Abstract
In Rhodobacter capsulatus, genes encoding enzymes of the Calvin-Benson-Bassham reductive pentose phosphate pathway are located in the cbbI and cbbII operons. Each operon contains a divergently transcribed LysR-type transcriptional activator (CbbRI and CbbRII) that regulates the expression of its cognate cbb promoter in response to an as yet unidentified effector molecule(s). Both CbbRI and CbbRII were purified, and the ability of a variety of potential effector molecules to induce changes in their DNA binding properties at their target promoters was assessed. The responses of CbbRI and CbbRII to potential effectors were not identical. In gel mobility shift assays, the affinity of both CbbRI and CbbRII for their target promoters was enhanced in the presence of ribulose-1,5-bisphosphate (RuBP), phosphoenolpyruvate, 3-phosphoglycerate, 2-phosphoglycolate. ATP, 2-phosphoglycerate, and KH2PO4 were found to enhance only CbbRI binding, while fructose-1,6-bisphosphate enhanced the binding of only CbbRII. The DNase I footprint of CbbRI was reduced in the presence of RuBP, while reductions in the CbbRII DNase I footprint were induced by fructose-1,6-bisphosphate, 3-phosphoglycerate, and KH2PO4. The current in vitro results plus recent in vivo studies suggest that CbbR-mediated regulation of cbb transcription is controlled by multiple metabolic signals in R. capsulatus. This control reflects not only intracellular levels of Calvin-Benson-Bassham cycle metabolic intermediates but also the fixed (organic) carbon status and energy charge of the cell.
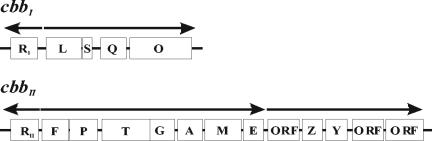
Rhodobacter capsulatus is a nonsulfur purple photosynthetic bacterium that possesses two cbb operons, cbbI and cbbII, encoding enzymes involved in the Calvin-Benson-Bassham (CBB) reductive pentose phosphate pathway of carbon dioxide fixation (24, 25) (Fig. 1). The cbbI operon of R. capsulatus contains cbbL and cbbS, encoding the large and small subunits of form I ribulose-1,5-bisphosphate (RuBP) carboxylase/oxygenase (RubisCO), respectively, as well as two genes, cbbQ and cbbO, of unknown function. Aside from cbbM, which encodes form II RubisCO, the cbbII operon also contains cbbF, encoding fructose-1,6/sedoheptulose-1,7-bisphosphatase; cbbP, encoding phosphoribulokinase; cbbT, encoding transketolase; cbbG, encoding glyceraldehyde 3-phosphate dehydrogenase; cbbA, encoding fructose-1,6-bisphosphate aldolase; cbbM, encoding the large subunit of form II RubisCO; cbbE, encoding ribulose-5-phosphate-3-epimerase; cbbZ, encoding 2-phosphoglycolate phosphatase; and cbbY, encoding an unknown function, as well as three unidentified open reading frames.
FIG. 1.
Gene organization of the R. capsulatus cbbI and cbbII operons. Gene designations: cbbR, positive transcriptional regulator; cbbL, large subunit, form I RubisCO; cbbS, small subunit, form I RubisCO; cbbQ and cbbO, unknown function; cbbF, fructose-1,6/sedoheptulose-1,7-bisphosphatase; cbbP, phosphoribulokinase; cbbT, transketolase; cbbG, glyceraldehyde 3-phosphate dehydrogenase; cbbA, fructose-1,6-bisphosphate aldolase; cbbM, large subunit, form II RubisCO; cbbE, ribulose-5-phosphate epimerase; cbbZ, 2-phosphoglycolate phosphatase; cbbY, unknown function. The direction of transcription and the extent of potential transcripts are indicated by arrows.
Divergently transcribed from the cbbI and cbbII operons are cbbRI and cbbRI, respectively, which encode regulators that positively affect transcription of their cognate operons. R. capsulatus cbbI and cbbII are regulated independently, and their expression levels have been shown to vary depending on the growth conditions (13, 14, 24, 25, 29, 43). The level of cbbI and cbbII expression is maximal under photoautotrophic conditions, when the CBB pathway is used to synthesize organic carbon from CO2 to support growth and maintain the redox balance of the cell (18, 46). During photoheterotrophic growth (i.e., anaerobic growth conditions in the presence of an organic carbon source), cbbII expression is reduced, while cbbI is not expressed. Under aerobic chemoheterotrophic conditions, when CO2 fixation is not needed, the level of cbb expression is lowest. Regulatory cross talk between the two operons also occurs, since inactivation of either of the two cbb operons in R. capsulatus leads to a compensatory increase in the expression of the remaining operon (25).
While the RegB-RegA two-component regulatory system is also involved in derepressing both cbbI and cbbII operon expression under photoautotrophic growth conditions (43), CbbRI and CbbRII for the most part specifically regulate their cognate operons. However, there is some indication that, in the absence of CbbRII, CbbRI may cross regulate cbbII operon expression under photoheterotrophic conditions (43). In addition, partial expression of the cbbII operon occurs under photoautotrophic conditions in the complete absence of CbbRI and CbbRII, suggesting that there may be additional regulators, as recently described for Rhodobacter sphaeroides (10).
The CbbR proteins belong to the LysR family of transcriptional regulators and have been shown to be involved in the regulation of cbb gene expression in many photosynthetic and chemoautotrophic bacteria (15, 25, 38, 41, 47), where they usually bind at the consensus DNA binding motif, T-N11-A, that is often located within 100 bp of the transcription start of the target gene or operon (16). In order to function as a transcriptional regulator, most members of this ubiquitous family of regulators must bind an effector or coinducer molecule that in turn alters the DNA binding properties of the protein (28). This effector/coinducer molecule is often an intermediate or end product of the physiological pathway that is regulated (28).
Differential regulation of the expression of the cbbI and cbbII operons by separate CbbR proteins has not been observed in other organisms. Thus, R. capsulatus is unusual in having two nonidentical CbbR proteins, CbbRI and CbbRII, that show 42.5% amino acid sequence identity, with many of the conserved residues in the putative coinducer/effector binding domain (25). The presence of two distinct yet homologous CbbR regulators with similar effector domains, combined with the observed differential regulation, suggests that the two proteins might respond to the same effector molecules but to different degrees. In phototrophic bacteria, it has long been speculated that the CBB pathway might be controlled by the redox state or the level of some key intracellular molecule(s) (1, 21, 33, 34). In addition, recent genetic and physiological studies point to potential effector molecules that might influence transcription of the cbb operons of both R. capsulatus and R. sphaeroides in vivo (25, 31, 36). With these past studies in mind, the current investigation was undertaken to determine the role of potential effector molecules in influencing the interactions of R. capsulatus CbbRI and CbbRII with the promoter-operator regions of their respective cbb operons in vitro.
