Abstract
Antibiotic resistance Staphylococcus aureus strains cause several life threatening infections. New drug treatment options are needed, but are slow to develop because 50% of the S. aureus genome is hypothetical. The goal of this is to aid in the annotation of the S. aureus NCTC 8325 genome by identifying hypothetical proteins related to the Major Facilitator Superfamily (MFS). The MFS is a broad protein group with members involved in drug efflux mechanisms causing resistance. To do this, sequences for three MFS proteins with x-ray crystal structures in E. coli were PSI-BLASTed against the S. aureus NCTC 8325 genome to identify homologs. Eleven identified hypothetical protein homologs underwent BLASTP against the non-redundant NCBI database to fit homologs specific to each hypothetical protein. ExPASy characterized the physiochemical features, CDD-BLAST and Pfam identified domains, and the SOSUI server defined transmembrane helices of each hypothetical protein. Based on size (300 – 700 amino acids), number of transmembrane helices (>7), CD06174 and MFS domains in CDD-BLAST and Pfam, respectively, and close relation to well-defined homologs, SAOUHSC_00058, SAOUHSC_00078, SAOUHSC_00952, SAOUHSC_02435, SAOUHSC_02752, and ABD31642.1 are members of the MFS. Further multiple-alignment and phylogeny analyses show SAOUHSC_00058 to be a quinolone resistance protein (NorB), SAOUHSC_00058 a siderophore biosynthesis protein (SbnD), SAOUHSC_00952 a glycolipid permease (LtaA), SAOUHSC_02435 a macrolide MFS transporter, SAOUHSC_02752 a chloramphenicol resistance (DHA1), and ABD31642.1 is a Bcr/CflA family drug resistance efflux transporter. These findings provide better annotation for the existing genome, and identify proteins related to antibiotic resistance in S. aureus NCTC 8325.
Keywords: Functional annotation, Hypothetical proteins, Superfamily, S.aureus
Background
Staphylococcus aureus is an opportunistic pathogen responsible for a wide variety of infections including superficial skin and surgical wound infections, toxic shock syndrome, and bacteremia [1]. Most are nosocomial infections, though there are increases in community acquired (CA) Methicillin-resistant Staphylococcus aureus (MRSA) infections, particularly among immunocompromised patients. Other health issues related to internalized infections are heart and lung diseases such as endocarditis and necrotizing pneumonia found in younger community populations rather than remaining solely a hospital acquired infection. Deaths from S. aureus caused heart and lung infections are reported [2]. In 2011, the Center for Disease Control estimate 80,000 invasive MRSA infections and 11,285 related deaths in the United States [3]. These deaths are primarily due to MRSA strains that are resistant to macrolides, monovalent cationic antimicrobials, quinolones, bivalent quaternary ammonium compounds, tetracycline, and all betalactam antibiotics including penicillin, amoxicillin, methicillin, and oxacillin [4]. Inactivation of antibiotics, reduction in cellular permeability, alteration of antibiotic target sites, and bacterial efflux pumps convey drug resistance [5]. Several multidrug efflux genes, such as the NorA, NorB, and NorC from the S. aureus chromosome, confer resistance to quinolones and other antibiotics [6,7]. Disturbingly, an increase in the variety of drug-resistant strains of S. aureus has been noted in the past years, with the most prevalent being vancomycin-resistant S. aureus (VRSA). Usually VRSA develops in MRSA patients treated with vancomycin, the frontline treatment to MRSA. While VRSA is rare with most S. aureus being vancomycin-intermediate meaning that large amounts of vancomycin still kill the organisms, this presents a new challenge to combat S. aureus infections. These superbugs are generally sensitive to intravenous medication, such as quinupristin– dalfopristin, that require slow infusion in a large fluid volume, making it unrealistic for administration to CA-MRSA patients in an outpatient setting. Quinupristin–dalfopristin can also cause disabling myopathy as a side effect. Due to documented increases of a global spread of CA-MRSA in just the past 20 years and the increases in antibiotic resistances, there is a need for new treatment options [2]
Most multidrug resistance efflux pumps eject various substrates regardless of structure, a common feature of the Major Facilitator Superfamily (MFS) they belong to [8]. In bacteria, about 25% of characterized membrane transport proteins come from the MFS [9]. This group of transporters contains 74 families that move a wide variety of substrates including sugar phosphates, nucleosides, ions, amino acids, peptides, and drugs across the cytoplasmic membrane [10]. X-ray crystallography established E. coli’s structure of three members of this conserved protein family: glycerol-3-phosphate transporter (GlpT), lactose permease (LacY), and multidrug transporter (EmrD). These structures, listed in the PDB as 1PW4, 1PV7, and 2GFP for GlpT, LacY, and EmrD, respectively, demonstrate that MFS proteins function via the substrate’s electrochemical potential. MFS proteins are usually 400 to 600 amino acids long, with most containing 11 to 14 transmembrane alpha helices connected by hydrophilic loops [10].
