Abstract
Representative strains of the Bacillus cereus group of bacteria, including Bacillus anthracis (11 isolates), B. cereus (38 isolates), Bacillus mycoides (1 isolate), Bacillus thuringiensis (53 isolates from 17 serovars), and Bacillus weihenstephanensis (2 isolates) were assigned to 59 sequence types (STs) derived from the nucleotide sequences of seven alleles, glpF, gmk, ilvD, pta, pur, pycA, and tpi. Comparisons of the maximum likelihood (ML) tree of the concatenated sequences with individual gene trees showed more congruence than expected by chance, indicating a generally clonal structure to the population. The STs followed two major lines of descent. Clade 1 comprised B. anthracis strains, numerous B. cereus strains, and rare B. thuringiensis strains, while clade 2 included the majority of the B. thuringiensis strains together with some B. cereus strains. Other species were allocated to a third, heterogeneous clade. The ML trees and split decomposition analysis were used to assign STs to eight lineages within clades 1 and 2. These lineages were defined by bootstrap analysis and by a preponderance of fixed differences over shared polymorphisms among the STs. Lineages were named with reference to existing designations: Anthracis, Cereus I, Cereus II, Cereus III, Kurstaki, Sotto, Thuringiensis, and Tolworthi. Strains from some B. thuringiensis serovars were wholly or largely assigned to a single ST, for example, serovar aizawai isolates were assigned to ST-15, serovar kenyae isolates were assigned to ST-13, and serovar tolworthi isolates were assigned to ST-23, while other serovars, such as serovar canadensis, were genetically heterogeneous. We suggest a revision of the nomenclature in which the lineage and clone are recognized through name and ST designations in accordance with the clonal structure of the population.
The Bacillus cereus group comprises closely related gram-positive bacteria that exhibit highly divergent pathogenic properties. Many bacteria classified as B. cereus are widely distributed in the environment, with probable reservoirs in the soil (57), and as commensal inhabitants of the intestines of insects (35). Occasionally they are associated with food poisoning (16) and with soft tissue infections, particularly of the eye (9). Other members of the group that are currently classified as Bacillus thuringiensis are primarily insect pathogens. These bacteria produce toxins in the form of parasporal crystal proteins that have been widely used for the biocontrol of insect pests (49). Occasionally, B. thuringiensis strains are responsible for human infections similar to those caused by strains of B. cereus (7, 25). A third pathogenic phenotype is exhibited by Bacillus anthracis, a pathogen of mammals and especially ungulates that can cause human disease (36). The principal virulence factors of B. anthracis are encoded by genes located on two plasmids: the tripartite toxin genes pag, lef, and cya are carried on plasmid pXO1, while the genes encoding the biosynthesis of the poly-d-glutamate capsule, capA, capB, and capC, are carried on a smaller plasmid, pXO2 (38). Similarly, the crystal protein genes responsible for the major features of insect toxicity of B. thuringiensis isolates are almost invariably plasmid encoded (49). The virulence genes of B. cereus, on the other hand, are chromosomal (17, 24, 43).
These three species of the B. cereus group were first described around the turn of the 19th century, yet despite this long history the relationships between these organisms have yet to be completely resolved (44, 46). Whole-genome DNA hybridization has been unhelpful (28, 37, 50), while conventional markers of chromosomal diversity, such as 16S and 23S rRNA genes, are essentially identical (2, 3). Comprehensive studies using a diverse range of techniques, including genomic mapping (5), pulsed-field gel electrophoresis of chromosomal DNA (4), multilocus enzyme electrophoresis (18, 19), variable number tandem repeat mapping, BOX-PCR fingerprinting (31), amplified fragment length polymorphism (AFLP) analysis (54), and multilocus sequence typing (MLST) (20), have revealed extensive genomic similarities and few consistent differences among isolates currently classified as B. anthracis, B. cereus, and B. thuringiensis. These studies have reinforced the phenotypic argument (15) that the three taxa should be considered a single bacterial species (15, 19).
Despite such biological arguments for unification, a separate species status for these bacteria has been maintained because of their distinctive pathogenic features. Virtually all B. cereus group isolates obtained from humans or animals exhibiting the symptoms of anthrax are very closely related to each other, and B. anthracis is very likely to be a clone, particularly if associated with the toxin-encoding plasmids pXO1 and pXO2 (29, 30). However, organisms classified as B. cereus and B. thuringiensis are more diverse, and the evolutionary relationships between all members of the group have yet to be definitively established (21). This is important, not only for understanding the evolution of virulence in the B. cereus group, but also for rapidly and accurately characterizing these organisms, a concern which has become of increasing scientific and political importance in recent years.
MLST studies that employ nucleotide sequence analysis to identify genetic variation have been highly successful for characterizing bacterial genetic variation and for developing evolutionary frameworks that interpret this diversity (56). Here we have employed this approach by determining the nucleotide sequences of seven housekeeping gene fragments for 105 representative members of the B. cereus group and related organisms. The results demonstrate a largely clonal population structure and indicate that the group comprises at least eight distinct lineages. Two of these lineages centered on B. anthracis and B. thuringiensis serovar sotto contain strains of a single species, but the remainder are mixed and contain isolates that are currently classified as different species.
MATERIALS AND METHODS
Bacterial isolates.
A total of 105 pure cultures, representing B. anthracis and related bacteria that were isolated globally over the period 1900-1999, were analyzed (Table 1). The collection comprised isolates that were classified as B. anthracis (11 isolates), B. cereus (38 isolates), Bacillus mycoides (1 isolate), B . thuringiensis (53 isolates representing 17 serovars), and Bacillus weihenstephanensis (2 isolates). Further details of the strain collection are available at http://pubmlst.org/bcereus/. B. thuringiensis isolates were classified on the basis of the presence of a crystal protein and/or insect toxicity, while B. cereus isolates lacked a crystal protein. Isolates were named as received and, if necessary, checked for the absence or presence of a crystal protein by light and/or scanning electron microscopy. Bacterial cultures for DNA isolation were grown in nutrient broth (5 ml) containing 0.5% glucose at 30°C until the late exponential phase, which required an incubation period of 16 h for most isolates.
TABLE 1.
