Abstract
Transferable antibiotic resistance in Haemophilus influenzae was first detected in the early 1970s. After this, resistance spread rapidly worldwide and was shown to be transferred by a large 40- to 60-kb conjugative element. Bioinformatics analysis of the complete sequence of a typical H. influenzae conjugative resistance element, ICEHin1056, revealed the shared evolutionary origin of this element. ICEHin1056 has homology to 20 contiguous sequences in the National Center for Biotechnology Information database. Systematic comparison of these homologous sequences resulted in identification of a conserved syntenic genomic island consisting of up to 33 core genes in 16 β- and γ-Proteobacteria. These diverse genomic islands shared a common evolutionary origin, insert into tRNA genes, and have diverged widely, with G+C contents ranging from 40 to 70% and amino acid homologies as low as 20 to 25% for shared core genes. These core genes are likely to account for the conjugative transfer of the genomic islands and may even encode autonomous replication. Accessory gene clusters were nestled among the core genes and encode the following diverse major attributes: antibiotic, metal, and antiseptic resistance; degradation of chemicals; type IV secretion systems; two-component signaling systems; Vi antigen capsule synthesis; toxin production; and a wide range of metabolic functions. These related genomic islands include the following well-characterized structures: SPI-7, found in Salmonella enterica serovar Typhi; PAP1 or pKLC102, found in Pseudomonas aeruginosa; and the clc element, found in Pseudomonas sp. strain B13. This is the first report of a diverse family of related syntenic genomic islands with a deep evolutionary origin, and our findings challenge the view that genomic islands consist only of independently evolving modules.
The evolutionary origins of transferable antibiotic resistance in Haemophilus influenzae, a strict human commensal, have not been explained. Prior to the 1970s H. influenzae was universally susceptible to ampicillin. In 1972, the first isolate with ampicillin resistance (Apr) that produced β-lactamase was detected (37). Over the following few years, β-lactamase-producing and ampicillin-resistant strains appeared worldwide, and their prevalence rapidly increased (42, 54). During the same period, strains resistant to tetracycline or chloramphenicol and strains multiply resistant to these antibiotics emerged (33, 42). In many countries, 20 to 30% of clinical isolates were resistant to at least one of these antibiotics (42, 47), and most people carried such antibiotic-resistant haemophili (35, 45).
It was recognized that this antibiotic resistance was transferred by conjugation and was encoded by 40- to 60-kb related mobile elements (10, 14, 15, 25, 31-33). Transposons, inserted at separate sites into the conjugative element, contained the resistance genes. The β-lactamase gene encoding Apr formed part of Tn3, and the genes encoding tetracycline and chloramphenicol resistance resided in a Tn10-like compound transposon (14, 15, 25, 26). Surprisingly, free plasmids could not be easily detected in clinical isolates (10, 52). However, extrachromosomal plasmids could be isolated from transconjugants (10, 52). It is now recognized that these related resistance elements not only conjugate but integrate site specifically into H. influenzae tRNALeu (11). This site-specific insertion into a tRNA gene was intriguingly similar to genomic islands (GIs) (13, 23) and raised the question of whether these large conjugative resistance elements had evolutionary relationships with GIs.
Genomic islands which are part of the horizontal or flexible gene pool often constitute a large part (more than 10%) of bacterial genomes (22, 23, 29, 49). These islands are typically characterized by a G+C content different from that of the host genome. In Proteobacteria, they are often integrated into tRNA genes (13, 22). Genes important in habitat-specific adaptation cluster on these islands (13, 22). Well-recognized examples include islands with genes involved in pathogenesis (23), avirulence (1), chemical degradation (58), antibiotic resistance (27), and metabolic functions (30). Hitherto, the evolutionary origin of these islands has not been elucidated (13). This is not surprising as the islands are thought to consist of modules with diverse origins joined together in a single structure, often referred to as a mosaic (5, 56). Such a structure would not have a unified evolutionary history. This mosaic type of structure is supported by studies of the organization of a number of well-characterized elements and closely related elements, such as SXT (2) and ICESt1 (6, 40). These elements have only short regions of homology with other distantly related elements that correlate with a single module. However, investigators have recently reported unexpectedly extensive homologies between the completed genome sequences of a wide range of Proteobacteria and the GIs PAP1 and pKLC102 found in Pseudomonas aeruginosa (24, 27), the clc element found in Pseudomonas sp. strain B13 (58), and the pathogenicity island SPI-7 found in Salmonella enterica serovar Typhi C18 and Ty2 (41). The extent of these homologies invites the thought that some GIs may have a coherent structure with a shared origin.
Here we report the findings of a systematic analysis of GIs exhibiting extensive homology. Homologies for most of these islands have recently been reported by other workers (24, 27, 41, 58), but the evolutionary implications of these homologies were not studied. The starting point of this investigation, however, was a bioinformatics study of the complete sequence of a large conjugative Haemophilus resistance element, ICEHin1056, which was previously referred to as p1056 (3, 10, 11, 35). Identification of homologous sequences in ICEHin1056 and elements or genomic islands found in β- and γ-Proteobacteria indicated the presence of a syntenic core element with a shared evolutionary origin. A wide range of accessory gene clusters with phylogenies independent of the phylogenies of the core genes nestled between the core genes of each related GI.
MATERIALS AND METHODS
Complete sequence of ICEHin1056.
Briefly, the whole sequence was derived from a random library generated from purified extrachromosomal closed circular ICEHin1056, an element harbored by the ampicillin-, tetracycline-, and chloramphenicol-resistant organism H. influenzae type b strain 1056 (3, 10). The sequences were assembled by using Pred and Phrap (17, 18, 21) and were viewed and edited with the Staden (50) suite of programs. Artemis was used to view and annotate the sequence (44).