Hypothetical proteins make up approximately 50% of reference strain S. aureus NCTC 8325 genome [11]. Nucleic acid sequence only predicts hypothetical proteins [12]. There is no experimental evidence for a hypothetical protein’s function exists; ergo a hypothetical protein may not actually be functional. Further, some hypothetical proteins do not follow conventional phylogenetic lineage. Usually the two groups of hypothetical proteins are uncharacterized protein families and domains of unknown function, or experimentally identified proteins with structural domains that do not relate to already established functions. However, databases frequently label a protein hypothetical if it comes from a newly deposited sequence and no annotation was available for it at that time [13]. This is likely the case with S. aureus NCTC 8325, whose genome was published in 2006 [14].
With approximately half of all S. aureus NCTC 8325 genomic protein sequences currently annotated as hypothetical proteins and 25% of all membrane transport proteins belonging to the MFS, ergo likely related to antibiotic resistance, there is great potential for the discovery of new drug targets here [15]. Since proper annotation of hypothetical proteins can lead to new therapeutic targets, a high demand to characterize hypothetical proteins is present. Ergo, this study uses in silico techniques to identify and characterize hypothetical proteins in S. aureus NCTC 8325 that are related to the protein MFS.
Methodology
Figure 1 illustrates the overall experimental design. To identify MFS-related hypothetical proteins, protein sequences for the three E. coli MFS crystal structures, 1PW4 (GlpT), 1PV7 (LacY), and 2GFP (EmrD), were downloaded from PDB. A Position-Specific Iterative Basic Local Alignment Search Tool (PSI-BLAST) revealed proteins of interest when searched with each sequence against the S. aureus NCTC 8325 genome. If the protein was hypothetical, it underwent protein-protein BLAST (BLASTP) against NCBI’s entire nonreductant protein database to identify top matching homologs in any species. NCBI provides both BLAST algorithms.
Figure 1.
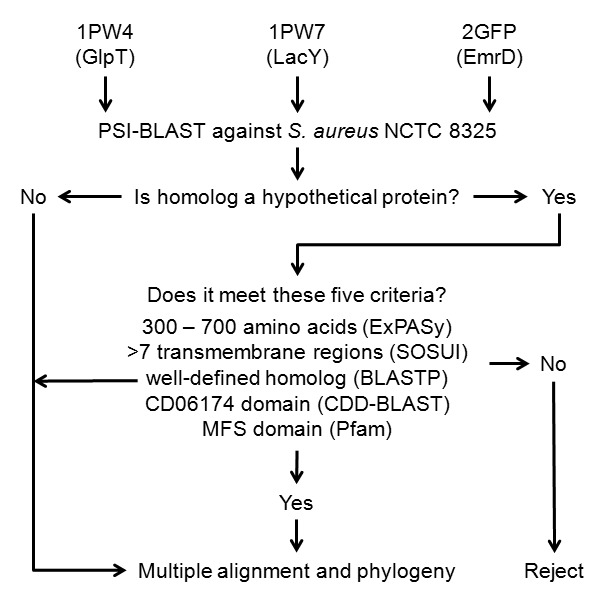
Experimental Overview. Protein sequences for three E. coli MFS proteins underwent PSI-BLAST. Hypothetical protein homologs meeting five parameters are likely functional MFS and then compared to homologs of predicted function.
Physicochemical Characterization
To characterize the proteins, the Expasy Protparam server computed several physicochemical characterizations. The number of amino acids, molecular weight, total number of charged residues (the addition of arginine and lysine for negatively charged and aspartic acid added to glutamic acid for positively charged) [16]. The algorithm determines the theoretical isoelectric point, the pH where a molecule carries no net electrical charge, from the number of charged residues. Further, the program calculates the extinction coefficient, the amount of light absorbed by the protein at a 280nm wavelength, which is helpful for protein purification procedures [17]. A protein’s stability in a test tube under physiological conditions is measured by its instability index [18]. The relative volume occupied by open side chain amino acids in a protein is the aliphatic index [19]. The grand average hydropathy (GRAVY) is the total of the hydropathy values of all amino acids in the protein divided by the number of resides a measure of hydrophobicity for a given molecule [20]. The SOSUI server also determines a protein’s hydrophobicity, though via solubility computations, and it further characterizes potential transmembrane regions [21].
