Abstract
The packaging of the eukaryotic genome into chromatin is likely to be important for the maintenance of genomic integrity. Chromatin structures are assembled onto newly synthesized DNA by the action of chromatin assembly factors, including anti-silencing function 1 (ASF1). To investigate the role of chromatin structure in the maintenance of genomic integrity, we examined budding yeast lacking the histone chaperone Asf1p. We found that yeast lacking Asf1p accumulate in metaphase of the cell cycle due to activation of the DNA damage checkpoint. Furthermore, yeast lacking Asf1p are highly sensitive to mutations in DNA polymerase alpha and to DNA replicational stresses. Although yeast lacking Asf1p do complete DNA replication, they have greatly elevated rates of DNA damage occurring during DNA replication, as indicated by spontaneous Ddc2p-green fluorescent protein foci. The presence of elevated levels of spontaneous DNA damage in asf1 mutants is due to increased DNA damage, rather than the failure to repair double-strand DNA breaks, because asf1 mutants are fully functional for double-strand DNA repair. Our data indicate that the altered chromatin structure in asf1 mutants leads to elevated rates of spontaneous recombination, mutation, and DNA damage foci formation arising during DNA replication, which in turn activates cell cycle checkpoints that respond to DNA damage.
The eukaryotic genome is packaged into a nucleoprotein structure known as chromatin. The basic repeating unit of chromatin, the nucleosome, consists of approximately two turns of DNA wrapped around an octamer of core histone proteins (34). This association plays a fundamental role in regulating gene expression (for recent reviews, see references 42 and 58). Chromatin structure modulates the access of proteins to DNA and is therefore likely to regulate other aspects of DNA processing, including the repair of double-strand DNA damage and DNA replication (15, 36, 40).
Cell survival and maintenance of genomic integrity are dependent on the efficient and accurate repair of DNA double-stand breaks. Double-strand breaks occur when DNA replication forks stall (7), in response to exogenous DNA-damaging agents or as a programmed event during growth or development (19, 47). The repair of double-strand breaks depends on the DNA damage checkpoint that detects and signals the presence of DNA damage and arrests cell cycle progression until the damage is repaired (67). In budding yeast Saccharomyces cerevisiae the DNA damage checkpoint is initiated by the independent localization of two checkpoint complexes to sites of DNA damage. Rad24p forms an RFC-like complex with Rfc2p-5p and loads the PCNA-like complex of Rad17p, Mec3p, and Ddc1p at the site of the DNA lesion (27, 39, 48). Independently, the PI3-family kinase ATR homolog Mec1p and its binding partner, the ATRIP homolog Ddc2p, are recruited to the DNA lesion in an RPA-dependent manner (27, 39, 69). Mec1p recruitment leads to the phosphorylation of histone H2A (or histone variant H2AX in mammals) on serine 129 in the chromatin flanking the lesion (10). Once recruited to the DNA, Mec1p phosphorylates the mediator kinase Mrc1p in response to DNA replication stress and Rad9p in response to double-strand DNA lesions (3, 14, 43). The key downstream target of Mrc1p and Rad9p is the effector kinase Rad53p (3, 14). Rad53p is important for maintaining nucleotide levels necessary for replication, stabilizing stalled replication forks and preventing the degradation of Pds1p (which leads to cell cycle arrest at the metaphase to anaphase transition) (44). Rad53p has also been implicated in maintaining proper histone levels during DNA replication, providing a link between chromatin assembly and DNA replication (17).
Replication defects are the major source of spontaneous genomic instability in the cell and the DNA damage checkpoint is the principal defense against such instability (44, 65). When a replication fork pauses or stalls, it is either stabilized and restarted by proteins involved in the DNA damage checkpoint response or the stalled replication fork reverses to form so-called “chicken-feet” structures that may lead to deleterious recombination events (44, 65). A balance exists between processing stalled replication forks via restarting or via recombination. This is seen by the fact that the mutation of proteins that stabilize stalled replication forks or that promote progression of replication forks leads to elevated levels of recombination, which is dependent on the RAD52 epistasis group of genes (2).
The chromatin assembly factors that package histones and DNA together into nucleosomes have recently been linked to DNA repair and the DNA damage response. The histone chaperones known as anti-silencing function 1 (ASF1) and chromatin assembly factor 1 (CAF-1) deposit histones H3 and H4 onto newly replicated DNA in vitro (51, 59). Yeast with ASF1 deleted are highly sensitive to double-strand DNA-damaging agents (5, 28, 59), whereas yeast with CAF-1 deleted are highly sensitive to UV irradiation (26). The increased sensitivity of chromatin assembly factor mutants to DNA-damaging agents may reflect a direct role for these factors in modulating chromatin structure during DNA repair. For example, human Asf1p cooperates with CAF-1 to assemble nucleosomes after nucleotide excision repair in vitro (38). In addition, a molecular connection between Asf1p and genomic stability has been provided by the identification of a dynamic interaction between Asf1p and the Rad53p DNA damage checkpoint protein (11, 24). The interaction between Asf1p and Rad53p downregulates the chromatin assembly activity of Asf1p. Upon phosphorylation of Rad53p by the DNA damage checkpoint, Asf1p is released, allowing it to bind histones and modify the chromatin structure (11, 24). These results suggest that the activation of Asf1p may be an important cellular response to DNA damage and replicational stress (11, 24).
Chromatin assembly factors have also been implicated in the maintenance of genomic integrity during normal growth. For example, expression of a dominant-negative form of the largest subunit of CAF-1 in human cells leads to DNA damage and activation of the S-phase checkpoint (64). Similarly, deletion of ASF1 or the genes encoding CAF-1 increases gross chromosomal rearrangements in yeast (41). These results taken together suggest that defects in chromatin assembly induce double-strand DNA damage. Despite the evidence linking the chromatin assembly factor Asf1p to genomic stability, the mechanism whereby chromatin structure affects genomic integrity is unclear. Here we show that the slow growth rate of yeast lacking Asf1p is due to activation of the DNA damage checkpoint as a consequence of increased levels of spontaneous DNA damage and recombination occurring during DNA replication. We propose that the altered chromatin structure in asf1 mutants causes genomic instability as a consequence of elevated levels of DNA repair events occurring during S phase.
MATERIALS AND METHODS
Yeast molecular genetics.
The yeast strains examined here are listed in Table 1. All yeast strains were haploid derivatives of W303-1 except strains BOB461 and JRY009, which are A364A, and BOB852 and NW020, which are S288C. The endogenous DDC2 gene product was epitope tagged at its C terminus with an enhanced green fluorescent protein (EGFP) cassette by using a previously described approach (33). Cells were grown in complete media at 30°C to mid log phase prior to all analyses unless indicated otherwise. Standard yeast genetic manipulations and media were used.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| BAT009 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 GAL ade3::Gal10:HO | This study |
| BAT022 | MATatrp1-1 ura3-1 can1-100 ADE bar::LEU2 his3-11 GAL rad52::TRP1 [pGAL-HO URA3] | This study |
| JKT0001 | MATa Δlys2 asf1::his5+ bar1::LEU2 TELVIIL::URA3 trp1-1 his3-11 leu2-3,112 can1-100 | This study |
| JKT0004 | MATarad52::TRP1 trp1-1 ura3-1 can1-100 ADE bar1::LEU2 his3-11 | This study |
| JKT0005 | MATaasf1::his5+rad52::TRP bar1::LEU2 leu2-3,112 his3-11 can1-100 Δlys2 TELVIIL::URA3 trp1-1 | This study |
| JKT0010 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 | This study |
| JKT0040 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP | This study |
| JKT0041 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP asf1::his5+ | This study |
| JKT0042 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP rad9::URA3 | This study |
| JKT0043 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP rad24::Kan | This study |
| JKT0044 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP asf1::his5+rad9::kan | This study |
| JKT0045 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP rad24::Kan asf1::his5+ | This study |
| JKT0181 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP asf1::his5+rad9::URA3 mrc1::kan | This study |
| JKT0182 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP rad9::URA3 mrc1::kan | This study |
| JKT0183 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP asf1::his5+mrc1::kan | This study |
| JKT0184 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 pds1::18MYC-TRP mrc1::kan | This study |
| JKT0185 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 sml1::HPH rad53::kan asf1::his5+ | This study |
| JKT0186 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 sml1::HPH mec1::kan | This study |
| JKT0187 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 sml1::HPH mec1::kan asf1::his5+ | This study |
| JKM179 | MATα Δhmla::ADE1 ΔHO::ADE1 Δhmr::ADE1 ade1 leu2-3,112 lys5 ura3-52 ade3::Gal10:HO | Jim Haber |
| JRY001 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 sml1::HPH | This study |
| JRY002 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 sml1::HPH asf1::his5+ | This study |
| JRY003 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 sml1::HPH rad53::kan | This study |
| JRY006 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 ddc2::GFP-kan | This study |
| JRY007 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 ddc2::GFP-kan asf1::his5+ | This study |
| JRY008 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 ddc2::GFP-kan rad52::TRP | This study |
| JRY009 | MATα leu2 trp1 lys2 pol1-17 asf1::kan | This study |
| JRY010 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 mrc1::13myc-Kan | This study |
| JRY011 | MATahis3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 mrc1::13myc-Kan asf1::his5+ | This study |
| JRY013 | MATatrp1::his3-Δ3′ his3Δ5′ URA3 asf1::KanMX | This study |
| JRY014 | MATahta1-htb1::LEU2 hta2-htb2::TRP1 leu2-1 ura3-52 his3-200 (hta1S129A-HTB1 CEN-HIS3) asf1::Kan | This study |
| JRY015 | MATahta1-htb1::LEU2 hta2-htb2::TRP1 leu2-1 ura3-52 his3-200 asf1::Kan (HTA1-HTB1 CEN-HIS3) | This study |
| JRY016 | MATα leu2 trp1 his7 cdc17-1 asf1::Kan | This study |
| JRY2334 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 | Rohinton Kamakaka |
| SRH014 | MATakanMX6-PGAL1-ASF1-3HA-PEST-HIS3 bar1::LEU2 can1-100 gal1::hisG his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 | This study |
| SRH015 | MATaASF1-3HA-kanMX6 bar1::LEU2 can1-100 gal1::hisG his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 | This study |
| SRH016 | MATaasf1::kanMX6 bar1::LEU2 can1-100 gal1::hisG his3-11 leu2-3,112 Δlys2 trp1-1 ura3-1 | This study |
| ROY1170 | MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 asf1::his5+ HMRa::ADE2 TELVIIL::URA3 can1-100 | Tyler et al. (59) |
| ROY1172 | MATα ade2-1 LYS2 leu2-3,112 his3-11 trp1-1 ura3-1 HMRa::ADE2 TELVIIL::URA3 can1-100 | Tyler et al. (59) |
| YM001 | MATα Δhmla::ADE1 ΔHO::ADE1 Δhmr::ADE1 ade1 leu2-3,112 lys5 ura3-52 ade3::Gal10:HO asf1::Kan | This study |
| YM003 | MATα Δhmla::ADE1 ΔHO::ADE1 Δhmr::ADE1 ade1 leu2-3,112 lys5 ura3-ade3::Gal10:HO msh2::Kan | This study |
| YM004 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 asf1::Kan | This study |
| BOB460 | MATα leu2 trp1 his7 cdc17-1 | Bob Sclafani |
| BOB461 | MATα leu2 trp1 lys2 pol1-17 | Bob Sclafani |
| BOB463 | MATacdc13 ura3 his3 can1 trp1 | Bob Sclafani |
| BOB852 | MATα met15 leu2 ura3 his3 | Bob Sclafani |
| NW020 | MATα met15 leu2 ura3 his3 asf1:kanMX6 | This study |
| YD122 | MATatrp1::his3-Δ3′ his3-Δ5′ URA3 | Michael Fasullo |
| 1825-1B | MATα cdc16-123 bar1 trp1-1 can1-100 his3-11,15 leu2-3,112 ura3 GAL | Paul Megee |
| YM004 | MATaade2-1 can1-100 his3-11 leu2-3,112 trp1-1 ura3-1 Δasf1::KanMX | This study |
| JKY001 | MATahta1-htb1::LEU2 hta2-htb2::TRP1 leu2-1 ura3-52 his3-200 (HTA1-HTB1 CEN-HIS3) | Jocelyn Krebs |
| JKY002 | MATahta1-htb1::LEU2 hta2-htb2::TRP1 leu2-1 ura3-52 his3-200 (hta1S129A-HTB1 CEN-HIS3) | Jocelyn Krebs |
Flow cytometry analysis.
