Abstract
Shewanella oneidensis MR-1 is a facultative Fe(III)- and Mn(IV)-reducing microorganism and serves as a model for studying microbially induced dissolution of Fe or Mn oxide minerals as well as biogeochemical cycles. In soil and sediment environments, S. oneidensis biofilms form on mineral surfaces and are critical for mediating the metabolic interaction between this microbe and insoluble metal oxide phases. In order to develop an understanding of the molecular basis of biofilm formation, we investigated S. oneidensis biofilms developing on glass surfaces in a hydrodynamic flow chamber system. After initial attachment, growth of microcolonies and lateral spreading of biofilm cells on the surface occurred simultaneously within the first 24 h. Once surface coverage was almost complete, biofilm development proceeded with extensive vertical growth, resulting in formation of towering structures giving rise to pronounced three-dimensional architecture. Biofilm development was found to be dependent on the nutrient conditions, suggesting a metabolic control. In global transposon mutagenesis, 173 insertion mutants out of 15,000 mutants screened were identified carrying defects in initial attachment and/or early stages in biofilm formation. Seventy-one of those mutants exhibited a nonswimming phenotype, suggesting a role of swimming motility or motility elements in biofilm formation. Disruption mutations in motility genes (flhB, fliK, and pomA), however, did not affect initial attachment but affected progression of biofilm development into pronounced three-dimensional architecture. In contrast, mutants defective in mannose-sensitive hemagglutinin type IV pilus biosynthesis and in pilus retraction (pilT) showed severe defects in adhesion to abiotic surfaces and biofilm formation, respectively. The results provide a basis for understanding microbe-mineral interactions in natural environments.
Microbe-mineral interactions are key steps in the biochemical cycles of elements. Of particular interest are microbe-mediated redox transformations of Fe and Mn oxide-containing minerals, which are abundantly present in soil as insoluble (oxyhydr)oxide minerals (e.g., goethite, ferrihydrite, and birnessite). In case of reductive interactions, microbially produced reactive Fe(II) and Mn(II) species dissolve from the mineral phase and can subsequently initiate geochemically important, abiotic reactions, such as the reductive transformation of Se, As, and Cr species (9, 22, 33). Shewanella oneidensis and Geobacter species are soil and sediment microbes that reduce insoluble Fe(III) and Mn(IV) in their catabolic metabolism under anaerobic conditions (24, 34, 35). Key to those microbe-mineral interactions, however, is the close association of the microbe with the mineral surface. In natural environments, such close contact is mediated by microbial biofilms, which develop on mineral particles and enable these biofilm microbes to metabolically interact with the metals of the mineral phase.
Despite intense research over the past decade on medically important microbial biofilms (e.g., see references 6, 11, 20, and 38), the molecular understanding of such microbe-mineral interactions, the biology of such biofilms, and the microorganisms involved is relatively poor. With this study, we provide the first insights into the molecular stages of biofilm formation by the group of Fe(III)- and Mn(IV)-reducing microbes and the factors involved. We focused on the facultative γ-proteobacterium Shewanella oneidensis MR-1. One of the most remarkable properties of this microbe is that it can use, under anaerobic conditions, an impressive variety of chemically diverse inorganic compounds as electron acceptors, including metals, such as Fe(III) and Mn(IV) (31, 32). Productive microbe-mineral interactions in S. oneidensis biofilms are initiated by cells adhering to iron or manganese oxide surfaces (25, 35), and electron transfer between the organism and the mineral surface can cause dissolution of insoluble Fe and Mn minerals (35) and alter the corrosion properties of steel (8).
In order to begin to understand the metabolic microbe-mineral interactions of S. oneidensis which occur in nature, we concentrated here on aerobic biofilms developing on glass surfaces, because this enabled us to use green fluorescent protein (GFP) as a single-cell reporter for a continuous, noninvasive monitoring of biofilms developing on glass surfaces in a hydrodynamic flow chamber system. We found that the biofilm biology of S. oneidensis resembles only to some extent that of other bacteria but exhibits several important differences. The results on molecular processes obtained from this study will facilitate the more complex investigations of anaerobic S. oneidensis biofilms developing on mineral surfaces.
MATERIALS AND METHODS
Growth conditions and media.