Bioinformatics analysis.
By interrogating the National Center for Biotechnology Information (NCBI) database with the TBLASTX algorithm, we identified GIs homologous to ICEHin1056. The input ICEHin1056 sequence consisted of the entire element with all the antibiotic resistance-associated sequences (i.e., Tn10-like and Tn3 sequences) removed. Sequences producing significant alignments (i.e., e value scores of <10−5) were individually interrogated by TBLASTX searching for contiguous sequences homologous to ICEHin1056. Twenty candidate GIs (including ICEHin1056) were identified for further investigation.
The Artemis comparison tool (ACT) (44) was used to visually compare GIs pairwise for homology by using the TBLASTX algorithm. Preliminary analysis of open reading frames of the homologous GIs indicated that four potential GIs contained only a short sequence with shared homology or were from incompletely sequenced genomes that limited or precluded investigation of a possible coherent GI. The remaining 16 GIs with extensive homology were investigated further. Initially, to determine possible phylogenetic relationships among these 16 GIs, the amino acid sequences encoded by two predicted genes present in all of them were aligned by using ClustalX for multiple-sequence alignment and tree estimation (55). This resulted in identification of four clusters. One well-known representative GI of each cluster (ICEHin1056, SPI-7 [41], PAP1 [24], and PAGI-3 [30]) was used to distinguish sequences shared by these related GIs and thereby identify potential core genes.
Core GI sequences were defined as GI sequences that were present (BLASTP e value, <10−5) in at least three of the four GIs. The presence of GI genes was visually scored by performing pairwise comparisons of each of the GIs with ACT and individually aligning potential homologues with the BLASTP algorithm. Thirty-three core genes fitting the definition were identified (designated genes 1 to 33) (Fig. 1). The sequence of each gene was used to construct four virtual core GIs representative of ICEHin1056, SPI-7, PAP1, and PAGI-3. Virtual core GIs consisting of either amino acid sequences or nucleotide sequences were constructed for each of the four GIs. For the genes missing from a GI, the homologue from PAP1 was used. The concatenated nucleotide sequences of core genes from each of these four representative GIs were used to systematically reinterrogate the NCBI databases with TBLASTX to identify any additional GIs missed in the original screening with ICEHin1056 (none were identified).
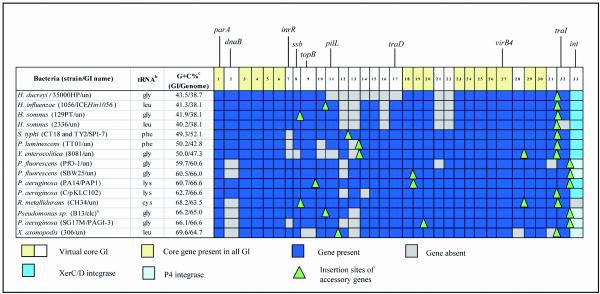
FIG. 1.
Array of predicted genes present in related coherent and syntenic GIs found in Proteobacteria. The clc element is also present in the complete sequence of B. fungorum. The tRNA data indicate the site of GI integration. The G+C contents of both the genomic island and the host genome are indicated. un, unnamed.
A virtual coherent GI (consisting of 33 core genes) was compared to each candidate GI by using ACT, as shown in Fig. 2. The presence of core genes was visually scored by a pairwise comparison of the element being studied and the virtual coherent element. A coherent evolutionarily related GI was defined as possessing >24 (∼75%) of the 33 core genes. Sequences of the virtual core GI genes are available at http://www.ndcls.ox.ac.uk/mohd-zain.html. The inferred attributes of the predicted noncore genes for all the GIs were determined by reannotation of the GIs by using Artemis and BLASTP available through NCBI. As a negative control, the virtual core element was compared to the following three well-characterized GIs (12) that had not been found to show homology during the TBLASTX interrogation of the NCBI database: PAI I536 (GenBank accession number AJ488511), PAI II536 (GenBank accession number AJ494981), and PAI III536 (GenBank accession number X16664).
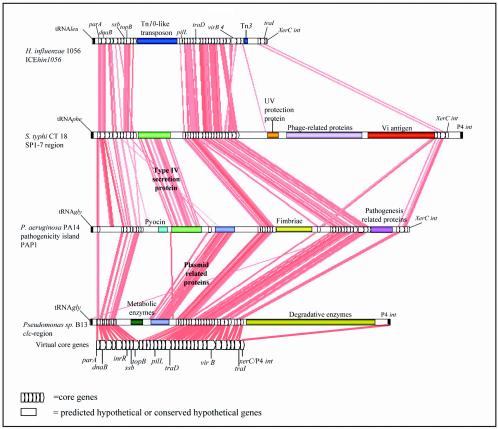
FIG. 2.
Modified Artemis Comparison Tool view of the homologous regions identified in four genomic islands (ICEHin1056, SPI-7, PAP1, and the clc element) and the core genes of the virtual genomic island. Homologous sequences (>30% amino acid identity) are indicated by red lines joining regions of the five schematic representations of the GIs.
Phylogenetic analysis.
The 15 core genes common to all 15 GIs (Fig. 1) were investigated for phylogenetic relationships. The relationships between amino acid sequences encoded by each of the 15 common genes individually or for the concatenated sequences were estimated by using ClustalX. For this analysis, we used the following settings: exclude position with gaps, correct for multiple substitutions, phylip format, and bootstrap neighbor-joining tree of 100. The output from the analysis was viewed by using TreeView (39). By using Artemis and the concatenated nucleotide sequences of the 15 common genes, the G+C content of the shared core sequences of each of the GIs was determined. The G+C content of the host's whole genome was accessible through GenBank accession numbers and linked web pages.
