1 and/or Rad9 as a mediator, as their combined elimination causes lethality (Table 2 and Fig. 4D). In accord with this, a recent study with Schizosaccharomyces pombe indicates that Tel1 and Rad3 (Mec1) can each play a role in Mrc1-mediated activation of Cds1 (Rad53) (44).
Significance of S-CDK regulation to tumorigenesis.
Proper regulation of CDK activity is critical for normal cell division and organismal development, whereas deregulation of CDK activity is frequently associated with tumorigenesis (reviewed in reference 32). The finding that diminished S-CDK function elicits DNA damage and replicative stress in S. cerevisiae suggests that a similar effect of decreased S-CDK function in metazoan cells is likely. The inability to inhibit CDK activity in G1 or G0 cells contributes significantly to the inappropriate proliferation of many cancer cells. Similarly, deregulation of S-CDK functions may contribute to the genesis of cancer cells by affecting the accurate execution of S phase, thus creating DNA damage and enhancing genomic instability. For example, recent studies in yeast have shown that deregulation of G1-CDK suppresses the assembly of functional pre-RCs, which increases replicon size and increases genomic instability (16, 36). Although the checkpoint status in these previous studies was not reported, diminished pre-RC activity as a result of mutations in ORC subunits does result in a DNA damage checkpoint response (12, 42; D. G. Gibson, F. Hu, and O. M. Aparicio, unpublished observation) Thus, mutations that affect pre-RC assembly are comparable to diminished S-CDK activity in that both have the effect of reducing origin usage and creating DNA damage. However, cells with a severely defective replication protein(s) (e.g., ORC) are unlikely to proliferate enough to contribute to tumorigenesis, whereas cells with deregulated CDK may retain strong replicative capacity, perhaps damage prone. Hence, reduction of S-CDK function may present another potential mechanism of genomic instability and tumorigenesis.
Acknowledgments
We thank N. Arnheim, S. Forsburg, and M. Goodman for sharing equipment; D. Arnold, W. Dolan, and J. Yuan for help with microscopy; J. Orr for excellent technical assistance; J. Bachant, F. Cross, S. Elledge, R. Rothstein, E. Schwob, S. Szyjka, D. Toczyski, and T. Weinert for strains and plasmids; M. Foiani for protocols; J. Bachant for critical reading of the manuscript; and S. Bell, J. Diffley, S. Elledge, and S. Haase for helpful discussions.
This work was supported by a Burroughs-Wellcome Career Award (992834) and NIH grant (1RO1GM-CA65494-01A1) to O.M.A.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, J. G., C. J. Viggiani, D. G. Gibson, and O. M. Aparicio. 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:4769-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 5.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 6.Cross, F. R., M. Yuste-Rojas, S. Gray, and M. D. Jacobson. 1999. Specialization and targeting of B-type cyclins. Mol. Cell 4:11-19. [DOI] [PubMed] [Google Scholar]
- 7.Deshpande, A. M., and C. S. Newlon. 1996. DNA replication fork pause sites dependent on transcription. Science 272:1030-1033. [DOI] [PubMed] [Google Scholar]
- 8.Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer, and W. L. Fangman. 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2:173-182. [DOI] [PubMed] [Google Scholar]
- 9.Epstein, C. B., and F. R. Cross. 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6:1695-1706. [DOI] [PubMed] [Google Scholar]
- 10.Foiani, M., G. Liberi, G. Lucchini, and P. Plevani. 1995. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B subunit. Mol. Cell. Biol. 15:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman, K. L., and B. J. Brewer. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 262:613-627. [DOI] [PubMed] [Google Scholar]
- 12.Garber, P. M., and J. Rine. 2002. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161:521-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunjan, A., and A. Verreault. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115:537-549. [DOI] [PubMed] [Google Scholar]
- 14.Ivessa, A. S., B. A. Lenzmeier, J. B. Bessler, L. K. Goudsouzian, S. L. Schnakenberg, and V. A. Zakian. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12:1525-1536. [DOI] [PubMed] [Google Scholar]
- 15.Kuhne, C., and P. Linder. 1993. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 12:3437-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lengronne, A., and E. Schwob. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 9:1067-1078. [DOI] [PubMed] [Google Scholar]
- 17.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 18.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 19.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 20.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miao, H., J. A. Seiler, and W. C. Burhans. 2003. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J. Biol. Chem. 278:4295-4304. [DOI] [PubMed] [Google Scholar]
- 22.Nakada, D., K. Matsumoto, and K. Sugimoto. 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17:1957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 25.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 27.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 29.Santocanale, C., and J. F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615-618. [DOI] [PubMed] [Google Scholar]
- 30.Schwob, E., and K. Nasmyth. 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7:1160-1175. [DOI] [PubMed] [Google Scholar]
- 31.Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6:648-655. [DOI] [PubMed] [Google Scholar]
- 32.Sherr, C. J. 1996. Cancer cell cycles. Science 274:1672-1677. [DOI] [PubMed] [Google Scholar]
- 33.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618-621. [DOI] [PubMed] [Google Scholar]
- 34.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, S., and J. F. Diffley. 2002. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev. 16:2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 38.Torres, J. Z., S. L. Schnakenberg, and V. A. Zakian. 2004. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24:3198-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usui, T., H. Ogawa, and J. H. Petrini. 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7:1255-1266. [DOI] [PubMed] [Google Scholar]
- 40.van Brabant, A. J., C. D. Buchanan, E. Charboneau, W. L. Fangman, and B. J. Brewer. 2001. An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint. Mol. Cell 7:705-713. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., M. Vujcic, and D. Kowalski. 2001. DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol. Cell. Biol. 21:4938-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe, K., J. Morishita, K. Umezu, K. Shirahige, and H. Maki. 2002. Involvement of RAD9-dependent damage checkpoint control in arrest of cell cycle, induction of cell death, and chromosome instability caused by defects in origin recognition complex in Saccharomyces cerevisiae. Eukaryot. Cell 1:200-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinreich, M., M. A. Palacios DeBeer, and C. A. Fox. 2004. The activities of eukaryotic replication origins in chromatin. Biochim. Biophys. Acta 1677:142-157. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, H., K. Tanaka, E. Nogochi, C. Nogochi, and P. Russell. 2003. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Mol. Cell. Biol. 23:8395-8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 46.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]