Strains used in this study are summarized in Table 1. Cultures of S. oneidensis MR-1 and Escherichia coli strains were grown in Luria-Bertani (LB) medium at 30 and 37°C, respectively. As necessary, the medium was supplemented with 25 μg of kanamycin, 10 μg of gentamicin, and/or 6 μg of chloramphenicol/ml. For plates, the medium was solidified using 1.5% (wt/vol) agar. Biofilm experiments were carried out in LM medium (0.02% [wt/vol] yeast extract, 0.01% [wt/vol] peptone, 10 mM [wt/vol] HEPES [pH 7.4], 10 mM NaHCO3) with a lactate concentration of 0.5 mM, if not indicated otherwise. Swimming motility was assayed in LM medium containing 15 mM lactate and solidified with 0.3% (wt/vol) agar.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| DH5α-λpir | φ80dlacZΔM15 Δ(lacZYA-argF) U196 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1/λpir | 30 |
| S17-λpir | thi pro recA hsdR [RP4-2Tc::Mu-Km::Tn7]λpir Tpr Smr | 44 |
| BW20767 | RP4-2-Tc::Mu-1 Kan::Tn7 integrant leu-63::IS10 recA1 zbf-5 creB510 hsdR17 endA1 thi uidA (ΔMluI)::pir+ | 29 |
| HB101 | Smr, recA thi pro leu hsdRM+ | 15 |
| Shewanella oneidensis | ||
| MR-1 | Shewanella oneidensis MR-1 wild type | 48 |
| AS92 | S. oneidensis MR-1, SO1282::pJP5603-Gm, Gmr | This study |
| AS93 | S. oneidensis MR-1 tagged with eGFPa in a mini-Tn7 construct, Gmr Cmr | This study |
| AS94 | fliK::pJP5603, Kmr Gmr Cmr in AS93 | This study |
| AS95 | flhB::pJP5603, Kmr Gmr Cmr in AS93 | This study |
| AS96 | pomA::pJP5603, Kmr Gmr Cmr in AS93 | This study |
| AS97 | mshA::pJP5603, Kmr Gmr Cmr in AS93 | This study |
| AS98 | pilT::pJP5603, Kmr Gmr Cmr in AS93 | This study |
| Plasmids | ||
| pJP5603 | KmrmobRP4+ori-R6K, suicide plasmid for mutation by plasmid integration | 39 |
| pJP5603Gm | GmrmobRP4+ ori-R6K | This study |
| pJP5603Gm-tp | fragment of SO1282 in pJP5603 | This study |
| pJP5603::flhB | fragment of flhB in pJP5603 | This study |
| pJP5603::fliK | fragment of fliK in pJP5603 | This study |
| pJP5603::pomA | fragment of pomA in pJP5603 | This study |
| pJP5603::mshA | fragment of mshA in pJP5603 | This study |
| pJP5603::pilT | fragment of pilT in pJP5603 | This study |
| pRL27 | Kmrori-R6K, Tn5 delivery vector | 23 |
| pBBR1MCS-5 | Gmr pBBR1-ori lacZ mob+ | 21 |
| pUX-BF13 | Aprmob+ori-R6K, helper plasmid, providing Tn7 transposition functions in trans | 2 |
| RK600 | Cmrori-ColE1 RK2-mob+ RK2-tra+, helper plasmid in matings | 15 |
| pBK-miniTn7-egfp2 | Gmr Cmr Apr mob+, delivery vector for mini-Tn7-efp2 | 19 |
eGFP, enhanced GFP.
DNA manipulations.
All restriction digests, ligations, cloning, and DNA electrophoresis were carried out according to standard techniques (42). Enzymes used were purchased from New England Biolabs (Beverly, Mass.). Kits for purifying DNA fragments and plasmid and chromosomal DNA were obtained from QIAGEN (Valencia, Calif.) and used according to the manufacturer's instructions.
Construction in S. oneidensis MR-1 strains.
Gene disruption mutations were generated by plasmid integration and carried out by using the suicide plasmids pJP5603 and pJP5603Gm (39; this study). pJP5603Gm was constructed by amplification of the gentamicin cassette from pBBR1MCS-5 with the primer pair Gm-fw-Nco and Gm-rv-Nco and ligating the product into the NcoI site of pJP5603, thereby disrupting kanamycin resistance (Table 2). To construct a gentamicin-resistant strain of S. oneidensis MR-1, an internal fragment of a transposase gene within the Shewanella genome (SO1282) was amplified with the primer pair tp-fw-bam and tp-rv-eco and ligated into pJP5603Gm after digestion with BamHI and EcoRI. The resulting suicide vector, pJP5603Gm-tp, was introduced into S. oneidensis MR-1 by mating, using E. coli S17I-λpir as the donor strain, yielding the gentamicin-resistant strain AS92, which was used for transposon mutagenesis.
TABLE 2.
Sequence of primers used in this study
| Primer name | Sequence |
|---|---|
| Gm-fw-Nco | GAACCTGAATCGCCAGCGGCATC |
| Gm-rv-Nco | AAGGCCATGGGCGAAAAGCTGCTGACGG |
| flhB-fw-V | TGCTGATATCGGTTGTGGCATTTATCGGTAACG |
| flhB-rv-V | TACCGATATCACATCCGCATTAGGCACTTCTGCC |
| flhB-check-fw | GTTGTTGGCAGCGAAATAGGTTGG |
| flhB-check-rv | CGGTAACGATGGCAATATTGTGCG |
| fliK-fw-eco | ATCCGAATTCTAGTGACGATAACCCCTCTCG |
| fliK-rv-bam | TGTTGGATCCCTTGGCTAACCATAGTCACCAG |
| fliK-check-fw | CAAAGAGCCTAATACGCTAGG |
| fliK-check-rv | GTGTCTGATCACCATGAACC |
| mshA-fw-V | AGGCGATATCCGTGCGTCTGCATTACAAGG |
| mshA-rv-V | TGTTGATATCTCTTAGCAGTACCTGGTGTAC |
| mshA-check-fw | TTGCTGTCACAGCAGCACCTAAG |
| mshA-check-rv | CATACTTAGGCAGTTCACCTGG |
| pilT-fw-V | AGTCGATATCTTAGCGCGTTTCCGTGTGAACG |
| pilT-fw-rv | GGTCGATATCCTTAGCTGCCGAGGTGGTATG |
| pilT-check-fw | GTGTACACAGCCTTGTGTACGAC |
| pilT-check-rv | ATTTCGTGGGCAGCAACACGAC |
| pom-fw-V | TTAGTTGATATCCTCGTCGACGGCCATG |
| pom-rv-V | ACGGGAATATCATTTCTTCACCCATTCG |
| pom-check-fw | CTTGAGGAAGCACAAATCTCC |
| pom-check-rv | CTATCACCCTAGGATTTTGGC |
| pPS-seq1 | AACAAGCCAGGGATGTAACG |
| tp-fw-bam | AAACGGATCCATTTGCCCAAGCTGTTCGAGC |
| tp-rv-eco | TTAAGAATTCGTCAACCTCTACGATGCCGG |
For biofilm studies, an S. oneidensis MR-1 strain was constructed that constitutively expressed GFP, using the Tn7 delivery system described by Koch et al. (19). Strain AS93 was constructed by four-parental mating of the S. oneidensis MR-1 wild type with E. coli HB101/RK600, E. coli HB101/pUX-BF13, and E. coli HB101 harboring pBK-mini-Tn7-egfp2. The resulting gentamicin-resistant strain, AS93, was not observed to have any growth or biofilm deficiency or any atypical physiology and was subsequently used as the S. oneidensis MR-1 wild type for biofilm studies. This strain also served as the host strain for introducing targeted gene disruptions.