Nucleotide sequence accession number.
The nucleotide sequence of ICEHin1056 has been deposited in the GenBank database under accession number AJ627386
RESULTS
Identification of related syntenic genomic islands.
Twenty potential GIs with homology to the non-resistance-associated sequences of ICEHin1056 were identified from interrogation of the NCBI database with the TBLASTX algorithm. Four of these GIs with extensive homology (ICEHin1056, SPI-7, PAP1, and PAGI-3) and representing the diversity of the GIs were selected to identify the predicted core element (i.e., genes present in at least three of four GIs) of these GIs. Thirty-three genes fulfilled this definition. Virtual core elements consisting of 33 genes, representing these four GIs, were constructed and used in a pairwise comparison with the 20 candidate and related GIs. Sixteen of the 20 GIs met the definition of a coherent element (24 of the 33 core genes [∼75%] were present). These GIs were restricted to β- and γ-Proteobacteria (two and nine species, respectively). The isolates included members of the Burkholderiaceae (Burkholderia fungorum and Ralstonia metallidurans), members of the Pasteurellaceae (one H. influenzae isolate, one Haemophilus ducreyi isolate, and two Haemophilus somnus isolates), three members of the Enterobacteriaceae (one S. enterica serovar Typhi isolate, one Yersinia enterocolitica isolate, and one Photorhabdus luminescens isolate), and seven members of the Pseudomonadaceae (three P. aeruginosa isolates, two Pseudomonas fluorescens isolates, one Xanthomonas axonopodis isolate, and Pseudomonas sp. strain B13). As the genomic island in B. fungorum (GenBank accession number NZ_AAAJ00000000) exhibits nucleotide sequence identity to the clc element of Pseudomonas sp. strain B13 (58), it was not analyzed as a separate GI. The species and strains harboring the syntenic and coherent GIs are listed in Fig. 1 and Table 1.
TABLE 1.
Species with syntenic GIs, their sizes, Genbank accession numbers, and major attributes of accessory genes
| Bacterium (strain/GI)a | Size (bp) | Accession no. | Major attributes of accessory genes |
|---|---|---|---|
| H. ducreyi (35000HP/un) | 49,075 | NC_002940 | Type I antirestriction, cytolethal distending toxin |
| H. influenzae (1056/ICEHin1056) | 59,393 | AJ627386 | Antibiotic resistance |
| H. somnus (129PT/un) | 56,442 | NZ_AAB002000009 | Type I antirestriction, virulence protein |
| H. somnus (2336/un) | 64,831 | NZ_AACJ00000000 | Antibiotic, metal, and antiseptic resistance |
| Y. enterocolitica (8081/un) | 65,285 | NC_003222 | Type IV secretion, arsenic resistance, hemagglutinin-related protein |
| S. enterica serovar Typhi (CT18 and TY2/SPI-7) | 134,000 | NC_003198 | Type IV secretion, Uv protection, Vi antigen |
| P. luminescens (TT01/un) | 140,163 | NC_005126 | Metabolic enzymes, type IV secretion |
| P. fluorescens (PfO-1/un) | 70,421 | NZ_AAAT00000000 | Biofilm regulators, fimbrial proteins, metabolic enzymes |
| P. fluorescens (SBW25/un) | 101,233 | NC_004507 | Metabolic enzymes |
| P. aeruginosa (PA14/PAP1) | 106,518 | AY273869 | Pyocin, type IV secretion, fimbrial protein, two-component signal systems |
| P. aeruginosa (C/pKLC102) | 103,532 | AY257538 | Type IV secretion, virulence protein |
| R. metallidurans (CH34/un) | >76,893 | NZ_AAA100000000 | Degradative enzymes, metabolic enzymes, pathogenesis-related protein |
| Pseudomonas sp. (B13/c/c)b | 105,027 | AJ617740 | Metabolic enzymes, degradative enzymes |
| P. aeruginosa (SG17M/PAG1-3) | 114,632 | AF440524 | Plasmid transfer proteins, type I restriction-modification, metabolic enzymes |
| X. axonopodis (306/un) | 86,019 | NC_003919 | Phage-related proteins, efflux pump, hemolysin |
un, unnamed.
The clc element is also present in B. fungorum.
The complete genomes of the following organisms contained contiguous sequences homologous to less than 24 of the 33 genes present in a virtual coherent GI: Xylella fastidiosa 9a5c (GenBank accession number NC_002488) (11 genes) and Pseudomonas syringae pv. tomato strain DC3000 (GenBank accession number NC_004578) (17 genes). Therefore, the GIs did not meet the definition of a coherent syntenic GI. An incomplete genome sequence of Azotobacter vinelandii (GenBank accession number NZ_AAAU00000000) contained sequences which suggested the presence of a homologous GI, but the unfinished sequence was unsuitable for determining whether it contained a coherent GI. Lastly, a P. fluorescens strain currently undergoing whole-genome sequencing contained a homologous GI, but it was not analyzed further due to publication restrictions. The three GIs which were investigated as negative controls (PAI I536, PAI II536, and PAI III536) exhibited no homology with a virtual core element as determined with ACT.
Features of the core genes.