For cell cycle analysis, ∼5 × 106 cells for each sample were stained with propidium iodide as described previously (52). 10,000 cells from each sample were scanned with a Beckman Coulter XL-MCL machine.
Micromanipulation of yeast.
Strains were grown overnight to mid-log phase in yeast extract-peptone-dextrose (YPD) media. Cultures were diluted 20-fold and spotted onto YPD plates. Individual cells representing various stages of the cell cycle were micromanipulated onto a grid. Growth and division were visualized microscopically over a period of 48 h at 23°C by using a ×20 magnification objective lens.
Western blot analysis of checkpoint proteins.
Western analysis of checkpoints proteins was performed as described previously (8). Antibodies for Rad53p and Rad9p were kindly provided by Noel Lowndes.
Growth curve analysis.
Yeast cultures were grown in yeast extract-peptone-raffinose (YEPR) plus 1% galactose to a Klett reading of 30 to induce expression of Asf1p in the conditional ASF1 and then switched to YPD medium to repress expression of Asf1p. Klett readings were taken every hour, and the cultures were diluted back to a Klett reading of 30 whenever any culture grew to a reading of more than 100 to keep the cells in mid-log phase. Dilution factors were multiplied back into the Klett readings to obtain the final Klett units. Growth readings were taken by using a Klett-Summersen photoelectric colorimeter (model 800-3).
Analysis of Ddc2p foci formation.
Logarithmically growing cells were fixed and processed for GFP fluorescence as described previously (39). Images were captured with a Nikon E800 epifluorescence microscope with a Gene-Snap cooled charge-coupled device camera and Metamorph imaging software.
Pulsed-field gel electrophoresis analysis.
Chromosome-sized DNA fragments were immobilized in low-melting-point agarose and processed for resolution by pulsed-field gel electrophoresis as described previously (18).
HO repair viability assay.
Induction of HO in liquid culture was achieved by growing cells in YEPR at 30°C and adding galactose (final concentration of 2%). Samples were removed at various times after galactose addition, sonicated and then counted on a hemocytometer to determine cell concentrations, followed by dilution and spreading of ca. 400 cells onto YEP-glucose to repress HO expression and determine the number of viable cells. The plates were scored after 3 days growth at 30°C.
PCR analysis of mating-type switching.
Primers flanking the HO site in the MAT locus were used to determine mating type by PCR amplification (primer sequences are available upon request). Cultures were grown overnight in raffinose-containing medium. Galactose and then glucose were added to 2% at the times indicated in the figure legends. Primers to the RAD27 gene were included in the multiplex PCR as an internal control. The number of PCR cycles to produce amplification in the linear range was determined empirically. The ratio of the MAT product to the control product was quantified by using Labworks (GelPro4.0; Media Cybernetics, LP).
Analysis of resection during homologous recombination.
Measurement of mating-type switching was performed as described previously (53). Briefly, purified genomic DNA was digested with StyI, and fragments were resolved on an alkaline denaturing gel. The Southern blot was probed with a fragment that overlaps the StyI fragment containing the HO cleavage site of MAT and the next most distal fragment.
Analysis of mutation frequency.
The rate of accumulation of Canr mutants in cell populations was determined by fluctuation analysis by using the method of the median (29) as described previously (35). Each fluctuation test was repeated independently at least two times. Independent Canr mutants were isolated by streaking strains JKM179 (WT), YM001 (asf1Δ), and YM003 (msh2Δ) to single colonies on YPD plates. Then, single colonies were resuspended in water and diluted as appropriate onto YPD and −ARG+canavanine plates; mutation of the CAN1 gene allows growth on −ARG+canavanine plates. The results are expressed as the mean ± the standard deviation of multiple independently derived mutation rates.
Recombination assay.
The rate of sister chromatid exchange (SCE) in cell populations was determined by fluctuation analysis by using the method of the median (29) as described previously (35). Each fluctuation test was repeated independently at least two times. A strain containing the SCE substrate of 3′Δ-his3 5′Δhis3 that forms a functional HIS3 upon recombination was kindly provided by Michael Fasullo. Yeast that had undergone independent recombination events were isolated by streaking strains YD122 (WT) and JRY013 (asf1Δ) to single colonies on YPD plates. Single colonies were then resuspended in water and diluted as appropriate on YPD and plates lacking histidine. The results are expressed as the mean ± the standard deviation of multiple independently derived mutation rates.
RESULTS
asf1 mutants have a metaphase-anaphase cell cycle defect.
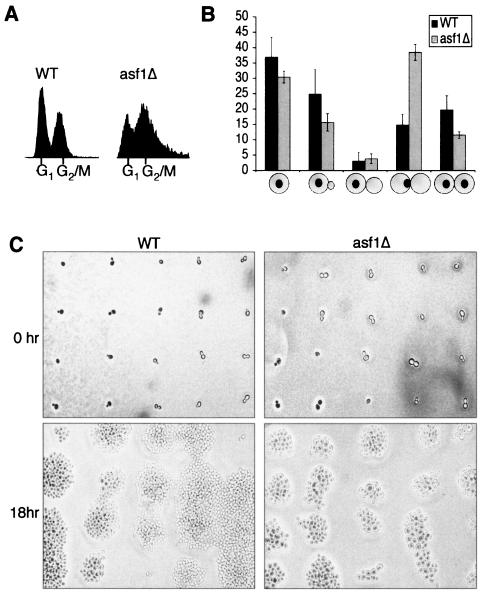
We set out to determine why yeast lacking Asf1p grow slowly (59). Flow cytometry analysis demonstrated that asynchronous cultures of yeast deleted for the ASF1 gene (asf1Δ) accumulate with a G2/M DNA content compared to an isogenic wild-type culture (Fig. 1A). To determine the stage in G2/M at which asf1 mutant cells arrest, we performed microscopic analysis of the DNA and bud size on a population basis. We found that deletion of ASF1 increases the proportion of cells in metaphase, defined as large budded cells with one mass of DNA at the bud neck, from 15% in wild-type cultures to ca. 40% of the population in asynchronous asf1Δ cultures (Fig. 1B). These cells also had short mitotic spindles pointing toward the bud neck, as determined by immunofluorescence analysis with an anti-tubulin antisera (data not shown), a finding consistent with being in metaphase. Taken together, these results demonstrate that asf1 mutants accumulate in metaphase of the cell cycle.
FIG. 1.
Yeast with ASF1 deleted have a metaphase-anaphase transition defect. (A) asf1 mutants accumulate in G2/M phase. Results from flow cytometry analysis of DNA content of JKT0040 (WT) and JKT0041 (asf1Δ) asynchronous cultures are shown. (B) asf1 mutants accumulate in metaphase. Budding indices of JKT0040 (WT) and JKT0041 (asf1Δ) asynchronous cultures are shown. The diagrams below each set of columns indicate the relative size of the bud and position of the nuclear DNA. The data shown are the averages and standard deviations of three independent blind counts of 100 cells for each strain. (C) asf1 mutant cells grow slowly. Individual cells from JKT0040 (WT) and JKT0041 (asf1Δ) strains were micromanipulated onto a grid of agar and photographed immediately and 18 h later. The magnification was identical for all panels.