For construction of targeted gene disruption mutants, internal fragments of the corresponding genes of 250 to 400 bp in length were amplified by PCR, using the primer pairs flhB-fw and -rv for flhB, fliK-fw and -rv for fliK, pom-fw and -rv for pomA, mshA-fw and -rv for mshA, and pilT-fw and -rv for pilT, respectively. The fliK fragment was cleaved with EcoRI and BamHI and ligated into pJP5603 treated with the same enzymes. All other fragments were digested with EcoRV and ligated into the SmaI site of pJP5603. The corresponding constructs were transferred into AS93 by conjugation from E. coli S17I-λpir, and plasmid integration mutants of S. oneidensis were selected on LB medium containing kanamycin and gentamicin. The correct insertion was verified by PCR, using primers flanking the location of insertion.
Transposon mutagenesis and screen for biofilm-deficient mutants.
Transposon mutagenesis was performed by mating AS92 with the donor strain E. coli BW20767 harboring suicide plasmid pRL27, which carries a hyperactive transposase and a Tn5-mini transposon with a kanamycin resistance cassette and an RK6 origin of replication (23). The mating was performed at a 1/1 donor-recipient ratio at room temperature for 5 h, and transconjugants were selected on LB medium containing kanamycin and gentamicin. The mutant screen was performed essentially as described earlier (37, 40). Briefly, the wells of 96-well microtiter dishes, filled with 175 μl of LM (15 mM lactate) medium, were inoculated with individual kanamycin-resistant colonies and allowed to grow at 30°C. After 16 h, cells were stained by addition of 10 μl of a 0.5% (wt/vol) crystal violet solution. Following an incubation for 10 min at room temperature, the supernatants were removed, the wells were rinsed with quartz-distilled water, and 200 μl of 96% (vol/vol) ethanol was added. Wells containing a reduced amount of cell material attached to the wall were indicated by a paler blue color than for the nonmutagenized strain as quantified spectrophotometrically at 570 nm.
All mutants putatively deficient in biofilm formation were retested for a biofilm phenotype in a second microtiter plate screen in triplicate and, subsequently, for their ability to grow planktonically in LM medium to eliminate mutants with severe growth deficiencies. Only biofilm mutants with a growth rate of greater than 80% of AS93's were retained for further studies. To map the location of a Tn5 insertion, chromosomal DNA was prepared from the mutants and digested with PstI. The resulting fragments were religated and used to transform E. coli DH5α-λpir. Religation products harboring Tn5 are maintained as stable “plasposons ” due to the presence of the R6K origin of replication and allowed selection on kanamycin-containing media. Subsequently, the plasposons were prepared from E. coli and subjected to sequencing of the regions flanking Tn5 with the primer pPS-seq1. Use of the obtained sequence information for databank searches (http://tigrblast.tigr.org/cmr-BLAST/) revealed the exact location of the transposon insertion.
Biofilm cultivation.
Biofilms were cultivated at 30°C in three-channel flow cells; the individual channel dimensions were 1 by 4 by 40 mm. Each flow chamber was prepared by gluing a microscope coverslip (Fisher Scientific, Pittsburgh, Pa.), which served as a substratum for microbial attachment, onto the flow chamber with silicone (GE Sealants & Adhesives, Hunterville, N.C.) and leaving it to dry for 24 h at room temperature prior to use. Assembly of the flow system was carried out essentially as described earlier (5) with the exception that the setup was sterilized by autoclaving before the inflow tubing was connected to the medium reservoir. After assembly, the system was equilibrated with medium at a low flow rate for at least 6 h before use.
LB overnight cultures of the appropriate S. oneidensis strain were diluted 1/100 in LB and grown until mid-logarithmic phase. The optical density at 600 nm was then adjusted to 0.01 in LM medium without lactate. A 0.5- to 1-ml volume of the diluted cell suspension was injected into each channel after the medium flow was arrested, and the chambers were turned upside down to enhance initial attachment. After a 40-min incubation at 30°C, the flow cells were inverted, and medium flow was started at a constant rate of 66 μl/min per channel, using a Watson-Marlow Bredel (Cornwall, United Kingdom) 205S peristaltic pump. All biofilm characterizations were conducted in triplicate in at least two independent experiments.
Image acquisition and processing.
Microscopic visualization of biofilms was carried out at the Stanford Biofilm Research Center using an upright Zeiss LSM510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany) equipped with the following objectives: ×10/0.3 Plan-Neofluar, ×20/0.5W Achroplan, and ×40/1.2W C-Apochromat. Biofilm parameters, such as surface coverage, biofilm mass, and average biofilm thickness, were quantified with the COMSTAT program (13). Image data obtained were further processed by using the IMARIS software package (Bitplane AG, Zürich, Switzerland) and Adobe Photoshop.