Domain Identification
Both Pfam and the conserved domain database (CDD) identified domains. The Pfam database is a collection of protein families represented by hidden Markov models and multiple sequence alignments [22,23]. Conserved domain database BLAST (CDDBLAST) uses a variant of PSI-BLAST to scan a set of pre-calculated position-specific scoring matrices from a protein query [24]. Researchers frequently use Pfam and CDD-BLAST together to examine protein domains to predict function prior to modeling the protein to evaluate its binding capability [15,25].
Protein relatedness
Hypothetical proteins between 300 and 700 amino acids long with well-defined BLASTP homologs, CD06174 MFS domain identified by CDD-BLAST, and an MFS domain found by Pfam underwent evolutionary analyses against non-redundant defined homologs identified from prior PSI-BLAST and BLASTP searches. Two programs, PROMALS3D and CLUSTALW, did multiple sequence alignment. PROMALS3D aligned each set of proteins based on homology to the E. coli protein with the established structure by constructing alignments using information from available homologs with 3D structures, sequence database searches, and secondary structure prediction [26]. CLUSTALW aligned all protein sequences together as one large dataset [27]. The PHYLIP package Protdist program used the output from CLUSTALW to produce a distance matrix using default settings such as the Jones- Taylor-Thornton matrix distance model [28]. Another program in the PHYLIP suite, Neighbor, used this matrix to construct a neighbor joining and unweighted pair group method with arithmetic mean trees. Again as verification, another phylogenetic tree building approach, the Fitch-Margoliash and Least-Squares Distance method, verified the results. DrawTree from the PHYLIP suite illustrated phylogenetic trees produced. All analyses used default settings.
Discussion:
This study aimed to identify and characterize hypothetical proteins in S. aureus NCTC 8325 that belong to the MFS of proteins. Three E. coli MFS proteins with established structure (GlpT, LacY, and EmrD) underwent PSI-BLAST against the S. aureus NCTC 8325 genome to identify homologs. Table 1, Table 2, Table 3 list homologs for GlpT, LacY, and EmrD, respectively. This method identified eleven hypothetical proteins. Researchers need further examination to determine if these hypothetical proteins belong to the MFS.
Table 1. Homologs in S. aureus NCTC 8325 to GlpT from E. coli.
| Protein | Accession | QC % | %ID |
| glycerol-3-phosphate transporter | WP_001010111.1 | 97 | 57 |
| antiporter | WP_001008722.1 | 96 | 32 |
| glucarate transporter | WP_000660709.1 | 90 | 22 |
| quinolone resistance protein NorB | WP_000381270.1 | 21 | 28 |
| multidrug efflux MFS transporter NorA | WP_001041272.1 | 15 | 27 |
| MFS transporter | WP_001154220.1 | 27 | 20 |
| probable glycolipid permease LtaA | Q2FZP8.2 | 27 | 20 |
| conserved hypothetical protein | ABD31816.1 | 7 | 27 |
| hypothetical protein SAOUHSC_00058 | YP_498663.1 | 56 | 23 |
| hypothetical protein SAOUHSC_00078 | YP_498678.1 | 50 | 22 |
| hypothetical protein SAOUHSC_00952 | YP_499505.1 | 41 | 20 |
| hypothetical protein SAOUHSC_02620 | WP_000436818.1 | 4 | 45 |
| QC, Query Cover; %ID, percentage of identity; MFS, major facilitator superfamily | |||
Table 2. Homologs in S. aureus NCTC 8325 to LacY from E. coli.
| Protein | Accession | QC % | %ID |
| multidrug efflux MFS transporter NorA | WP_001041272.1 | 45 | 28 |
| hydroxyethylthiazole kinase | WP_000610051.1 | 17 | 25 |
| siderophore biosynthesis protein SbnD | WP_000610051.1 | 17 | 25 |
| MFS transporter | WP_000610884.1 | 35 | 27 |
| glucarate transporter | WP_000660709.1 | 46 | 29 |
| proline/betaine transporter | WP_000347061.1 | 14 | 27 |
| nitrate transporter NarT | WP_000278558.1 | 17 | 32 |
| nickel ABC transporter permease | WP_000584765.1 | 19 | 34 |
| antibiotic MFS transporter | WP_000675401.1 | 11 | 29 |
| hypothetical protein SAOUHSC_02866 | YP_501322.1 | 12 | 31 |
| hypothetical protein SAOUHSC_02307 | YP_500786.1 | 23 | 23 |
| hypothetical protein SAOUHSC_02309 | WP_001287088.1 | 11 | 31 |
| QC, Query Cover; %ID, percentage of identity; MFS, major facilitator super-family | |||
Table 3. Homologs in S. aureus NCTC 8325 to EmrD from E. coli.