To gain further evidence of a cell cycle defect in cells lacking Asf1p, we examined the growth of individual cells. Cells were micromanipulated onto a grid on an agar plate, and cell growth and division were observed (Fig. 1C). Wild-type cells had progressed through multiple divisions by 18 h. In contrast, asf1Δ cells had only progressed through three to five divisions by 18 h. Notably, many of the asf1Δ cells were greatly enlarged compared to the wild type (Fig. 1C), where enlarged cells reflect continued growth during cell cycle arrest. The enlarged size of cells lacking Asf1p is also apparent from the broadening of the DNA peaks in our flow cytometry analyses (Fig. 1A). Taken together, these data clearly indicate that loss of Asf1p leads to cell cycle defects.
Loss of Asf1p activates the DNA damage checkpoint.
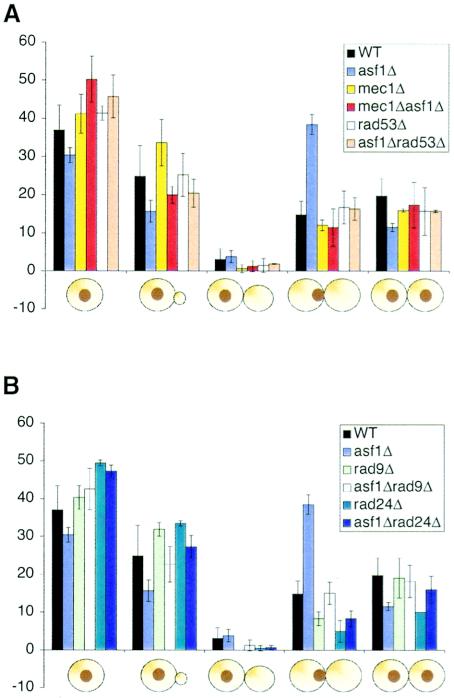
We had previously observed that asf1 mutants are highly sensitive to double-strand DNA-damaging agents (59). To determine whether the metaphase-anaphase delay of yeast lacking Asf1p is due to activation of the DNA damage checkpoint, we measured the bud index for yeast deleted for ASF1 in combination with DNA damage checkpoint mutants. Deletion of MEC1 and RAD53, the central kinases in all genomic integrity checkpoint pathways, rescued the accumulation of asf1Δ cells in metaphase (Fig. 2A). Note that SML1 is also deleted in these strains to suppress the requirement for Rad53p and Mec1p in regulating nucleotide levels (66). Deletion of the gene encoding the Rad24p DNA damage checkpoint protein from asf1Δ cells also prevented the accumulation in metaphase (Fig. 2B). Similarly, deletion of the RAD9 gene encoding, the mediator kinase that is activated in response to double-strand lesions, from asf1Δ cells prevented the accumulation in metaphase (Fig. 2B). These results indicate that the delay in the metaphase to anaphase transition in asf1 mutants is dependent on DNA damage checkpoint proteins.
FIG. 2.
Loss of Asf1p activates the DNA damage checkpoint. (A) Deletion of MEC1 or RAD53 fixes the metaphase defect of asf1Δ cells. Budding index of JKT0040 (WT), JKT0041 (asf1Δ), JKT0186 (mec1Δ), JKT0187 (asf1Δ mec1Δ), JRY003 (rad53Δ), and JKT0185 (asf1Δ rad53Δ) strains was determined as described in Fig. 1B. (B) Deletion of RAD24 or RAD9 fixes the metaphase defect of asf1Δ cells. Budding index of JKT0040 (WT), JKT0041 (asf1Δ), JKT0042 (rad9Δ), JKT0044 (asf1Δ rad9Δ), JKT0043 (rad24), and JKT0045 (asf1Δ rad24Δ) were determined as described for Fig. 1B.
The accumulation of yeast lacking Asf1p in metaphase appears to be wholly due to activation of the DNA damage checkpoint. Activation of the spindle assembly checkpoint can also leads to metaphase arrest (30), and yet deletion of the MAD2 component of the spindle assembly checkpoint failed to rescue the metaphase accumulation defect of asf1Δ cells (data not shown). Furthermore, by using a GFP-tagged minichromosome system (37), we found no increase in minichromosome missegregation in asf1Δ cells compared to wild-type cells (data not shown). Consistent with our results, yeast that are deleted for both ASF1 and the gene encoding the large subunit of the CAF-1 chromatin assembly factor have been reported to show a G2/M arrest independent of spindle assembly checkpoint activation and segregate a minichromosome at frequencies similar to wild-type cells (50).
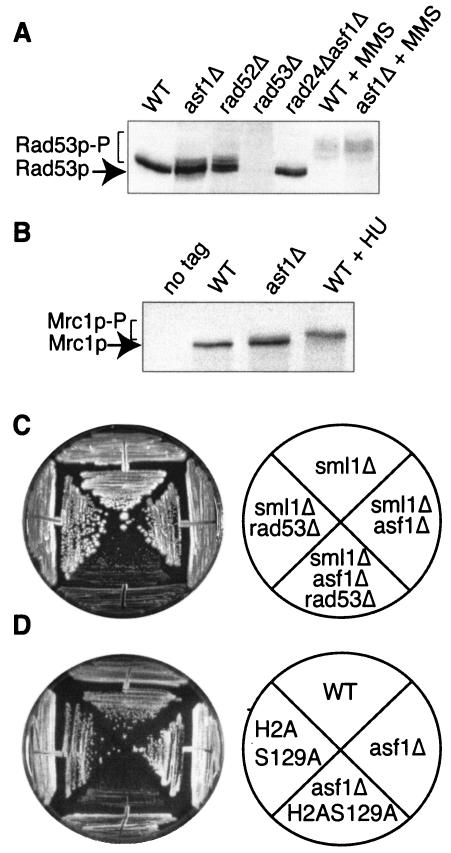
To gain molecular proof that the mitotic delay in cells lacking Asf1p is due to activation of the DNA damage checkpoint pathway, we looked for phosphorylation of DNA damage checkpoint kinases. We confirmed that the Rad53p DNA damage checkpoint protein is activated in the absence of DNA-damaging agents in asf1Δ cells but not in wild-type cells (Fig. 3A), as published previously (24). As a reference for the amount of Rad53p activation that occurs if the spontaneous DNA damage occurring during DNA replication is not repaired, we compared Rad53p activation in asf1Δ and rad52Δ strains. We found that the extent of Rad53p activation in asf1Δ cells is very similar to that observed previously in asf1 mutant cells (24) and is similar to the extent of Rad53p activation that occurs in a rad52Δ strain that is unable to perform any homologous recombination (Fig. 3A). This result indicates that Rad53p is activated in asf1Δ cells at levels similar to cells that are unable to repair spontaneous double-strand DNA damage. As expected, we found that deletion of RAD24, MEC1, or RAD9, but not deletion of MRC1, prevented activation of Rad53p due to endogenous DNA damage in asf1Δ cells (Fig. 3A and data not shown). This lack of requirement for Mrc1p for Rad53p activation reflects the fact that Rad9p is recruited to stalled replication forks in mrc1 mutants, resulting in Rad53p activation (25). To address whether replicational stress is activating the DNA damage checkpoint, we examined whether Mrc1p is phosphorylated in asf1 mutants, where Mrc1p phosphorylation is a specific response to replicational stress. We observed a shift upward of the Mrc1p band corresponding to phosphorylation in the asf1Δ strains that was absent in the wild-type strain (Fig. 3B). This result indicates that the DNA damage checkpoint is activated in response to increased replicational stress in cells lacking Asf1p.
FIG. 3.
The loss of Asf1p activates the DNA damage checkpoint. (A) Rad53p is activated in asf1 mutants. Western blot analysis of the DNA damage checkpoint kinase Rad53p was done in total protein extracts derived from JKT0040 (WT), JKT0041 (asf1Δ), JKT0004 (rad52Δ), JRY003 (rad53Δ), and JKT0045 (rad24Δ asf1Δ) and strains, and WT and asf1Δ strains treated with 0.2% MMS for 1 h. Activation of Rad53p is determined by phosphorylation and is apparent by the higher-molecular-weight bands that are indicated as “Rad53p-P.” (B) Mrc1p is activated in asf1 mutants. Western blot analysis of the 13-myc tagged DNA replication checkpoint protein Mrc1p was done in total protein extracts derived from strains JRY010 (WT), JRY011 (asf1Δ), a wild-type strain treated with 200 mM hydroxyurea for 2 h, and strain JKT0004 (no tag). Activation of Mrc1p was determined by phosphorylation and is apparent by the higher-molecular-weight bands that are indicated as “Mrc1p-P.” (C) Deletion of both RAD53 and ASF1 leads to severe growth defects. Strains JRY001 (sml1Δ), JRY003 (sml1Δ rad53Δ), JRY002 (sml1Δ asf1Δ), and JKT0185 (sml1Δ asf1Δ rad53Δ) were streaked onto YPD and grown for 3 days before photographing them. (D) Mutation of serine-129 of H2A, together with deletion of ASF1, leads to severe growth defects. Strains JKY001 (WT), JKY002 (H2AS129A), JRY015 (asf1Δ), and JRY014 (H2AS129A asf1Δ) were streaked onto YPD and grown for 3 days before being photographed.