RESULTS AND DISCUSSION
Characterization of S. oneidensis MR-1 biofilm development.
In order to obtain the first basic insights into the molecular determinants of biofilm formation in Fe(III)- and Mn(IV)-reducing S. oneidensis MR-1, we investigated the formation and dynamics of biofilms developing under hydrodynamic conditions on glass surfaces in flow chambers, using gfp-labeled cells in conjunction with confocal laser scanning microscopy. In contrast to frequently used static systems, this experimental approach provides steady conditions of nutrient and oxygen supply and of product removal and allows continuous, noninvasive monitoring of biofilm progression (5). So far, biofilm formation of only very few microorganisms has been studied thoroughly by this approach, which reflects the natural environment of the organism.
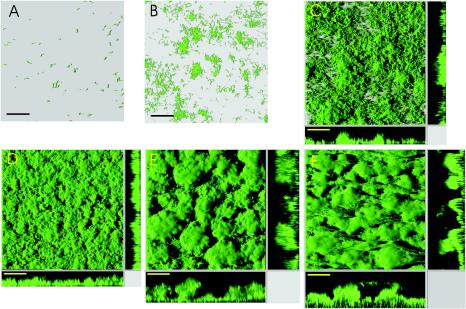
We used strain AS93 as the wild type strain for all of our biofilm studies. After injection into the flow chambers, the cells were allowed to adhere for 40 min, and flow of medium, containing 0.5 mM lactate, was started. Figure 1 displays representative images of the time course of S. oneidensis biofilm formation. Following initial attachment, cells divided rapidly at an approximate doubling time of 2 h (between t = 8 h and t = 16 h) and formed isolated microcolonies that appear to result from clonal growth. In addition to these early microcolonies, a few new microcolonies could be observed to form after 6 to 8 h, which probably originated from detached, free-swimming cells that attached to uncovered surface regions. Small three-dimensional structures (up to 10 μm in thickness) appeared after 12 h, and the previously isolated microcolonies began to fuse with each other, thereby increasing the overall surface coverage. After about 20 h, almost complete and confluent coverage of the surface was observed, with a height of an average biofilm of about 20 μm. From this time on, biofilms developed into extensive, three-dimensional structure containing mushroom-like protrusions and valley-shaped indents. The progression reached a quasisteady state after about 5 days, when the biofilms were dominated by towering, mushroom-like structures, which extended more than 200 μm into the bulk liquid phase. This observed development of S. oneidensis MR-1 biofilms is reminiscent of the biofilm progression described for Pseudomonas aeruginosa (17, 43, 47). Similar initial phases were also observed in strains of E. coli K-12 (41).
FIG. 1.
Biofilm formation of S. oneidensis MR-1 (strain AS93). Images display shadow projections of AS93 biofilms; x-z and y-z sagittal images at selected positions in the biofilm are shown at the bottom and right side of images C-F, respectively. Biofilms were grown in LM medium containing 0.5 mM lactate. Images displayed were recorded after 1, 8, 16, 24, 48, and 120 h (A-F). Bar = 80 μm.
Cross-section imaging of mature (>4 days) AS93 biofilms revealed that the brightest GFP fluorescence was found in cell layers that were in closest contact with the flowthrough medium (Fig. 1F, cross section). Cells located in those regions are exposed to the highest nutrient and oxygen concentrations, which are expected to lead to higher levels of metabolic activity and an associated enhanced production of maturated GFP. For Pseudomonas putida biofilms, it was shown that cells in close proximity to the nutrient medium have a higher growth rate than more distantly located cells (45). However, the absence of GFP fluorescence in deeper layers of mature S. oneidensis biofilms correlated with the absence of cells in those layers, as revealed by bright-field microscopy, suggesting that the decreased fluorescence is predominantly due to cell loss or lysis (data not shown). Cell lysis was demonstrated for P. aeruginosa by Webb and colleagues (52), who proposed that prophage-induced cell death provides a mechanism for Pseudomonas biofilms to differentiate. Notably, the S. oneidensis MR-1 genome sequence revealed the presence of three putative prophages, although none has been shown so far to be active (12).
Influence of nutrient concentration on biofilm formation.
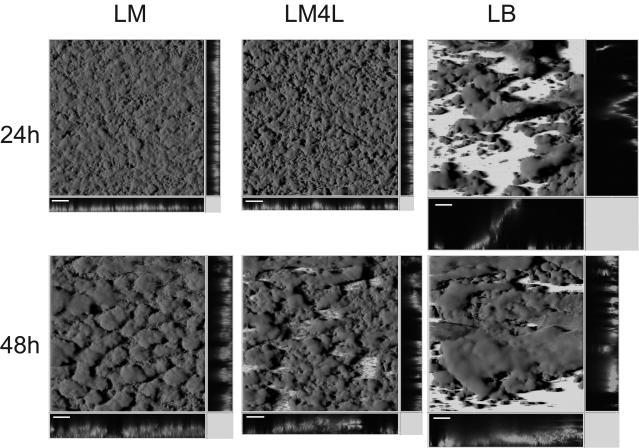
Studies of Pseudomonas and Vibrio species showed that the type of carbon source and the nutrient concentration strongly influence the development and architecture of the respective biofilms. P. aeruginosa was found to form flat, unstructured biofilm layers when citrate, Casamino Acids, or benzoate served as the carbon source, while the formation of pronounced, three-dimensional structures occurred only in medium containing glucose (17). Mutagenesis of Pseudomonas fluorescens WCS365 yielded mutants that exhibited an attachment phenotype only when grown in a minimal glucose medium but not in a citrate or glutamate medium (37). Vibrio cholerae biofilms show distinct differences in architecture when grown in fresh or seawater medium (16). In order to investigate the effect of the carbon source on S. oneidensis MR-1 biofilm formation, flow-chamber experiments were carried out, using AS93 with our standard LM (500 μM lactate) medium, with LM medium containing 4 mM lactate (LM4L), and with LB medium. After 24 and 48 h of incubation, biofilms grown in LM4L medium were less compact and had a different surface structure than LM biofilms (Fig. 2). This observation was similar to that for P. aureofaciens grown with different concentrations of citrate (13). The architectures of 5-day-old S. oneidensis biofilms grown with either LM or LM4L were indistinguishable, which could be due to increasing substrate limitation in the densely grown LM biofilms (data not shown).