| Protein | Accession | QC % | %ID |
| Bcr/CflA family drug resistance efflux transporter | WP_000999131.1 | 62 | 25 |
| drug:proton antiporter | WP_000038961.1 | 42 | 27 |
| multidrug MFS transporter | WP_001820335.1 | 40 | 26 |
| nitrate transporter NarT | WP_000278558.1 | 52 | 24 |
| multidrug efflux MFS transporter NorA | WP_001041272.1 | 84 | 20 |
| chloramphenicol resistance protein DHA1 | WP_000026194.1 | 41 | 20 |
| quinolone resistance protein NorB | WP_001066546.1 | 34 | 23 |
| conserved hypothetical protein | ABD31642.1 | 62 | 25 |
| hypothetical protein SAOUHSC_02435 | YP_500904.1 | 34 | 22 |
| hypothetical protein SAOUHSC_02752 | YP_501211.1 | 51 | 20 |
| QC, Query Cover; %ID, percentage of identity; MFS, major facilitator super-family | |||
To distinguish that, homolog identification, physiochemical characterization, transmembrane enumeration, and domain identification compared hypothetical proteins to established MFS proteins. Table 4 lists the top BLASTP homolog for each hypothetical protein regardless of origin. A hypothetical protein that has a homolog with a well-defined function is more likely related. SAOUHSC_02307, SAOUHSC_02309, and ABD31816.1 hit several general membrane proteins. SAOUHSC_02620 and SAOUHSC_02866 matched several hypothetical proteins as well as general membrane proteins. This indicates these hypothetical proteins may not be in the MFS.
Table 4. Homologs to hypothetical proteins.
| Hypothetical Protein | Top Match |
| SAOUHSC_00058 | quinolone resistance protein NorB |
| SAOUHSC_00078 | siderophore biosynthesis protein SbnD |
| SAOUHSC_00952 | glycolipid permease LtaA |
| SAOUHSC_02307 | putative membrane spanning protein |
| SAOUHSC_02309 | membrane protein |
| SAOUHSC_02435 | macrolide MFS transporter |
| SAOUHSC_02620 | membrane protein |
| SAOUHSC_02752 | chloramphenicol resistance protein DHA1 |
| SAOUHSC_02866 | membrane protein |
| ABD31816.1 | membrane protein |
| ABD31642.1 | Bcr/CflA family drug resistance efflux transporter |
| MFS, major facilitator super-family | |
Table 5 lists the physiochemical parameters calculated by ExPASy. Since MFS proteins are usually 400 to 600 amino acids, any hypothetical protein outside the 300 to 700 amino acid range is unlikely to be functional. Four of the five hypothetical proteins that had ambiguous membrane protein BLASTP hits, SAOUHSC_02307, SAOUHSC_02309, SAOUHSC_02620, and SAOUHSC_02866, came up outside this size range. Examination of other physiochemical calculations failed to be consistent in predicting MFS proteins among hypotheticals though some proteins outside this size range had varied theoretical index (SAOUHSC_02620 and SAOUHSC_02866) and GRAVY values (‹0.05 for SAOUHSC_02307, SAOUHSC_02309, SAOUHSC_02866, and ABD31816.1) compared to proteins with expected size. This indicates that these specific hypothetical proteins may not be in the MFS.
Table 5. Physiochemical properties.