To investigate whether activation of the DNA damage checkpoint is necessary to protect asf1Δ cells from exiting mitosis with catastrophic DNA damage, we examined microcolony formation. We found that deletion of both ASF1 and the DNA damage checkpoint gene RAD53, MEC1, or RAD9 greatly reduces viability compared to each individual mutation. An example of the reduced viability of cells deleted for both ASF1 and a DNA damage checkpoint component is shown in Fig. 3C for the Rad53p checkpoint kinase. We also observed greatly reduced viability when we deleted ASF1 in combination with a mutation in histone H2A that prevents its phosphorylation in response to DNA damage (Fig. 3D). The low viability of the asf1Δ H2AS129A double mutant indicates that phosphorylation of H2A in response to DNA damage is important in the absence of Asf1p. Our results indicate that the absence of Asf1p increases the requirement for the DNA damage checkpoint and that failure to arrest at the DNA damage checkpoint leads to increased cell death in asf1 mutants.
asf1 mutants are sensitive to drugs and DNA polymerase mutants that induce replicational stress.
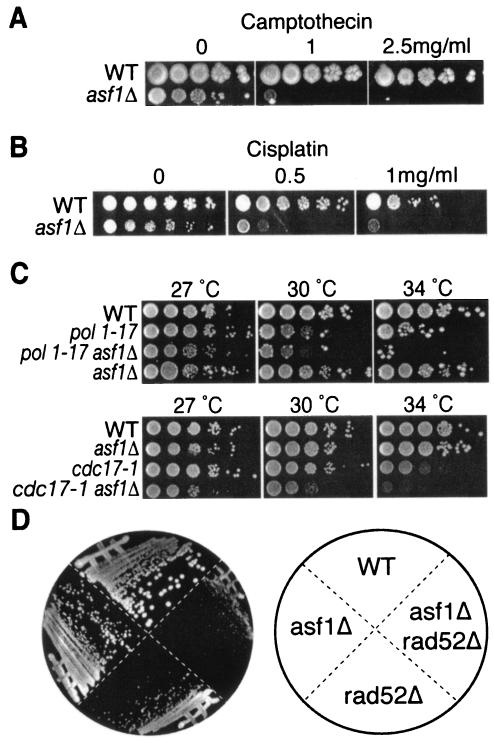
To investigate whether the activation of the DNA damage checkpoint in asf1 mutants is a consequence of DNA replication stress, we examined their sensitivity to agents that induce replicational stress. Consistent with this idea, we have previously observed that asf1 mutants are sensitive to hydroxyurea, which depletes the deoxynucleoside triphosphate pools and leads to stalling of replication forks (59). We found that loss of Asf1p also greatly increases sensitivity to camptothecin (Fig. 4A). Camptothecin inhibits the ability of topoisomerase 1 to religate the single-strand breaks that it creates to relieve tension ahead of the replication fork, leading to double-strand DNA damage during DNA replication (23). Similarly, asf1 mutants are extremely sensitive to cisplatin (Fig. 4B), which generates intrastrand DNA cross-links that cause the replication fork to stall (13). The problems generated by hydroxyurea, camptothecin, and cisplatin during DNA replication can be fixed eventually by DNA repair mechanisms. Therefore, to address directly whether asf1 mutants have DNA replication problems rather than DNA repair problems, we examined mutations of DNA polymerase. We found that deletion of ASF1 lowered the restrictive temperature of the DNA polymerase alpha mutants, pol1-1 and cdc17-1 (Fig. 4C). These results indicate that the absence of Asf1p exasperates DNA replication problems.
FIG. 4.
Cells lacking Asf1p are sensitive to replicational stress. (A) asf1 mutants are sensitive to camptothecin. Cultures of JKT0040 (WT) and JKT0041 (asf1Δ) were serial diluted (10-fold), spotted onto YPD and YPD plus 1 or 2.5 mg of camptothecin/ml, and grown for 3 days at 30°C. (B) asf1 mutants are sensitive to cisplatin. Cultures of JKT0040 (WT) and JKT0041 (asf1Δ) were serial diluted (10-fold), spotted onto YPD and YPD plus 0.5 or 1 mg of cisplatin/ml, and grown for 3 days at 30°C. (C) Deletion of ASF1 reduces the permissive temperature of polymerase alpha mutants. Cultures of JKT0040 (WT), BOB461 (pol1-17ts), JRY009 (asf1Δ pol1-17), BOB460 (cdc17-1), JRY016 (asf1Δ cdc17-1), and JKT0041 (asf1Δ) were serial diluted (10-fold), spotted onto YPD, and incubated at 27, 30, or 34°C for 3 to 4 days. (D) Synthetic sickness upon deletion of both ASF1 and RAD52. Strains JKT0010 (WT), JKT0001 (asf1Δ), JKT0005 (asf1Δ rad52Δ), and JKT0004 (rad52Δ) were struck out, grown at 30°C for 3 days, and then photographed.
During the course of our analyses, we generated a mutant with both ASF1 and RAD52 deleted, where the Rad52 protein is an essential component of the homologous recombination machinery. We found that this double mutant was very sick compared to either single mutant (Fig. 4D). This result suggests that in the absence of Asf1p, cells have an increased requirement for the homologous recombinational machinery. This result is particularly interesting because it is the homologous recombinational machinery that rescues cells that have experienced replication problems.
The cell cycle defect of yeast lacking Asf1p is not due to incomplete DNA replication.
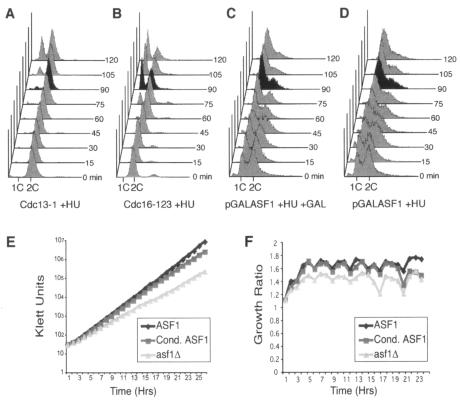
Next, we sought to determine whether the metaphase-anaphase transition defect in cells lacking Asf1p is a consequence of incomplete DNA replication. This question was relevant because we have now shown that asf1Δ cells are sensitized to DNA replication problems, and it was possible that the accumulation of asynchronous asf1Δ cells with a 2C DNA content may reflect late-S-phase cells rather than metaphase cells. To determine whether asf1 mutants have completed DNA replication, we added the DNA synthesis inhibitor hydroxyurea to a yeast strain that had its endogenous copy of ASF1 under the control of the galactose-inducible GAL1 promoter. The addition of hydroxyurea to cells that had been arrested before completion of replication, such as cdc13-1 mutants, causes immediate arrest with a 2C DNA content upon switching to the permissive temperature (62) (Fig. 5A). In contrast, the addition of hydroxyurea to metaphase-arrested cells with completely replicated chromosomes, such as cdc16-123 mutants, allows progression through mitosis to the subsequent S phase upon switching to the permissive temperature (Fig. 5B). Addition of hydroxyurea to cells lacking Asf1p at the same time that Asf1p was induced by the addition of galactose permitted the cells that had accumulated with a 2C DNA content to exit mitosis and arrest as they entered the subsequent S phase (Fig. 5C). This experiment indicated that the accumulation of cells lacking Asf1p with a 2C DNA content is not due to incomplete DNA replication and therefore reflects arrest in metaphase rather than late S phase. No significant difference was seen whether Asf1p was induced or not at the time of hydroxyurea addition (Fig. 5C and D) or in cells where ASF1 was deleted (data not shown), demonstrating that Asf1p is not required for passage from metaphase to the subsequent S phase. By a process of elimination, these data indicate that the functions of Asf1p during S phase and/or G2 phase of the cell cycle leads to metaphase arrest.
FIG. 5.
Cells lacking Asf1p complete DNA replication. (A) cdc13-1 mutation prevents the completion of replication. An asynchronous culture of logarithmically growing strain BOB463 with a temperature-sensitive cdc13-1 allele was shifted to the nonpermissive temperature (37°C) for 2 h, leading to arrest in late S phase. Hydroxyurea was added to a 200 mM final concentration at the same time the cells were shifted to a permissive temperature (23°C) to induce functional Cdc13p. A sample was taken for flow cytometry analysis of the cell cycle distribution immediately (“0 min”) and at every 15 min thereafter, as shown. (B) A cdc16-123 mutation allows completion of replication. The identical manipulations were performed in parallel as for panel A, with strain 1825-1B carrying the temperature-sensitive cdc16-123 allele. (C) The 2C accumulation of asf1 mutants is not due to incomplete replication. Hydroxyurea was added to 200 mM to strain SRH014 carrying the ASF1 gene under control of the pGAL1 promoter growing in raffinose medium at 23°C. At the same time as the hydroxyurea addition, galactose was added (1%) to induce Asf1p expression. No Asf1p is detected during growth of this strain in raffinose, and 1% galactose was empirically determined to induce Asf1p to the same levels as the endogenous protein. The induction of Asf1p was assayed during this experiment by Western analysis, in which synthesis of Asf1p was detected within 15 min, and was fully induced by 45 min (data not shown). A sample was taken for flow cytometry analysis of the cell cycle distribution immediately (“0 min”) and at every 15 min thereafter. (D) Asf1p is not required for passage from metaphase to S phase. The experiment was performed as described for panel C but without the inclusion of 1% galactose. The flow cytometry profiles of DNA content are shown, and the black profiles indicate a time point where the result is most apparent between the strains. (E) The growth defect in asf1 mutants is a result of an accumulation of defects. Strains SRH015 (WT), SRH014 (Cond. ASF1), and SRH016 (asf1Δ) were grown in YEPR plus 1% galactose to a Klett reading of 30 and then switched to YPD medium to repress ASF1 transcription in the Cond. ASF1 strain. The Asf1 protein was not detectable 5 min after the switch to YPD medium. Klett readings were taken every hour, and the cultures were diluted back to a Klett reading of 30 whenever any culture grew to a reading of >100 to keep the cells in mid-log phase. Dilution factors were multiplied back into the Klett readings to obtain the Klett units plotted above. (F) Growth ratio switch analysis. To determine the time at which the growth rate after degradation of Asf1p (Cond. ASF1) switches from the growth rate of wild-type yeast (ASF1) to that of asf1 mutant yeast (asf1Δ), the growth ratio for each culture was plotted every hour after Asf1p degradation in panel E. The growth ratio is the Klett reading at any given time divided by the Klett reading at the previous time, reflecting how much the culture grew in each 1-h period. The growth ratio is not absolutely constant, since the cultures spend a short amount of time outside their ideal growth conditions during the hour that dilution occurred (the time points with the lowest growth ratios). It is clear that the growth rate after degradation of Asf1p in the Cond. ASF1 strain very closely followed that of the wild-type (ASF1) strain, until 20 h after degradation of Asf1p, wherein the growth ratio drops to that of the asf1 mutant (asf1Δ).