MATERIALS AND METHODS
Bacterial strains and plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown aerobically at 37°C in Luria broth (LB) medium unless specified (2). Antibiotic concentrations were 50 μg/ml for kanamycin and 200 μg/ml for ampicillin. R. capsulatus was cultured as previously described (24, 43).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli ER 2566 | F− λ−fhuA2[lon] ompT lacZ::T7 genel gal sulAII Δ(mcrC-mrr)114::IS10 R(mcr-73::miniTn10)2 R(zgb-210::Tn10)1 (TeTS) endA1 [dem] | 6 |
| Plasmids | ||
| pK18 | Kmr, derivative of cloning vector pUC18 | 26 |
| pK19 | Kmr, derivative of cloning vector pUC19 | 26 |
| pTYB1 | Apr, T7 RNA polymerase-based expression vector | New England Biolabs |
| pGEM-7Zf(+/−) | Apr | Promega |
| PCR-ScriptSK(+) | Apr | Stratagene |
| pK18FIIS4.4 | Kmr, pK18 with a 4.4-kb SalI fragment containing R. capsulatus cbbRill, cbbF, and part of cbbP | 24 |
| pK18FIIB2.3 | Kmr, pK18 with a 2.3-kb BamHI fragment containing R. capsulatus cbbF and part of cbbP and cbbRill | 24 |
| pEULA4 | Kmr, pK19 with a 4-kb EcoRI fragment containing R. capsulatus cbbL, cbbRI, part of anfA, and uncharacterized sequence between cbbRI and anfA | 23 |
| pEULPE1.2 | Kmr, pK19 with a 1.2-kb PstI-HindIII fragment containing R. capsulatus cbbRI-cbbI promoter | G. C. Paoli and F. R. Tabita (unpublished data) |
| RIscript6 | Apr, PCR script with an amplified fragment containing cbbRI from the first to the last codons plus NdeI site at the 5′- end and SapI site at the 3′ end | 43 |
| pTYBRI | Apr, pTYB1 with an NdeI-SapI fragment from pRIscript6 | 43 |
| pGEM-7RII | Apr, pGEM-7 with an amplified fragment containing cbbRII from the first to the last codons plus NdeI site at the 5′ -end and the SapI site at the 3′ end | 43 |
| pTYBRII | Apr, pTYB1 with an NdeI-SapI fragment from pGEM-7RII | 43 |
DNA manipulations.
Standard protocols were used for routine DNA techniques such as plasmid preparation, restriction enzyme digestion, DNA ligation, agarose gel electrophoresis, and bacterial transformation (2).
Construction of R. capsulatus cbbRI and cbbRII overexpression plasmids in Escherichia coli.
R. capsulatus cbbR overexpression plasmids were constructed by insertion of an NdeI-SapI fragment containing only the coding portion of either the cbbRI or the cbbRII gene from the translational start site to the last amino acid codon (serine) (excluding the stop codon), into the NdeI and SapI sites of the expression vector pTYB1. The N-terminal NdeI and C-terminal SapI sites were introduced into cbbRI and cbbRII during PCR amplification. The forward primers, complementary to the noncoding strands of cbbRI and cbbRII at the 5′ ends, included an NdeI site plus an extra AAA at the 5′ ends. The nucleotide sequences of the forward primers for cbbRI and cbbRII are 5′-AAACATATGCGTTGCACGCTTCGCCAGTTGC-3′ and 5′-AAACATATGGTCCGGCTGGACGGGATCACG-3′, respectively. The reverse primers, complementary to the coding strands of cbbRI and cbbRII at the 3′ ends, included a SapI site plus an extra AAA at the 5′ ends. The nucleotide sequences of the reverse primers for cbbRI and cbbRII are 5′-AAAGCTCTTCCGCAGCTCGGGCCCGCGCTGCCCGCCGCC-3′ and 5′-AAAGCTCTTCCGCACGACGCGTCAATCATCGGAATCG-3′, respectively.
Plasmids pEULA4 and pK18FIIS4.4 were used as the plasmid templates to amplify the cbbRI and the cbbRII genes, respectively. Amplification was performed at 94°C for 7 min and then at 94°C for 30 s, followed by 3 s at 60°C and 30 s at 72°C for 30 cycles, with a final extension at 72°C for 10 min with Pfu Turbo polymerase (Stratagene, La Jolla, Calif.). The blunt-ended PCR product containing cbbRI was cloned into the PfuI site of PCR-Script SK(+) to yield plasmid pRIscript6. The blunt-ended cbbRII PCR product was cloned into the SmaI site of pGEM-7Zf(+/−) yielding plasmid pGEM-7RII. The NdeI-SapI fragments from pRIscript6 and pGEM-7RII were subsequently inserted into the NdeI and SapI sites of the expression vector pTYB1, yielding plasmids pTYBRI and pTYBRII, respectively. The DNA sequence of each insert was determined with a Thermosequenase II kit (Amersham, Piscataway, N.J.) and an ABI Prism 310 genetic analyzer.
CbbRI and CbbRII purification.
E. coli strain ER2566, containing plasmid pTYBRI or pTYBRII, was grown in LB medium to an optical density at 600 nm of 0.8 at 37°C. The culture was then shifted to 42°C for 30 min in order to increase the proportion of soluble CbbRI and CbbRII since the majority of the recombinant protein was found in inclusion bodies. Cultures were then induced overnight with 1 mM isopropylthiogalactopyranoside (IPTG) at room temperature with low aeration (100 rpm), harvested, and washed with a buffer containing 20 mM HEPES-NaOH (pH 8.0 and 0.1 mM EDTA, and stored at −70°C. Cell pellets were resuspended (4.5 ml/g of wet cell pellet) in column buffer (20 mM HEPES-NaOH, pH 8.0, 1 M NaCl, 0.1 mM EDTA, 0.3% Triton X-100) and sonicated (model 550, Fisher Scientific, Pittsburgh, Pa.). Crude cell extracts were cleared by centrifugation (17,000 × g for 90 min), filtered through a 0.45 μm filter (Gelman Sciences, Ann Arbor, Mich.), and loaded on a 15-ml chitin column (Impact T7 kit, New England Biolabs, Beverly, Mass.) that had been equilibrated with column buffer at 4°C.
On-column cleavage of the intein-chitin domain (6, 7) was accomplished by incubation in column buffer containing 40 mM dithiothreitol without Triton X-100 for 40 h at 4°C as described by the manufacturer. CbbRI and CbbRII were eluted from the column with 45 ml of column buffer. Since cbbRI and cbbRII were cloned into the SapI site of plasmid pTYB1, the eluted proteins contained no additional C-terminal residues (6). The fractions were pooled, concentrated (Ultrafree-15 concentrator, Millipore, Billerica, Mass.), dialyzed against storage buffer (50 mM HEPES, pH 7.8, 200 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 0.05 mM phenylmethylsulfonyl fluoride, 50% glycerol), and stored at −70°C. The protein concentration was determined with a Bradford dye binding assay kit (Bio-Rad, Hercules, Calif.).
DNase I footprint analysis.