| Protein | # AA | MW | pI | # neg | # pos | EC | II | AI | GRAVY |
| GlpT | 451 | 50204.2 | 8.69 | 28 | 33 | 115660 | 36.12 | 99.73 | 0.505 |
| LacY | 417 | 46457.1 | 9.02 | 17 | 24 | 54240 | 29.54 | 109.93 | 0.906 |
| EmrD | 375 | 39995.1 | 9.1 | 12 | 18 | 61800 | 37.9 | 121.23 | 0.942 |
| SAOUHSC_00058 | 462 | 48999.6 | 9.49 | 17 | 29 | 59610 | 22.37 | 128.74 | 0.948 |
| SAOUHSC_00078 | 418 | 44835.3 | 9.54 | 13 | 27 | 53570 | 46.13 | 120.65 | 0.786 |
| SAOUHSC_00952 | 402 | 45457.6 | 9.54 | 16 | 26 | 63830 | 36.9 | 128.28 | 0.936 |
| SAOUHSC_02307 | 163 | 19404.9 | 9.62 | 16 | 22 | 32430 | 48.51 | 123.8 | 0.134 |
| SAOUHSC_02309 | 159 | 18972.3 | 9.18 | 13 | 18 | 38850 | 36.78 | 108.43 | 0.14 |
| SAOUHSC_02435 | 397 | 44337.3 | 9.44 | 16 | 31 | 47830 | 24.38 | 127.68 | 0.868 |
| SAOUHSC_02620 | 215 | 24956.7 | 6.1 | 16 | 15 | 34630 | 56.72 | 111.91 | 0.573 |
| SAOUHSC_02752 | 375 | 40245.1 | 9.68 | 12 | 20 | 43430 | 29.41 | 132.51 | 0.918 |
| SAOUHSC_02866 | 822 | 90409.3 | 6.46 | 90 | 86 | 58790 | 31.15 | 107.93 | 0.159 |
| ABD31816.1 | 416 | 47645 | 9.19 | 31 | 43 | 39810 | 26.46 | 107.62 | 0.282 |
| ABD31642.1 | 312 | 34467.5 | 10.05 | 11 | 20 | 34950 | 28.19 | 125.61 | 0.897 |
SOSUI calculates the average hydrophobicity of a protein. If hydrophobicity exists, the server labels that portion of the protein as a transmembrane region. Table 6 details transmembrane regions as described by the SOSUI server. MFS proteins typically have 11 to 13 transmembrane regions, so SAOUHSC_02307, SAOUHSC_02309, SAOUHSC_02620, and ABD31816.1 are unlikely functioning MFS proteins because they have half or less of the necessary number of transmembrane regions. Since GlpT and LacY had 10 transmembrane regions per SOSUI (data not shown), it is still possible that SAOUHSC_02752 and ABD31642.1 are related to MFS proteins based on this analysis.
Table 6. Transmembrane Regions Identified by SOSUI.
| Locus Tag | Number | Base-Pair Position |
| GlpT | 10 | 26 – 43, 102 – 124, 160 – 181, 187 – 208, 251 – 273, 290 – 312, 320 – 342, 350 – 372, 384 – 406, 414 – 435 |
| LacY | 12 | 11 – 33, 44 – 66, 75 – 97, 106 – 128, 144 – 166, 174 – 196, 215 – 237, 260 – 282, 288 – 310, 313 – 335, 346 – 368, 379 – 401 |
| EmrD | 10 | 4 – 26, 32 – 54, 69 – 91, 127 – 149, 153 – 175, 205 – 227, 235 – 256, 263 – 285, 291 – 313, 346 – 368 |
| SAOUHSC_00058 | 13 | 12 – 34, 45 – 67, 88 – 110, 134 – 156, 163 – 184, 198 – 220, 226 – 247, 262 – 284, 301 – 323, 330 – 349, 355 – 373, 403 – 425, 431 – 453 |
| SAOUHSC_00078 | 12 | 11 – 33, 45 – 67, 80 – 102, 104 – 126, 145 – 167, 171 – 192, 224 – 246, 256 – 277, 291 – 313, 317 – 339, 351 – 371, 376 – 398 |
| SAOUHSC_ 00952 | 11 | 15 – 37, 42 – 64, 85 – 107, 113 – 134, 144 – 166, 173 – 195, 218 – 240, 251 – 273, 280 – 302, 307 – 329, 374 – 396 |
| SAOUHSC_02307 | 2 | 17 – 39, 50 – 72 |
| SAOUHSC_02309 | 2 | 20 – 42, 48 – 70 |
| SAOUHSC_02435 | 11 | 5 – 27, 36 – 58, 66 – 88, 92 – 114, 133 – 155, 162 – 184, 212 – 234, 248 – 270, 286 – 308, 333 – 355, 361 – 382 |
| SAOUHSC_02620 | 4 | 38 – 60, 73 – 95, 108 – 130, 143 – 165 |
| SAOUHSC_02752 | 8 | 12 – 34, 53 – 75, 80 – 101, 110 – 132, 139 – 161, 193 – 215, 224 – 246, 260 – 282 |
| SAOUHSC_02866 | 12 | 10 – 32, 170 – 192, 198 – 220, 225 – 246, 267 – 289, 303 – 324, 348 – 370, 510 – 532, 538 – 560, 579 – 601, 624 – 646, 655 – 677 |
| ABD31816.1 | 6 | 75 – 97, 119 – 141, 143 – 165, 169 – 191, 195 – 216, 389 – 411 |
| ABD31642.1 | 9 | 13 – 35, 53 – 74, 80 – 102, 107 – 129, 138 – 160, 167 – 189, 221 – 243, 254 – 276, 285 – 307 |
Finally, potential MFS hypothetical proteins should have similar domains to those found in GlpT, LacY, and EmrD. Table 7 lists the CDD-BLAST results and Table 8 the Pfam results for domain identification. Most hypothetical proteins with the appropriate size and well-defined PSI-BLAST homologs had the CD06174 MFS domain identified by CDD-BLAST and the MFS_1 domain identified by Pfam. GlpT and EmrD have these domains too.