Strains used for this study and their allocation to lineages
| Clade or lineage | ST | Strain | Country of origin | Yr isolated | Original designationa | Reference or sourceb |
|---|---|---|---|---|---|---|
| Clade 1 (B. cereus) | ||||||
| Anthracis | ST-1 | Ames | United States | 1981 | B. anthracis | |
| ST-1 | Ames (cured strain) | United States | B. anthracis | 46 | ||
| ST-1 | K0610/A0034 | China | B. anthracis | 30 | ||
| ST-1 | K4834/A0039 | Australia | 1994 | B. anthracis | 30 | |
| ST-1 | K1340/A0062 | Poland | 1962 | B. anthracis | 30 | |
| ST-1 | K1694/A0462 | United States | 1932 | B. anthracis | 30 | |
| ST-1 | K5135/A0463 | Pakistan | 1978 | B. anthracis | 30 | |
| ST-1 | K4596/A0488 | United Kingdom | 1997 | B. anthracis | 30 | |
| ST-2 | K3700/A0267 | United States | 1937 | B. anthracis | 30 | |
| ST-3 | K2478/A0102 | Mozambique | 1944 | B. anthracis | 30 | |
| ST-3 | K2762/A0465 | France | 1997 | B. anthracis | 30 | |
| Cereus I | ST-5 | m1545 | Brazil | 1987 | B. cereus | MADM |
| ST-6 | m1564 | Brazil | 1987 | B. cereus | MADM | |
| ST-7 | M21 | Finland | 1998 | B. cereus | 40 | |
| ST-32 | ATCC 10987 | Canada | 1930 | B. cereus | ATCC | |
| Cereus II (emetic) | ST-26 | F4810/72 | United States | 1972 | B. cereus | 55 |
| ST-26 | S710 | United Kingdom | 1979 | B. cereus | 41 | |
| ST-26 | F3080B/87 | United Kingdom | 1987 | B. cereus | 40 | |
| ST-26 | F3942/87 | United Kingdom | 1987 | B. cereus | 40 | |
| ST-31 | S366 | North Sea | B. cereus | 41 | ||
| ST-45 | m1293 | Brazil | 1987 | B. cereus | MADM | |
| ST-47 | m1576 | Brazil | 1987 | B. cereus | MADM | |
| Cereus III | ST-27 | F4370/75 | United Kingdom | 1975 | B. cereus | 41 |
| ST-57 | T10024 | Pakistan | 1975 | B. thuringiensis serovar darmstadiensis | IP | |
| ST-60 | T18004 | Iraq | 1984 | B. thuringiensis serovar kumamotoensis | IP | |
| Clade 2 (B. thuringiensis) | ||||||
| Kurstaki | ST-8 | S57 | United States | 1975 | B. cereus | BA |
| ST-8 | S58 | United States | 1975 | B. cereus | 41 | |
| ST-8 | S59 | United States | 1975 | B. cereus | BA | |
| ST-8 | T03a001 | France | 1961 | B. thuringiensis serovar kurstaki | IP | |
| ST-8 | T03a075 | Iraq | 1976 | B. thuringiensis serovar kurstaki | IP | |
| ST-8 | T03a172 | Pakistan | 1982 | B. thuringiensis serovar kurstaki | IP | |
| ST-8 | T03a287 | Kenya | 1988 | B. thuringiensis serovar kurstaki | IP | |
| ST-8 | T03a361 | Australia | 1990 | B. thuringiensis serovar kurstaki | IP | |
| ST-13 | T04b001 | Kenya | 1962 | B. thuringiensis serovar kenyae | IP | |
| ST-13 | T04b054 | Iraq | 1986 | B. thuringiensis serovar kenyae | IP | |
| ST-13 | T04b060 | Iraq | 1987 | B. thuringiensis serovar kenyae | IP | |
| ST-13 | T04b073 | Chile | 1993 | B. thuringiensis serovar kenyae | IP | |
| ST-15 | T07033 | Japan | 1975 | B. thuringiensis serovar aizawai | IP | |
| ST-15 | T07058 | France | 1983 | B. thuringiensis serovar aizawai | IP | |
| ST-15 | T07180 | Spain | 1992 | B. thuringiensis serovar aizawai | IP | |
| ST-18 | T13028 | Chile | 1993 | B. thuringiensis serovar pakistani | IP | |
| ST-25 | T05005 | United States | 1964 | B. thuringiensis serovar galleriae | IP | |
| ST-25 | T05033 | United States | 1975 | B. thuringiensis serovar galleriae | IP | |
| ST-25 | T05144 | France | 1985 | B. thuringiensis serovar galleriae | IP | |
| ST-33 | ATCC 10876 | 1945 | B. cereus | ATCC | ||
| ST-39 | SPS 2 | 1999 | B. cereus | 40 | ||
| ST-40 | TSP 11 | 1999 | B. cereus | 40 | ||
| ST-44 | m1292 | Brazil | 1987 | B. cereus | MADM | |
| ST-51 | T05a015 | United States | 1977 | B. thuringiensis serovar canadensis | IP | |
| ST-54 | T07196 | Brazil | 1993 | B. thuringiensis serovar aizawai | IP | |
| ST-59 | T18001 | Japan | 1980 | B. thuringiensis serovar kumamotoensis | IP | |
| ST-59 | T18002 | United States | 1980 | B. thuringiensis serovar kumamotoensis | IP | |
| Sotto | ST-9 | NCTC 6474 | United Kingdom | B. cereus | 41 | |
| ST-12 | T04002 | Canada | 1965 | B. thuringiensis serovar sotto | IP | |
| ST-12 | T04016 | Pakistan | 1980 | B. thuringiensis serovar sotto | IP | |
| ST-12 | T04024 | Pakistan | 1981 | B. thuringiensis serovar sotto | IP | |
| ST-12 | T15001 | United States | 1983 | B. thuringiensis serovar dakota | IP | |
| ST-16 | CCCT 2259 | Brazil | 1993 | B. thuringiensis serovar israelensis | 27 | |
| ST-16 | T08025 | France | 1988 | B. thuringiensis serovar morrisoni | IP | |
| ST-23 | T08001 | United States | 1963 | B. thuringiensis serovar morrisoni | IP | |
| ST-23 | T08009 | United States | 1979 | B. thuringiensis serovar morrisoni | IP | |
| ST-23 | T08012 | Pakistan | 1980 | B. thuringiensis serovar morrisoni | IP | |
| ST-23 | T08023 | Brazil | 1987 | B. thuringiensis serovar morrisoni | IP | |
| ST-23 | T08031 | Brazil | 1991 | B. thuringiensis serovar morrisoni | IP | |
| ST-49 | T04236 | Indonsesia | 1991 | B. thuringiensis serovar sotto | IP | |
| ST-55 | T10016 | United States | 1982 | B. thuringiensis serovar darmstadiensis | IP | |
| ST-56 | T10003 | Germany | 1967 | B. thuringiensis serovar darmstadiensis | IP | |
| ST-56 | T10018 | Japan | 1982 | B. thuringiensis serovar darmstadiensis | IP | |
| Thuringiensis | ST-10 | T01001 | Canada | 1958 | B. thuringiensis serovar thuringiensis | IP |
| ST-10 | T01015 | Bulgaria | 1962 | B. thuringiensis serovar thuringiensis | IP | |
| ST-10 | T01022 | United States | 1964 | B. thuringiensis serovar thuringiensis | IP | |
| ST-10 | T01326 | Chile | 1993 | B. thuringiensis serovar thuringiensis | IP | |
| ST-20 | WSBC 10312 | Thailand | 1999 | B. cereus | 43 | |
| ST-43 | m1278 | Brazil | 1987 | B. cereus | MADM | |
| ST-58 | T15006 | South Korea | 1993 | B. thuringiensis serovar dakota | IP | |
| Tolworthi | ST-4 | ATCC 14579T | United States | 1916 | B. cereus | ATCC |
| ST-14 | T06007 | Pakistan | 1983 | B. thuringiensis serovar entomocidus | IP | |
| ST-14 | T06010 | Pakistan | 1983 | B. thuringiensis serovar entomocidus | IP | |
| ST-17 | T13001 | Pakistan | 1976 | B. thuringiensis serovar pakistani | IP | |
| ST-17 | T13004 | Pakistan | 1980 | B. thuringiensis serovar pakistani | IP | |
| ST-19 | WSBC 10249 | Denmark | 1999 | B. cereus | 43 | |
| ST-19 | m1280 | Brazil | 1987 | B. cereus | MADM | |
| ST-22 | T09010 | United States | 1979 | B. thuringiensis serovar tolworthi | IP | |
| ST-22 | T09011 | Iraq | 1987 | B. thuringiensis serovar tolworthi | IP | |
| ST-22 | T09024 | Indonesia | 1991 | B. thuringiensis serovar tolworthi | IP | |
| ST-22 | T09034 | Brazil | 1992 | B. thuringiensis serovar tolworthi | IP | |
| ST-24 | NCIB 6349 | B. cereus | 41 | |||
| ST-24 | Cal3 | Finland | 1998 | B. cereus | 40 | |
| ST-24 | WSBC 10028 | Germany | 1999 | B. cereus | 43 | |
| ST-29 | S86 | B. cereus | 41 | |||
| ST-34 | ATCC 11778 | United States | B. cereus | ATCC | ||
| ST-46 | m1550 | Brazil | 1987 | B. cereus | MADM | |
| ST-48 | T01246 | Iraq | 1984 | B. thuringiensis serovar thuringiensis | IP | |
| ST-50 | T05a001 | Canada | 1968 | B. thuringiensis serovar canadensis | IP | |
| ST-52 | T05a019 | Pakistan | 1980 | B. thuringiensis serovar canadensis | IP | |