The cell cycle defect of asf1 mutants is due to the accumulation of defects.
To gain further insight into Asf1p function, we investigated how rapidly or slowly the problems associated with loss of Asf1p lead to the cell cycle defect. To do this, we used a yeast strain in which an unstable copy of Asf1p is expressed from the galactose-inducible and glucose-repressible pGAL1 promoter that results in the disappearance of any detectable Asf1p within minutes of its transcriptional repression (S. Zabaronick and J. Tyler, submitted for publication). After repression of ASF1, the passage of 20 h was required before the growth rate of yeast lacking Asf1p switched from the growth rate of wild-type yeast to that of asf1Δ yeast (Fig. 5B). This distinct switch in growth rate at 20 h is apparent from comparison of the culture growth between each time point (Fig. 5C). These data indicate that a threshold of molecular defects has to be reached before the slow-growth phenotype occurs after removal of Asf1p.
Loss of Asf1p leads to genomic instability.
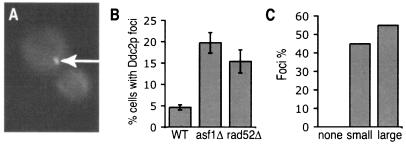
To better understand the role of Asf1p in chromosomal integrity, we wanted to distinguish between activation of the DNA damage checkpoint in asf1Δ cells as a consequence of altered chromatin structure or by DNA lesions. Activation of the DNA damage checkpoint by disrupted chromatin structure in human cells is not accompanied by the formation of DNA repair foci (4). Therefore, we examined whether asf1 mutants have more spontaneous DNA repair foci than wild-type cells by looking for foci of Ddc2p tagged with GFP (39). DNA repair foci are “factories” in which multiple DNA lesions, DNA damage checkpoint proteins, and DNA repair proteins come together to mediate repair (31, 32). We observed that 5% of wild-type cells have spontaneous Ddc2p-GFP foci, a finding consistent with published data (39) (Fig. 6A and B). In contrast, 20% of asf1Δ yeast had spontaneous Ddc2p-GFP foci (Fig. 6B). For reference, 15% of rad52Δ cells that are incapable of repairing damage by homologous recombination had Ddc2p-GFP foci (Fig. 6B). The Ddc2p foci in asf1Δ cells were found primarily in small budded (S-phase) (45%) or large budded (G2/M-phase) (55%) cells (Fig. 6C). These data demonstrate that asf1Δ cells have elevated amounts of DNA lesions due to problems during DNA replication.
FIG. 6.
Loss of Asf1p increases Ddc2p-GFP foci formation. (A) A typical metaphase asf1Δ cell with the Ddc2p-GFP focus indicated by the arrow. A single focus is the site of repair of multiple DNA lesions and the majority of cells with Ddc2p-GFP foci in this analysis contained a single focus. (B) Proportion of asynchronous cells containing Ddc2p-GFP foci in strain JRY006 (WT), JRY007 (asf1Δ), and JRY008 (rad52Δ). (C) Cell cycle distribution of asf1Δ cells containing Ddc2p-GFP foci, as determined by bud morphology, where “none,” “small,” and “large” refer to the bud size.
Asf1p is not required for repair of double-strand DNA damage.
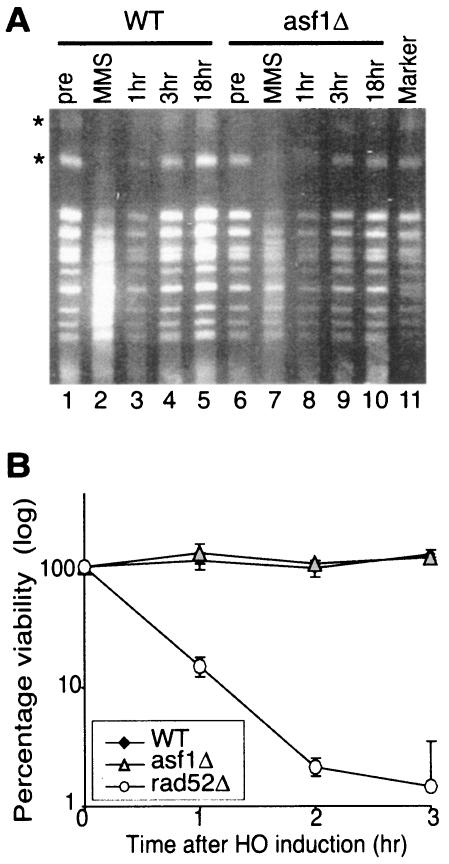
To determine whether the increased number of DNA damage foci in asf1Δ cells is due to the failure of asf1Δ cells to repair DNA damage or due to increased incidence of DNA lesions, we examined the ability of asf1Δ cells to perform double-strand DNA repair. First, we analyzed the integrity of chromosomes isolated from asf1Δ and wild-type cells at increasing times after removal of methyl methanesulfonate (MMS). MMS is a DNA alkylating agent that results in both single and double-strand DNA breaks (49). After a short treatment (5 min) with MMS, double-strand DNA damage was apparent from the smearing of the chromosomes, as detected by pulsed-field electrophoresis (Fig. 7A). The chromosomes of both wild-type and asf1Δ cells were distinct bands again after 1 and 3 h after the removal of MMS, indicating repair of double-strand DNA lesions (Fig. 7A). In parallel, we measured the viability of the cells that were used for chromosome isolation and found no significant changes in viability (as determined by growth on YPD) for each of the wild-type and asf1Δ samples before, during, or after MMS treatment (data not shown). This result indicates that the reappearance of the intact chromosomes (Fig. 7A) was indeed due to DNA repair. Therefore, we conclude that Asf1p is not required for religation of double-strand breaks on a chromosomal scale.
FIG. 7.
Asf1p does not appear to be required for double-strand break repair. (A) asf1 mutants are competent at repairing gross chromosomal damage. Cells from cultures of ROY1172 (WT) and ROY1170 (asf1Δ) were arrested for 2 h with nocodazole, followed by a 5-min treatment with 0.048% MMS. The MMS was then removed, and the cells were recovered for 1, 3, or 18 h at 30°C. Chromosomes were isolated after the arrest (pre), after MMS treatment (MMS), and at 1, 3, or 18 h after removal of the MMS (1, 3, and 18 h, respectively) and were resolved by pulsed-field agarose gel electrophoresis, followed by ethidium bromide staining. The characteristic pattern of S. cerevisiae chromosomes is seen in the marker lane. Induction and repair of double-strand breaks is most apparent by monitoring the disappearance and reappearance of the two largest chromosomes (indicated by the asterisks), the majority of which appears to be damaged at this level of MMS exposure. Double-strand breaks are seen as a reduction in the intensity of a chromosome and an increase in chromosome ladder smearing. Identical results were obtained with or without nocodazole arrest (data not shown). (B) asf1 mutants are competent for repair of a defined HO lesion by homologous recombination. Plasmid yECP50 carrying the pGAL1:HO gene was introduced into strains JRY2334 (WT), YM004 (asf1Δ), and JKT0004 (rad52Δ), and HO endonuclease was induced by the addition of galactose to the medium. Identical amounts of dilutions were plated onto glucose medium at increasing times after HO induction, grown for 3 days at 30°C, and viability was determined by colony counting. Viability for each strain prior to induction of the HO endonuclease was designated 100%. The data are the average and standard deviation of three independent experiments.
In order to examine the repair of a defined double-strand break, we sought to determine whether Asf1p was required for the repair of an HO lesion at the MAT locus by homologous recombination (Fig. 7B). At increasing times after induction of the HO endonuclease (under the control of the GAL1 promoter), cells were plated onto glucose to repress the HO endonuclease and their viability was determined. Viability is dependent on the repair of the HO lesion by homologous recombination. As a control for a strain that is defective for repair of the HO lesion, the viability of rad52Δ cells was greatly reduced (Fig. 7B). We found no significant difference in cell viability between asf1 mutants and wild-type cells after HO induction, suggesting that asf1Δ cells are competent to repair the HO lesion (Fig. 7B).