Table 7. CDD-BLAST domain data.
| Hypothetical | Protein Domains |
| Hypothetical | Protein Domains |
| GlpT | cd06174 |
| PRK11273 | |
| LacY | cd06174 |
| pfam01306 | |
| EmrD | cd06174 |
| PRK11652 | |
| SAOUHSC_00058 | cd06174 |
| pfam07690 | |
| SAOUHSC_00078 | cd06174 |
| PRK09874 | |
| SAOUHSC_00952 | cd06174 |
| pfam07690 | |
| SAOUHSC_02307 | COG3402 |
| SAOUHSC_02309 | cl01348 |
| SAOUHSC_02435 | cd06174 |
| TIGR00900 | |
| SAOUHSC_02620 | COG3152 |
| SAOUHSC_02752 | cd06174 |
| COG2814 | |
| SAOUHSC_02866 | pfam03176 |
| cl21543 | |
| COG2409 | |
| ABD31816.1 | COG1289 |
| ABD31642.1 | cd06174 |
| cd06174 and pfam07690, Major Facilitator Superfamily; PRK11273, glycerol- 3-phosphate transporter; pfam01306, LacY proton/sugar symporter; PRK11652, multidrug resistance protein D; PRK09874, drug efflux system protein MdtG; COG3402, YdbS; cl01348, bPH_2 super family; TIGR00900, H+ Antiporter protein; COG3152, yhaH; COG2814, arabinose efflux permease; pfam03176 and cl21543, MMPL super family; COG2409, YdfJ; COG1289, YccC. | |
Table 8. Pfam domain data.
| Hypothetical | Protein Domains |
| GlpT | MFS_1 |
| LacY | LacY_symp |
| EmrD | MFS_1 |
| SAOUHSC_00058 | MFS_1 |
| SAOUHSC_00078 | MFS_1 |
| sugar_tr | |
| SAOUHSC_00952 | MFS_1 |
| SAOUHSC_02307 | bPH_2 |
| SAOUHSC_02309 | No Pfam-A matches |
| SAOUHSC_02435 | MFS_3 |
| SAOUHSC_02620 | DUF805 |
| SAOUHSC_02752 | MFS_1 |
| SAOUHSC_02866 | MMPL family |
| ABD31816.1 | No Pfam-A matches |
| ABD31642.1 | MFS_1 |
| MFS_1, Major Facilitator Superfamily; LacY_symp, LacY proton/sugar symporter; sugar_tr, sugar and other transporter; bPH_2, bacterial Pleckstrin homology domain; MFS_3, Transmembrane secretion effector; DUF805, Protein of unknown function; MMPL family, putative integral membrane proteins. | |
Based on these data collectively, the following hypothetical proteins are likely MFS proteins due to their size (300 – 700 amino acids), well-defined BLAST homologs (no generalized membrane or hypothetical proteins), more than seven transmembrane regions, and CD06174 and MFS domains from CDD-BLAST and Pfam, respectively, underwent evolutionary analyses: SAOUHSC_00058, SAOUHSC_00078, SAOUHSC_00952, SAOUHSC_02435, SAOUHSC_02752, and ABD31642.1. Since either the GlpT or EmrD proteins identified the six hypothetical proteins most likely to belong to the MFS, the study removed LacY and its homologs in Table 2 from further study.