| ST-53 | T07146 | Indonesia | 1991 | B. thuringiensis serovar aizawai | IP | |
| Unassigned | ST-28 | F4431/3 | Indonesia | 1973 | B. cereus | 41 |
| ST-30 | S363 | North Sea | B. cereus | 41 | ||
| ST-38 | ATCC 4342 | United States | 1900 | B. cereus | 15 | |
| Other | ST-21 | WSBC 10277 | Germany | 1999 | B. mycoides | 43 |
| ST-35 | AH621 | Norway | B. cereus | 18 | ||
| ST-36 | AH647 | Norway | B. cereus | 18 | ||
| ST-37 | AH684 | Norway | B. cereus | 18 | ||
| ST-41 | WSBC 10202 | Germany | 1999 | B. weihenstephanensis | 43 | |
| ST-42 | WSBC 10364 | Germany | 1999 | B. weihenstephanensis | 43 |
B. thuringiensis strains are given serovar designations when they are known.
IP, Collection of Bacillus thuringiensis and Bacillus sphaericus, Institut Pasteur, Paris, France; BA, Brian Austin, Heriot Watt University, Edinburgh, United Kingdom; and MAMD, Marilena Aquino de Muro, CABI Biosciences, Egham, United Kingdom.
Molecular methods.
Chromosomal DNAs were prepared from 1.0-ml aliquots of the cultures by the use of a PureGene DNA isolation kit (Gentra Systems) in accordance with the manufacturer's instructions, with the exception that the lyticase solution was increased to 2.5 μl. Seven genes distributed around the chromosome of B. anthracis Ames were chosen for MLST. Four loci, glpF, gmk, pta, and tpi, were derived from those used for MLST of Staphylococcus aureus, a low-G+C gram-positive bacterium (11). The nucleotide sequences of these loci were obtained from the B. anthracis complete genome sequence by BLAST searches and were used to design PCR amplification and nucleotide sequencing oligodeoxyribonucleotide primer sequences. The PCR amplification and nucleotide sequencing primers for the remaining loci, ilvD, pur, and pycA, were designed from the sequences described by Økstad et al. (39). The primers used (annealing temperatures are in parentheses) were as follows: Glp-F, 5′-GCGTTTGTGCTGGTGTAAGT; Glp-R, 5′-CTGCAATCGGAAGGAAGAAG (59°C); Gmk-F, 5′-ATTTAAGTGAGGAAGGGTAGG; Gmk-R, 5′-GCAATGTTCACCAACCACAA (56°C); Gmk2-F (an alternative forward primer for gmk that was sometimes necessary), 5′-ATCGTTCTTTCAGGACCTTC (56°C); IlvD-F, 5′-CGGGGCAAACATTAAGAGAA; and IlvD-R, 5′-GGTTCTGGTCGTTTCCATTC (58°C). For emetic strains of B. cereus, the following alternative primers were necessary: IlvD2, 5′-AGATCGTATTACTGCTACGG; IlvD2-R, 5′-GTTACCATTTGTGCATAACGC (58°C); Pta-F, 5′-GCAGAGCGTTTAGCAAAAGAA; Pta-R, 5′-TGCAATGCGAGTTGCTTCTA (58°C); Pur-F, 5′-CTGCTGCGAAAAATCACAAA; Pur-R, 5′-CTCACGATTCGCTGCAATAA (56°C); PycA-F, 5′-GCGTTAGGTGGAAACGAAAG; PycA-R, 5′-CGCGTCCAAGTTTATGGAAT (57°C); Tpi-F, 5′-GCCCAGTAGCACTTAGCGAC; and Tpi-R, 5′-CCGAAACCGTCAAGAATGAT (58°C). The same primers were used for DNA sequencing, and the methods used are available at http://pubmlst.org/bcereus/.
Each locus was amplified by PCR and purified by polyethylene glycol precipitation as described previously (10). Nucleotide sequence extension reactions were performed on the purified amplicons by the use of BigDye Ready Reaction mix (ABI Corp), and reaction products were separated and detected on a Prism 3700 or a Prism 310 automated DNA analyzer (ABI Corp.). Nucleotide sequences were determined at least once for each DNA strand and were assembled with the STADEN software package (52). All sequences are available from http://pubmlst.org/bcereus/, while representative sequences have been submitted to GenBank (Table 2). Each unique sequence was assigned an arbitrary allele number by reference to the B. cereus group MLST database (http://pubmlst.org/bcereus/), which employed MLSTdbnet software (26). The combination of allele numbers for all seven loci of a given isolate was assigned an arbitrary sequence type (ST); each ST was equivalent to a unique haplotype.
TABLE 2.
Genetic loci analyzed in this study and their characteristics
| Locus | Encoded protein | Genomic positiona | Fragment length (bp) | Total length of gene (bp) | No. of alleles | Avg p distance | dN/dS | Representative accession no. |
|---|---|---|---|---|---|---|---|---|
| glpF | Glycerol uptake facilitator protein | 1014815 | 381 | 822 | 37 | 0.023 | 0.108 | AY729746-AY729753 |
| gmk | Guanylate kinase (putative) | 3688226 | 504 | 618 | 19 | 0.044 | 0.022 | AY729753-AY729761 |
| ilvD | Dihydroxyacid dehydratase | 1736221 | 393 | 1674 | 30 | 0.067 | 0.017 | AY729762-AY729769 |
| pta | Phosphate acetyltransferase | 5122669 | 414 | 972 | 31 | 0.023 | 0.019 | AY729770-AY729777 |
| pur | Phosphoribosylaminoimidazole carboxamide formyltransferase | 306074 | 348 | 1,536 | 30 | 0.046 | 0.010 | AY729778-AY729785 |
| pycA | Pyruvate carboxylase | 3809749 | 363 | 3,447 | 33 | 0.065 | 0.028 | AY729786-AY729793 |
| tpi | Triosephosphate isomerase | 4861379 | 435 | 756 | 34 | 0.015 | 0.110 | AY729794-AY729801 |
Based on the B. anthracis Ames genome (46).
Analysis of sequence diversity.
The nucleotide sequences were analyzed with the MEGA (32) and DnaSP (48) packages, which were used to calculate the (uncorrected) p distances, mean numbers of nonsynonymous (dN) and synonymous (dS) substitutions per site, numbers of differences among various groups of sequences, and numbers of fixed differences and shared polymorphisms among lineages. Distances among concatenated sequences were visualized by split decomposition analysis implemented in the SPLITSTREE program (23), using Hamming distances, which were equivalent to p distances.
Phylogenetic analysis.
Maximum likelihood (ML) phylogenetic trees were reconstructed by using the general time-reversible model of DNA substitution, with a nucleotide substitution matrix and a shape parameter (α) of a discrete approximation (with four categories) to a gamma distribution of rate heterogeneity among sites, the proportion of invariant sites (I), and the base composition estimated from the empirical data during tree reconstruction. For the ML tree of the concatenated data (see below), these parameter values were as follows: for the general time-reversible substitution model, A→C = 0.60228, A→G = 4.24750, A→T = 0.91374, C→G = 0.32520, C→T = 6.28401, G→T = 1.00000, α = 1.70796, and I = 0.81537. The base compositions were as follows: A = 0.33003, C = 0.16656, G = 0.23004, and T = 0.27307. The parameter values for the individual loci are available upon request. To assess the phylogenetic support for groupings on the tree, we performed a bootstrap resampling analysis (1,000 replications). This analysis was run by using 1,000 replicate neighbor-joining trees estimated by the maximum likelihood substitution model described above. To obtain a general measure of the overall degree of incongruence between trees of each of the seven loci, we compared, for each locus in turn, the likelihood of the ML tree for that locus to those of the ML topologies obtained for the other loci and to 200 randomly generated trees of the same size, with the branch lengths being re-estimated in each case. If the ML trees for each locus were congruent, then all of them would have likelihoods that were higher than those of the random trees (12). All of these analyses were undertaken by using the PAUP* 4.0 software package (53).
