Abstract
The MLL gene is a frequent target for leukemia-associated chromosomal translocations that generate dominant-acting chimeric oncoproteins. These invariably contain the amino-terminal 1,400 residues of MLL fused with one of a variety of over 30 distinct nuclear or cytoplasmic partner proteins. Despite the consistent inclusion of the MLL amino-terminal region in leukemia oncoproteins, little is known regarding its molecular contributions to MLL-dependent oncogenesis. Using high-resolution mutagenesis, we identified three MLL domains that are essential for in vitro myeloid transformation via mechanisms that do not compromise subnuclear localization. These include the CXXC/Basic domain and two novel domains of unknown function. Point mutations in the CXXC domain that eliminate myeloid transformation by an MLL fusion protein also abolished recognition and binding of nonmethylated CpG DNA sites in vitro and transactivation in vivo. Our results define a critical role for the CXXC DNA binding domain in MLL-associated oncogenesis, most likely via epigenetic recognition of CpG DNA sites within the regulatory elements of target genes.
A distinctive subset of transcriptional regulators exerts its functions via epigenetic modifications of chromatin surrounding the regulatory elements of target genes. These are typified by the trithorax and polycomb groups of chromatin-associated proteins, which function antagonistically as upstream transcriptional regulators of the homeotic genes in Drosophila spp. and mammals (31). The MLL protein (10, 15, 37) is a mammalian orthologue of trithorax and is required for embryonic development via the maintenance but not initiation of expression of target genes such as the Hox genes (2, 38, 41, 42). MLL exists in a multicomponent complex and mediates its epigenetic transcriptional effector functions via a SET domain-dependent histone methyltransferase activity (23, 24). MLL specifically methylates lysine 4 (K4) present on the N-terminal tail of histone H3, a modification typically associated with transcriptionally active regions of chromatin (36).
The MLL gene is a frequent target for chromosomal translocations associated with aggressive human acute leukemias, generating novel chimeric genes between MLL and 1 of over 30 distinct partner genes (3). These genetic events lead to the production of dominant-acting oncogenic fusion proteins that invariably consist of the N-terminal 1,400 residues of MLL fused to variable portions of the partner protein. The results of structure-function studies of a subset of MLL fusion proteins suggest a common theme whereby transcriptional effector domains of the partner proteins make critical contributions to the oncogenicity of MLL fusion proteins (8, 9, 18, 22, 32-34, 43). The consistent inclusion of the N-terminal 1,400 residues in all described MLL fusion proteins suggests that it contributes critical common functions to leukemogenesis, although the mechanistic basis for such functions remains poorly defined. Similarly, while much progress has been made in defining the epigenetic transcriptional effector functions of MLL, the mechanism by which either wild-type or oncogenic mutants of MLL specifically recognize and selectively discriminate chromatin surrounding target genes remains unknown. Recently a Fugu MLL homologue has been described that exhibits extensive sequence homology throughout its length, including the N-terminal region of MLL retained in its oncogenic fusion protein derivatives (7). The identification of Fugu MLL allowed the putative assignment of numerous novel conserved functional domains within the N terminus of MLL that have the potential to contribute to MLL-associated oncogenesis.
In this report, we undertook extensive high-resolution mutagenesis mapping to delineate domains throughout the N terminus that are essential for MLL-associated transformation. We identified three regions of MLL that are essential for transformation, two of which have no known function but do not compromise nuclear localization. The third required region is the CXXC/basic domain, whose contributions to myeloid transformation correlate with its ability to mediate binding to nonmethylated CpG DNA sites in vitro and transcriptional activation in vivo. We conclude that the CXXC/basic domain within MLL fusion proteins may facilitate essential recognition of regulatory elements within MLL target genes in vivo in a methylation-sensitive manner.
MATERIALS AND METHODS
Plasmids.
MLL deletion and site-directed mutants (Table 1) were created by PCR with PfuTurbo DNA polymerase (Stratagene). PCR products flanked by convenient restriction sites were subcloned into either an SP72 vector containing a EcoRI-BglII cDNA fragment encoding a FLAG epitope at its 5′ end followed by MLL nucleotides (nt) 1 to 2249 or a pBluescript vector containing an AccI-BamHI cDNA fragment encoding MLL nt 1334 to 3754. All junctions were sequenced, and the mutant inserts were cloned in-frame into the MSCV FLAG MLL-ENL (Delta SB) vector, encoding a FLAG-tagged MLL-ENL fusion cDNA (FlagME) in which vector SalI and BamHI sites had been destroyed. For expression in COS-7 cells, FlagME and deletion mutants were excised from the MSCV vector by use of EcoRI-XhoI and ligated into the EcoRI-SalI sites of the expression plasmid pCMV5. For bacterial expression, polyhistidine-tagged MLL CXXC mutants (nt 3406 to 3754) were cloned in-frame into the NcoI-HindIII sites of the prokaryotic expression plasmid pRSETB.
TABLE 1.