The elements were all predicted to or are known to integrate into tRNA genes, as shown in Fig. 1. There were five different tRNA genes that were sites of insertion for these GIs, and 6 of 15 GIs inserted into tRNAGly. The core genes of each of the GIs were largely in the same order (i.e., synteny) (Fig. 2). The level of amino acid sequence homology between GIs at the extremes of homology was as low as 25 to 30% for many of the shared core genes, indicating that there was wide evolutionary divergence. Of the 33 core genes, 10 had homology to genes with known functions. These included four genes (parA, dnaB, ssb, and topB) among genes 1 to 10 (Fig. 1 and 2). Homologues of these genes are recognized to play a role together in plasmid replication (16, 20). One of the genes in this region, inrR, has recently been shown to regulate integrase function in the clc element and controls excisive and integrative recombination of that element with tRNAGly (48). For the most part the functions of genes 11 to 32 are unknown, but four of these genes exhibit homology to pilL (core gene 11), virB4 (core gene 27), traD (core gene 17), and a relaxase or traI (core gene 32); homologues of these four genes are recognized to play a role in conjugative DNA transfer (60).
Integrases were found in all GIs other than that in R. metallidurans. As the DNA sequence of this organism is not complete, it is premature to conclude that an integrase associated with this GI is absent. The remaining GIs possess an integrase of either the P4 or XerC/D lineage. SPI-7, the pathogenicity island found in S. enterica serovar Typhi, possesses copies of both types of integrase (Fig. 2). The xerCD integrase gene that was immediately adjacent to the innermost tRNAPhe sequence present at the right end of the GI (the putative attR) was selected as the integrase gene shown in Fig. 2, because only this integrase gene was consistently present in SPI-7-like GIs identified in a range of Salmonella enterica serovars (46).
Fifteen of the core GI genes were conserved in all the GIs (Fig. 1). The G+C contents of the GIs were determined and ranged widely, from 40.2% for H. somnus 2336 to 69.6% for X. axonopodis. Compared to the average G+C content of the host's genome, nine GIs had higher G+C contents. These GIs consisted of all the GIs found in members of the Pasteurellaceae and Enterobacteriacae (except SPI-7 in S. enterica serovar Typhi, which had a lower G+C content than its host genome). Similarly, the GIs found in Pseudomonas sp. strain B13, B. fungorum, (genomic G+C content, 61.8%), R. metallidurans, and X. axonopodis had higher G+C contents than their hosts' genomes. However, the GIs found in both P. fluorescens isolates and the GIs PAGI-3, PAP1, and pKLC102 found in P. aeruginosa had lower G+C contents than their hosts.
Phylogenetic relationships between GIs.
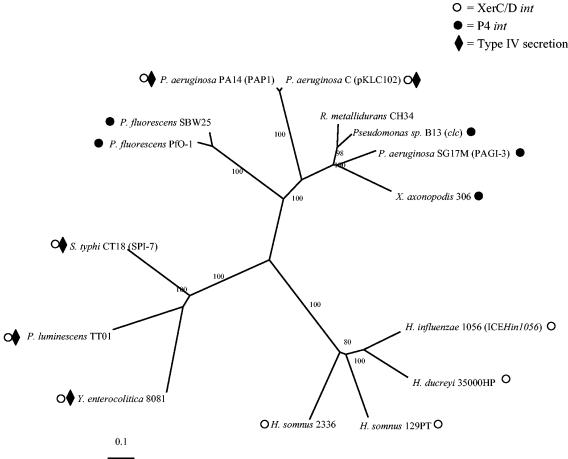
The phylogenetic relationships of each of the 15 genes common to all the GIs exhibited a congruent structure, with a few minor exceptions. The trees for four representative core genes (genes 1, 6, 18, and 27) are available as supplemental material (http://www.ndcls.ox.ac.uk/mohd-zain.html). The phylogenetic relationships of the GIs as determined by alignment of the concatenated amino acid sequences of all 15 conserved genes are shown in Fig. 3 and are congruent with the phylogenetic relationships of each of the individual genes.
FIG. 3.
SplitTree tree based on ClustalX analysis of the 15 GIs. The amino acid sequences of the 15 predicted genes common to all 15 GIs were concatenated and aligned by using ClustalX. The alignment of each of the genes alone was consistent with the alignment illustrated. Each strain containing a GI is identified, and where available, the designation of the GI (e.g., ICEHin1056) is given in parentheses. S. typhi, S. enterica serovar Typhi.
Accessory genes, including antibiotic resistance gene content.
The size of the coherent virtual element was approximately 18 kb, while the sizes of the whole GIs varied from 49 kb for H. ducreyi to 140 kb for P. luminescens (Table 1). The variation in the sizes of these related GIs was attributable largely to differences in accessory gene content. Nestled between conserved core genes of each GI were a wide range of different accessory genes (Fig. 1 and 2). The sites of insertion were largely clustered in two regions, between core genes 9 and 14 and between core genes 31 and 33. The major attributes of the accessory genes are listed in Table 1 and include antibiotic, metal, and antiseptic resistance; degradation of chemicals; type IV secretion systems; two-component signaling systems; biofilm regulation; Vi antigen capsule synthesis; cytolethal toxin production; antirestriction systems; and a wide range of metabolic functions. Type IV secretion system genes were present in 5 of the 15 GIs (Fig. 3). The genes homologous to type IV secretion system genes contained in these five GIs were homologous and in synteny (see the supplemental material [http://www.ndcls.ox.ac.uk/mohd-zain.html]). Many of the accessory genes do not exhibit homology to genes with known functions (Fig. 2). The resistance genes found in ICEHin1056 were inserted at two different sites. A Tn10-like transposon that contains the tetA and cat genes lies between core genes 10 and 11. Tn3 that contains the bla gene is inserted between core genes 31 and 32 (Fig. 1 and 2).
DISCUSSION
Syntenic core GI.