RESULTS
Sequence diversity.
The MLST gene fragments varied in length from 381 to 504 bp, with average p distances of 0.015 (tpi gene fragment) to 0.067 (ilvD gene fragment). The ratio of nonsynonymous to synonymous mutations (dN/dS) was less than one for all loci, from 0.01 (pur gene fragment) to 0.110 (tpi gene fragment), revealing strong purifying selection in each case. All seven loci examined exhibited base compositions in the range of 38.6 to 44.4 mol% G+C. The most diverse locus in terms of numbers of unique sequences was glpF, with 37 MLST alleles, and the least diverse was gmk, with 19 MLST alleles (Table 2). The strains were recovered in 59 unique allelic profiles, or STs, which were numbered sequentially, with the exception that the ST-11 designation was not used. Only three STs were represented more than four times in the data set: they were ST-1 (associated with eight B. anthracis isolates), ST-8 (associated with three B. cereus and five B. thuringiensis isolates), and ST-23, which comprised five isolates of B. thuringiensis serovar morrisoni. Five STs were present four times in the data set, 3 STs were present three times, 7 STs were present twice, and the remaining 41 STs occurred only once.
Population structure.
ML trees were constructed for the single sequence of 2,838 bp of concatenated loci (Fig. 1) and individually for all seven loci (Fig. 2). To test for the presence of similar phylogenetic signals in the eight trees obtained, we performed an ML randomization test by which the similarities in tree topologies among loci were compared to those expected by chance alone. This analysis confirmed that while the topologies of the eight trees had different likelihoods, indicating that the signals present in these data were not completely congruent and therefore that the population was not entirely clonal, they were far more similar than would be expected by chance alone (Fig. 3). As such, there is a clear signal of phylogenetic history present in these MLST data so that they can be used to reconstruct an evolutionary history of the B. cereus group.
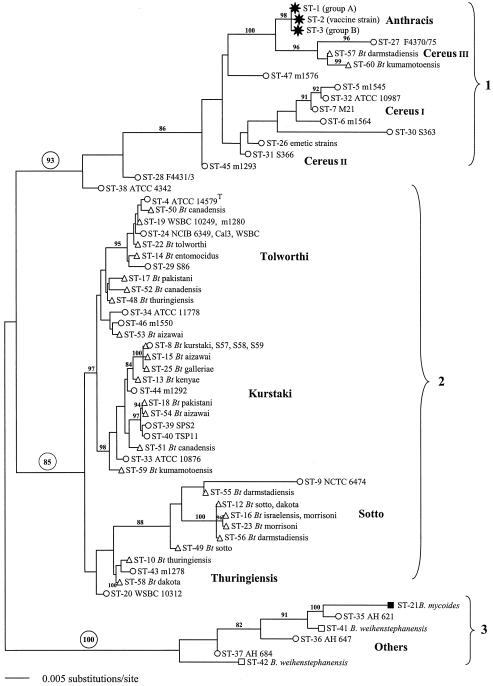
FIG. 1.
ML phylogenetic tree for the concatenated gene sequences for the 59 STs included in the study. Strain identifications:  , B. anthracis; ○, B. cereus; ▵, B. thuringiensis; ▪, B. mycoides; □, B. weihenstephanensis. All horizontal branch lengths were drawn to a scale of substitutions per site, and the tree was rooted at the midpoint for the purpose of clarity only. All bootstrap support values of >80% are shown next to the appropriate nodes. The 85% bootstrap value associated with clade 2 excludes the highly divergent ST-9 type.
, B. anthracis; ○, B. cereus; ▵, B. thuringiensis; ▪, B. mycoides; □, B. weihenstephanensis. All horizontal branch lengths were drawn to a scale of substitutions per site, and the tree was rooted at the midpoint for the purpose of clarity only. All bootstrap support values of >80% are shown next to the appropriate nodes. The 85% bootstrap value associated with clade 2 excludes the highly divergent ST-9 type.
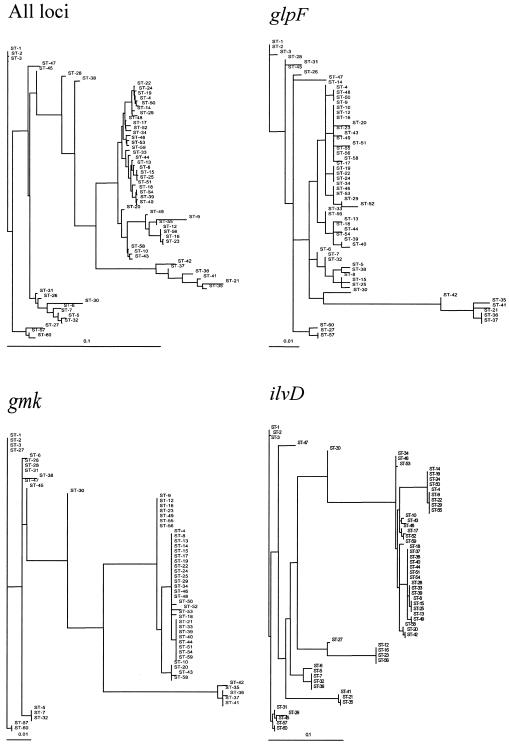
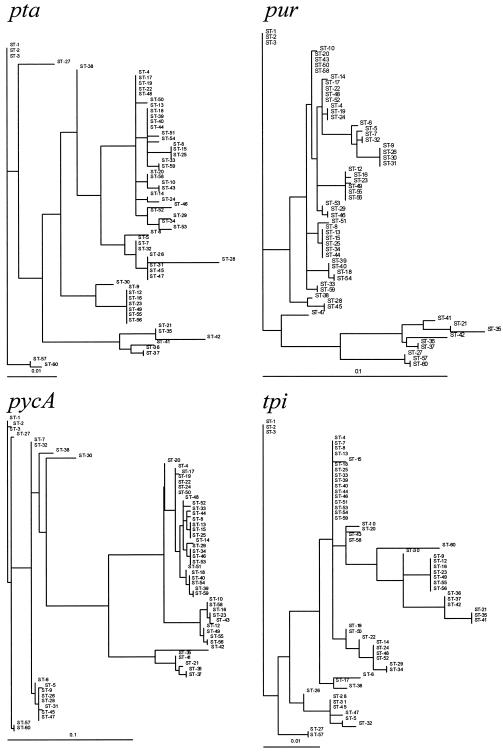
FIG. 2.
Maximum likelihood phylogenetic trees obtained for the concatenated sequence and the seven loci. ST designations are given in Table 1. All horizontal branch lengths were drawn to scale.
FIG. 3.
Maximum likelihood analysis of phylogenetic congruence in the B. cereus group. An ML tree that was reconstructed from the data for the concatenated loci and each of the seven loci was compared to each of the eight ML trees, with the branch lengths optimized for each analysis. The differences in likelihood (Δ−ln L) are shown for each tree (open symbols) and for 200 random trees (closed symbols).
The gmk tree conformed to the concatenated tree most consistently, and glpF, pycA, and tpi provided strain assignments that were reasonably well correlated with those in the concatenated tree. The ilvD, pta, and pur trees, however, failed to resolve the STs into the major monophyletic groups described below.