To evaluate how related these hypothetical proteins were, proteins of interest underwent multiple sequence alignment and phylogenetic tree construction. For these analyses, all defined homologs from Tables 1 and Table 3 combined with hypothetical proteins fitting the study’s criteria completed multiple sequence alignment as displayed in Figure 2 and Figure 3, respectively. These analyses included top BLASTP hits from Table 4 not already included in Tables 1 and Table 3 Both alignments found over 10 alpha helices in the consensus sequences, as expected from MFS members. Hypothetical proteins aligned with their top BLASTP hits from Tables 4 best, with SAOUHSC_00952 also closely aligning with the MFS transporter. To visualize how closely related these proteins are phylogenetic trees were constructed. Though all similar, a representative tree of all the proteins and their homologs is shown in Figure 4. The same NorA multidrug efflux MFS transporter came up in all three E. coli PSI-BLASTs while GlpT (1PW4) and EmrD (3GFP) identified two separate NorB quinolone resistance proteins. SAOUHSC_00058 nuzzled between the two NorB proteins. As expected, phylogeny confirmed the multiple sequence alignment. Hypothetical proteins related closely with their top PSI-BLAST hits from Tables 4, with SAOUHSC_00952 being closely related to the MFS transporter also. Floyd’s illustration of the established proteins shown in the phylogenetic tree presented here is similarly arranged [4].
Figure 2.
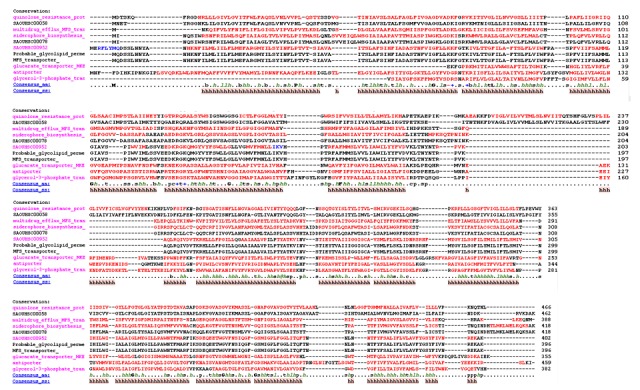
Alignment of GlpT homologs aligned by PROMALS3D. Magenta names are representative sequences colored red to identify predicted alpha-helix secondary structures. The black names belonging to the same alignment group as the magenta name above it, indicating a strong relationship between the two. Consensus_aa, consensus amino acid sequence; Consensus_ss, consensus predicted secondary structures; h, consensus predicted secondary structure alpha-helix.
Figure 3.

Alignment of EmrD homologs aligned by PROMALS3D. Magenta names are representative sequences colored red to identify predicted alpha-helix secondary structures. The black names belonging to the same alignment group as the magenta name above it, indicating a strong relationship between the two. Consensus_aa, consensus amino acid sequence; Consensus_ss, consensus predicted secondary structures; h, consensus predicted secondary structure alpha-helix.
Figure 3.
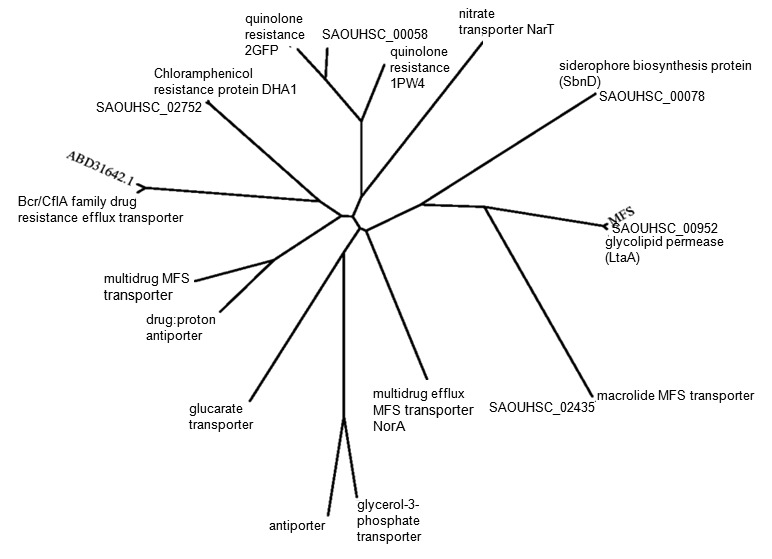
Representative phylogenetic tree of proteins produced via PHYLIP package programs showing six hypothetical proteins belong evolutionarily to the major facilitator superfamily (MFS). SAOUHSC_00058, SAOUHSC_02435, SAOUHSC_02752 and ABD31642.1 are related to drug efflux proteins. SAOUHSC_00078 is closely related to a siderophore biosynthesis protein as SAOUHSC_00952 is confirmed to be a glycolipid permease.