A diverse family of related and syntenic GIs with a common evolutionary origin in proteobacterial hosts has been recognized for the first time. These GIs have diverged widely, as demonstrated by as much as 30% divergence (40 to 70%) in the G+C contents of the GIs and amino acid homologies as low as 20 to 30% for proteins encoded by homologous GI genes. These GIs have acquired diverse accessory genes with habitat-specific adaptive functions typical of GIs or pathogenicity islands (13, 22). Other features typical of proteobacterial GIs are insertion into tRNA genes and G+C contents different from those of their hosts' genomes (13).
It has been hypothesized that GIs are mosaics of independently evolving genes or clusters of genes (5, 56). This is referred to as modular evolution. In the reports of Burrus et al., it was demonstrated that heterogeneous modules were assorted in different GIs (5, 6, 40). Such a lack of coherent structure is probable following repeated recombination arising from frequent horizontal spread of GIs. This would be expected to erase coherent structures in different GIs over time. The findings reported here, at least, identify one family of related GIs that challenges this hypothesis. A coherent syntenic element with a common evolutionary origin has been found. The accessory genes of these related GIs are inserted in a manner typical of modular evolution. However, variable modules consisting of accessory genes nestle in a conserved syntenic core GI. The presence of a syntenic core GI suggests a fitness property of the conserved core genes acting collectively that has ensured their survival as a coherent syntenic whole. It is unclear what property this may be, yet it is reasonable to surmise that it is the ability of these GIs to transfer and propagate better as a coherent whole. The role of accessory genes in the survival of this family of elements appears to be interwoven with allowing organism-specific adaptation to habitats. This necessarily involves complex interactions among the core syntenic GI, accessory genes, and the survival of the host bacterium in new hostile environments.
Bioinformatics analysis of the core genes and review of the functional properties of some of the well-characterized examples of this family of GIs provide insight into the possible functions of the core GI genes. An integrase gene is present in all but one (R. metallidurans) of these GIs. The integrase genes are likely to encode integration with tRNA genes, and all belong to the family of tyrosine integrase genes. The only GI in which this has been conclusively shown is the clc element found in Pseudomonas sp. strain B13, which contains a homologue of the P4 integrase gene (43). A curious aspect of the tyrosine integrase genes found in these GIs is that they belong to two different lineages, a P4 lineage and a XerC/D lineage. Both types of integrase genes are known to encode recombination with tRNA genes in other mobile elements (36). This is the only core gene of these syntenic elements which has an evolutionary history more typical of modular evolution. How members of the GIs have acquired these two divergent lineages of integrase genes is intriguing, and the available data provide no obvious explanation. The observation that SPI-7 found in S. enterica serovar Typhi contains both integrase gene lineages, while SPI-7-like GIs found in other S. enterica serovars do not (46), may suggest that there are ready opportunities for exchanging integrase gene lineages.
How GIs transfer horizontally has not been clearly explained (13), although it has been recognized that the clc element is a genomic island and is capable of conjugative transfer (58). The observation made here that another conjugative element, ICEHin1056, found in H. influenzae, is also a related GI strengthens this observation. Hitherto, only the clc element and ICEHin1056 have been shown to conjugate. Both the clc element and ICEHin1056 transfer by conjugation at frequencies of 10−6 to 10−7 (transconjugants/donors) (10, 43). The genes encoding this property have not been identified in either GI. However, the genes encoding this function are likely to be present among the core genes as none of the accessory genes present in either GI is shared. Also, none of their accessory genes is known to encode conjugative functions. Furthermore, four of the core GI genes are homologous to pilL, traD, virB4, and traI and are interspersed along a contiguous 22-gene segment of the GI. These four genes are known to be involved in DNA conjugative transfer systems (60). Therefore, it can be inferred that core genes are likely to encode transfer functions that would explain how this family of GIs transfers between hosts. Before this can be firmly concluded, however, this possibility needs to be formally investigated and demonstrated experimentally.
Autonomous replication would be an unexpected property of GIs (13). However, 4 of the first 10 genes are homologues of parA, dnaB, ssb, and topB, which are genes known to be associated with plasmid replication (16, 20). Furthermore, both ICEHin1056 found in H. influenzae and pKLC102 found in P. aeruginosa C appear to exist as extrachromosomal closed circular plasmids under some conditions (9, 10, 27, 35). ICEHin1056 or other related Haemophilus conjugative elements are found largely in plasmid form in transconjugants immediately following conjugation (9, 10). This conclusion is supported by two observations: first, ready isolation of plasmids from transconjugants and not from parent donor clinical isolates (10); and second, the appearance of Southern blot hybridization patterns consistent with a closed circular form in transconjugants and a pattern indicating chromosomal integration in parent donor strains (9, 10, 35). pKLC102 has not been formally shown to replicate autonomously; however, Klockgether et al. reported the presence of an extrachromosomal form and sequences close to the ssb gene with features typical of an oriV (27). These data suggest that some, if not all, of these GIs may be capable of autonomous replication under some conditions, but this needs to be formally determined experimentally. If this was found to be true, it would be a new, hitherto unrecognized dimension of genomic islands (13, 22) or the integrating and conjugating elements (ICEs) reviewed by Burrus et al. (5). Furthermore, an element that was capable of integrative and excisive recombination, conjugative transfer, and autonomous replication would require finely coordinated regulation of these functions to ensure that their timing was appropriate to the relevant state of the GI.