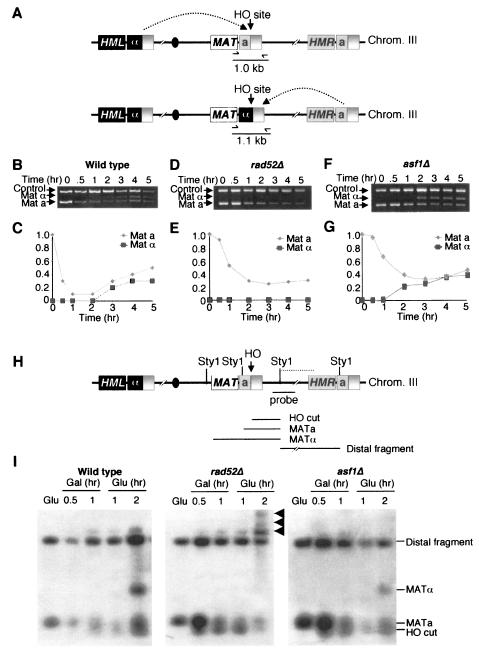
To evaluate the ability of yeast lacking Asf1p to perform homologous recombination at the molecular level, we monitored repair of the HO lesion. To do this, we used a quantitative PCR-based analysis of the MAT region to monitor the cutting and repair of the HO lesion (Fig. 8A) (B. Tamburini and J. Tyler, submitted for publication). We used a primer pair that would produce a product unique to MAT that would differ by 0.1 kb between MATa and MATα cells (Fig. 8A). A MAT locus with an HO lesion will yield no MAT PCR product. In general, MATa cells are repaired by using the information at HMLα to yield MATα yeast, whereas MATα yeast are repaired by using the information at HMRa to yield MATa yeast (Fig. 8A) (63). This PCR-based approach generated the same results as Southern analysis (data not shown) but was more convenient and easier to internally control. To analyze HO cleavage and repair, HO endonuclease was induced at 0 h by addition of galactose, followed by the addition of glucose at 2 h to repress HO endonuclease. HO cleavage and repair was apparent in the wild-type strain (Fig. 8B and C). Both mating types are generated during repair because immediate recleavage and repair occurs on some templates during the 2 h induction of HO endonuclease. As a control for a strain that cannot perform homologous recombination, we examined repair in a rad52Δ strain (Fig. 8D and E). Minimal repair occurred in the rad52Δ strain and presumably the slight increase in repaired MATα product is a result of repair by simple ligation via nonhomologous end joining. We found that the asf1Δ strain was able to repair its HO lesion with similar kinetics to the wild-type strain and with no less than 50% the efficiency of repair seen in the wild-type strain (Fig. 8F and G). This finding indicates that Asf1p is not required for homologous recombination, since homologous recombination can clearly occur in the absence of Asf1p.
FIG. 8.
Molecular analysis of DNA repair in asf1 mutants. (A) Schematic of the mating-type loci in yeast. The locations of PCR primers are shown. If the mating type is “a,” then the PCR product is 1.0 kb; if it is “α,” then the product is 1.1 kb. (B) Cutting and repair of the HO lesion in wild-type yeast. HO endonuclease was induced at . = 0 in strain BAT009 (WT) by the addition of galactose and was repressed at 2 h by the addition of glucose. Samples were analyzed throughout the time course with the primers shown in panel A and control primers. (C) Quantitation of DNA cutting and repair in WT yeast. The MATa and α products were quantified from the gel in panel B and were normalized to the control product. The amount of MAT product at . = 0 was normalized to 1. (D) As in panel B, but with strain BAT022 (rad52Δ). (E) Same as in panel C, but with strain BAT022 (rad52Δ). (F) Same as in panel B, but with strain YM004 (asf1Δ). (G) Same as in panel C, but with strain YM004 (asf1Δ). (H) Schematic of the mating-type loci used to investigate resection during repair of an HO induced double-strand break. (I) Analysis of resection by Southern hybridization after HO induction. Genomic DNA from mating type a strains JRY2334 (WT), YM004 (asf1Δ) and JKT004 (rad52Δ) was isolated from asynchronous cultures before induction of the HO nuclease “Glu” at 0.5 and 1 h after the addition of galactose and then at 1 and 2 h after the subsequent addition of glucose to repress the HO endonuclease and allow repair of the HO lesion by mating-type switching. Genomic DNA was digested with StyI, and the resulting Southern blots were hybridized by using the probe indicated in panel A. The HO-cut fragment appeared between 0.5 and 1 h after HO induction with galactose, followed by the appearance of the MATα product in wild-type and asf1Δ strains, 2 h after repression of the HO endonuclease by glucose addition. The MATα product cannot be observed in the rad52Δ strain due to the inability to perform homologous recombinational repair of the HO lesion. Arrows indicate the formation of single-stranded DNA tails that result from the inability of StyI to cleave the single-stranded DNA, which is due to resection beyond one or more distal StyI sites.
The accessibility of the chromatin structure at the donor locus is known to influence the strand invasion step of homologous recombination (53). To investigate whether the chromatin assembly factor Asf1p influences the chromatin structure at the donor locus, we analyzed the efficiency and intermediates of HO mating type switching (Fig. 8H). The HO site was efficiently cut in asf1Δ, wild-type, and rad52Δ strains (Fig. 8I). After transcriptionally repressing the HO endonuclease with glucose, we found that the asf1Δ and wild-type cells were equally capable of mating-type switching (by repairing the HO lesion at MATa with the HMLα sequences), where ca. 50% of the cells of each strain had switched to MATα by 2 h. As a positive control for a gene required for mating-type switching, the rad52Δ strain failed to detectably switch to MATα (Fig. 8I). Instead, the rad52Δ strain exhibited extensive formation of single-strand DNA, where the double-strand break end is resected. The asf1Δ strain exhibited no greater resection than the wild-type strain. These results show that Asf1p does not noticeably affect the rate or extent of mating-type switching and indicate that there is no enhancement or reduction in the ability of the broken end to invade the chromatin of the donor sequences in asf1Δ cells. All of these analyses taken together demonstrate that Asf1p is not required for the repair of double-strand breaks via homologous recombination and, by inference, the increased DNA damage foci in asf1Δ cells reflect an increased incidence of DNA damage rather than a failure to repair.
Yeast lacking Asf1p have an elevated rate of spontaneous mutation and SCE.
To determine whether Asf1p and consequently, chromatin structure, are required for genomic stability, we determined the spontaneous mutation rates for yeast disrupted or wild type for ASF1. The approach was to measure the mutation rate of the endogenous CAN1 gene that encodes an arginine permease. The product of the wild-type CAN1 gene allows uptake of arginine and the closely related metabolic poison canavanine. Therefore, only cells that have deleted or inactivated their CAN1 gene by mutation will be viable on canavanine plates (16, 35). We found that CAN1 was consistently inactivated in asf1Δ yeast at a 2.5-fold-higher rate (1.58 × 10−7 ± 1.24 × 10−7) than CAN1 inactivation in wild-type yeast (6.39 × 10−8 ± 2.24 × 10−8). Therefore, there is a significant (. = 2.03 × 10−4) increase in mutation rate upon loss of Asf1p. As a positive control for an extreme example of an elevated spontaneous mutation rate, we determined in parallel the mutation rate of yeast deleted for the MSH2 gene that is required for mismatch repair. The msh2Δ strain exhibited a spontaneous mutation rate of 1.04 × 10−5—163 times higher than wild-type cells. To determine what types of mutations had occurred to increase the mutation rate of asf1Δ strains, Canr colonies were assayed for the presence of the CAN1 locus by PCR. We found that none of the Canr asf1Δ colonies were due to deletion of the CAN1 locus. Sequencing of the entire CAN1 gene from multiple independent Canr asf1Δ colonies indicated that the spontaneous mutations were nucleotide substitutions and single nucleotide deletions (data not shown). From this analysis, we conclude that loss of Asf1p results in an increase in the rate of spontaneous DNA mutation.
To gain further evidence for a requirement for Asf1p in maintaining genomic stability after DNA replication, we measured the rate of spontaneous recombination occurring in asf1Δ cells. We used an assay to measure SCE, a recombination event that is initiated in S phase (2, 9). Wild-type and asf1 mutant strains containing a 5′ fragment of HIS3 next to a 3′ fragment of HIS3 were used to quantitate spontaneous SCE by the formation of a functional HIS3 gene, as described previously (12). We consistently found that asf1 mutants performed SCE 2.5 times more frequently (1.71 × 10−7 ± 7.42 × 10−9) than wild-type yeast (6.96 × 10−8 ± 2.5 × 10−8). Therefore, there is a significant (. = 1.20 × 10−4) increase in recombination rate upon loss of Asf1p. These data indicate that asf1 mutants have an elevated rate of recombination, suggesting that abnormal chromatin structures lead to increased recombination during replication.
DISCUSSION
We have established that the slow growth of yeast lacking the Asf1p chromatin assembly factor is due to the activation of their DNA damage checkpoint. The activation of the DNA damage checkpoint in asf1 mutants is a consequence of increased genomic instability, as indicated by elevated probability of DNA damage foci, elevated recombination rates, and elevated mutation rates; events that are all initiated during S phase. We have found that Asf1p is not required for DNA repair per se, and therefore our data indicate that the altered chromatin structure in asf1 mutants leads to increased genomic instability after DNA replication.
Cells lacking Asf1p have increased DNA damage, even though they can repair DNA damage.
Spontaneous DNA damage occurs during DNA replication and is repaired by homologous recombination. There are two possible models to explain the striking increase in spontaneous DNA repair foci seen in asf1 mutants (Fig. 6). First, asf1 mutants may not be capable of repairing endogenous damage as seen in rad52 mutants or, second, the loss of Asf1p may increase the occurrence of endogenous DNA damage. Initially, the first possibility seemed likely because yeast lacking Asf1p are clearly inviable when plated onto medium containing double-strand DNA-damaging agents (28, 59). However, we have not been able to detect a significant requirement for Asf1p in the extent or efficiency of repair of a defined HO lesion at the MAT locus by homologous recombination (Fig. 8). It is possible that the recombinational repair of the HO lesion is not truly representative of general double-strand break repair since the donor loci at HMR/HML are present in an inaccessible chromatin structure (19). The cell may have evolved specialized machinery to gain access to the buried donor DNA sequences at HMR and HML, such that repair of the HO lesion at MAT may be less affected by the loss of Asf1p than double-strand breaks elsewhere in the genome. However, this does not appear to be the case, since the repair of MMS-induced double-strand breaks is indistinguishable between wild-type cells and cells with ASF1 deleted on the chromosomal scale in our pulsed-field gel electrophoresis analyses (Fig. 7).