Mutants of MLL-ENL employed for these studies
| Designation(s) | Deleted amino acids | Description |
|---|---|---|
| ΔN1 | 35-289 | Large N-terminal deletion |
| ΔN2 | 35-170 | SAG+NCS deletion |
| AT+NTS | 172-317 | Contiguous deletion of all AT hooks and NTS1 |
| NTS1 | 242-288 | Deletion of NTS1 |
| AT1-2, -3 | 172-226, 303-306 | Deletion of AT1 through AT2 and AT3 |
| AT1 | 176-179 | Deletes RGRP sequence of AT1 |
| AT1 | 176-179 | Deletes RGRP sequence of AT1 |
| AT2 | 222-225 | Deletes RGRP sequence of AT2 |
| AT3 | 303-306 | Deletes RGRP sequence of AT3 |
| NTS3-SNL2 | 727-1106 | Deletion of NTS3 through SNL2 |
| U1 snRNP-1 | 869-1014 | Large deletion of RERE/U1 snRNP and flanking sequences |
| U1 snRNP-2 | 869-897 | Small deletion of RERE/U1 snRNP |
| SNL1+2 | 400-443, 1008-1106 | Deletion of SNL1 and SNL2 |
| SNL1 | 400-443 | Deletion of SNL1/NTS2/Trx1 homology domain |
| NTS3 | 727-748 | Deletion of NTS3 |
| SNL2 | 1008-1106 | Deletion of SNL2/Trx2 homology domain |
| CXXC/basic | 1153-1250 | Deletion of CXXC/DNA methyltransferase homology domain and flanking basic region |
| CXXC | 1151-1197 | Deletion of CXXC domain |
| Basic | 1200-1250 | Deletion of basic region |
| CC/AA | 1155, 1158 | Substitution of cysteines with alanines at 1155 and 1158 in CXXC domain |
| E1165A | 1165 | Substitution of glutamate with alanine at 1165 in CXXC domain |
| KFGG/AAAA | 1178-1181 | Substitution of KFGG with AAAA in CXXC domain |
Myeloid immortalization assay.
Transduction of murine bone marrow cells and methylcellulose replating assays were performed as previously described (19).
Western blotting.
Cos-7 cells grown in six-well plates were transfected with 2 μg of pCMV5 FlagME or its derivatives by use of Lipofectamine Plus (Invitrogen). At 2 days later, cells were lysed in sodium dodecyl sulfate (SDS) sample buffer, boiled, sheared, and frozen. Dithiothreitol and bromophenol blue were added prior to electrophoresis through SDS-6% polyacrylamide gels. Proteins were electroblotted onto Hybond ECL (Amersham) in Tris-glycine-0.05% SDS-10% methanol for 3 h. Membranes were probed with monoclonal antibody FLAG M2 (Sigma) diluted 1:2,000 followed by a secondary antibody conjugated to horseradish peroxidase (Accurate Antibodies). Bands were visualized using ECL reagent (Amersham).
Immunofluorescence.
Cos-7 cells were plated onto glass slides and transfected with pCMV5 FlagME plasmids as described above. After 24 h, cells were washed three times with phosphate-buffered saline (PBS), permeabilized, and blocked with 0.5% Triton X-100 and 10% normal goat serum in PBS for 30 min. FLAG M5 antibody (10 μg/ml) diluted in PBS-10% goat serum was added to the slides and kept overnight at 4°C in a humidified chamber. Slides were then washed three times with 0.1% Triton in PBS and incubated with anti-mouse immunoglobulin G conjugated to Texas Red (Jackson Laboratories) diluted 1:100 in PBS-10% goat serum. After 90 min of incubation, slides were washed three times and overlain with Vectashield (Vector Laboratories, Burlingame, Calif.).
Reporter assays.
The REH human pro-B cell line was grown to log phase and washed with RPMI medium. Cells were resuspended at a concentration of 3 × 106cells/0.8 ml in RPMI medium plus 5 μg of DEAE-dextran/ml (30) together with 1 μg each of herpes simplex virus (HSV) thymidine kinase (TK) luciferase reporter and various pCMV5-FlagME effector plasmids. Cells were electroporated at 250 V and 960 μF, transferred into 4.2 ml of complete medium, and harvested 2 days later for determination of luciferase activities (Luciferase Assay System; Promega) and protein concentrations (Bio-Rad).
Gel-shift assays.
pRSET CXXC mutants were expressed in BL21(DE3)pLys bacteria. Histidine-tagged proteins were purified using Ni-nitrilotriacetic acid agarose beads (QIAGEN) according to the manufacturer's instructions. Proteins were concentrated in Centricon 3 filters (Amicon, Beverly, Mass.), and concentrations were determined by a Bradford assay. Gel shifts were performed as previously described (6) with double-stranded oligonucleotide probes including probe 1 (GGGCCGTGCTAGTGCGTCGTACCCGCCGA) or the HSV TK promoter-proximal CG box (GGGATGCAGTTCGGGGCGGCGCGGTCCGAGGT).
RESULTS
A structure-function analysis was conducted to determine which regions of MLL are required by the oncogenic fusion protein MLL-ENL to induce the enhanced replating of myeloid progenitors in vitro, a phenotype representative of its transformation properties in primary bone marrow cells (19). In view of the large portion of MLL retained in MLL-ENL (1,400 amino acids [aa]), it was divided into three distinct subdomains (aa 1 to 400, 401 to 1150, and 1151 to 1400) for the purposes of our analyses. Details of the various MLL-ENL deletion mutants are summarized in Table 1.
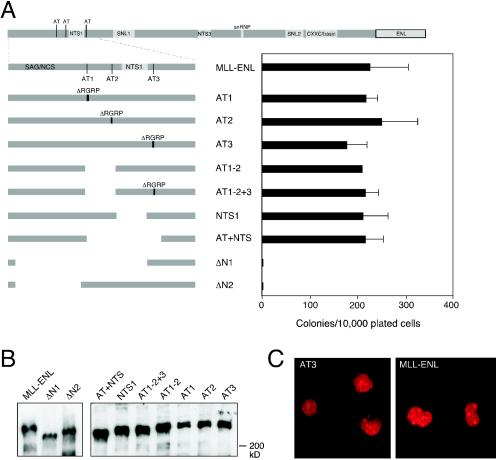
The AT hook DNA binding motifs are dispensable for myeloid transformation by MLL-ENL.