The origin and host range of this family of GIs are not apparent from the data obtained in this investigation. The wide range of G+C contents (40 to 70%) for the shared core GI genes is not substantially different from the range found for β- and γ-Proteobacteria (28). This could be interpreted to suggest that there is a longstanding relationship with this phylum. This interpretation is based on the proposition that GIs are constrained to this phylum and the proposition that horizontally acquired DNA with a different G+C equilibrates over time with its host's genomic G+C content through a process referred to as amelioration (34). It follows that for this family of GIs to have evolved such divergent G+C contents, substantial time in hosts with similarly divergent G+C contents would have been necessary. It would also require limited horizontal transfer and recombination between divergent members of this family of GIs, as this would tend to homogenize the G+C contents. The presence of two GIs with G+C contents of 68.2 and 69.6%, which are markedly higher than the corresponding host genomic G+C contents (namely, the G+C contents of R. metallidurans [63.5%] and X. axonopodis [64.7%]) is curious. The fact that these G+C contents are at the upper limit of the values for Proteobacteria (28) raises the question of whether these GIs originated from nonproteobacterial hosts with higher G+C contents than Proteobacteria. Conclusive evidence of this may become apparent from the growing number of whole genomic sequences available for analysis and improved algorithms for interrogating the databases for large contiguous and weakly homologous sequences.
Antibiotic resistance accessory genes and this family of GIs.
The observation of evolutionarily related coherent and syntenic GIs provides new insight into how the recent emergence and spread of β-lactamase-positive Apr and/or tetracycline and chloramphenicol resistance in H. influenzae occurred. The phylogenetic relationship of ICEHin1056 to other distantly related GIs indicates that transferable resistance in H. influenzae has deep evolutionary origins. The resistance genes, conveyed by transposons (e.g., Tn10 or Tn3) (15, 26), form clusters of accessory genes in the core element that have apparently evolved stable relationships. No other accessory genes with different properties were apparent from an analysis of the whole sequence. The emergence of this resistance element in pathogenic H. influenzae only became readily detectable in the early 1970s (4, 14, 42, 53, 54). Curiously, resistance then rapidly emerged over the next few years worldwide among pathogenic H. influenzae strains and became very prevalent (20 to 30%) in many countries (42, 47). Evidence indicates that this resistance is accounted for by the appearance of ICEs (GIs) that are highly related to ICEHin1056 (9-11, 35, 42, 47). The epidemic expansion of this family of ICEs among pathogenic H. influenzae and the high prevalence among other commensal haemophili (35, 45, 46) suggest that this GI is well adapted to these bacterial hosts and provides a survival advantage under antibiotic exposure conditions. An intriguing question is whether the acquisition of resistance transposons and their intimate and apparent stable relationship with the GIs in haemophili evolved recently or have deeper origins preceding the antibiotic era. The latter possibility would support the notion that antibiotic resistance is an ancient adaptive response that pre-evolved in bacteria. Modern-day intensive use of antibiotics only serves to change the distribution or organization of the pre-evolved structures.
The association of resistance genes with this family of GIs is not unique to H. influenzae. GIs found in both H. somnus and P. aeruginosa contain accessory genes with homology to resistance genes. In the case of H. somnus 2336, homologues of tetA and romA (a multidrug resistance gene) are present, and the strain is tetracycline resistant (the MIC of tetracycline is 8 μg/ml [Inzana, unpublished data]). In P. aeruginosa strain C, the GI, pKLC102, contains aadB that encodes tobramycin and gentamicin resistance (27).
Major properties of other accessory genes.
The accessory genes conveyed by the family of GIs described here have a wide range of attributes other than antibiotic resistance that contribute to the habitat-specific survival of their host organisms. The next most common associated attribute is type IV secretion found in five of the GIs, two of which, PAP1 (24) and SPI-7 (41), are well-described pathogenicity islands. The genes of all five GIs were homologous and in synteny, suggesting that they share a common evolutionary origin. The type IV secretion system in SPI-7 is involved in the adhesion of S. enterica serovar Typhi to macrophages and is important to S. enterica serovar Typhi's intracellular life cycle and in disease causation (38, 57, 61). The role of genes encoding the type IV secretion system in PAP1 is less clear. These genes were not reported as contributing to the virulence of P. aeruginosa PA14 (24). However, the most likely role of type IV secretion found in these GIs is adherence of the host organisms to specific targets in their habitats.
Other genes important in virulence are present in these GIs. The Vi antigen encoded by SPI-7 is a well-known capsular antigen characteristic of S. enterica serovar Typhi that contributes to the pathogenesis of typhoid fever and is a valuable vaccine antigen (41). PAP1 contains a number of virulence-associated accessory genes, including two-component regulators that may be involved in regulating pathogenicity activities (24). The island found in H. ducreyi contains the cytolethal distending toxin genes, which have an uncertain role in disease pathogenesis (19, 51, 59).
The accessory genes important in the survival of organisms in environmental habitats include a range of genes encoding metabolic functions. These genes are found in organisms that live in potentially nutrient-deficient environments, such as water or soil, including P. aeruginosa, Pseudomonas sp. strain B13, B. fungorum, and R. metallidurans. Another attribute of the accessory genes of some of these environmental species is highly processive degradative enzymes. In particular, the clc element found in Pseudomonas sp. strain B13 and B. fungorum encodes a number of genes that inactivate toxic chemicals, such as chlorocatechols (58). Organisms possessing these enzymes and related DNA elements have become widely distributed, presumably through selection, in environments that are heavily contaminated with toxic waste (7, 8, 58).
Conclusions.
There is a very large and diverse population of GIs in bacteria (13), and most of these GIs do not exhibit detectable homology with the family of related GIs described here. However, the methods used here to identify and characterize one family of GIs may be used in principle to identify other families of coherent GIs among the many apparently unrelated GIs. It seems possible that there are examples of both mosaics without evidence of a coherent syntenic core GI structure and other GIs which have syntenic core genes and a common evolutionary origin. This will be increasingly possible to investigate with the improving bioinformatics techniques and the rapidly increasing sequence data available from complete bacterial genomes. Such a systematic classification of GIs is likely in turn to necessitate a commensurate naming system.