Conclusion:
This study identified eleven hypothetical proteins homologous to E. coli MFS proteins with known structure. Six of those hypothetical proteins, SAOUHSC_00058, SAOUHSC_00078, SAOUHSC_00952, SAOUHSC_02435, SAOUHSC_02752, and ABD31642.1, were between 300 and 700 amino acids, had well-defined BLASTP homologs, over seven transmembrane regions, CD06174 domain from CDD-BLAST, and an MFS domain from Pfam. Based on these results alongside multiple sequence alignment and phylogenetic trees, SAOUHSC_00058 is likely a quinolone resistance protein (NorB) due to its close relation to two NorB proteins identified by either GlpT or EmrD. SAOUHSC_00058 may be a siderophore biosynthesis protein (SbnD). SAOUHSC_00952, a glycolipid permease (LtaA), and another MFS transporter closely are related. Further, UniProt has SAOUHSC_00952 labeled as a glycolipid permease LtaA with experimental evidence at the protein level, unlike all the other hypothetical proteins studied here labeled as uncharacterized [29]. SAOUHSC_02435 hits a macrolide MFS transporter while SAOUHSC_02752 matches a chloramphenicol resistance (DHA1), and ABD31642.1 may be a Bcr/CflA family drug resistance efflux transporter. These data manually verifies the in silico identity of six hypothetical proteins from reference strain S. aureus NCTC 8325 in public databases.
Edited by P Kangueane
Citation:Marklevitz & Harris, Bioinformation 12(4): 254-262 (2016)
References
- 1.Holden MT, et al. Proc Natl Acad Sci. 2004;101:26. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stinear TP, et al. Genome Biol Evol. 2014;6:2. doi: 10.1093/gbe/evu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. http://www.cdc.gov/drugresistance/pdf/carb_national_strategypdf.
- 4.Floyd JL, et al. Antimicrob Agents Chemother. 2010;54:12. doi: 10.1128/AAC.00580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikaido H. Annu Rev Biochem. 2009;78 doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fournier B, et al. J Bacteriol. 2000;182:3. doi: 10.1128/jb.182.3.664-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding Y, et al. J Bacteriol. 2008;190:21. doi: 10.1128/JB.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pao SS, et al. Microbiol Mol Biol Rev. 1998;62:1. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Law CJ, et al. Annu Rev Microbiol. 2008;62 doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy VS, et al. FEBS J. 2012;279:11. doi: 10.1111/j.1742-4658.2012.08590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.School K, et al. Bioinformation. 2016;12:3. doi: 10.6026/97320630012209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bharat Siva Varma P, et al. J Infect Public Health. 2015;8:6. doi: 10.1016/j.jiph.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Ijaq J, et al. Front Genet. 2015;6:119. doi: 10.3389/fgene.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillaspy AF, et al. The Staphylococcus aureus NCTC 8325 genome. Washington (DC): 2006. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors Gram - positive pathogens 2nd ed ASM Press p. 381–412. [Google Scholar]
- 15.Islam MS, et al. Genomics Inform. 2015;13:2. doi: 10.5808/GI.2015.13.2.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkins MR, et al. Methods Mol Biol. 1999;112 doi: 10.1385/1-59259-584-7:445. [DOI] [PubMed] [Google Scholar]
- 17.Gill SC, von Hippel PH. Anal Biochem. 1989;182:2. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 18.Guruprasad K, et al. Protein Eng. 1990;4:2. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 19.Ikai AJ. J Biochem. 1980;88:6. [PubMed] [Google Scholar]
- 20.Kyte J, Doolottle RF. J Mol Biol. 1982;157:1. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Hirokawa T, et al. Bioinformatics. 1998;14:4. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 22.Sonnhammer E, et al. Proteins. 1997;28:3. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, et al. Nucleic Acids Res. 2006;34:D247. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, et al. Nucleic Acids Res. 2015;43:D222-6. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohan R, Venugopal S. Bioinformation. 2012;8:15. doi: 10.6026/97320630008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei J, et al. Nucleic Acids Res. 2008;36:7. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, et al. Nucleic Acids Res. 1994;22:22. doi: 10.1093/nar/22.13.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Felsenstein J, et al. PHYLIP -- Phylogeny Inference Package (Version 3.2) 1989. Cladistics 5: 163-166. [Google Scholar]
- 29.Gründling A, Schneewind O. Journal of bacteriology. 2007;189:6. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