FIG. 2.
Architecture of S. oneidensis MR-1 (AS93) biofilms grown in different media. Displayed are shadow projections of confocal laser scanning microscopy images of 24- and 48-h-old AS93 biofilms grown in flow chambers irrigated with LM (0.5 mM lactate), LM4L (2.0 mM), or LB medium. Bar = 100 μm.
LB-grown biofilms of S. oneidensis MR-1 were found to have a fundamentally different appearance (Fig. 2). The cells formed densely packed but fragile clusters with large biomasses anchored to the surface only at a very small region. Some of the structures were found to span the entire distance between the coverslip and the bottom of the flow chamber (>1 mm). Minor agitation resulted in rapid separation of large portions of the attached cell mass. After 72 h, LB-irrigated channels were totally overgrown (data not shown). As demonstrated here, the S. oneidensis biofilm structure and development depends highly on the medium used, indicating that biofilm dynamics is probably metabolically controlled. It also suggests that for this strain, poor nutrient conditions result in more-robust biofilms with enhanced microbe-surface interactions.
Identification of mutants with defects in early biofilm formation.
To identify genes and gene products required for the initial phases of S. oneidensis MR-1 biofim formation, we performed a transposon mutagenesis and screened Tn5 mutants for loss of biomass in microtiter plates (37). This microtiter plate assay has been used previously with other microorganisms, such as E. coli, V. cholerae, and P. fluorescens, to identify genes involved in initial attachment and early stages of biofilm formation (37, 40, 50, 51).
The transposon mutagenesis was carried out as described in Materials and Methods with S. oneidensis AS92. Mutants identified as defective in the microtiter biofilm assay were retested in triplicate and examined for growth defects in LM medium. Out of approximately 15,000 mutants screened, 173 were identified that were impaired in forming biofilms on the polyvinyl chloride surface in the microtiter plate assay (less than 85% crystal violet retention compared to the parent strain) but exhibited no change in the growth rate. None of the mutants had a delayed biofilm phenotype, since the mutants were not observed to form wild-type biofilms upon prolonged incubation of the microtiter plates (up to 48 h) or when LB was used in the microtiter biofilm assay. All mutants were tested for swimming motility in a soft agar assay, and 71 were found to be defective. The positions of the Tn5 insertion were determined for seven randomly chosen mutants of this group, and all were found to be in genes annotated as encoding flagellum-related proteins (fliA, fliE, fliP, fliD, flhA, and flgK).
Analysis of motility- and pilus-defective mutants in the hydrodynamic biofilm assay.
Since the static microtiter biofilm assay does not provide much temporal information or any architectural information, we examined the biofilm features of selected mutants in more detail in the hydrodynamic flow chamber. We focused on the predominant mutant group identified (i.e., the motility mutants) and also included two mutants defective in genes (mshA and pilT) putatively related to type IV pilus-mediated twitching motility. These mutants as well as all swimming motility mutants were reconstructed from the wild type, AS93, by targeted gene disruption and used in all further analysis. Such reconstructions are necessary, since IS sequences in S. oneidensis MR-1 are known to cause spontaneous mutations, suggesting that a transposon insertion per se might not be the unequivocal cause of an observed phenotype (3). The gene disruption mutants were constructed in gfp-labeled wild-type AS93 by integration of a suicide plasmid carrying an internal sequence of the target.
The role of swimming motility on initial attachment and maturation of S. oneidensis MR-1 biofilms.
To study the roles of the flagellum, motility, and pili in the early phases of biofilm formation, two mutants defective in flagellum assembly, flhB and fliK mutants, designated AS95 and AS94, respectively, were constructed. Both mutants were unable to swim according to soft agar assay results (data not shown). It has been demonstrated for P. aeruginosa and Clostridium difficile that the flagellum itself is involved in surface attachment in natural environments (1, 46). Therefore, we constructed additional mutants with an intact but paralyzed flagellum. S. oneidensis MR-1 contains two similar gene clusters that might be involved in flagellar rotation and are designated motA/B and pomA/B (SO4286/87 and SO1529/30, respectively). In analogy to Vibrio species, motA/B is likely to be responsible for proton-driven swimming, while pomA/B potentially uses a Na+ gradient to drive the rotation of the polar flagellum as shown for Vibrio parahaemolyticus (28). Both gene clusters were inactivated by targeted gene disruption, and only interruption of the SO1529 (pomA) gene yielded a nonswimming mutant, AS96, as revealed by the soft agar assay. A deletion of SO4287 (motA) did not affect swimming under the conditions tested, suggesting that rotation of the polar flagellum of S. oneidensis MR-1 is sodium dependent. None of the introduced mutations caused a growth defect.