Further evidence for a lack of requirement for Asf1p in the repair of an HO lesion has been provided by Qin and Parthun (46). Consistent with our results showing that homologous recombinational repair is intact in asf1 mutant cells, the study of Qin and Parthun showed at worst a 50% efficiency of HO repair in an asf1 mutant compared to wild-type cells upon inspection of their data. Clearly, the subtle, if any, defect in homologous recombination in asf1 mutants cannot explain the elevated occurrence of DNA damage foci in the absence of Asf1p, because even higher levels of DNA damage foci were observed in an asf1 mutant compared to a rad52 mutant that is totally defective in homologous recombination (Fig. 6). In support of this finding, a recent report demonstrated that asf1 mutant cells have an elevated spontaneous rate of occurrence of Rad52 DNA repair foci in budded cells (45). Therefore, the elevated occurrence of DNA damage foci in the absence of Asf1p is likely to reflect a higher incidence of DNA damage rather than the failure to repair DNA damage. The fact that the DNA damage checkpoint is activated in cells lacking Asf1p also supports the idea that these cells have a higher incidence of DNA damage. As such, the reduced viability of asf1Δ yeast upon inactivation of the DNA damage checkpoint (Fig. 3) is probably a consequence of precocious progression through mitosis with damaged DNA.
Yeast lacking Asf1p grow slowly as a consequence of activating the DNA damage checkpoint; however, this slow growth rate is not apparent until 20 h after Asf1p removal (Fig. 5). This rules out the possibility that the loss of Asf1p disregulates transcription of genes important for cell cycle progression because we would expect to see a growth defect upon degradation of Asf1p sooner than 20 h since the average half-life of mRNA is a mere 19 min (22). Also, there is no disregulation of obvious candidates for alteration of the cell cycle in our microarray analyses of asf1 mutants (Zabaronick and Tyler, submitted). A more likely model to explain the delayed growth defect upon degradation of Asf1p is that chromatin structure gradually becomes more perturbed upon loss of Asf1p until a threshold is reached that affects cellular processes enough to activate the DNA damage checkpoint. Taken together, the available data indicate that Asf1p is not required for DNA repair per se but, in its absence, the accumulation of altered chromatin structures eventually triggers the DNA damage checkpoint. Consistent with this idea, Asf1p is dispensable for the recovery from DNA damage checkpoint-induced metaphase arrest (Fig. 5). This result is important because it indicates that Asf1p is not required for the repair events that the DNA damage checkpoint is stalling the cell cycle to allow. Rather, the absence of Asf1p is causing more damage events that provide opportunities for inaccurate repair.
If asf1 mutants can repair double-strand DNA damage, why are they inviable on plates containing double-strand DNA-damaging agents? An important clue to the answer comes from the fact that transient exposure to high doses of DNA-damaging agents does not reduce the viability of asf1 mutants. We have found that an event after religation of the DNA lesion, perhaps the repair of the chromatin structure, does not occur efficiently in asf1 mutant cells, leading to a delay in turning off or recovery from the DNA damage checkpoint (S. Howar and J. Tyler, unpublished data). Constant exposure of asf1 mutants to DNA-damaging agents therefore would never give the cells an opportunity to grow because of the delayed recovery from the DNA damage checkpoint.
Yeast lacking Asf1p are mutators and have increased rates of recombination.
Consistent with the idea that asf1 mutants have elevated rates of spontaneous DNA damage foci, the loss of Asf1p leads to a moderate but consistently elevated rate of spontaneous mutation and recombination. This finding was confirmed in a recent report that also found elevated rates of sister chromatid exchange in asf1 mutant cells (45). If Asf1p were actively involved in the repair of DNA damage, we would expect the mutation rate to increase in proportion to the amount of lesions induced in cells lacking Asf1p. However, treatment of asf1Δ cultures with MMS did not increase the mutation rate any more than treatment of wild-type cells with MMS (data not shown), a finding consistent with an indirect influence of Asf1p on the fidelity of DNA repair. This indirect influence is most likely via an affect of Asf1p during or after DNA replication, since mutations result when errors made by cellular replicases go unrepaired or are repaired in an error-prone fashion. Similarly, the loss of Asf1p leads to elevated rates of SCE, which is also initiated during DNA replication. Therefore, we propose that the elevated mutation rate and recombination rate upon deletion of ASF1 is a consequence of the loss of Asf1p's function in maintaining normal chromatin structure (59), leading to problems during or after DNA replication.
Asf1p protects against genomic instability during DNA replication.
The weight of evidence suggests that the absence of Asf1p leads to problems during S phase that result in genomic instability. This evidence includes the fact that asf1 mutants activate the Mrc1p kinase that responds to replication problems and are sensitive to both DNA replication stresses and mutation of DNA polymerase alpha. Deletion of ASF1 also leads to gross chromosomal rearrangements (41), increased spontaneous DNA damage foci during S phase (45; the present study), and increased recombination (45; the present study) and mutation rates—events that are all triggered during DNA replication. There are also many genetic interactions that strongly suggest a mechanistic link between Asf1p and the processing of stalled replication forks. For example, deletion of ASF1 is synthetically lethal with deletion of the SGS1 Rec-Q helicase that regulates recombination at stalled replication forks (56). Similarly, deletion of ASF1 is synthetically lethal with deletion of the MGS1 helicase that facilitates fork progression through alternative damage avoidance mechanisms (57). Deletion of ASF1 is also synthetically lethal with deletion of MMS4, which encodes part of an endonuclease complex involved in processing of stalled replication forks (57). We noted a similar relationship between Rad52p and Asf1p in that rad52Δasf1Δ cells have a greatly reduced viability compared to either single mutant (Fig. 3). Rad52p is required for the formation of Holliday junctions during S phase, an intermediate of the rescue of stalled DNA replication forks (68). Similarly, deletion of both ASF1 and the RAD50 gene, whose product is required for the processing of stalled replication forks by homologous recombination, results in inviability (57).
There are several potential mechanisms whereby loss of Asf1p may function to influence genomic stability during S phase: (i) the altered chromatin structure in asf1 mutants may lead to elevated rates of replicational stalling, which results in elevated rates of homologous recombinational repair to resolve the stalled forks; or (ii) replication forks stall at the normal frequency in asf1 mutants, but the abnormal chromatin structure in asf1 mutants leads to preferential processing of the stalled forks by homologous recombination rather than restarting. Neither model would absolutely require Asf1p to be functioning to assemble chromatin at the DNA replication fork, although this certainly is possible. There is evidence that Asf1p is involved in both replication-independent and replication-dependent chromatin assembly, as well as replication-independent chromatin disassembly in vivo (1). Evidence for a replication-independent chromatin assembly role for Asf1p in vivo includes the fact that Asf1 exists in a complex with the histone variant H3.3 that is assembled into highly transcribed genes and the histone chaperone HirA that is required for replication-independent chromatin assembly (55). Evidence for a replication-dependent chromatin assembly role for Asf1p in vivo includes the facts that Asf1 was discovered in a complex with newly synthesized histones that are assembled onto newly replicated DNA (59), that Asf1 exists in a complex with the histone variant H3.1 that is assembled only onto newly replicated DNA (55), and that Asf1 interacts with the replication-specific histone chaperone CAF-1 (55, 59). As such, the altered global chromatin structure that exists in asf1 mutants (1a) that is leading to genomic instability during S phase could result from either the replication-dependent or replication-independent functions of Asf1p in maintaining normal chromatin structure because half of the chromatin on each newly replicated daughter DNA strand is inherited from the old chromatin and half is newly assembled chromatin.
We do not favor the idea that all our results are due to indirect effects of Asf1p on histone levels. Asf1p is known to contribute to the cell cycle regulation of histone gene transcription (54); however, the levels and stability of histone proteins are not detectably different in wild-type and asf1 mutant cells (1a, 17). Also, overexpression of histones H3 and H4 failed to restore the cell cycle and growth defects of asf1 mutants (data not shown). Furthermore, there have not been any reports of histone levels affecting genomic stability, and altered histone levels do not appear to hamper the completion of DNA replication (20).
There is previous evidence that chromatin structure can regulate the movement and timing of the initiation of replication forks. For example, deletion of the histone deacetylase Rpd3p creates a more open chromatin structure that leads to an earlier firing of replication origins (61). It has also been reported that mec1-4 temperature-sensitive mutants show preferential break sites termed replication slow zones (RSZs) that may occur due to specialized chromatin structures that are difficult for replication forks to traverse, such as at the ribosomal DNA (6). By analogy to the situation at RSZs, we have found that asf1 mutants have a more compact or overassembled chromatin structure than normal (1a) that may provide an obstacle for the replication machinery leading to replicational stalls. Strikingly, deletion of ASF1 results in an absolute requirement for the product of the RRM3 gene that mediates efficient replication through the compact chromatin structure of ribosomal DNA (57). Conversely, overexpression of Asf1p may generate a more open or underassembled chromatin structure that is easier to replicate through, as suggested by the observation that overexpression of Asf1p can suppress the requirement for Rad53p in stabilizing replication forks during hydroxyurea treatment (24). It is possible that Asf1p may be directly influencing the processivity of DNA replication forks rather than indirectly through altered chromatin structures. For example, Asf1-mediated reassembly of chromatin behind the DNA replication fork (59) may facilitate proper timing of chromatin assembly to allow for the correct processing of Okazaki fragments, preventing DNA damage. In support of this idea, deletion of both ASF1 and the gene encoding Rad27p, a protein required for Okazaki fragment processing, leads to synthetic lethality (57). Another possibility is that Asf1-mediated disassembly of chromatin (1) ahead of the DNA replication fork may facilitate smooth passage of the replication machinery. However, if Asf1p is affecting the processivity of replication forks, the effect is subtle because, despite our extensive efforts, we have been unable to find any evidence for increased replicational stalling in asf1 mutants by either pulsed-field gel electrophoresis or two-dimensional replicational gels (data not shown). Therefore, we favor the idea that the altered chromatin structure in the absence of Asf1p does not lead to more replication stalls per se but increases the probability that the stalled replication forks will be resolved by homologous recombination rather than be stabilized and restarted. As such, our findings are consistent with the idea that the chromatin structures formed by Asf1p stabilize stalled replication forks, facilitating their restarting.