Acknowledgments
We thank The Wellcome Sanger Institute, Hinxton, United Kingdom, for sequence data and support in annotation of DNA sequences. A preliminary DNA sequence was obtained from the H. ducreyi sequencing project, a collaborative effort between the Institute for Systems Biology, Seattle Wash., and the laboratory of Robert Munson at the Children's Research Institute and The Ohio State University.
The H. ducreyi sequencing project was funded by NIH grant R01-AI45091. The H. somnus sequencing project was funded by USDA/CSREES grant 2001-52100-11314. D.W.C. was funded by Wellcome Trust Research Leave Fellowship 057366/Z/99/Z. Z.M.Z. was funded through a scholarship awarded by the Government of Malaysia.
REFERENCES
- 1.Alfano, J. R., A. O. Charkowski, W. L. Deng, J. L. Badel, T. Petnicki-Ocwieja, K. van Dijk, and A. Collmer. 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97:4856-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brightman, C. A., D. W. Crook, W. A. Kraak, I. D. Dimopoulou, E. C. Anderson, W. W. Nichols, and M. P. Slack. 1990. Family outbreak of chloramphenicol-ampicillin resistant Haemophilus influenzae type b disease. Lancet 335:351-352. [DOI] [PubMed] [Google Scholar]
- 4.Brunton, J., D. Clare, and M. A. Meier. 1986. Molecular epidemiology of antibiotic resistance plasmids of Haemophilus species and Neisseria gonorrhoeae. Rev. Infect. Dis. 8:713-724. [DOI] [PubMed] [Google Scholar]
- 5.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 6.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77-97. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, D. K., and A. M. Chakrabarty. 1983. Genetic homology between independently isolated chlorobenzoate-degradative plasmids. J. Bacteriol. 153:532-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, D. K., S. T. Kellogg, S. Hamada, and A. M. Chakrabarty. 1981. Plasmid specifying total degradation of 3-chlorobenzoate by a modified ortho pathway. J. Bacteriol. 146:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimopoulou, I. D., J. Z. Jordens, N. J. Legakis, and D. W. Crook. 1997. A molecular analysis of Greek and UK Haemophilus influenzae conjugative resistance plasmids. J. Antimicrob. Chemother. 39:303-307. [DOI] [PubMed] [Google Scholar]
- 10.Dimopoulou, I. D., W. A. Kraak, E. C. Anderson, W. W. Nichols, M. P. Slack, and D. W. Crook. 1992. Molecular epidemiology of unrelated clusters of multiresistant strains of Haemophilus influenzae. J. Infect. Dis. 165:1069-1075. [DOI] [PubMed] [Google Scholar]
- 11.Dimopoulou, I. D., J. E. Russell, Z. Mohd-Zain, R. Herbert, and D. W. Crook. 2002. Site-specific recombination with the chromosomal tRNALeu gene by the large conjugative Haemophilus resistance plasmid. Antimicrob. Agents Chemother. 46:1602-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobrindt, U. H., B. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 14.Elwell, L. P., J. De Graaff, D. Seibert, and S. Falkow. 1975. Plasmid-linked ampicillin resistance in Haempohilus influenzae type b. Infect. Immun. 12:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elwell, L. P., J. R. Saunders, M. H. Richmond, and S. Falkow. 1977. Relationships among some R plasmids found in Haemophilus influenzae. J. Bacteriol. 131:356-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinosa, M. C., S. Couturier, M. del Solar, G. Diaz-Orejas, R. Giraldo, R. Janniere, L. Miller, C. Osborn, and C. M. Thomas. 2000. Plasmid replication and copy number control, p. 1-47. In C. M. Thomas (ed.), The horizontal gene pool bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 17.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 18.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Gelfanova, V., E. J. Hansen, and S. M. Spinola. 1999. Cytolethal distending toxin of Haemophilus ducreyi induces apoptotic death of Jurkat T cells. Infect. Immun. 67:6394-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerdes, K. A., S. Carnosa, I. Ceglowski, P. Diaz-Orejas, R. Franch, T. Gultyarev, A. P. Gugge Jensen, R. Kobayashi, I. Macpherson, C. Summers, C. M. Thomas, and U. Zielenkiewicz. 2000. Plasmid maintenance systems, p. 49-85. In C. M. Thomas (ed.), The horizontal gene pool bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 21.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 22.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 24.He, J., R. L. Baldini, E. Deziel, M. Saucier, Q. Zhang, N. T. Liberati, D. Lee, J. Urbach, H. M. Goodman, and L. G. Rahme. 2004. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. USA 101:2530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jahn, G., R. Laufs, P. M. Kaulfers, and H. Kolenda. 1979. Molecular nature of two Haemophilus influenzae R factors containing resistances and the multiple integration of drug resistance transposons. J. Bacteriol. 138:584-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaulfers, P. M., R. Laufs, and G. Jahn. 1978. Molecular properties of transmissible R factors of Haemophilus influenzae determining tetracycline resistance. J. Gen. Microbiol. 105:243-252. [DOI] [PubMed] [Google Scholar]
- 27.Klockgether, J., O. Reva, K. Larbig, and B. Tummler. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J. Bacteriol. 186:518-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krieg, N. R., and J. G. Holt (ed.). 1984. Bergey's manual of systemic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 29.Lan, R., and P. R. Reeves. 1996. Gene transfer is a major factor in bacterial evolution. Mol. Biol. Evol. 13:47-55. [DOI] [PubMed] [Google Scholar]