Within its first 310 aa, MLL contains three AT hook DNA binding motifs that are highly conserved with Fugu MLL. A previous study implicated a large amino-terminal region containing these three motifs as being required for myeloid transformation (32). To assess their relative contributions to MLL-associated oncogenesis, small internal deletions of the RGRP core sequence were generated within each AT hook motif. The solution structure of the AT hook predicts that such mutations would abolish association with the minor groove of DNA (17). MLL-ENL constructs lacking single AT hook motifs (constructs AT1, AT2, and AT3) induced efficient myeloid transformation in vitro (Fig. 1A), indicating that each motif is individually dispensable. Therefore, a more extensive set of mutants was constructed that lacked two or more AT hooks. These included mutants with a deletion of AT hooks 1 and 2, with or without additional deletion of the RGRP of AT hook 3 (AT1-2 and AT1-2+3, respectively), and a larger deletion spanning AT hooks 1 to 3 (AT+NTS). All of these MLL-ENL mutants induced enhanced replating of myeloid progenitors (Fig. 1A), indicating that AT hook function was not required for in vitro transformation. The nuclear targeting signal 1 (NTS-1), which resides between AT hooks 2 and 3 (39), was also shown by this analysis to be unnecessary for transformation.
FIG. 1.
Amino-terminal MLL sequences are essential for myeloid transformation by MLL-ENL. At the top of the figure is a scale map of conserved and/or functional MLL domains in the MLL-ENL chimeric oncoprotein. Vertical lines indicate AT hook motifs (AT1, AT2, and AT3). Shaded boxes indicate nuclear translocation sequences (NTS1, NTS2, and NTS3), subnuclear localization domains (SNL1 and SNL2), homology with U1 snRNP (U1 snRNP), and similarity to DNA methyltransferase (CXXC and basic). (A) Murine bone marrow cells enriched for stem-progenitor cells following 5-fluorouracil treatment were transduced with retroviral constructs expressing proteins that are schematically shown on the left side of the panel. The bar graph (right) indicates numbers of colonies with BLAST-like morphology obtained in the third round of serial replating. (B) Detection of MLL-ENL fusion protein expression in Cos-7 cells by Western blot analysis. Faint lower bands represent processed, prematurely terminated, or degraded protein. (C) Representative immunofluorescence analysis of transfected Cos-7 cells shows the typical localization to nuclear speckles manifested by all MLL-ENL proteins analyzed in this mutant series. Transfected proteins were detected with M2 monoclonal antibody that recognizes an amino-terminal FLAG epitope in all constructs.
Conversely, deletion of MLL residues 35 to 289 (construct ΔN1) completely eliminated myeloid transformation. Although AT hooks 1 and 2 were deleted by this mutation, a smaller deletion spanning residues 35 to 170 (construct ΔN2) implicated a novel upstream region as necessary for MLL-ENL transformation activity. This essential region contains two notable subdomains. One consists of 60 residues (aa 43 to 102), which we term the SAG domain due to its high content of serine (32%), alanine (30%), glycine (20%), and proline (13%), which is not conserved with Fugu MLL. It is flanked by a conserved sequence (aa 103 to 171) termed the N-terminal conserved sequence (NCS), which is 48.5% identical to the Fugu MLL. Both the SAG domain and NCS are unique to the MLL protein family, but their sequences offer no clues regarding their contributions to MLL function. Thus, MLL oncogenic activity is AT hook independent but requires novel uncharacterized sequences in the MLL amino terminus.
All mutants of MLL-ENL were efficiently expressed and migrated at their predicted sizes in Western blot analysis following transfection into Cos-7 cells (Fig. 1B), indicating that lack of myeloid transformation by select mutants was not due to lack of expression. All MLL-ENL mutants localized to the nucleus and displayed a speckled subnuclear distribution in transiently transfected Cos-7 cells (Fig. 1C). Notably, the NTS-1 domain, previously identified to be sufficient for targeting a cytoplasmic protein into the nucleus (39), was not required for either nuclear localization or myeloid transformation by MLL-ENL. These results suggest that the ability of amino-terminal mutants of MLL-ENL to eliminate myeloid transformation occurs via a mechanism independent of subnuclear localization.
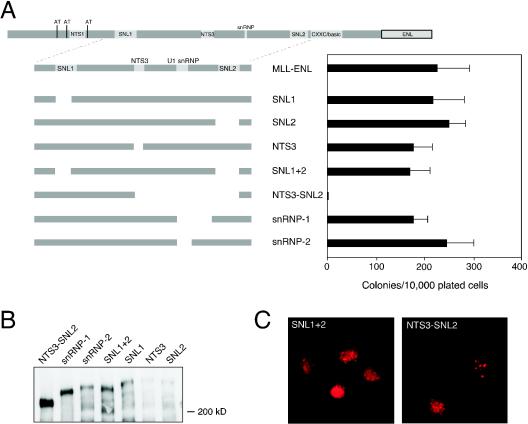
Conserved subnuclear localization motifs are dispensable for nuclear localization and myeloid transformation.