The importance of proper chromatin modulation during DNA replication has been demonstrated in human cells (64). Overexpression of a dominant-negative form of the large subunit (p150) of the chromatin assembly factor CAF-1 slowed S-phase progression, induced double-strand breaks, and activated components of the DNA damage checkpoint that sensed both DNA replication problems and DNA damage (64). However, depletion of CAF-1 p150 by siRNA only activated the DNA damage checkpoint components that respond to DNA replication problems and not to DNA damage (21). It is possible that the dominant-negative version of CAF-1 p150 in the earlier study was acting by sequestering hAsf1 via its interaction with the p60 subunit of CAF-1 (38, 60). As such, the DNA damage and the activation of the DNA damage checkpoint that result from overexpressing the dominant-negative CAF-1 (64) may be a consequence of disrupting the function of Asf1 in humans. We are investigating this possibility and the role of Asf1 in genome stability in human cells.
In summary, Asf1p is required for the formation of proper chromatin structures that are likely to be important for the processing of stalled replication forks. The absence of Asf1p leads to a greater requirement for resolution of stalled replication forks by homologous recombination, providing opportunities for inaccurate repair. As such, proper chromatin structures mediated by Asf1p have a novel and important role in maintaining genomic integrity.1a
Acknowledgments
We thank Paul Megee for critical reading of the manuscript. We are very grateful to Jim Haber, Rohinton Kamakaka, Paul Megee, Jocelyn Krebs, Michael Fasullo, and Bob Sclafani for yeast strains; David Toczyski for the Ddc2p-GFP plasmid; Noel Lowndes for antibodies to Rad53 and Rad9; and Steven Jackson and Jocelyn Krebs for antibodies to phosphorylated H2A. We thank Miguel Ferreria for stimulating discussions and help and advice with pulsed-field gel electrophoresis and Michelle Pham, Josh Carson, and Jack Milwid for technical assistance. We are particularly grateful to the University of Colorado Cancer Center Flow Cytometry Facility for flow cytometry analyses.
This study was supported by an NIH award CA95641-01 to J.K.T. J.K.T. is a scholar of the Leukemia and Lymphoma Society. C.J.R. was supported by a predoctoral training grant in molecular biology NIH T32GM 08730.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 1a.Adkins, M. W., and J. K. Tyler. The histone chaperone Asf1p mediates global chromatin dissembly in vivo. J. Biol. Chem., in press. [DOI] [PubMed]
- 2.Aguilera, A., S. Chavez, and F. Malagon. 2000. Mitotic recombination in yeast: elements controlling its incidence. Yeast 16:731-754. [DOI] [PubMed] [Google Scholar]
- 3.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 4.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 6.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 7.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 8.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, Z., and M. Fasullo. 2003. Multiple recombination pathways for sister chromatid exchange in Saccharomyces cerevisiae: role of RAD1 and the RAD52 epistasis group genes. Nucleic Acids Res. 31:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 11.Emili, A., D. M. Schieltz, J. R. Yates III, and L. H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7:13-20. [DOI] [PubMed] [Google Scholar]
- 12.Fasullo, M., J. Koudelik, P. AhChing, P. Giallanza, and C. Cera. 1999. Radiosensitive and mitotic recombination phenotypes of the Saccharomyces cerevisiae dun1 mutant defective in DNA damage-inducible gene expression. Genetics 152:909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuertesa, M. A., J. Castillab, C. Alonsoa, and J. M. Perez. 2003. Cisplatin biochemical mechanism of action: from cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Curr. Med. Chem. 10:257-266. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 15.Gontijo, A. M., C. M. Green, and G. Almouzni. 2003. Repairing DNA damage in chromatin. Biochimie 85:1133-1147. [DOI] [PubMed] [Google Scholar]
- 16.Grenson, M., M. Mousset, J. M. Wiame, and J. Bechet. 1966. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. I. Evidence for a specific arginine-transporting system. Biochim. Biophys. Acta 127:325-338. [DOI] [PubMed] [Google Scholar]
- 17.Gunjan, A., and A. Verreault. 2003. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in Saccharomyces cerevisiae. Cell 115:537-549. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie, C. F., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press, New York, N.Y..
- 19.Haber, J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 20.Han, M., M. Chang, U. J. Kim, and M. Grunstein. 1987. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48:589-597. [DOI] [PubMed] [Google Scholar]
- 21.Hoek, M., and B. Stillman. 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100:12183-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 23.Hryciw, T., M. Tang, T. Fontanie, and W. Xiao. 2002. MMS1 protects against replication-dependent DNA damage in Saccharomyces cerevisiae. Mol. Genet. Genomics 266:848-857. [DOI] [PubMed] [Google Scholar]
- 24.Hu, F., A. A. Alcasabas, and S. J. Elledge. 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078-1083. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman, P. D., R. Kobayashi, and B. Stillman. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11:345-357. [DOI] [PubMed] [Google Scholar]
- 27.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 28.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029-1042. [DOI] [PubMed] [Google Scholar]
- 29.Lea, D. E., and C. A. Coulson. 1948. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264-268. [DOI] [PubMed] [Google Scholar]
- 30.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 31.Lisby, M., A. Antunez de Mayolo, U. H. Mortensen, and R. Rothstein. 2003. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle 2:479-483. [PubMed] [Google Scholar]
- 32.Lisby, M., U. H. Mortensen, and R. Rothstein. 2003. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 5:572-577. [DOI] [PubMed] [Google Scholar]
- 33.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 34.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 35.Marsischky, G. T., N. Filosi, M. F. Kane, and R. Kolodner. 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10:407-420. [DOI] [PubMed] [Google Scholar]
- 36.McNairn, A. J., and D. M. Gilbert. 2003. Epigenomic replication: linking epigenetics to DNA replication. Bioessays 25:647-656. [DOI] [PubMed] [Google Scholar]
- 37.Megee, P. C., and D. Koshland. 1999. A functional assay for centromere-associated sister chromatid cohesion. Science 285:254-257. [DOI] [PubMed] [Google Scholar]
- 38.Mello, J. A., H. H. Sillje, D. M. Roche, D. B. Kirschner, E. A. Nigg, and G. Almouzni. 2002. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modesti, M., and R. Kanaar. 2001. DNA repair: spot(light)s on chromatin. Curr. Biol. 11:R229-R232. [DOI] [PubMed] [Google Scholar]
- 41.Myung, K., V. Pennaneach, E. S. Kats, and R. D. Kolodner. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc. Natl. Acad. Sci. USA 100:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 43.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasero, P., K. Shimada, and B. P. Duncker. 2003. Multiple roles of replication forks in S phase checkpoints: sensors, effectors and targets. Cell Cycle 2:568-572. [PubMed] [Google Scholar]
- 45.Prado, F., F. Cortes-Ledesma, and A. Aguilera. 2004. The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep. 5:497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin, S., and M. R. Parthun. 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 22:8353-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roth, D. B., and S. Y. Roth. 2000. Unequal access: regulating V(D)J. recombination through chromatin remodeling. Cell 103:699-702. [DOI] [PubMed] [Google Scholar]
- 48.Rouse, J., and S. P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19:5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz, J. L. 1989. Monofunctional alkylating agent-induced S-phase-dependent DNA damage. Mutat. Res. 216:111-118. [DOI] [PubMed] [Google Scholar]
- 50.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in Saccharomyces cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, S., and B. Stillman. 1989. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58:15-25. [DOI] [PubMed] [Google Scholar]
- 52.Stone, E. M., and L. Pillus. 1996. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 135:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugawara, N., E. L. Ivanov, J. Fishman-Lobell, B. L. Ray, X. Wu, and J. E. Haber. 1995. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373:84-86. [DOI] [PubMed] [Google Scholar]
- 54.Sutton, A., J. Bucaria, M. A. Osley, and R. Sternglanz. 2001. Yeast asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 56.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 57.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303:808-813. [DOI] [PubMed] [Google Scholar]
- 58.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 59.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 60.Tyler, J. K., K. A. Collins, J. Prasad-Sinha, E. Amiott, M. Bulger, P. J. Harte, R. Kobayashi, and J. T. Kadonaga. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Biol. Cell 21:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogelauer, M., L. Rubbi, I. Lucas, B. J. Brewer, and M. Grunstein. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10:1223-1233. [DOI] [PubMed] [Google Scholar]
- 62.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 63.Wu, X., J. K. Moore, and J. E. Haber. 1996. Mechanism of MAT alpha donor preference during mating-type switching of Saccharomyces cerevisiae. Mol. Cell. Biol. 16:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye, X., A. A. Franco, H. Santos, D. M. Nelson, P. D. Kaufman, and P. D. Adams. 2003. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11:341-351. [DOI] [PubMed] [Google Scholar]
- 65.Zegerman, P., and J. F. Diffley. 2003. Lessons in how to hold a fork. Nat. Struct. Biol. 10:778-779. [DOI] [PubMed] [Google Scholar]
- 66.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 68.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]
- 69.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]