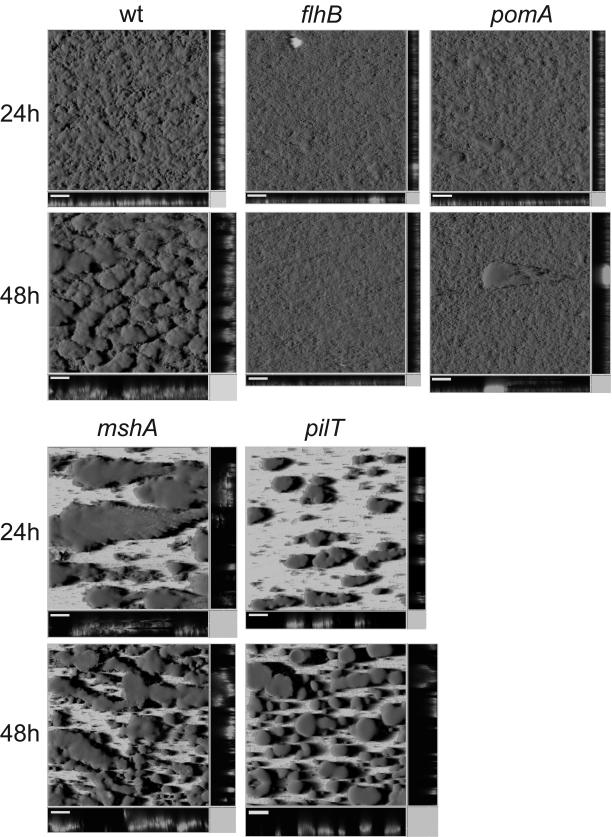
Biofilms of flhB, fliK, and pomA mutants were grown in LM medium in the hydrodynamic flow-chamber system and monitored for 48 h. In each experiment, strain AS93 was grown as a wild-type reference in parallel (Fig. 3). The extent of initial attachment of the flhB, fliK, and pomA mutants was indistinguishable from that of the wild type. The numbers of cells attached to the glass surface after 1 and 4 h (data not shown) were the same for the swimming mutants and for the wild type, and almost complete surface coverage was achieved after 24 h by all strains (Fig. 3). However, major differences in biofilm architecture were noticeable. By 24 h, the wild-type biofilms had undergone the transition from flat to structured, while motility mutant biofilms remained flat with only sporadically occurring thicker clusters of cells (Fig. 3). COMSTAT analysis (13) revealed that both flhB and fliK mutants accumulate only about half of the amount of biomass (6.4 ± 0.6 and 8.93 ± 0.8 μm3/μm2) and displayed a significant lower average thickness (11.2 ± 1.3 and 14.1 ± 1.5 μm) compared to the wild type (16.7 ± 0.5 μm3/μm2 and 19.5 ± 1.9 μm, respectively). Additionally, both mutants showed a distinct phenotype by forming long chains of cells that were aligned in the direction of the flow, which could be observed only in cells grown in flow chambers but not in batch liquid culture (data not shown). In later-stage biofilms, pomA mutant cells also formed chains of cells, but these were shorter than those of the flagella mutants. pomA biofilms formed slightly thicker layers than flhB and fliK biofilms (16.2 ± 0.9 μm) and contained little more biomass (10.6 ± 0.5 μm3/μm2) than the flagellum mutants.
FIG. 3.
Biofilm architecture of S. oneidensis MR-1 mutants in flagellum assembly (flhB; AS95), flagellum rotation (pomA; AS96), MSHA pilus (mshA; AS97), or pilus retraction (pilT; AS98), taken at 24 h or 48 h. The wild-type reference is strain AS93. Bar = 100 μm.
After 48 h, the difference between these motility mutants and the wild type became more severe. Cessation of development or a slight decrease of biomass and thickness was observed with all three nonmotile mutants, while wild-type biofilm thickness almost doubled in that time period (Fig. 3). The motility mutants biofilms remained flat. No further structural development or biomass accumulation occurred (observed until day 5; data not shown). Obviously, swimming motility has no impact on initial attachment of S. oneidensis MR-1 cells to a glass surface, but contributes significantly to the development of biofilm architecture. Notably, the presence of a paralyzed flagellum might lead to slightly enhanced biomass accumulation and biofilm thickness compared to mutants without flagellum.
A role of swimming motility in bacterial biofilm formation has been reported previously, but the exact contribution remains unknown. Observations in earlier studies of E. coli, P. aeruginosa, and V. cholerae El Tor mutants in static systems suggested that flagellum-driven motility enhances initial cell-surface contact and might also contribute to the spreading of cells on the substratum (36, 40, 49). Our studies did not confirm this observation and suggest that flagella and flagellum rotation are not critical for initial attachment but rather for structural development. More-recent studies with P. aeruginosa and V. cholerae O136 have shown that flagella are apparently involved in building or maintaining the architecture of biofilms. In flow chamber experiments with P. aeruginosa, it was found that a fliM mutation leads to a more structured biofilm with a higher biomass accumulation than that for a wild-type strain tested under the same conditions (17). In static systems, the absence of a flagellum in V. cholerae O136 was shown to result in a so-called rugose phenotype, characterized by an overproduction of extracellular polymeric substances, as indicated by an altered colony morphology and biofilm structure (51). However, the phenotype found for these two microorganisms is strikingly different from that observed for S. oneidensis here and, so far, has not been described for any other microorganism. The possible role of the flagella and/or flagellum rotation is the subject of ongoing work in our laboratory.