The central region of N-terminal MLL (aa 400 to 1100) contains several domains conserved with Fugu MLL, including nuclear (NTS-3) and subnuclear (SNL1 and SNL2) targeting motifs and a region of similarity with U1 snRNP (11, 39). The short SNL1 and SNL2 motifs are the only sequences conserved between Drosophila trithorax and MLL that are consistently retained by oncogenic MLL fusion proteins. To determine the role of nuclear and/or subnuclear targeting in the oncogenic action of MLL-ENL, mutants were generated that lacked the SNL1, SNL2, or NTS-3 domains and were assayed for their oncogenic potential. All efficiently transformed myeloid progenitors (Fig. 2A), indicating that the targeted domains are individually dispensable. Furthermore, a mutant of MLL-ENL lacking both SNL1 and SNL2 (SNL1+2) also retained full oncogenic activity (Fig. 2A), thus excluding a redundant SNL function in oncogenesis. Similarly, deletion of the U1 snRNP homology domain (constructs snRNP1 and snRNP2) did not eliminate myeloid transformation (Fig. 2A). Thus, no single motif in this region of MLL was individually necessary for in vitro transformation. However, larger deletions spanning multiple motifs (e.g., NTS3 to SNL2) eliminated transformation.
FIG. 2.
Subnuclear localization and nuclear translocation sequences 2 and 3 are dispensable for MLL-ENL-mediated myeloid immortalization. (A) The bar graph (right) indicates numbers of colonies with BLAST-like morphology obtained in the third round of serial replating (averages of triplicate determinations) in methylcellulose cultures following retroviral transduction of the mutant proteins shown on the left. (B) Detection of MLL-ENL fusion protein expression in Cos-7 cells by Western blot analysis. (C) Photomicrographs show typical localization in nuclear speckles for mutants in this series.
When transiently transfected into Cos-7 cells, all deletion mutants in the central region of MLL were appropriately expressed at predicted sizes (Fig. 2B). All deletion mutants also efficiently localized to characteristic MLL subnuclear domains (Fig. 2C), including the NTS3-SNL2 mutant, which lacked oncogenic activity. Thus, a large central region spanning residues 727 to 1106 is essential for MLL-associated oncogenesis via a mechanism independent of subnuclear localization; however, we were unable to define a single conserved motif responsible for this requirement.
The CXXC/basic domain is essential for myeloid transformation.
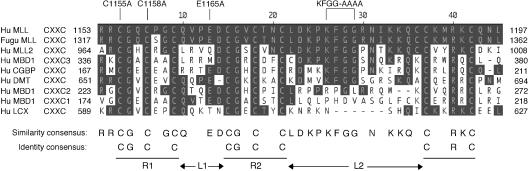
The CXXC DNA binding domain of MLL is highly conserved with Fugu MLL (7) and also shares similarity with domains present in a number of transcriptional regulators, several of which are implicated in modulation of DNA methylation. These can be aligned to reveal numerous conserved residues, which predict two repeats of the CGXCX2C sequence (R1 and R2) flanked by two linker sequences (L1 and L2) and followed by a conserved CX2RXC sequence (Fig. 3). A short basic region is located immediately carboxy terminal to the CXXC domain (Fig. 4A) and exhibits a high lysine content (27% over 56 residues) that is not extensively conserved with Fugu MLL or other mammalian MLL homologues.
FIG. 3.
Alignment of CXXC domains demonstrating highly conserved residues in various proteins. Shading denotes amino acid residues that are identical with human MLL. Site-directed mutations created in MLL-ENL are indicated above the alignment. Identity and similarity consensus sequences are indicated at the bottom of the figure.
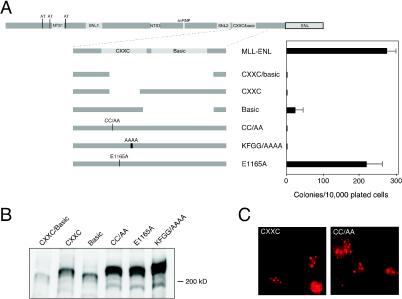
FIG. 4.
The CXXC domain is essential for MLL-ENL transformation. (A) The bar graph (right) indicates numbers of colonies with BLAST-like morphology obtained in the third round of serial replating (averages of triplicate determinations) in methylcellulose cultures following retroviral transduction of the mutant proteins shown on the left. (B) Detection of MLL-ENL fusion protein expression in Cos-7 cells by Western blot analysis. (C) The photomicrographs show typical localization in nuclear speckles for MLL-ENL mutants in this series.
MLL-ENL constructs lacking the CXXC and/or basic domains were examined for their ability to transform myeloid progenitors (Fig. 4A). Lack of the CXXC/basic or the CXXC domain alone completely eliminated in vitro replating potential, indicating that the CXXC domain is essential for MLL-associated transformation. Deletion of the basic domain alone compromised, but did not eliminate, transformation. Clonogenic activity was observed, but colonies were decreased in number (Fig. 4A) and size and were less compact with a more dispersed morphology (data not shown).