A probe for DNase I footprint analyses was selectively labeled at the 5′ end with 32P as previously described (43). The primers for PCR amplification of the probes used to determine the CbbRI binding site were 5′-GCGTCATAGGTCTTGGCG-3′ and 5′-GCAATTCCTCGGCGGCGC-3′. The primers for PCR amplification of the labeled probes used to determine the CbbRII binding sites were 5′-CCGAGACCTTCAAGCTCG-3′ and 5′-CCGCAGTCAGCGAGCCCC-3′ for the top strand and 5′-CCAGCCGGGTCATCACATCC-3′ and 5′-CCGCAGTCAGCGAGCCCC-3′ for the bottom strand. Plasmids pEULPE1.2 and pK18FIIB2.3 were used as templates for PCR amplification of the cbbI and cbbRII promoter probes, respectively. The labeled probes were purified by electroelution from an 8% nondenaturing polyacrylamide gel. Probes were resuspended in a buffer containing 50 mM HEPES (pH 8.0) and 100 mM sodium acetate (pH 7.9).
DNase I footprint assays were performed in a buffer of 50 mM HEPES, pH 8.0, containing 200 mM KCl and 1 mM dithiothreitol, as described previously (3). Metabolites were added to the reaction mix containing CbbR prior to addition of the probe. Equal amounts of each reaction were loaded onto an 8% polyacrylamide-7 M urea gel along with a Maxam and Gilbert chemical cleavage G+A ladder generated from the same labeled probe as described elsewhere (2). The gel was dried onto 3MM Whatman paper and exposed to either X-ray film or a phosphoscreen for visualization with a Molecular Dynamics Storm 840 imaging system (Molecular Dynamics, Sunnyvale, Calif.).
Gel mobility shift assays.
The 32P-labeled probes used in gel mobility shift assays were generated by PCR amplification of fragments containing the cbbI and cbbII promoters. The sets of oligonucleotide primers, templates, and reaction conditions used to amplify the fragments were the same as in the DNase I footprinting experiments.
Gel mobility shift assays were performed as previously described (9). Binding reactions (50 μl total volume) were comprised of 0.08 nmol of CbbRI or 0.71 nmol of CbbRII, radiolabeled DNA fragment (15,000 to 30,000 cpm), and 1.5 μg of dI/dC in a buffer of 10 mM Tris, pH 8.5, containing 300 mM potassium glutamate, 1 mM dithiothreitol, and 30% glycerol. CbbR was incubated in the presence of competitor poly(dI::dC)-poly(dI::dC) DNA for 5 min at room temperature prior to addition of the radiolabeled probe. The reaction was incubated for 20 min after the addition of the probe. Metabolites were added before the addition of CbbR. The pH of the gel mobility shift reaction mixtures, with or without added metabolites, remained at approximately pH 8.0. Samples were separated with a Tris-glycine gel system as described previously (2, 9). Gels were dried onto 3MM Whatman paper, exposed on a phosphoscreen, and visualized with a Storm 840 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Overexpression of cbbRI and cbbRII and purification of recombinant R. capsulatus CbbRI and CbbRII from extracts of E. coli.
R. capsulatus CbbRI and CbbRII overexpression plasmids contained either cbbRI or cbbRII translationally fused to the intein-chitin binding domain at the carboxy terminus.
Purification on a chitin column and excision of the intein through intraprotein self-splicing in the presence of thiols yielded CbbRI and CbbRII proteins that did not contain extraneous C-terminal amino acids (6, 7). Purified CbbRII and CbbRI had predicted molecular masses of approximately 34 and 32 kDa, respectively, (25). A single minor contaminating protein of approximately 64 kDa was detected in both preparations (data not shown). This contaminant was likely to be GroEL because this protein often copurifies with misfolded proteins in the E. coli host (27). R. capsulatus CbbRI and CbbRII were not insoluble in low-salt buffers as are a number of other LysR family regulatory proteins (4, 9, 20) and were used directly in DNA binding experiments.
Effect of metabolites on CbbRI and CbbRII binding in gel mobility shift assays.
Most (but not all) LysR family regulators require a coinducer or effector molecule to mediate or influence gene expression at target sequences (28). This coinducer is often a specific metabolite or product of an enzyme whose gene is regulated by the LysR family regulator in question. In several cases, binding of the coinducer molecule causes a change in the DNA binding characteristics of the LysR family regulator protein to its target promoter (5, 12). With this in mind, CbbRI and CbbRII were used to determine the ability of phosphorylated CBB pathway and other intermediates to influence DNA binding as measured by DNase I footprint and gel mobility shift assays.
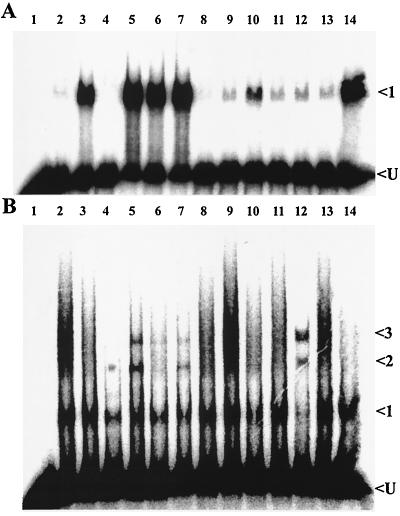
The results of gel mobility shift assays testing the ability of potential effector molecules to alter the DNA binding properties of CbbRI and CbbRII are shown in Fig. 2 and 3. The amounts of CbbRI and CbbRII used in the experiments were adjusted so that a minimum of binding was detected in positive control reactions containing no metabolites. This allowed easier detection of enhanced DNA binding. The binding of CbbRI to its cognate promoter was visibly enhanced in the presence of 2-phosphoglycerate, 2-phosphoglycolate, 3-phosphoglycerate, phosphoenolpyruvate, KH2PO4 and, to a lesser degree, ATP (Fig. 2A, lanes 3, 5, 6, 7, 14, and 10, respectively). This enhanced binding was manifested by a large increase in the intensity of a single protein-DNA complex that is normally observed in the absence of effectors (42) (Fig. 2A, complex 1).
FIG. 2.
Binding of CbbRI and CbbRII to their cognate promoters in the presence of various metabolites. A phosphorimage of a gel mobility shift assay is shown with CbbRI (0.08 nmol) and a cbbI probe (A) and CbbRII (0.71 nmol) and a cbbII probe (B). Lane 1, probe only; lane 2, CbbR and probe with no metabolite added; lane 3, 2-phosphoglycerate; lane 4, RuBP; lane 5, 2-phosphoglycolate; lane 6, 3-phosphoglycerate; lane 7, phosphoenolpyruvate; lane 8, NADPH; lane 9, NADH; lane 10, ATP; lane 11, fructose-6-phosphate; lane 12, fructose-1,6-bisphosphate; lane 13, ribose-5- phosphate; lane 14, KH2PO4. All metabolites were present at a concentration of 1 mM. All reactions contained 3.7 μg of poly(dI-dC)::poly(dI-dC) and 15,000 cpm of 32P-labeled probe. Arrows indicate unbound probe (U) and shifted protein-DNA complexes.
FIG. 3.