The role of the MSHA pilus and pilT.
Two of the genes identified during transposon mutagenesis were identified as members of the mannose-sensitive hemagglutinin type IV pilus (MSHA) gene cluster, mshN (SO4210) and mshO (SO4102). Type IV pili are known to represent a critical factor for biofilm formation on abiotic surfaces. The MSHA pilus was first described and characterized for V. cholerae (14, 26) and was shown to play a critical role in the initial attachment of V. cholerae El Tor to abiotic surfaces but not to nutritive substrates such as chitin (49, 50, 51). More than half of all V. cholerae mutants found in a screen for mutants defective in initial attachment mapped to genes in the two clusters encoding the gene products for transport and pili subunits (50). Notably, the MHSA pilus is not involved in the same process in another V. cholerae strain, strain O139 (51). In order to identify a possible role of this pilus in biofilm formation of S. oneidensis MR-1, a gene disruption mutant in the putative main pilus subunit, mshA, was constructed, resulting in strain AS97. Characterization of the mshA mutant in flow chambers revealed a severe defect in initial adhesion, as observed after 1 and 4 h. Only a very few cells were found to be attached to the glass surface (data not shown). After 24 h, large but well-isolated suspended structures had formed, presumably by clonal growth of the few mutant cells that initially adhered firmly. After 48 h, more colonies appeared, and the overall coverage of the surface increased, but in contrast to wild-type biofilms (Fig. 3), large areas remained uncovered. This observation is different from data for V. cholerae El Tor (50), where mshA mutant cells attached sparsely to form isolated colonies but apparently did not form such extensive structures. However, the results were obtained from different biofilm systems (static for V. cholerae versus hydrodynamic for S. oneidensis) and thus are difficult to compare. Despite this difference, it is obvious that the MSHA pilus represents a critical component for initial attachment to abiotic surfaces in both organisms. The similarity between the biofilms formed by the mshA mutant and the LB-grown AS93 strain is interesting. The poor surface coverage and the large amount of biomass may suggest a metabolic control of MSHA gene cluster expression, which might be down-regulated under high-nutrient conditions.
The group of biofilm-defective S. oneidensis Tn5 mutants with inactivated motility genes also included a pilT insertion (SO3351). This gene is involved in twitching motility in other microbes, presumably by mediating retraction of the type IV pili (27). Mutants of pilT of Myxococcus xanthus, Neisseria gonorrhoeae, P. aeruginosa, and V. cholerae are nontwitching and hyperpiliated (4, 50, 53, 54). We constructed a pilT mutant of S. oneidensis MR-1, strain AS93, and characterized its biofilm in flow chamber experiments (Fig. 3). In contrast to the mshA mutant phenotype, initial attachment was unaffected in the pilT mutant cells compared to that in wild-type cells, while significant differences in biofilm architecture could be observed in later stages. After 24 and 48 h, pilT mutants formed well-isolated, highly symmetrical and pronounced structures, up to 100 μm in height. In contrast to wild-type biofilms, most of the surface area of pilT biofilms remained uncovered, except for the spots where the towering structures developed. The number of colonies increased slightly with time, though. In a sense, this mutant phenotype resembled that of the mshA mutant. However, while the latter one gave rise to structures comparable to early stages of the wild-type biofilm with cells being rather loosely attached to each other, pilT mutants formed very densely packed, tower-like structures. Enhanced biofilm formation of P. aeruginosa has been associated with hyperpiliated mutants (7).
Similar to the case with S. oneidensis, a pilT mutant of P. aeruginosa was found to form more-densely packed cell clusters (4). However, in contrast to our observations, in microtiter plate assays as well as in hydrodynamic biofilm systems, the P. aeruginosa mutant accumulated larger amounts of biomass than the wild type. Similar to our observations, a pilT mutant in V. cholerae was reported to exhibit decreased biofilm formation in microtiter plate assays (50).
The distinct phenotype of the pilT mutant suggests a possible role for pili-mediated cell-cell contact in S. oneidensis MR-1. For E. coli it was shown that conjugative pili can promote biofilm formation by acting as an adhesion factor (10) and appear to be sufficient for extensive structural development, even in the background of strains with mutations that normally impair biofilm formation in the E. coli K-12 wild type (41). In order to test whether or not type IV pili exert a structural function within S. oneidensis MR-1 wild-type biofilms beyond a role in mediating contact to the substratum, mutations were introduced into genes designated pilM and pilC. Both gene products are thought to be essential for type IV pilus assembly (27). However, the introduced mutations did not diminish initial attachment or structural development, either in the microtiter plate assay or in flow chamber experiments (data not shown). This suggests that type IV pili mediated cell-cell contact is not a dominant determinant for biofilm architecture under the conditions tested. Notably, with the exception of mshA, genes encoding proteins with high homologies to pilin subunits are absent.
While for some Vibrio strains, the MSHA type IV pilus is crucial for attachment to surfaces, such as polysterene or glass (50), type IV pilus-mediated twitching motility seems to have an important function in development of biofilm architecture. Unlike in S. oneidensis, in P. aeruginosa biofilms, type IV pili are not essential for the initial attachment process. Results obtained from static and hydrodynamic systems suggested that twitching motility is critical for microcolony and structure formation of biofilm cells (17, 18, 36). Although our studies did not demonstrate directly that S. oneidensis MR-1 cells move by twitching motility, we conclude from the observed pilT phenotype and by analogy to other bacterial systems that pili and pilT-dependent dynamic processes might contribute to the biofilm dynamics of S. oneidensis. In addition to mediating cell-cell contacts, such roles may include cell movement along the substratum or within the biofilm.