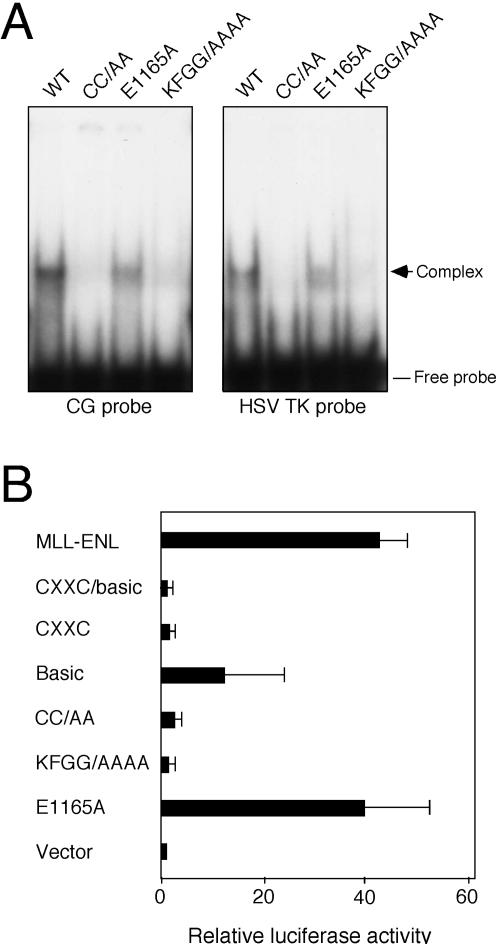
Site-directed missense mutations were introduced into the CXXC domain to investigate its molecular requirement in greater detail. Double alanine substitutions were introduced in conserved cysteine residues C1155/C1158 within the amino-terminal end of CGXCX2C repeat 1 (Fig. 3). Alanine substitutions were also introduced into one of two conserved acidic residues (E1165) in L1 and the central conserved KFGG block (aa 1178 to 1181) positioned within the larger L2 region. Replating assays showed that the CC/AA and KFGG/AAAA site-directed mutants of MLL-ENL were completely defective for myeloid transformation (Fig. 4A). In contrast, the E1165A mutant was indistinguishable from wild-type MLL-ENL in its ability to immortalize myeloid progenitors. When transiently transfected into Cos-7 cells, all CXXC deletion and site-directed mutants were appropriately expressed at their predicted sizes and localized in discrete subnuclear domains (Fig. 4B and C). Thus, highly conserved residues in the R1 and L2 regions are essential for CXXC function and MLL-associated oncogenesis via a mechanism independent of subnuclear localization.
Residues within the CXXC domain that eliminate MLL-associated myeloid transformation are required for binding to nonmethylated CpG DNA and for transactivation.
The CXXC domain of MLL mediates binding to nonmethylated CpG DNA (6). We therefore determined the relationship between mutations within the CXXC domain of MLL that eliminate myeloid transformation and their effect on binding to nonmethylated CpG DNA. Purified, recombinant proteins containing the wild-type or mutant MLL CXXC/basic domains were utilized for gel shift assays. DNA probes contained five CpG sites corresponding to clone 1 derived from the binding site selection (6) or the HSV TK promoter-proximal CG box (Fig. 5A). In agreement with previous studies, the wild-type CXXC/basic domain bound efficiently to both probes. Binding was CXXC dependent and did not require the basic domain (data not shown). For mutants CC/AA and KFGG/AAAA, binding to both CpG probes was completely eliminated. Conversely, the E1165A mutant was unimpaired in its binding ability. Thus, residues within the CXXC domain that eliminate MLL-associated myeloid transformation are required for binding to nonmethylated CpG DNA sites in vitro.
FIG. 5.
Point mutations in the CXXC domain abolish transcriptional activation and DNA binding by MLL-ENL. (A) Gel-shift assay of CXXC mutants. Bacterially expressed proteins (indicated above the gel lanes) were incubated with a radiolabeled CG dinucleotide-containing probe or the proximal CG box of the HSV TK promoter. Protein-DNA complexes (arrow) were electrophoresed on nondenaturing polyacrylamide gels and exposed to film overnight. WT, wild type. (B) MLL-ENL constructs containing mutations in the CXXC domain were transfected into REH cells along with an HSV TK luciferase reporter gene. Relative luciferase values were normalized to protein concentrations. Bars represent means and standard deviations of three experiments.
Several MLL fusion proteins, including MLL-ENL, have been shown to transactivate a variety of promoters, and their transcriptional effector properties are essential for myeloid oncogenesis (8, 9, 30, 32, 33). Therefore, the role of the CXXC/basic domain in transcriptional activation by MLL-ENL was also investigated. MLL-ENL expression plasmids were transiently transfected together with a luciferase reporter construct under the control of the HSV TK promoter into the REH pro-B cell line (Fig. 5B). MLL-ENL strongly activated transcription of the luciferase reporter, whereas mutants lacking either the CXXC/basic domains or the CXXC domain alone were unable to activate transcription above background. MLL-ENL lacking the basic region alone exhibited reduced transactivating properties, suggesting that while the basic domain is not required for binding to CpG sites in vitro, it does contribute to transactivation in vivo. MLL-ENL proteins harboring the CC/AA or KFGG/AAAA mutations were unable to activate transcription. Conversely, the E1165A mutant behaved in an manner identical to that of wild-type MLL-ENL in its ability to activate transcription. These data support a model whereby MLL-associated oncogenesis requires CXXC domain-dependent recognition and binding to nonmethylated CpG sites within the regulatory elements of target genes, leading to their constitutively maintained expression via the transcriptional effector properties of the MLL partner protein.
DISCUSSION
Despite the prevalent involvement of MLL in a variety of mutations in human leukemias, little is known regarding its critical molecular contributions to the transforming properties of MLL oncoproteins. In this study, we have utilized a variety of functional assays to determine the essential regions required for target recognition, transactivation, and myeloid transformation by a representative MLL fusion protein, MLL-ENL. Unexpectedly, most conserved and/or previously defined functional domains were individually dispensable. These include the three AT hook motifs that are highly conserved in vertebrate MLL orthologues and thought to facilitate binding to AT-rich DNA in the minor groove. This property of MLL, therefore, appears to be nonessential for transformation. Nevertheless, previous studies implicated sequences near or overlapping the AT hook motifs as being required for myeloid transformation (32), and our present analysis assigns this function to a novel region flanking the AT hook motifs herein described as the SAG and NCS domains. The SAG sequence is not found in other mammalian MLL homologues or orthologues, while the NCS is well conserved with Fugu MLL, suggesting this region may play an important functional role. The SAG and NCS regions do not exhibit any sequence homology to other proteins, and the mechanism by which the SAG/NCS contributes to MLL-associated oncogenesis is presently unclear.