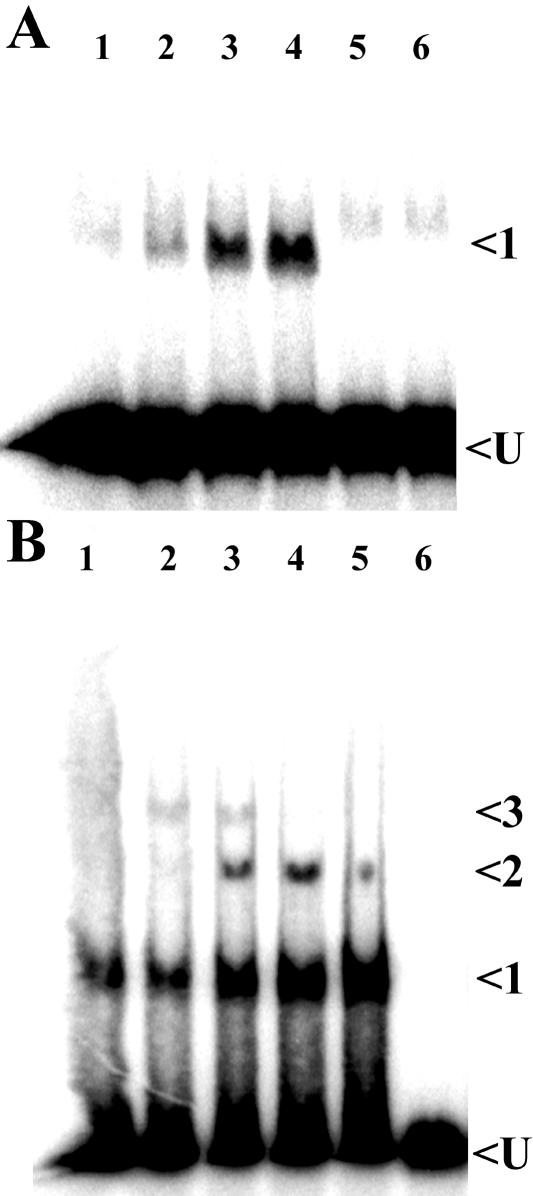
Concentration dependence of RuBP on the binding of CbbRI and CbbRII to their cognate promoters. Phosphorimage of gel mobility shift assays are shown with CbbRI (0.08 nmol) and a cbbI probe (A) and CbbRII (0.71 nmol) and a cbbII probe (B). Lane 1, CbbR with no metabolite added; lane 2, 1 μM RuBP; lane 3, 10 μM RuBP; lane 4, 100 μM RuBP; lane 5, 1.0 mM RuBP; lane 6, probe only. All reactions contained 3.7 μg of poly(dI-dC)::poly(dI-dC) and 15,000 cpm of 32P-labeled probe. Arrows indicate unbound probe (U) and shifted protein-DNA complexes.
It was somewhat surprising that the addition of 1 mM RuBP to a CbbRI binding reaction appeared to inhibit CbbRI DNA binding (Fig. 2A, lane 4 compared with lane 2). This was confirmed in further gel mobility shift experiments with a range of RuBP concentrations (Fig. 3A, lane 5). However, these experiments also showed that RuBP at concentrations of less than 1 mM stimulated CbbRI DNA binding (Fig. 3A, lanes 1 to 4). No effect on CbbRI DNA binding was observed in the presence of either NADPH (Fig. 2A, lane 8), a molecule that had been shown previously to alter the DNA binding of CbbRs from two other bacteria (35, 38); NADH (Fig. 2A, lane 9); fructose-6-phosphate (Fig. 2A, lane 11); fructose-1,6-bisphosphate (Fig. 2A, lane 12); or several concentrations of ribose-5-phosphate (Fig. 2A, lane 13, and data not shown).
The binding of CbbRII to its cognate promoter was enhanced in the presence of RuBP, 2-phosphoglycolate, 3-phosphoglycerate, phosphoenolpyruvate, and fructose-1,6-bisphosphate (Fig. 2B, lanes 4, 5, 6, 7, and 12, respectively). Addition of these metabolites to CbbRII binding reactions resulted in the appearance and/or change in signal intensity of one or more of three protein DNA complexes (Fig. 2B, complexes 1, 2, and 3). While complex 1 is normally observed in the absence of effectors (Fig. 2B, lane 2), complexes 2 and 3 presumably represent either higher orders of CbbR oligomerization or various degrees of CbbRII-induced DNA bending. CbbRII DNA binding was not appreciably affected by the presence of either 2-phosphoglycerate (Fig. 2B, lane 3), NADPH (Fig. 2B, lane 8), NADH (Fig. 2B, lane 9), ATP (Fig. 2B, lane 10), fructose-6-phosphate (Fig. 2B, lane 11), ribose-5-phosphate (Fig. 2B, lane 13, and results not shown), or KH2PO4 (Fig. 2B, lane 14).
Additional gel mobility shift assays examined the effect of a range of different concentrations of RuBP to influence the ability of CbbRII to bind to target DNA sequences; the results showed that low concentrations of RuBP (1 and 10 μM) in a CbbRII binding reaction stimulated the formation of protein-DNA complexes 1, 2, and 3 (Fig. 3B, lanes 2 and 3). Incremental increases in the RuBP concentration above 10 μM resulted in the disappearance of complex 3 along with increases in the intensity of complexes 1 and 2 (Fig. 3B, lanes 3, 4, and 5). In the case of both CbbRI and CbbRII, the degree to which the metabolites other than RuBP influenced DNA binding in gel mobility shift assays was also concentration dependent (in all cases 1 mM metabolite was stimulatory) and resulted in similar changes in the observed banding patterns (data not shown). The amount of CbbRII added to binding reactions was nearly ninefold greater than that of CbbRI due to reduced binding activity of the CbbRII sample. It was assumed that this was due to either different DNA affinities of the two proteins or differences in the activities of the two protein samples (43).
Effect of metabolites on CbbRI and CbbRII binding in DNase I protection assays.
It has been shown previously that CbbRs generally protect the cbb promoter-operator region within −76 bp to −10 bp relative to the cbb transcription start site in the absence of an effector molecule (8, 14, 19, 40, 43). DNA binding studies on a number of LysR family regulators have shown that the presence of a coinducer/effector molecule can result in changes in the size of the DNase I-protected region (22, 32, 44). We tested the effect of glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, sedoheptulose-7-phosphate, xylulose-5-phosphate, ribose-5-phosphate, ribulose-5-phosphate, RuBP, erythrose-4-phosphate, 6-phosphogluconate, acetyl-coenzyme A, ribose, phosphoenolpyruvate, 3-phosphoglycerate, 2-phosphoglycolate, ATP, ADP, AMP, cyclic AMP, NADPH, NADH, K2HPO4, ribulose-5-phosphate plus ADP (1 mM), and ATP plus ADP (1 mM) on the binding of CbbRI and CbbRII to their cognate promoters in DNase I protection assays.