Phylogenetic groupings.
The overall structure of the ML tree generated from concatenated sequences (Fig. 1) revealed three major phylogenetic groups, with each defined by high bootstrap support values of 85 to 100%. One heterogeneous group based on B. mycoides (ST-21) and B. weihenstephanensis (ST-41 and ST-42) is referred to here as “others” (Table 1) and was not further considered. A group including B. anthracis, numerous B. cereus strains, and rare B. thuringiensis isolates, notably ST-57 and ST-60, is referred to as clade 1 and labeled B. cereus since that was the predominant organism of the cluster (Table 1). Finally, a large cluster that was mostly composed of B. thuringiensis strains but that included some B. cereus isolates is described as clade 2 and labeled B. thuringiensis. The only ambiguous strain in this clade was recovered as ST-9, and the bootstrap value for this clade excluded this highly divergent strain (see below).
The ML tree of concatenated sequences was used as the basis for grouping the allelic profiles into lineages. The validity of these assignments was augmented by examinations of the individual gene trees (Fig. 2) and split decomposition analyses of clades 1 and 2 (see Fig. S1 in the supplemental material). In this way, 50 of the 59 STs were grouped into eight lineages which were assigned names that were as consistent as possible with previous microbiological and serological designations but that were given a unique format (capitalized, nonitalic) to avoid confusion with valid taxonomic labels. The lineage compositions, with the exception of Cereus II, were supported by bootstrap values of >87%. Cereus II was the only lineage for which strain allocation did not correlate with a monophyletic group in the ML tree. Its composition was determined by split decomposition analysis, individual alleles that isolates had in common, and the preponderance of fixed differences compared to shared polymorphisms.
Variation among and within lineages.
There was an excess of fixed differences compared to shared polymorphisms in pairwise comparisons of all but two of the lineages (Kurstaki and Tolworthi), with the highest numbers of shared polymorphisms occurring between lineages that were more closely positioned in the ML tree of the concatenated sequences (Table 3). Compared with the overall diversity of the data set, there were generally fewer sequence differences within each of the lineages at each locus. It was noteworthy that in many cases, multiple sequence differences were due to single genes that were present in individual STs (Table 4), probably as a consequence of lateral gene transfer.
TABLE 3.
Numbers of fixed differences across all seven loci and shared polymorphisms among the B. cereus group lineages
| Lineage | No. of fixed differences (no. of shared polymorphisms)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Cereus I | Cereus II | Cereus III | Kurstaki | Tolworthi | Sotto | Thuringiensis | Others | |
| Anthracis | 54 (0) | 28 (0) | 27 (0) | 127 (0) | 120 (0) | 92 (1) | 126 (0) | 100 (0) |
| Cereus I | 11 (7) | 53 (4) | 95 (3) | 88 (3) | 52 (1) | 94 (2) | 78 (5) | |
| Cereus II | 37 (8) | 95 (4) | 91 (4) | 64 (15) | 94 (2) | 87 (13) | ||
| Cereus III | 124 (2) | 120 (3) | 85 (9) | 127 (2) | 79 (11) | |||
| Kurstaki | 3 (5) | 27 (12) | 13 (0) | 64 (9) | ||||
| Tolworthi | 24 (16) | 11 (6) | 67 (10) | |||||
| Sotto | 21 (13) | 55 (33) | ||||||
| Thuringiensis | 70 (6) | |||||||
| Others | ||||||||
TABLE 4.
Numbers of nucleotide sequence differences within subdivisions of the B. cereus group
| Subdivision | No. of variable sites in gene
|
|||||||
|---|---|---|---|---|---|---|---|---|
| All loci | glpF | gmk | ilvD | pta | pur | pycA | tpi | |
| All STs | 395 | 50 | 52 | 81 | 50 | 63 | 74 | 25 |
| Clade 1 (B. cereus) | 158 | 16 | 8 | 44a | 17 | 42 | 14 | 17 |
| Anthracis | 3 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| Cereus I | 35 | 4 | 3 | 8 | 3 | 4 | 3 | 11 |
| Cereus II | 54 | 8 | 1 | 17b | 0 | 24c | 1 | 3 |
| Cereus III | 45 | 2 | 1 | 24d | 0 | 1 | 0 | 8e |
| Clade 2 (B. thuringiensis) | 195 | 20 | 11 | 49 | 24 | 28 | 49 | 14 |
| Kurstaki | 45 | 9 | 1 | 10 | 5 | 12 | 7 | 1 |
| Tolworthi | 61 | 6 | 4 | 19 | 9 | 9 | 9 | 5 |
| Sotto | 92 | 0 | 0 | 39 | 0 | 16f | 37g | 0 |
| Thuringiensis | 27 | 4 | 1 | 6 | 1 | 1 | 12h | 2 |
| Others | 146 | 9 | 33 | 50 | 11 | 22 | 19 | 2 |
| Unassigned | 114 | 11 | 11 | 29 | 14 | 22 | 17 | 10 |
Includes 11 variable sites contributed only from ST-27.
Includes 11 variable sites contributed only from ST-47.
Includes 11 variable sites contributed only from ST-45.
Includes 24 variable sites contributed only from ST-27.
Includes eight variable sites contributed only from ST-60.
Includes 14 variable sites contributed only from ST-9.
Includes 32 variable sites contributed only from ST-9.
Includes 10 variable sites contributed only from ST-20.
Relationships of lineages to previous designations and phenotypic properties.
All of the B. anthracis strains exhibited STs that were assigned to the Anthracis lineage (1, 2, and 3); indeed, there were only three polymorphisms identified among the 11 isolates examined, highlighting how closely related these isolates are. Although the two major phylogenetic groups (A and B) of B. anthracis (29) were evident as ST-1 and ST-3, respectively, the subdivisions within group A could not be resolved. The vaccine strain used in the United States (V770-NPI-R) was assigned to ST-2, with unique alleles at ilvD and pycA, both of which were the result of single nucleotide polymorphisms that do not appear elsewhere in the data set and probably represent mutational changes.
Strains designated B. cereus were distributed among several of the lineages, indicating that the characteristics used to identify this species do not necessarily reflect the phylogenetic origins of the strains. For example, several strains of B. cereus, including the type strain ATCC 14579, were included in the B. thuringiensis-rich clade 2 in lineages Tolworthi, Kurstaki, and Thuringiensis, while most strains were assigned to clade 1. The Cereus I lineage included B. cereus ATCC 10987, an atypical xylose-positive strain isolated from cheese, and three other isolates from foods. B. cereus strains associated with the emetic form of food poisoning constitute a recognized clone (40) and were recovered here as ST-26 within the Cereus II lineage. The ST-26 strains were isolated from cases of food poisoning, except strain S710, which was isolated from soil. The latter has since been shown to synthesize the emetic toxin cereulide (1.6 ng/ml of culture fluid), consistent with its clonal root with other emetic toxin-forming strains. We included three other STs representing nonemetic B. cereus isolates in this lineage, with the lineage being distinguished by a unique pta5 allele.
The only two strains of B. thuringiensis recovered in clade 1 were allocated to the Cereus III lineage together with one strain of B. cereus that was isolated from a case of diarrheal food poisoning (Table 1). This lineage was the closest relative of the Anthracis lineage in our collection. Indeed, B. cereus F4370/75 was the only strain in the collection that shared an allele with B. anthracis, specifically, the gmk1 allele.