The central region of MLL (aa 401 to 1150) that is retained in oncogenic fusion proteins contains multiple motifs previously implicated in nuclear and/or subnuclear localization. Our analysis showed that these were dispensable individually or in combination for myeloid transformation. However, a large internal deletion removing multiple motifs (NTS3 to SNL2), including the snRNP homology domain, eliminated myeloid transformation. The NTS3-SNL2 mutant localized correctly to subnuclear domains, suggesting that the central region requirement was independent of nuclear localization. However, we were unable to attribute any single domain to this essential activity and therefore conclude that such large deletions may be sufficient to compromise basic structural conformation of the amino terminus of MLL or that the deleted domains cooperate to provide an essential function in transformation. Furthermore, our present study suggests that the central snRNP homology domain of MLL exhibits a differential requirement for myeloid transformation by MLL-ENL versus a heterologous MLL fusion with an artificial dimerization domain (35). One potential explanation for this functional discrepancy may lie in the mechanisms of action of distinct classes of MLL fusion proteins that possess intrinsic transcriptional effector functions such as MLL-ENL or dimerization domains. Alternatively, select mutants with the particularly weak myeloid-transforming activity of a heterologous MLL fusion with an artificial dimerization domain, such as that lacking the RERE/snRNP homology domain, may exhibit only moderately reduced activity and yet be scored as a complete loss of transforming function.
Our results demonstrate a critical role for the CXXC domain of MLL in myeloid transformation that correlates with its recognition of CpG sites in vitro and transactivation in vivo. Subtle mutations within the CXXC domain that eliminated myeloid transformation highlighted conserved residues that were also required for transactivation and recognition of CpG DNA sites in vitro. On the basis of sequence alignments, the CXXC domain can be subdivided into five components, including two CGXCXXC repeats, each flanked at their carboxy terminus by linker regions of 5 and 14 residues, respectively, followed by a CXXRXC motif. Our mutational analysis revealed essential contributions of conserved residues within one of the CGXCXXC repeats, as well as the KFGG motif within the extended second linker region. Conversely, E1165 within the short first linker region was not required for transactivation, CpG recognition, or myeloid transformation, suggesting that either the charge of this conserved residue or a more general role of the first linker region may not be critical for CXXC function. We note that the two adjacent acidic residues, E1165 and D1166, have the potential to form an acidic pocket within the linker 1 region of the CXXC domain. Thus, mutation of only one of these residues, E1165, may not be sufficient to compromise the charge of the putative acidic pocket. Cysteine residues located within the CGXCXXC repeats may be required for coordination of zinc atoms and correct conformation, as has been reported for the CXXC protein CGBP (21). We also note that the KFGG motif within the second linker region of the CXXC domain has been previously associated with CXXC-dependent recognition of nonmethylated CpG sites. This association is most clearly illustrated by the MBD1 protein, which possesses three copies of the CXXC domain, of which only the CXXC3 domain that contains a KFGG motif within its second linker region is capable of recognizing nonmethylated promoters (14). In contrast, CXXC domains in MBD1 lacking the KFGG motif bind only to methylated promoters. Thus, the presence of the KFGG motif seems to discriminate CXXC domains that recognize nonmethylated CpG sites. Moreover, KFGG motif-dependent myeloid transformation by an MLL oncoprotein underscores the critical importance of an appropriate epigenetic state of target sites to facilitate MLL recruitment and subsequent transcriptional deregulation.
We also identified a role for the flanking basic region in myeloid transformation and transactivation in vivo but not for binding to CpG sites in vitro. The close proximity of the basic domain to the CXXC domain suggests that it may contribute to CXXC function, either by ensuring optimal conformation or by modulating the sequence-specific binding to nucleotides flanking the CpG core site in vivo. Interestingly, analysis of all other presently identified CXXC domain proteins reveals that the flanking basic region is unique to MLL and could therefore provide the means to discriminate MLL versus non-MLL targets in the context of CpG sites. However, recent in vitro binding site selection studies using the CXXC/basic region did not reveal any sequence specificity flanking the CpG site (6). Nevertheless, our transcriptional reporter assays clearly demonstrated a requirement for the basic region for efficient MLL-ENL-mediated transactivation. Furthermore, the possibility that CXXC function is modulated by a flanking sequence is supported by the recent analysis of the CXXC protein CGBP, which requires additional residues immediately carboxy terminal to the CXXC domain for binding to CpG sites in vitro (21).
Recently, a number of related MLL proteins have been described that possess variable numbers of structural domains conserved with MLL and/or TRX. These include MLL2 (13, 16), ALR (26), MLL3 (28), and MLL5 (12). It is presently unclear whether any of these structural MLL homologues exhibit overlapping functions during embryonic development, hematopoiesis, or oncogenesis. However, of the three MLL regions identified here as required for transformation, only the CXXC domain is conserved with another MLL family member, MLL2 (16). Based on the crucial role of the CXXC domain for MLL-associated oncogenesis, we speculate that MLL2 represents the only other candidate MLL family member that may harbor oncogenic potential via CpG site recognition. Future domain swap experiments are required to address functional compensation between the CXXC domains of MLL and MLL2.