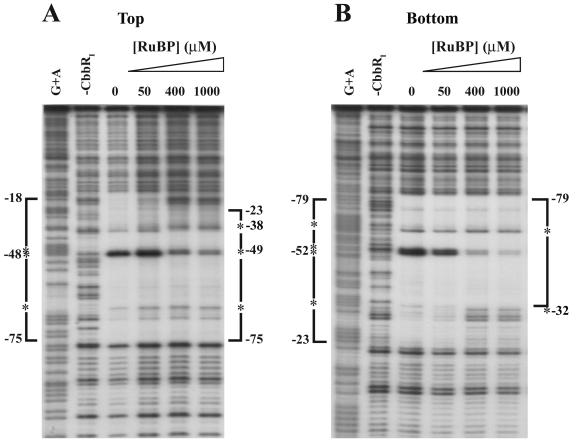
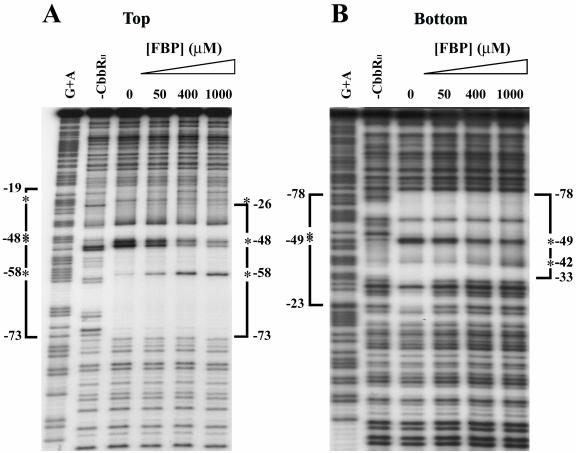
It was found that the region of the cbbI promoter protected from DNase I digestion by the binding of CbbRI was reduced upon the addition of 1 mM RuBP to the binding reaction (Fig. 4A and B). In the presence of 1 mM RuBP, the CbbRI-protected region on the bottom strand was reduced from nucleotides −79 to −23 to nucleotides −79 to −33 (Fig. 4B). The appearance of an additional hypersensitive site at nucleotide −32 was observed along with the disappearance of the strong hypersensitive sites at nucleotides −53 and −52 (Fig. 4B). On the top strand the extent of the CbbRI-protected region was also reduced from nucleotides −75 to −18 to nucleotides −75 to −23, and the intensity of the hypersensitive sites at nucleotides −48 and −47 was drastically reduced, while a new hypersensitive site appeared at −38 and the intensity of the hypersensitive site at −67 was increased (Fig. 4A). This shortening of the CbbRI-protected region was dependent on the concentration of RuBP (Fig. 4A and 4B); however, the addition of RuBP did not enhance the binding affinity of CbbRI to the cbbI promoter (data not shown).
FIG. 4.
Effect of RuBP on the DNase I-protected region of CbbRI binding to the cbbI promoter. The phosphorimage of a DNase I footprint is shown. The probe fragment used spans nucleotides −156 to +58 relative to the cbbL transcription start and is labeled on the top (A) and bottom (B) strands. Brackets indicate regions of protection, and asterisks indicate DNase I-hypersensitive sites. A control lane to which no CbbRI was added to the binding reaction is shown along with a lane containing a standard Maxim-Gilbert A+G sequencing ladder of the probe. The concentration of RuBP present in each reaction is indicated. Unless otherwise indicated, all reactions contained 88 nM CbbRI.
A similar shrinkage in the CbbRII-protected region was also observed in the presence of 1 mM fructose-1,6-bisphosphate. The CbbRII-protected region decreased from nucleotides −73 to −19 to nucleotides −73 to −27 on the top strand and from nucleotides −78 to −23 to nucleotides −78 to nucleotides −33 on the bottom strand (Fig. 5B). The intensity of the hypersensitive sites at nucleotides −48 to −45 of the top strand was significantly reduced, while the intensity of the hypersensitive site at −58 was increased. On the bottom strand, a reduction in the intensity of the strong hypersensitive sites at nucleotides −51 to −49 was observed, with an appearance of an additional hypersensitive site at −42 (Fig. 5A).
FIG. 5.
Effect of fructose-1,6-bisphosphate on the DNase I-protected region caused by CbbRII binding to the cbbII promoter. The phosphorimage of a DNase I footprint is shown. The probe fragment used spans nucleotides −151 to +46 relative to the cbbF transcription start for the top strand (A) and −151 to +79 on the bottom strand (B) is labeled on the top and bottom strands. Brackets indicate regions of protection, and asterisks indicate DNase I-hypersensitive sites. A control lane that does not contain CbbRII is shown along with a lane containing a standard Maxim-Gilbert A+G sequencing ladder of the probe. The concentration of fructose-1,6-bisphosphate present in each reaction is indicated. Unless otherwise indicated, all reactions contained 3.7 μM CbbRII.
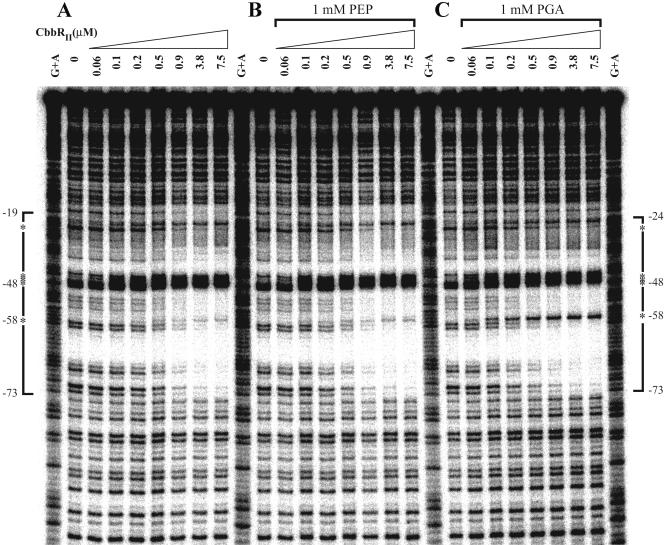
These changes in the CbbRII DNase I-protected regions were dependent on the concentration of fructose-1,6-bisphosphate in the binding reactions (Fig. 5A and B). Moreover, the presence of 1 mM fructose-1,6-bisphosphate enhanced the binding affinity of CbbRII for the cbbII promoter, since the concentration of CbbRII required to protect the cbbII promoter region at nucleotides −73 on the top strand and −78 on the bottom strand was reduced by approximately fivefold in the presence of 1 mM fructose-1,6-bisphosphate (data not shown). The CbbRII-protected region was also reduced in the presence of KH2PO4; the addition of 1 mM KH2PO4 to the binding reaction caused a change in the CbbRII protection pattern similar to that seen in the presence of 1 mM fructose-1,6-bisphosphate (data not shown). The presence of 1 mM 3-phosphoglycerate in the binding reaction reduced the CbbRII-protected region in the cbbII promoter from nucleotides −73 to −19 to nucleotides −73 to −24 on the top strand and induced the appearance of a hypersensitive site at nucleotide −58 without any change in the intensity of the strong hypersensitive sites at −48 to −45 (Fig. 6C). A lesser effect on the CbbRII-protected region was observed in the presence of 1 mM RuBP, which caused an increase in the intensity of the hypersensitive site at −58 (data not shown). The presence of 1 mM phosphoenolpyruvate did not affect the CbbRII binding pattern (Fig. 6B), even though this metabolite affected CbbRII binding in gel mobility shift assays (Fig. 2B, lane 7).
FIG. 6.
Effect of 3-phosphoglycerate and phosphoenolpyruvate on the pattern of DNase I protection caused by CbbRII binding to the cbbII promoter. The phosphorimage of a DNase I footprint of CbbRI binding to a cbbII promoter probe fragment alone (A) and in the presence of 1 mM phosphoenolpyruvate (PEP) (B) or 1 mM 3-phosphoglycerate (PGA) (C). The probe is labeled on the top strand and spans nucleotides −151 to +46 relative to the cbbF transcription start. Brackets indicate regions of protection, and asterisks indicate DNase I hypersensitive sites. The concentration of CbbRII in each reaction is indicated above each lane. A standard lane containing a Maxim-Gilbert A+G sequencing ladder of the probe is also shown.