Within clade 2, the Sotto lineage was comprised almost exclusively of B. thuringiensis isolates, including both dipteran (serovar israelensis)- and various lepidopteran-active strains. The only B. cereus isolate in the Sotto lineage was an outlier (ST-9) which had atypical pur and pycA alleles, both of which were more commonly associated with the B. cereus strains of clade 1, suggesting a chimeric B. cereus/B. thuringiensis genome. Indeed, the position of ST-9 could not be resolved in the bootstrap analysis. STs in this lineage correlated loosely with previous serovar designations. For example, three strains of B. thuringiensis serovar sotto from Pakistan and Canada that were isolated over a 16-year period formed a discrete clone (ST-12), but a strain of B. thuringiensis serovar dakota from the United States was also included in this clone, while a fourth strain of serovar sotto was allocated to the unique ST-49 type. Similarly, five strains of B. thuringiensis serovar morrisoni were identical (ST-23), but a sixth strain joined a B. thuringiensis serovar israelensis isolate in ST-16. Nevertheless, the Sotto lineage was unified by four common alleles, glp15, gmk7, pta2, and tpi13, and formed a coherent (with the exception of ST-9) monophyletic group (Fig. 1).
The Kurstaki lineage was the largest in the study, including 12 STs, 8 of which comprised exclusively or predominantly B. thuringiensis strains. Several B. thuringiensis serovars in this lineage correlated with discrete clones; notably, three strains of serovar aizawai were ST-15, three strains of serovar galleriae were ST-25, and four strains of serovar kenyae were ST-13. ST-8, comprising three strains of B. cereus and five strains of B. thuringiensis serovar kurstaki, was the only example in the study of B. cereus and B. thuringiensis strains being allocated to the same ST.
Four strains of B. thuringiensis serovar tolworthi from different continents formed the basis of the Tolworthi lineage as ST-22. They were associated with numerous other clones and strains of B. thuringiensis representing various serovars and some strains of B. cereus (Table 1; Fig. 1). The distinction between lineages Kurstaki and Tolworthi was slight, with only three fixed differences, but the splits graph (see Fig. S1 in the supplemental material) confirmed the divergence that was evident in the ML tree (Fig. 1).
The remaining lineage in clade 2 comprised a clone of four B. thuringiensis serovar thuringiensis strains (ST-10) together with a strain of B. thuringiensis serovar dakota and two isolates of B. cereus (Fig. 1; also see Fig. S1 in the supplemental material).
DISCUSSION
The definition of bacterial species is a continual source of debate (6, 47). While largely pragmatic definitions have been invaluable throughout the history of bacteriology, this has led to a situation in which the degree of genetic diversity seen within different bacterial species varies widely, as does the reproducibility and accuracy of bacteriological identification to the species level. In addition, these definitions may be misleading when a major characteristic used for classification purposes is encoded by a mobile element such as a phage or plasmid. Here we have described a sequence-based multilocus analysis of chromosomally encoded housekeeping genes to explore the relationships among members of the B. cereus group of bacteria, which have proved to be particularly refractory to traditional taxonomic investigations. The use of nucleotide sequences has several advantages. The data generated are definitive and reproducible among laboratories, and with the wide availability of complete genome sequences, genetic diversity in any part of the chromosome can be accessed rapidly and inexpensively. Furthermore, the data can be analyzed by a variety of phylogenetic and population genetic approaches to establish the nature of the variation under examination and to investigate possible evolutionary models for how this variation has arisen.
A preliminary analysis of the variation detected for the seven housekeeping genes used in this study indicated that the genes were similar in their diversity and were all under strong purifying selection. The clonality that is inherent in bacterial populations as a consequence of asexual reproduction can be broken down by recombination, and it is the extent of this lateral gene transfer that sets the degree of clonality in a given bacterial population (51). An analysis of congruence performed on ML trees generated from the concatenated loci and from each of the loci individually indicated that the B. cereus group was largely clonal, with evidence for some recombination (Table 4). However, the extent of this recombination is not sufficient to erode the phylogenetic signal in the data, as seen for some other bacterial species (12). Previous estimates of the degrees of association and recombination between alleles (IA) of strains of the B. cereus group similarly concluded that the population structure was clonal with limited recombination (20). With clonal organisms, it is possible to exploit conventional phylogenetic analyses to determine the population structure and evolution as we have done here, although it is necessary to be aware of recombination events because they will compromise the analysis. Since mutation will be more important than recombination in clonal organisms, it is preferable to use nucleotide sequences rather than allelic profiles as the basis for classification and evolutionary analysis of these organisms because allelic profiles do not retain the magnitude of changes between alleles. This contrasts with the case for essentially nonclonal organisms, such as Neisseria meningitidis, for which recombination invalidates phylogenetic approaches and for which allelic profiles are a more appropriate basis for such investigations (34).
The phylogenetic tree generated from the concatenated sequences (Fig. 1), together with the individual gene trees, resolved the isolates into eight distinct groups or lineages distributed between two major clades: clade 1 comprises B. anthracis and predominantly B. cereus strains, and clade 2 comprises largely B. thuringiensis strains with sporadic B. cereus isolates. A third major clade comprising other species of the B. cereus group was also observed. Four of the seven loci (glpF, gmk, pycA, and tpi) supported this primary division, while the remaining loci did not assign the STs of clade 1 to a monophyletic group, extending the finding from comparative genome sequences that lateral gene transfer has played a role in metabolic specialization in these bacteria (45). This primary division has been noted in several other population studies of these organisms (20, 21, 44, 57), supporting the contention that the phylogenetic signal is intact despite recent recombinational exchanges, although this will need to be confirmed through the analysis of more loci. In particular, an extensive AFLP analysis of these organisms recognized three major clusters, of which AFLP clusters 1 and 2 map almost perfectly to MLST clades 2 and 1, respectively, while representatives of AFLP cluster 3 were not included in this study (21).
The B. cereus group comprises bacteria that have most likely evolved from a saprophyte or insect gut commensal common ancestor, principally by asexual processes. At least eight distinct lineages have arisen, each of which appears to have attained global distribution, although the presence of the unassigned B. cereus genotypes represented by ST-28, ST-30, and perhaps ST-38 suggests that more exhaustive sampling would probably identify further lineages and add definition to the extant ones. However, it is notable that B. cereus ATCC 4342 (ST-38) was unassigned in both this and a previous MLST study (20) as well as the more extensive AFLP survey (21), suggesting that it is a true atypical strain rather than a representative of a poorly sampled lineage.
The eight lineages correlate closely with the phylogenetic branches of the AFLP analysis described by Hill et al. (21). The mammalian pathogens present in the Anthracis lineage are similar to the insecticidal pathogens in that they form a distinct lineage that has presumably evolved as a consequence of its association with particular plasmids. Despite its wide geographic representation, the Anthracis lineage contains only three sequence genotypes and three polymorphic nucleotides, two of which are only present in a laboratory vaccine strain. It is possible that the latter polymorphisms may indicate further mutational changes in the genome of this strain that could compromise its efficacy as a vaccine. Nevertheless, the high degree of clonality among these strains is consistent with previous reports of the very low genetic diversity of this organism (29, 30), contrasting with the multiple variants observed for the insecticidal lineages. This suggests that the Anthracis lineage is much younger than the insect pathogenic lineages. Hill et al. defined the Anthracis lineage more broadly in their AFLP analysis and included strains of B. cereus such as F4431/73 (ST-28), which was unassigned in our study, as well as strains of B. thuringiensis in their B. anthracis branch F (21). However, in view of the distinctive allelic profiles of B. anthracis strains, their strong bootstrap support, and their isolation in a splits graph (see Fig. S1 in the supplemental material), we consider it appropriate to define this lineage more strictly. The Cereus I lineage includes B. cereus ATCC 10987, for which there is now a complete genome sequence (45) that confirms its closer phylogenetic affinity to B. anthracis than to the B. cereus type strain located in clade 2.