The amino-terminal regions of MLL studied here in the context of an MLL fusion protein are also present in two other classes of oncogenic MLL mutants, including partial tandem duplications (PTD) and internal PHD finger 1 deletions (PHD1) (2). Thus, our results are likely to have important mechanistic implications for the functions of most if not all MLL oncoproteins as well as those of normal MLL. We suggest that the CXXC/basic domain may also mediate target recognition by wild-type MLL, with at least some overlap in the spectrum of target genes regulated by wild-type and oncogenic mutants of MLL. Indeed, MLL-associated human and murine leukemias express numerous HOXA cluster genes (1, 3, 27, 40) whose expression is perturbed in MLL-deficient embryos. Furthermore, using a genetic analysis, recent studies have shown that MLL-associated myeloid transformation in vitro and in vivo is dependent on select 5′ Hoxa genes (3), known targets of wild-type MLL during embryogenesis. Future studies may define a similar critical role for the CXXC/basic domain for the oncogenic functions of PTD and PHD1 mutants.
The CXXC/basic domain of MLL epigenetically binds to CpG DNA sites in a methylation-sensitive manner (6). During hematopoietic differentiation, 5′ HOXA gene expression is maintained in stem cell and committed progenitor compartments but is subsequently down-regulated as progenitors differentiate towards mature effector cells of the myeloid lineage (20, 25, 29). The molecular mechanisms that control Hox down-regulation during differentiation are presently unknown. Global patterns of DNA methylation are normally established early during embryonic development and then faithfully replicated throughout the lifespan of the adult (5). Our results suggest that one potential mechanism for Hox gene silencing during normal hematopoietic differentiation may involve promoter CpG methylation, which would functionally prevent CXXC-dependent MLL binding and transcriptional regulation. This concept offers a novel rationale for therapeutic intervention, namely, induction of CpG methylation as a potential means to blocking continued MLL fusion protein occupancy of target gene regulatory elements after DNA replication occurs.
Acknowledgments
We thank Cita Nicolas and Maria Ambrus for excellent technical assistance and Caroline Tudor for graphics support.
E. H. Chen was supported by Public Health Service grant 5T32-CA09151 from the National Cancer Institute. This work was supported by funds from the National Cancer Institute (CA55029) and the Children's Health Initiative.
REFERENCES
- 1.Armstrong, S. A., J. E. Staunton, L. B. Silverman, R. Pieters, M. L. den Boer, M. D. Minden, S. E. Sallan, E. S. Lander, T. R. Golub, and S. J. Korsmeyer. 2002. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 30:41-47. [DOI] [PubMed] [Google Scholar]
- 2.Ayton, P., S. F. Sneddon, D. B. Palmer, I. R. Rosewell, M. J. Owen, B. Young, R. Presley, and V. Subramanian. 2001. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis 30:201-212. [DOI] [PubMed] [Google Scholar]
- 3.Ayton, P. M., and M. L. Cleary. 2001. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 20:5695-5707. [DOI] [PubMed] [Google Scholar]
- 4.Ayton, P. M., and M. L. Cleary. 2003. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 17:2298-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 6.Birke, M., S. Schreiner, M. P. Garcia-Cuellar, K. Mahr, F. Titgemeyer, and R. K. Slany. 2002. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 30:958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldas, C., M. H. Kim, A. MacGregor, D. Cain, S. Aparicio, and L. M. Wiedemann. 1998. Isolation and characterization of a pufferfish MLL (mixed lineage leukemia)-like gene (fMll) reveals evolutionary conservation in vertebrate genes related to Drosophila trithorax. Oncogene 16:3233-3241. [DOI] [PubMed] [Google Scholar]
- 8.DiMartino, J. F., P. M. Ayton, E. H. Chen, C. C. Naftzger, B. D. Young, and M. L. Cleary. 2002. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL-AF10. Blood 99:3780-3785. [DOI] [PubMed] [Google Scholar]
- 9.DiMartino, J. F., T. Miller, P. M. Ayton, T. Landewe, J. L. Hess, M. L. Cleary, and A. Shilatifard. 2000. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood 96:3887-3893. [PubMed] [Google Scholar]
- 10.Djabali, M., L. Selleri, P. Parry, M. Bower, B. D. Young, and G. A. Evans. 1992. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nat. Genet. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 11.Domer, P. H., S. S. Fakharzadeh, C. S. Chen, J. Jockel, L. Johansen, G. A. Silverman, J. H. Kersey, and S. J. Korsmeyer. 1993. Acute mixed-lineage leukemia t(4;11)(q21;q23) generates an MLL-AF4 fusion product. Proc. Natl. Acad. Sci. USA 90:7884-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerling, B. M., J. Bonifas, C. P. Kratz, S. Donovan, B. R. Taylor, E. D. Green, M. M. Le Beau, and K. M. Shannon. 2002. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene 21:4849-4854. [DOI] [PubMed] [Google Scholar]
- 13.FitzGerald, K. T., and M. O. Diaz. 1999. MLL2: a new mammalian member of the trx/MLL family of genes. Genomics 59:187-192. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, N., N. Shimotake, I. Ohki, T. Chiba, H. Saya, M. Shirakawa, and M. Nakao. 2000. Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1. Mol. Cell. Biol. 20:5107-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu, Y., T. Nakamura, H. Alder, R. Prasad, O. Canaani, G. Cimino, C. M. Croce, and E. Canaani. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71:701-708. [DOI] [PubMed] [Google Scholar]