DISCUSSION
The results presented in this study identified a number of molecules that have the ability to alter the in vitro DNA binding properties of one or both of the CbbR proteins of R. capsulatus. The binding of both CbbRI and CbbRII to their cognate promoters was altered in the presence of RuBP, phosphoenolpyruvate, 3-phosphoglycerate, 2-phosphoglycolate, and KH2PO4. ATP and 2-phosphoglycerate were found to affect only CbbRI binding, while fructose-1,6-bisphosphate altered the binding properties of only CbbRII. The fact that the pattern of responses to the molecules tested was different between CbbRI and CbbRII, as well as the fact that certain molecules (NADPH, NADH, fructose-6-phosphate, and ribose-5-phosphate) had little effect on either CbbRI or CbbRII DNA binding, suggested that the observed responses were not due to the nonspecific effects of phosphorylated compounds.
The observation that RuBP affected CbbR binding in vitro is of particular interest because it reinforces earlier physiological and genetic studies in R. capsulatus and R. sphaeroides that indicate that RuBP, and possibly another CBB cycle intermediate(s), acts as a positive inducer of CbbR-mediated cbb gene expression (25, 31, 36). Evidence supporting RuBP as a positive inducer of cbb expression arises from in vivo studies with form II RubisCO and phosphoribulokinase knockout strains (25) and form I/form II RubisCO/phosphoribulokinase mutant strains of R. capsulatus (36) and RubisCO deletion strains of R. sphaeroides (31). In R. capsulatus, form I RubisCO (encoded by cbbLS) is not expressed in the wild-type strain grown under photoheterotrophic conditions on malate (25). However, in a form II RubisCO mutant strain (cbbM), cbbLS expression was induced under photoheterotrophic conditions. This induction of cbbLS expression was shown to be dependent on the presence of a functional cbbP, encoding phosphoribulokinase.
Phosphoribulokinase catalyzes the synthesis of the RubisCO substrate RuBP, and it is thus conceivable that an accumulation of RuBP in the form II RubisCO mutant caused the induction of form I RubisCO synthesis in this strain. Studies with double RubisCO deletion strains (cbbLS/cbbM) of R. capsulatus (31, 36) and R. sphaeroides (31) reinforced this initial finding, because cbb promoters were substantially induced but absolutely dependent on cbbP expression under photoheterotrophic growth conditions in the double RubisCO deletion strains of both organisms.
The in vitro results reported here are consistent with the physiological evidence implicating RuBP, the specific product of phosphoribulokinase activity, as a physiological positive effector of regulators that influence cbb transcription. Thus, RuBP affected the DNA binding properties of CbbRI and CbbRII in both gel mobility shift and DNase I footprint assays. However, it should be noted that 1 mM RuBP did not stimulate CbbRI DNA binding in gel mobility shifts but did induce binding site contraction in the DNase I footprinting assays. The reason for this is not known. It may be due to the different reaction conditions between the two assays; i.e., 50 mM HEPES, pH 8.0, containing 200 mM KCl for the footprints and 10 mM Tris, pH 8.5, containing 300 mM potassium glutamate in the gel mobility shifts. Only CbbRI showed a strong reduction of its DNase I footprint, on the side proximal to the cbbI transcription start in the presence of RuBP (Fig. 4 and Fig. 7A).
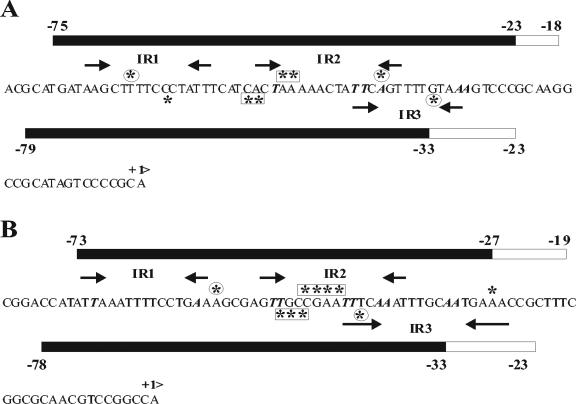
FIG. 7.
Summary and model of DNase I footprinting results for CbbRI binding to the cbbI promoter in the presence and absence of the effector RuBP (A) and CbbRII binding to the cbbII promoter in the presence and absence of fructose-1,6-bisphosphate (B). Bars indicate regions of protection on the top (upper) and bottom (lower) DNA strands in the presence and absence of effector. The black portion of each bar represents the region of protection in the presence of effector. Asterisks indicate DNase I-hypersensitive sites. Asterisks in boxes indicate hypersensitive sites that disappear or diminish in intensity in the presence of effector. Circled asterisks indicate hypersensitive sites that appear only in the presence of effector. Arrows indicate conserved inverted repeat (IR) sequences. A and T residues within the LysR consensus binding motif (T-N11-A) sequences are in bold italics.
This effector- and activation-induced contraction in the DNase I footprint is characteristic of group II LysR family transcriptional regulators (28, 22) such as OccR (44), OxyR (32), and ClcR (22). It has recently been proposed that the mechanism of CbbR-mediated activation of cbb transcription in Xanthobacter flavus (39) follows a “sliding dimer model” proposed for the LysR family transcriptional regulators OxyR (37) and OccR (45). The cbb promoter operator contains two CbbR binding sites, denoted R and A, that are conserved in a number of cbb promoters (30). The promoter-distal R site contains one conserved consensus CbbR binding motif (IR1), while the promoter-proximal A site spans two of these motifs that partially overlap (IR2 and IR3) (Fig. 7). The model suggests that in the absence of inducer, CbbR dimers are bound to IR1 in the R site and IR3 of the A site. Exposure to the inducer causes a shift in the position of the CbbR dimer occupying the A site from IR3 to IR2, leading to a reduction in CbbR-induced DNA bending and exposure of the −35 region of the cbb promoter.
The RuBP-induced reduction in the DNase I footprint of CbbRI bound to the cbbI promoter was consistent with a shift of CbbR dimer binding from IR3 to IR2, as predicted by this model. In addition, the RuBP-induced loss of the central DNase I hypersensitive sites at −52 bp and −53 bp was indicative of a change in DNA bending. RuBP had a slight effect on the DNase I footprint of CbbRII bound to its cognate promoter, implying that either the sensitivity of the two CbbRs to RuBP was different or the mechanism of RuBP activation may be different for the two proteins. The CbbRII footprint was reduced in the presence of fructose-1,6-bisphosphate, K2HPO4 and 3-phosphoglycerate, in a manner similar to the RuBP effect on the CbbRI footprint, but a decrease in the strongly hypersensitive sites was observed only in the presence of fructose-1,6-bisphosphate and K2HPO4 (Fig. 5, 6, and 7B and data not shown).