Of the lineages in clade 2, the Sotto lineage was comprised almost exclusively of B. thuringiensis isolates. This group was recognized as branch A by AFLP analysis and was similarly composed exclusively of B. thuringiensis strains, with serovars darmstadiensis, israelensis, morrisoni, and sotto in common between the two studies (21). Serovar assignments did not correlate perfectly with STs in this group (Table 1). However, there is evidence that the ST designation may relate more to insect toxicity than does the serovar. For example, B. thuringiensis serovar israelensis (ST-16) constitutes a large, globally widespread clone of highly active, mosquito-pathogenic strains with similar or identical crystal proteins (Cry4Aa, Cry4Ba, Cry10Aa, and Cry11Aa) (1, 27). Interestingly, the only strain of B. thuringiensis serovar morrisoni included in ST-16 is also a dipteran pathogen and contains Cry4 toxins (data not shown), making this ST exclusive to Cry4-containing mosquito pathogens. Most B. thuringiensis serovar morrisoni strains, on the other hand, are lepidopteran pathogens containing crystals composed of Cry1Aa and Cry1Bc and were assigned to ST-23. The preponderance of crystalliferous bacteria in this lineage is unique among the four lineages of clade 2. The relatively high numbers of fixed differences and rare shared polymorphisms clearly delineate it from other lineages (Table 3), suggesting that this line of descent represents a particularly successful association between crystal-encoding plasmids and the host genotype.
The Kurstaki lineage corresponds to branch C of the AFLP analysis (21). B. thuringiensis serovar assignments in common between the two studies include serovars aizawai, kenyae, kumamotoensis, kurstaki, and galleriae. However, the Tolworthi lineage was not recognized in the AFLP study, and isolates of B. thuringiensis serovars canadensis, entomocidus, pakistani, and tolworthi were included with Kurstaki lineage strains in branch C by AFLP (21). The ability to distinguish between lineages Kurstaki and Tolworthi may reflect the higher resolution of MLST, although the few fixed differences and an excess of shared polymorphisms between the two lineages suggest that the division is weak. Nevertheless, there was strong bootstrap support for this division (Fig. 1).
Most serovars in the Kurstaki and Tolworthi lineages were represented by cognate STs, reinforcing the clonal structure of B. thuringiensis noted in other studies (13, 14, 42). B. thuringiensis serovar kurstaki strains are widespread lepidopteran pathogens containing Cry1 and Cry2 toxins and are often used for biocontrol in agriculture. B. thuringiensis serovars aizawai, galleriae, kenyae, and kumamotoensis similarly contain Cry1 toxins, although the exact compositions of the crystals in these bacteria have not been determined (14). This is a group that apparently undergoes extensive sharing of plasmids since all are Cry1-containing types and yet discrete clones are apparent. It seems likely that some purging of diversity gave rise to the extant clones while plasmid promiscuity counters this by enhancing diversity through the generation of novel Cry proteins by recombination (8). The result is a balance represented by numerous clones (ST-8, ST-13, ST-15, and ST-25) distributed among unique STs within the confines of the lineage. ST-8 was the only ST in the study that comprised both B. thuringiensis (Cry+) and B. cereus (Cry−) strains. There was no sign of crystals in sporulated cultures of the B. cereus ST-8 strains that were examined by light and electron microscopy, and Western blots using an anti-Cry1 antiserum revealed a trace of crystal protein in strain S58 but none in the other two strains (data not shown). Presumably, these are strains that have lost most or all of the Cry plasmids, supporting the concept of the fluidity of plasmid-borne crystal protein synthesis in this lineage (22).
The Thuringiensis lineage was based on the clone of B. thuringiensis serovar thuringiensis isolates from four different countries (ST-10) together with some strains of B. cereus. It correlated with branch B defined by AFLP, which also largely comprised B. thuringiensis serovar thuringiensis strains (21).
The lineage assignments were confirmed and refined by analyses of the data by split decomposition and examinations of the sequence variation within and among the assigned lineages. On the basis of these results, a redefinition of the nomenclature of the B. cereus group was suggested. Each of the eight lineages was considered to be sufficiently distinct to warrant a separate label, and names for these lineages were chosen that were, as far as was possible, consistent with taxonomic designations but distinct in format to avoid confusion with the current system of nomenclature. Such a classification, in which the clone or phylogenetic lineage is given recognition, has been suggested for other clonal taxa, such as the four “species” of the Mycobacterium tuberculosis complex, which would become clones Africanum, Bovis, Tuberculosis, and Microti (33). It provides for an effective taxonomy in which, for example, Anthracis can be recognized as a pathogenic lineage but other lineages will contain both entomopathogens (B. thuringiensis strains) and nonpathogens (B. cereus strains). While it may seem incongruous to retain a separate species status for bacteria that are to be included in a coherent phylogenetic taxon such as a lineage, the implications of renaming B. thuringiensis strains as B. cereus would be severe for the biocontrol industry. Therefore, for pragmatic reasons, we retained the current species identifications. Nevertheless, clones can be named or coded within lineages and associated where appropriate with specific species and pathogenic traits, such as the emetic clone (ST-26) of the Cereus II lineage or the Morrisoni clone (ST-23) of the Sotto lineage. The MLST scheme described in this study provides the basis for more extensive sampling of the B. cereus group such that the population diversity can be more fully estimated and assigned to existing and new lineages and clones in due course.
Supplementary Material
Acknowledgments
M.C.J.M. is a Wellcome Trust Senior Research Fellow in Basic Biological Sciences.
We are grateful to various colleagues, in particular M. Aquino de Muro, A.-B. Kolstø, M. Salkinoja-Salonen, and S. Scherer, for the provision of strains and to N. Agata for an estimation of cereulide synthesis by B. cereus S710. We thank K. Jolley for assistance with preparation and maintenance of the web site.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ankarloo, J., D. A. Caugant, B. M. Hansen, A. Berg, A.-B. Kolstø, and A. Lovgren. 2000. Genome stability of Bacillus thuringiensis subsp. israelensis isolates. Curr. Microbiol. 40:51-56. [DOI] [PubMed] [Google Scholar]
- 2.Ash, C., and M. D. Collins. 1992. Comparative analysis of 23S ribosomal RNA gene sequences of Bacillus anthracis and emetic Bacillus cereus determined by PCR-direct sequencing. FEMS Microbiol. Lett. 73:75-80. [DOI] [PubMed] [Google Scholar]
- 3.Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, C. R., D. A. Caugant, and A.-B. Kolstø. 1994. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 60:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, C. R., T. Johansen, and A.-B. Kolstø. 1996. The chromosome map of Bacillus thuringiensis subsp. canadensis HD224 is highly similar to that of Bacillus cereus type strain ATCC 14579. FEMS Microbiol. Lett. 141:163-167. [DOI] [PubMed] [Google Scholar]
- 6.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 7.Damgaard, P. H., P. E. Granum, J. Bresciani, M. V. Torregrossa, J. Eilenberg, and L. Vealentino. 1997. Characterization of Bacillus thuringiensis isolated from infections in burn wounds. FEMS Immunol. Med. Microbiol. 18:47-53. [DOI] [PubMed] [Google Scholar]
- 8.de Maagd, R. A., A. Bravo, and N. Crickmore. 2001. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 17:193-199. [DOI] [PubMed] [Google Scholar]
- 9.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Embley, T. M. 1991. The linear PCR reaction: a simple and robust method for sequencing amplified rRNA genes. Lett. Appl. Microbiol. 13:171-174. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feil, E., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaviria Rivera, A., and F. G. Priest. 2003. Molecular typing of Bacillus thuringiensis serovars by RAPD-PCR. Syst. Appl. Microbiol. 26:254-261. [DOI] [PubMed] [Google Scholar]
- 14.Gaviria Rivera, A., and F. G. Priest. 2003. Pulsed field gel electrophoresis of chromosomal DNA reveals a clonal population structure to Bacillus thuringiensis that relates in general to crystal protein content. FEMS Microbiol. Lett. 223:61-66. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, R. E., W. C. Haynes, and C. H.-N. Pang. 1973. The genus Bacillus. Agriculture handbook no. 427. United States Department of Agriculture, Washington, D.C.