- 30.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laufs, R., and R. Fock. 1979. Characterization of genes specifying ampicillin resistance in bacterial isolates using a single-strand specific nuclease for analysis of plasmid DNA-DNA duplexes. J. Gen. Microbiol. 111:233-237. [DOI] [PubMed] [Google Scholar]
- 32.Laufs, R., and P. M. Kaulfers. 1977. Molecular characterization of a plasmid specifying ampicillin resistance and its relationship to other R factors from Haemophilus influenzae. J. Gen. Microbiol. 103:277-286. [DOI] [PubMed] [Google Scholar]
- 33.Laufs, R., F. C. Riess, G. Jahn, R. Fock, and P. M. Kaulfers. 1981. Origin of Haemophilus influenzae R factors. J. Bacteriol. 147:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leaves, N. I., I. Dimopoulou, I. Hayes, S. Kerridge, T. Falla, O. Secka, R. A. Adegbola, M. P. Slack, T. E. Peto, and D. W. Crook. 2000. Epidemiological studies of large resistance plasmids in Haemophilus. J. Antimicrob. Chemother. 45:599-604. [DOI] [PubMed] [Google Scholar]
- 36.Mantri, Y., and K. P. Williams. 2004. Islander: a database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 32(Database Issue):D55-D58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathies A. W. J. 1972. Penicillins in the treatment of bacterial meningitis. J. R. Coll. Physicians Lond. 6:139-146. [PMC free article] [PubMed] [Google Scholar]
- 38.Morris, C., C. M. Yip, I. S. Tsui, D. K. Wong, and J. Hackett. 2003. The shufflon of Salmonella enterica serovar Typhi regulates type IVB pilus-mediated bacterial self-association. Infect. Immun. 71:1141-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 40.Pavlovic, G., V. Burrus, B. Gintz, B. Decaris, and G. Guedon. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759-774. [DOI] [PubMed] [Google Scholar]
- 41.Pickard, D., J. Wain, S. Baker, A. Line, S. Chohan, M. Fookes, A. Barron, P. O. Gaora, J. A. Chabalgoity, N. Thanky, C. Scholes, N. Thomson, M. Quail, J. Parkhill, and G. Dougan. 2003. Composition, acquisition, and distribution of the Vi exopolysaccharide-encoding Salmonella enterica pathogenicity island SPI-7. J. Bacteriol. 185:5055-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powell, M. 1988. Antimicrobial resistance in Haemophilus influenzae. J. Med. Microbiol. 27:81-87. [DOI] [PubMed] [Google Scholar]
- 43.Ravatn, R., S. Studer, A. J. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 45.Scheifele, D. W., and S. J. Fussell. 1981. Ampicillin-resistant Haemophilus influenzae colonizing ambulatory children. Am. J. Dis. Child. 135:406-409. [DOI] [PubMed] [Google Scholar]
- 46.Scheifele, D. W., S. J. Fussell, and M. C. Roberts. 1982. Characterization of ampicillin-resistant Haemophilus parainfluenzae. Antimicrob. Agents Chemother. 21:734-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schito, G. C., E. A. Debbia, and A. Marchese. 2000. The evolving threat of antibiotic resistance in Europe: new data from the Alexander Project. J. Antimicrob. Chemother 46(Suppl. T1):3-9. [DOI] [PubMed] [Google Scholar]
- 48.Sentchilo, V., A. J. Zehnder, and J. R. van der Meer. 2003. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol. 49:93-104. [DOI] [PubMed] [Google Scholar]
- 49.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 51.Stevens, M. K., J. L. Latimer, S. R. Lumbley, C. K. Ward, L. D. Cope, T. Lagergard, and E. J. Hansen. 1999. Characterization of a Haemophilus ducreyi mutant deficient in expression of cytolethal distending toxin. Infect. Immun. 67:3900-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuy, J. H. 1980. Chromosomally integrated conjugative plasmids are common in antibiotic-resistant Haemophilus influenzae. J. Bacteriol. 142:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syriopoulou, V., D. Scheifele, V. Howie, J. Ploussard, J. Sloyer, and A. L. Smith. 1976. Incidence of ampicillin-resistant Hemophilus influenzae in otitis media. J. Pediatr. 89:839-841. [DOI] [PubMed] [Google Scholar]
- 54.Syriopoulou, V., D. Scheifele, A. L. Smith, P. M. Perry, and V. Howie. 1978. Increasing incidence of ampicillin resistance in Hemophilus influenzae. J. Pediatr. 92:889-892. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toussaint, A., and C. Merlin. 2002. Mobile elements as a combination of functional modules. Plasmid 47:26-35. [DOI] [PubMed] [Google Scholar]
- 57.Tsui, I. S., C. M. Yip, J. Hackett, and C. Morris. 2003. The type IVB pili of Salmonella enterica serovar Typhi bind to the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 71:6049-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Meer, J. R., and V. Sentchilo. 2003. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 14:248-254. [DOI] [PubMed] [Google Scholar]
- 59.Young, R. S., K. R. Fortney, V. Gelfanova, C. L. Phillips, B. P. Katz, A. F. Hood, J. L. Latimer, R. S. Munson, Jr., E. J. Hansen, and S. M. Spinola. 2001. Expression of cytolethal distending toxin and hemolysin is not required for pustule formation by Haemophilus ducreyi in human volunteers. Infect. Immun. 69:1938-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zechner, E. L., F. Eisenbrandt, R. Grahn, A. M. Koraimann, G. Lanka, E. Muth, G. Pansegrau, W. Thomas, C. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 61.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]