The existence of a CbbRII-specific effector molecule(s) other than RuBP was previously indicated by the fact that an R. capsulatus cbbL-cbbPII double mutant strain still displays CbbRII-dependent regulated expression of the cbbII promoter (36), suggesting that fructose-1,6-bisphosphate and 3-phosphoglycerate may indeed be effectors for CbbRII. The available evidence derived from effector studies of CbbRs from other sources indicates that the effector molecules to which they respond can be organism specific. For instance, NADPH has been proposed to be a positive effector of CbbR-mediated cbb transcription in Xanthobacter flavus (40) and Hydrogenophilus thermoluteolus (35), while phosphoenolpyruvate has been implicated as a negative effector of CbbR activity in the chemoautotroph Ralstonia eutropha (Alcaligenes eutrophus), where it has been proposed to function as an indicator of the fixed carbon status of the cell (17). This organism-specific variation in effector molecules could explain the different patterns of in vitro responses to potential effectors observed between CbbRI and CbbRII because phylogenetic analyses indicate that the R. capsulatus cbbI operon and cbbRI were acquired through a horizontal gene transfer event, probably from a chemoautotrophic ancestor (23).
It should be noted that phosphoenolpyruvate affected the DNA binding of both CbbRI and CbbRII in a manner similar to that observed in R. eutropha, i.e., an increase in DNA binding affinity in gel mobility shift assays but no effect on the DNase I footprint (17). Thus, phosphoenolpyruvate and possibly one or more of the other metabolites that affected CbbR binding affinity without altering the DNase I footprint could function as negative effectors of CbbR activity in R. capsulatus. All of the compounds identified in this study that affect the DNA binding of CbbR are logical candidates for effectors of CbbR-mediated CBB cycle gene regulation. While ATP and K2HPO4 are indicators of the energy available to the pathway, each of the other compounds is either a CBB cycle intermediate (RuBP, fructose-1,6-bisphosphate, and 2-phosphoglycerate), end product (3-phosphoglycerate and 2-phosphoglycolate), or derived from CBB cycle end products (phosphoenolpyruvate).
One might speculate that phosphoenolpyruvate and the CBB cycle end product 3-phosphoglycerate would be indicators of the fixed (organic) carbon status of the cell and therefore may serve as negative effectors of CbbR activity. However, the 3-phosphoglycerate-induced shrinkage of the CbbRII DNase I footprint is more consistent with the role of a positive effector. This could be rationalized by the fact that a portion of the 3-phosphoglycerate pool is fed back into the CBB cycle to regenerate RuBP, the CO2 acceptor molecule. K2HPO4 and 2-phosphoglycolate might also negatively affect CbbR activity, because high levels of K2HPO4 would signal high rates of ATP consumption, while the product of the energetically wasteful oxygenase reaction of RubisCO, 2-phosphoglycolate, would indicate high O2 levels. While the case for RuBP as a positive effector of CbbR activity is strong, the precise role that the other potential effector molecules play in CbbR-mediated regulation of cbb transcription is less clear. However, the accumulated data suggest that CbbR-mediated activation of cbb transcription in R. capsulatus is influenced by multiple metabolic signals that reflect not only the levels of CBB cycle intermediates but also the fixed (organic) carbon status and energy charge of the cell.
Finally, future studies in R. capsulatus should consider whether there might be cross regulation by one CbbR protein to the promoter-operator region of the opposite operon. Although CbbRI and CbbRII bind their noncognate cbb promoters in gel mobility shift assays (data not shown), it is not conclusive at this time from in vitro studies whether such interactions are efficient enough to have a significant physiological effect. Available DNase I footprinting experiments suggested that the binding affinity to the noncognate promoter was very low, because the same amount of CbbRI and CbbRII used to bind the cognate promoter did not result in binding to the noncognate promoter (data not shown). Yet there are several indications that such interactions might be significant in vivo. For example, earlier physiological studies showed that CbbRI and CbbRII might positively regulate each other's expression (43). Some evidence also suggests that CbbRI may positively cross-regulate cbbM because cbbM transcripts were detected in a photoheterotrophically grown cbbRII strain but not in a cbbRI cbbRII strain (43). Such interactions and the potential role of other regulator proteins (10, 11, 13) might have a profound influence on the overall regulatory mechanism.
Acknowledgments
These studies were supported by Public Health Service grant GM45404 from the National Institutes of Health.
REFERENCES
- 1.Anderson, L., and R. C. Fuller. 1967. Photosynthesis in Rhodospirillum rubrum. III. Metabolic control of reductive pentose phosphate and tricarboxylic acid cycle enzymes. Plant Physiol. 42:497-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology, vol. 1. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 3.Bird, T. H., S. Du, and C. E. Bauer. 1999. Autophosphorylation, phospho-transfer and DNA binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J. Biol. Chem. 274:16343-16348. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, R. E., and J. H. Weiner. 1993. Overproduction, solubilization, purification and DNA binding properties of AmpR from Citrobacter freundii. Eur. J. Biochem. 213:405-412. [DOI] [PubMed] [Google Scholar]
- 5.Chang, M., and I. P. Crawford. 1991. In vitro determination of the effect of indoleglycerol phosphate on the interaction of purified TrpI with its DNA binding sites. J. Bacteriol. 173:1590-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong, S., F. B. Mersha, D. G. Comb, M. E., Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus, and M. Q. Xu. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192:271-281. [DOI] [PubMed] [Google Scholar]
- 7.Chong, S., and M.-Q. Xu. 1996. Protein splicing of the Saccharomyces cerevisiae VMA intein without the endonuclease motifs. J. Biol. Chem. 272:15587-15590. [DOI] [PubMed] [Google Scholar]
- 8.Dubbs, J. M., T. H. Bird, C. E. Bauer, and F. R. Tabita. 2000. Interaction of CbbR and RegA* transcription regulators with the Rhodobacter sphaeroides cbbI promoter-operator region. J. Biol. Chem. 275:19224-19230. [DOI] [PubMed] [Google Scholar]
- 9.Dubbs, J. M., and F. R. Tabita. 1998. Two functionally distinct regions upstream of the cbbI operon of Rhodobacter sphaeroides regulate gene expression. J. Bacteriol. 180:4903-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubbs, J. M., and F. R. Tabita. 2003. Interactions of the cbbII promoter-operator region with CbbR and RegA (PrrA) regulators indicate distinct mechanisms to control expression of the two cbb operons of Rhodobacter sphaeroides. J. Biol. Chem. 278:16443-16450. [DOI] [PubMed] [Google Scholar]
- 11.Dubbs, J. M., and F. R. Tabita. 2004. Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28:353-376. [DOI] [PubMed] [Google Scholar]