- 16.Granum, P. E., and T. Lund. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 17.Guttmann, D. M., and D. J. Ellar. 2000. Phenotypic and genotypic comparisons of 23 strains from the Bacillus cereus complex for a selection of known and putative B. thuringiensis virulence factors. FEMS Microbiol. Lett. 188:7-13. [DOI] [PubMed] [Google Scholar]
- 18.Helgason, E., D. A. Caugant, M.-M. Lecadet, Y. Chen, J. Mahillon, A. Lovgren, I. Hegna, K. Kvaloy, and A.-B. Kolstø. 1998. Genetic diversity of Bacillus cereus/B. thuringiensis isolates from natural sources. Curr. Microbiol. 37:80-87. [DOI] [PubMed] [Google Scholar]
- 19.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A.-B. Kolstø. 2004. Multilocus sequence typing for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, K. K., L. O. Ticknor, R. T. Okinaka, M. Asay, H. Blair, K. A. Bliss, M. Laker, P. E. Pardington, A. P. Richardson, M. Tonks, D. J. Beecher, J. D. Kemp, A.-B. Kolstø, A. C. Lee Wong, P. Keim, and M. A. Jackson. 2004. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 70:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, X., B. M. Hansen, J. Eilenberg, N. B. Hendriksen, L. Smidt, Z. Yuan, and G. B. Jensen. 2004. Conjugative transfer, stability and expression of a plasmid encoding a cry1Ac gene in Bacillus cereus group strains. FEMS Microbiol. Lett. 231:45-52. [DOI] [PubMed] [Google Scholar]
- 23.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 24.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chung, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 25.Jackson, S. G., R. B. Goodbrand, R. Ahmed, and S. Kasatiya. 1995. Bacillus cereus and Bacillus thuringiensis isolated in gastroenteritis outbreak investigations. Lett. Appl. Microbiol. 21:103-105. [DOI] [PubMed] [Google Scholar]
- 26.Jolley, K. A., M.-S. Chan, and M. C. J. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaji, D. A., Y. B. Rosato, V. P. Canhos, and F. G. Priest. 1994. Characterization by polyacrylamide gel electrophoresis of whole cell proteins of some strains of Bacillus thuringiensis subsp. israelensis isolated in Brazil. Syst. Appl. Microbiol. 17:104-107. [Google Scholar]
- 28.Kaneko, T., R. Nozaki, and K. Aizawa. 1978. Deoxyribonucleic acid relatedness between Bacillus anthracis, Bacillus cereus and Bacillus thuringiensis. Microbiol. Immunol. 22:639-641. [DOI] [PubMed] [Google Scholar]
- 29.Keim, P., A. Kalif, J. M. Schupp, K. K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. E. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. Appl. Environ. Microbiol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, W., Y.-P. Hong, J.-H. Yoo, W.-B. Lee, C.-S. Choi, and S.-I. Chung. 2001. Genetic relationships of Bacillus anthracis and closely related species based on variable-number tandem repeat analysis and BOX-PCR genomic fingerprinting. FEMS Microbiol. Lett. 207:21-27. [DOI] [PubMed] [Google Scholar]
- 32.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1255. [DOI] [PubMed] [Google Scholar]
- 33.Lan, R., and P. R. Reeves. 2001. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 9:419-424. [DOI] [PubMed] [Google Scholar]
- 34.Maiden, M. C. J., J. A. Bygraves, J. A. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, and D. A. Caugant. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margulis, L., J. Z. Jorgensen, S. Dolan, R. Kolchinsky, F. A. Rainey, and S. C. Lo. 1998. The Arthromitus stage of Bacillus cereus: intestinal symbionts of insects. Proc. Natl. Acad. Sci. USA 95:1236-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, L. K. 1994. DNA relatedness among Bacillus thuringiensis serovars. Int. J. Syst. Bacteriol. 44:125-129. [DOI] [PubMed] [Google Scholar]
- 38.Okinaka, R. T., K. Cloud, O. Hampton, A. Hoffmaster, K. Hill, P. Keim, T. Koehler, G. Lamke, S. Kumano, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. Jackson. 1999. Sequence, assembly and analysis of pXO1 and pXO2. J. Appl. Bacteriol. 87:261-262. [DOI] [PubMed] [Google Scholar]
- 39.Økstad, O. A., I. Hegna, T. Lindback, A.-L. Rishovd, and A.-B. Kolstø. 1999. Genome organization is not conserved between Bacillus cereus and Bacillus subtilis. Microbiology 145:621-631. [DOI] [PubMed] [Google Scholar]
- 40.Pirttijarvi, T. S. M., M. A. Andersson, A. C. Scoging, and M. S. Salkinoja-Salonen. 1999. Evaluation of methods for recognising strains of the Bacillus cereus group with food poisoning potential among industrial and environmental contaminants. Syst. Appl. Microbiol. 22:133-144. [DOI] [PubMed] [Google Scholar]
- 41.Priest, F. G., M. Goodfellow, and C. Todd. 1988. A numerical classification of the genus Bacillus. J. Gen. Microbiol. 134:1847-1882. [DOI] [PubMed] [Google Scholar]
- 42.Priest, F. G., D. A. Kaji, Y. B. Rosato, and V. P. Canhos. 1994. Characterization of Bacillus thuringiensis and related bacteria by ribosomal RNA gene restriction fragment length polymorphisms. Microbiology 140:1015-1022. [DOI] [PubMed] [Google Scholar]
- 43.Prüss, B. M., R. Dietrich, B. Nibler, E. Martlbauer, and S. Scherer. 1999. The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl. Environ. Microbiol. 65:5436-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radnedge, L., P. G. Agron, K. K. Hill, M. A. Jackson, L. O. Ticknor, P. Keim, and G. L. Andersen. 2003. Genome differences that distinguish Bacillus anthracis from Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 69:2755-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasko, D. A., J. Ravel, O. A. Økstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourrasse, S. V. Angiuoli, J. Kolonay, K. E. Nelson, A.-B. Kolstø, C. M. Fraser, and T. D. Read. 2004. The genome of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to B. anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baille, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Økstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanen, R. J. Dodson, L. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, S. C. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, P. C. Hanna, A.-B. Kolstø, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 47.Rossello-Mora, R. 2003. Opinion: the species problem, can we achieve a universal concept? Syst. Appl. Microbiol. 26:323-326. [DOI] [PubMed] [Google Scholar]
- 48.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 49.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Somerville, H. J., and M. L. Jones. 1972. DNA competition studies within the Bacillus cereus group of bacilli. J. Gen. Microbiol. 73:257-265. [DOI] [PubMed] [Google Scholar]
- 51.Spratt, B. G., W. P. Hanage, and E. J. Feil. 2001. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Curr. Opin. Microbiol. 4:602-606. [DOI] [PubMed] [Google Scholar]
- 52.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 53.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 54.Ticknor, L. O., A. B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turnbull, P. C. B., J. M. Kramer, K. Jorgensen, and R. J. Gilbert. 1979. Properties and production characteristics of vomiting, diarrheal, and necrotizing toxins of Bacillus cereus. Am. J. Clin. Nutr. 32:219-228. [DOI] [PubMed] [Google Scholar]
- 56.Urwin, R., and M. C. J. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 57.Vilas-Boas, G., V. Sanchis, D. Lereclus, M. F. L. Lemos, and D. Bourguet. 2002. Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 68:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.