- 16.Huntsman, D. G., S. F. Chin, M. Muleris, S. J. Batley, V. P. Collins, L. M. Wiedemann, S. Aparicio, and C. Caldas. 1999. MLL2, the second human homolog of the Drosophila trithorax gene, maps to 19q13.1 and is amplified in solid tumor cell lines. Oncogene 18:7975-7984. [DOI] [PubMed] [Google Scholar]
- 17.Huth, J. R., C. A. Bewley, M. S. Nissen, J. N. Evans, R. Reeves, A. M. Gronenborn, and G. M. Clore. 1997. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 4:657-665. [DOI] [PubMed] [Google Scholar]
- 18.Lavau, C., C. Du, M. Thirman, and N. Zeleznik-Le. 2000. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO J. 19:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavau, C., S. J. Szilvassy, R. Slany, and M. L. Cleary. 1997. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 16:4226-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, H. J., G. Sauvageau, N. Ahmadi, A. R. Lopez, M. M. LeBeau, M. Link, K. Humphries, and C. Largman. 1995. Stage- and lineage-specific expression of the HOXA10 homeobox gene in normal and leukemic hematopoietic cells. Exp. Hematol. 23:1160-1166. [PubMed] [Google Scholar]
- 21.Lee, J. H., K. S. Voo, and D. G. Skalnik. 2001. Identification and characterization of the DNA binding domain of CpG-binding protein. J. Biol. Chem. 276:44669-44676. [DOI] [PubMed] [Google Scholar]
- 22.Luo, R. T., C. Lavau, C. Du, F. Simone, P. E. Polak, S. Kawamata, and M. J. Thirman. 2001. The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol. Cell. Biol. 21:5678-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10:1107-1117. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 25.Pineault, N., C. D. Helgason, H. J. Lawrence, and R. K. Humphries. 2002. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 30:49-57. [DOI] [PubMed] [Google Scholar]
- 26.Prasad, R., A. B. Zhadanov, Y. Sedkov, F. Bullrich, T. Druck, R. Rallapalli, T. Yano, H. Alder, C. M. Croce, K. Huebner, A. Mazo, and E. Canaani. 1997. Structure and expression pattern of human ALR, a novel gene with strong homology to ALL-1 involved in acute leukemia and to Drosophila trithorax. Oncogene 15:549-560. [DOI] [PubMed] [Google Scholar]
- 27.Rozovskaia, T., E. Feinstein, O. Mor, R. Foa, J. Blechman, T. Nakamura, C. M. Croce, G. Cimino, and E. Canaani. 2001. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4: 11) abnormality. Oncogene 20:874-878. [DOI] [PubMed] [Google Scholar]
- 28.Ruault, M., M. E. Brun, M. Ventura, G. Roizes, and A. De Sario. 2002. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene 284:73-81. [DOI] [PubMed] [Google Scholar]
- 29.Sauvageau, G., P. M. Lansdorp, C. J. Eaves, D. E. Hogge, W. H. Dragowska, D. S. Reid, C. Largman, H. J. Lawrence, and R. K. Humphries. 1994. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc. Natl. Acad. Sci. USA 91:12223-12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiner, S. A., M. P. Garcia-Cuellar, G. H. Fey, and R. K. Slany. 1999. The leukemogenic fusion of MLL with ENL creates a novel transcriptional transactivator. Leukemia 13:1525-1533. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher, A., and T. Magnuson. 1997. Murine Polycomb- and trithorax-group genes regulate homeotic pathways and beyond. Trends Genet. 13:167-170. [PubMed] [Google Scholar]
- 32.Slany, R. K., C. Lavau, and M. L. Cleary. 1998. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol. Cell. Biol. 18:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.So, C. W., and M. L. Cleary. 2002. MLL-AFX requires the transcriptional effector domains of AFX to transform myeloid progenitors and transdominantly interfere with forkhead protein function. Mol. Cell. Biol. 22:6542-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.So, C. W., and M. L. Cleary. 2003. Common mechanism for oncogenic activation of MLL by forkhead family proteins. Blood 101:633-639. [DOI] [PubMed] [Google Scholar]
- 35.So, C. W., M. Lin, P. M. Ayton, E. H. Chen, and M. L. Cleary. 2003. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell 4:99-110. [DOI] [PubMed] [Google Scholar]
- 36.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71:691-700. [DOI] [PubMed] [Google Scholar]
- 38.Yagi, H., K. Deguchi, A. Aono, Y. Tani, T. Kishimoto, and T. Komori. 1998. Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92:108-117. [PubMed] [Google Scholar]
- 39.Yano, T., T. Nakamura, J. Blechman, C. Sorio, C. V. Dang, B. Geiger, and E. Canaani. 1997. Nuclear punctate distribution of ALL-1 is conferred by distinct elements at the N terminus of the protein. Proc. Natl. Acad. Sci. USA 94:7286-7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeoh, E. J., M. E. Ross, S. A. Shurtleff, W. K. Williams, D. Patel, R. Mahfouz, F. G. Behm, S. C. Raimondi, M. V. Relling, A. Patel, C. Cheng, D. Campana, D. Wilkins, X. Zhou, J. Li, H. Liu, C. H. Pui, W. E. Evans, C. Naeve, L. Wong, and J. R. Downing. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 1:133-143. [DOI] [PubMed] [Google Scholar]
- 41.Yu, B. D., R. D. Hanson, J. L. Hess, S. E. Horning, and S. J. Korsmeyer. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 95:10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, B. D., J. L. Hess, S. E. Horning, G. A. Brown, and S. J. Korsmeyer. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505-508. [DOI] [PubMed] [Google Scholar]
- 43.Zeisig, B. B., S. Schreiner, M. P. Garcia-Cuellar, and R. K. Slany. 2003. Transcriptional activation is a key function encoded by MLL fusion partners. Leukemia 17:359-365. [DOI] [PubMed] [Google Scholar]