This study provided a comprehensive, high-optical-resolution analysis of S. oneidensis biofilms developing on glass surfaces in a hydrodynamic flow chamber system. Despite the active research on biofilms over the last decade, only for a few microorganisms (e.g., see references 17, 41, and 47) have similar single-cell-resolution biofilm studies been reported. Our studies included biofilms of S. oneidensis mutants defective in flagellum, swimming motility and type IV pili. Although equivalent studies were performed with other microbes as well, the actual roles and functions of flagella, motility, and pili are still largely unknown and might vary between different species. The noticeable differences that we observed in the S. oneidensis mutant biofilms will serve as a useful basis for developing more-specific hypotheses for their physiological functions, such as the potential role of Na+ metabolism in biofilm formation. Biofilm formation and dynamics are particularly relevant to the study of S. oneidensis because of its capacity to metabolically interact with insoluble electron acceptors (24, 31, 32, 35) and for investigating how the motility-related and other components identified could be involved in mediating catabolic electron transfer to Fe(III) and Mn(IV) oxide-containing mineral surfaces under anoxic conditions.
Acknowledgments
We are grateful to Søren Molin for kindly providing the Tn7 delivery system for the construction of the gfp-expressing S. oneidensis MR-1 strains.
This work was supported by grants from the NSF and a Powell Foundation faculty award to A.M.S.
REFERENCES
- 1.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, Y., D. P. Lies, H. Fu, and G. P. Roberts. 1991. An improved Tn7-based system for single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene 109:167-168. [DOI] [PubMed] [Google Scholar]
- 3.Bordi, C., C. Iobbi-Nivol, V. Méjean, and J.-C. Patte. 2003. Effects of ISSo2 insertions in structural and regulatory genes of the trimethylamine oxide reductase of Shewanella oneidensis. J. Bacteriol. 185:2042-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang, P., and L. L. Burrows. 2003. Biofilm formation by hyperpilated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Giskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Déziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubiel, M., C. H. Hsu, C. C. Chien, F. Mansfeld, and D. K. Newman. 2002. Microbial iron respiration can protect steel from corrosion. Appl. Environ. Microbiol. 68:1440-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fendorf, S. E., B. W. Weilinga, and C. M. Hansel. 2000. Chromium transformations in natural environments: the role of biological and abiological processes in chromium(VI) reduction. Int. Geol. Rev. 42:691-701. [Google Scholar]
- 10.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 11.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 12.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. M. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1093-1094. [DOI] [PubMed] [Google Scholar]
- 13.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Giskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 14.Jonson, G., M. Lebens, and J. Holmgren. 1994. Cloning and sequencing of Vibrio cholerae mannose-sensitive haemagglutinin pilin gene: localization of mshA within a cluster of type 4 pilin genes. Mol. Microbiol. 13:109-118. [DOI] [PubMed] [Google Scholar]
- 15.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram negative eubacteria: regulation of the Pm promoter in the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 16.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 18.Klausen, M., A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 50:61-68. [DOI] [PubMed] [Google Scholar]
- 19.Koch, B., L. E. Jensen, and O. Nybroe. 2001. A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45:187-195. [DOI] [PubMed] [Google Scholar]
- 20.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 21.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 22.Langner, H. W., and W. P. Insleep. 2000. Microbial reduction of arsenate in the presence of ferrihydrite. Environ. Sci. Technol. 34:3131-3136. [Google Scholar]
- 23.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-200. [DOI] [PubMed] [Google Scholar]
- 24.Lovley, D. R. 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47:263-290. [DOI] [PubMed] [Google Scholar]
- 25.Lower, S. K., M. F. Hochella, Jr., and T. J. Beveridge. 2001. Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and alpha-FeOOH. Science 292:1360-1363. [DOI] [PubMed] [Google Scholar]
- 26.Marsh, J. W., and R. K. Taylor. 1999. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J. Bacteriol. 181:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 28.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 30.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers, C., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1320. [DOI] [PubMed] [Google Scholar]
- 32.Myers, C. R., and K. H. Nealson. 1990. Respiration-linked proton translocation coupled to anaerobic reduction of manganese(IV) and iron(III) in Shewanella putrefaciens MR-1. J. Bacteriol. 172:6232-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myneni, S. B. C., T. K. Tokunaga, and G. E. Brown, Jr. 1997. Abiotic selenium redox transformations in the presence of Fe(II,III) hydroxides. Science 278:1106-1109. [Google Scholar]
- 34.Neal, L. A., K. M. Rosso, G. G. Geesey, Y. A. Gorby, and B. J. Little. 2003. Surface structure effects on direct reduction of iron oxides by Shewanella oneidensis. Geochim. Cosmochim. Acta 67:4489-4503. [Google Scholar]
- 35.Nealson, K. H., and D. Saffarini. 1994. Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48:311-343. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 37.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 38.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 39.Penfold, R. J., and J. M. Pemberton. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145-146. [DOI] [PubMed] [Google Scholar]
- 40.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 41.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1:784-791. [Google Scholar]
- 45.Sternberg, C., B. B. Christensen, T. Johansen, A. Toftgaard-Nielsen, J. B. Andersen, M. Giskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tasteyre, A., M.-C. Barc, A. Collignon, H. Boureau, and T. Karjalainen. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and dynamics of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 49.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Giskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfgang, M., P. Lauer, H. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]
- 54.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]