- 12.Gao, J., and G. N. Gussin. 1991. Mutations in TrpI binding site II that differentially affect activation of the trpBA promoter of Pseudomonas aeruginosa. EMBO J. 10:4137-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, J. L., J. M. Dubbs, and F. R. Tabita. 2002. Differential expression of the CO2 fixation operons of Rhodobacter sphaeroides by the Prr/Reg two-component system during chemoautotrophic growth. J. Bacteriol. 184:6654-6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, J. L., and F. R. Tabita. 1977. Isolation and preliminary characterization of two forms of ribulose 1,5-bisphosphate carboxylase from Rhodopseudomonas capsulatus. J. Bacteriol. 132:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, J. L., and F. R. Tabita. 1993. Nucleotide sequence and function analysis of CbbR, a positive regulator of the Calvin cycle operons of Rhodobacter sphaeroides. J. Bacteriol. 175:5778-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goethals, K., M. Van Montagu, amd M. Holsters. 1992. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc. Natl. Acad. Sci. USA 89:1646-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grzeszik, C., T. Jeffke, J. Schaferjohann, B., Kusian, and B. Bowein. 2000. Phosphoenolpyruvate is a signal metabolite in transcriptional control of the cbb CO2 fixation operons in Ralstonia eutropha. J. Mol. Microbiol. Biotechnol. 2:311-320. [PubMed] [Google Scholar]
- 18.Hallenbeck, P. L., R. Lerchen, and S. Kaplan. 1990. Roles of CfxA, CfxB, and external electron acceptors in regulation of ribulose 1,5-bisphosphate carboxylase/oxygenase expression in Rhodobacter sphaeroides. J. Bacteriol. 172:1736-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusano, T., and K. Sugawara. 1993. Specific binding of Thiobacillus ferrooxidans RbcR to the intergenic sequence between the rbc operon and the rbcR gene. J. Bacteriol. 175:1019-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusian, B., and B. Bowein. 1995. Operator binding of the CbbR protein, which activates the duplicate cbb CO2 assimilation operons of Alcaligenes eutrophus. J. Bacteriol. 177:6568-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lascelles, J. 1960. The formation of ribulose 1,5-diphosphate carboxylase by growing cultures of Athiiorhodaceae. J. Gen. Microbiol. 23:499-510. [DOI] [PubMed] [Google Scholar]
- 22.McFall, S. M., M. R. Parsek, and A. M. Chakrabarty. 1997. 2-Chloromuconate and ClcR-mediated activation of the clcABD operon: in vitro transcriptional and DNase I footprint analyses. J. Bacteriol. 179:3655-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paoli, G. C., F. Soyer, J. Shively, and F. R. Tabita. 1998. Rhodobacter capsulatus genes encoding form I ribulose-1,5-bisphosphate carboxylase/oxygenase (cbbLS) and neighboring genes were acquired by a horizontal gene transfer. Microbiology 144:219-227. [DOI] [PubMed] [Google Scholar]
- 24.Paoli, G. C., N. Strom-Morgan, F. R. Tabita, and J. M. Shively. 1995. Expression of the cbbLcbbS and cbbM genes and distinct organization of the cbb Calvin cycle structural genes of Rhodobacter capsulatus. Arch Microbiol. 164:396-405. [PubMed] [Google Scholar]
- 25.Paoli, G. C., P. Vichivanives, and F. R. Tabita. 1998. Physiological control and regulation of the Rhodobacter capsulatus cbb operons. J. Bacteriol. 180:4258-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 27.Rohman, M., and K. J. Harrison-Lavoie. 2000. Separation of copurifying GroEL from glutathione-S-transferase fusion proteins. Protein Expr. Purif. 20:45-47. [DOI] [PubMed] [Google Scholar]
- 28.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 29.Shively, J. M., E. Davidson, and B. L. Marrs. 1984. Derepression of the synthesis of the intermediate and large forms of ribulose-1,5-bisphosphate carboxylase/oxygenase in Rhodobacter capsulatus. Arch. Microbiol. 138:233-236. [DOI] [PubMed] [Google Scholar]
- 30.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 31.Smith, S. A., and F. R. Tabita. 2002. Up-regulated expression of the cbbI and cbbII operons during photoheterotrophic growth of a ribulose 1,5-bisphosphate carboxylase-oxygenase deletion mutant of Rhodobacter sphaeroides. J. Bacteriol. 184:6721-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storz, G., L. A. Tartaglia, and B. N. Ames. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189-194. [DOI] [PubMed] [Google Scholar]
- 33.Tabita, F. R. 1988. Molecular and cellular regulation of autotrophic carbon dioxide fixation in microorganisms. Microbiol. Rev. 52:155-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabita, F. R. 1995. The biochemistry and metabolic regulation of carbon metabolism and CO2 fixation in purple bacteria, p. 885-914. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 35.Terazono, K., N. R. Hayashi, and Y. Igarashi. 2001. CbbR, a LysR-type transcriptional regulator from Hydrogenophilus thermoluteolus bind two cbb promoter regions. FEMS Microbiol. Lett. 198:151-157. [DOI] [PubMed] [Google Scholar]
- 36.Tichi, M. A., and F. R. Tabita. 2002. Metabolic signals that lead to control of CBB gene expression in Rhodobacter capsulatus. J. Bacteriol. 184:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 38.van den Bergh, E. R. E., L. Dijkhuizen,, and W. G. Meijer. 1993. CbbR, a LysR-type transcriptional activator, is required for transcription of the autotrophic CO2 fixation enzymes of Xanthobacter flavus. J. Bacteriol. 175:6097-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Keulen, G., L. Girbal, A. N. J. A. Ridder, L. Dijkhuizen, L., and W. G. Meijer. 2003. Analysis of DNA binding and transcriptional activation by the LysR-type transcriptional regulator CbbR. J. Bacteriol. 185:1245-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Keulen, G., L. Girbal, E. R. E. van den Bergh, E., L. Dijkhuizen, and W. G. Meijer. 1998. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J. Bacteriol. 180:1411-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viale, A. M., H. Kobayashi, T. Akazawa, and S. Henikoff. 1991. rcbR, a gene coding for a member of the LysR family of transcriptional regulators, is located upstream of the expressed set of ribulose 1,5-bisphosphate carboxylase/oxygenase genes in the photosynthetic bacterium Chromatium vinosum. J. Bacteriol. 173:5224-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vichivanives, P. 2000. Ph.D. dissertation. Ohio State University, Columbus, Ohio.
- 43.Vichivanives, P., T. H. Bird, C. E. Bauer, and F. R. Tabita. 2000. Multiple regulators and their interactions in vivo and in vitro with the cbb regulons of Rhodobacter capsulatus. J. Mol. Biol. 300:1079-1099. [DOI] [PubMed] [Google Scholar]
- 44.Wang, L., K. Cho, J. D. Helmann, and S. C. Winans. 1992. The A. tumefaciens transcriptional activator OccR causes a bend at a target promoter, which is partially relaxed by a plant tumor metabolite. Cell 69:659-667. [DOI] [PubMed] [Google Scholar]
- 45.Wang, L., and S. C. Winans. 1995. The sixty nucleotide OccR operator contains a subsite essential and sufficient for OccR binding and a second subsite required for ligand responsive DNA bending. J. Mol. Biol. 253:691-702. [DOI] [PubMed] [Google Scholar]
- 46.Wang, X., D. L. Falcone, and F. R. Tabita. 1993. Reductive pentose phosphate-independent CO2 fixation in Rhodobacter sphaeroides and evidence that ribulose bisphosphate carboxylase/oxygenase activity serves to maintain the redox balance of the cell. J. Bacteriol. 175:3372-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Windhovel, U., and B. Bowien. 1991. Identification of cfxR, an activator gene of autotrophic CO2 fixation in Alcaligenes eutrophus. Mol. Microbiol. 5:2695-2705. [DOI] [PubMed] [Google Scholar]