Abstract
Background
Molecular biomarkers assisting risk-group assignment and subsequent treatment stratification are urgently needed for patients with squamous cell cancer of the head and neck region (HNSCC). Aberrant methylation is a frequent event in cancer and, therefore, a promising source for potential biomarkers. Here, the methylation status of the paired-like homeodomain transcription factor 3 (PITX3) was evaluated in HNSCC.
Methods
Using a quantitative real-time PCR, PITX3 methylation was assessed in a cohort of 326 HNSCC patients treated for localized or locally advanced disease (training cohort). The results were validated with Infinium HumanMethylation450 BeadChip data from a 528 HNSCC patient cohort (validation cohort) generated by The Cancer Genome Atlas (TCGA) Research Network.
Results
PITX3 methylation was significantly higher methylated in tumor compared to normal adjacent tissue (NAT; training cohort: median methylation NAT 32.3%, tumor 71.8%, p < 0.001; validation cohort: median methylation NAT 16.9%, tumor 35.9%, p < 0.001). PITX3 methylation was also significantly correlated with lymph node status both in the training (p = 0.006) and validation (p < 0.001) cohort. PITX3 methylation was significantly higher in HPV-associated (p16-positive) tumors compared to p16-negative tumors (training cohort: 73.7 vs. 66.2%, p = 0.013; validation cohort: 40.0 vs. 33.1%, p = 0.015). Hypermethylation was significantly associated with the risk of death (training cohort: hazard ratio (HR) = 1.80, [95% confidence interval (CI) 1.20–2.69], p = 0.005; validation cohort: HR = 1.43, [95% CI 1.05–1.95], p = 0.022). In multivariate Cox analyses, PITX3 added independent prognostic information. Messenger RNA (mRNA) expression analysis revealed an inverse correlation with PITX3 methylation in the TCGA cohort.
Conclusions
PITX3 DNA methylation is an independent prognostic biomarker for overall survival in patients with HNSCC and might aid in the process of risk stratification for individualized treatment.
Keywords: PITX3, Biomarker, Head and neck squamous cell carcinoma, HNSCC, DNA methylation, HPV, Prognosis
Background
With an estimated incidence of more than 60,000 cases in the USA alone, cancer of the head and neck region is a common malignant disease. In male patients, cancer of the oral cavity and pharynx is the eighth most common cancer type. For 2016, 13,190 patients are estimated to succumb to the disease [1]. The most prevalent histological subtype in the head and neck region is squamous cell carcinoma (HNSCC), and common risk factors are the concurrent abuse of tobacco and alcohol as well as infections with a high-risk human papillomavirus (HPV) like HPV16 and HPV18. Depending on the causative agent, distinct differences have been recorded with regard to molecular alterations, tumor site, and prognosis of the tumors. While TP53 mutations are the most common alterations in smoking-related cancers, HPV-associated tumors predominantly harbor PIK3CA mutations [2]. Moreover, patients with HPV-associated tumors tend to be younger, and tumors are more likely to develop in the oropharynx. Notably, these patients also have a better overall survival [3].
Most patients present in an advanced stage of disease. Surgery or radiation therapy with concurrent chemotherapy are first-line treatment options for HNSCC patients, but local and distant failure is common [4]. Newer drugs like the anti-EGFR antibody cetuximab have been successfully implemented into the therapeutic portfolio of HNSCC [5]. Combined with standard platinum-based chemotherapy, they provide a significant overall survival benefit for patients with recurrent or metastatic disease. For the same patient group, immunotherapy agents like the immune checkpoint inhibitor pembrolizumab have recently shown high efficacy [6]. Based on these results, pembrolizumab has recently been approved by the US Food and Drug Administration (FDA) for patients with recurrent or metastatic HNSCC. Considering the dismal prognosis in the event of metastatic or recurrent disease, such advances in drug therapy are urgently needed [7].
As yet, no prognostic molecular biomarkers have been established to assist clinicians in stratifying treatment or surveillance options after initial curative treatment. Currently, clinicopathological parameters like lymph node metastasis or tumor stage serve to identify high-risk patients. Additional biomarkers would be of utmost value to influence treatment strategy and surveillance intensity. With regard to emerging immunotherapy options that have an improved safety and side-effect profile, molecular biomarkers could distinguish patient subgroups that would benefit from adjuvant or neoadjuvant therapy. These biomarkers could further be evaluated in conjunction with other newly identified adverse risk factors like delayed time to treatment [8].
Tissue-based biomarkers have already been established for other tumor entities. In breast carcinoma, the commercially available multi-gene panel assay Oncotype DX© estimates risk of recurrence after first-line treatment for early stage breast cancer by analyzing the expression of 21 genes [9]. Patients who have been identified as high risk based according to the subsequently calculated recurrence score may profit from intensified surveillance or adjuvant chemotherapy. Another promising biomarker approach is the investigation of the methylation status in a tumor sample. Epigenetic regulation is conferred through methylation, and malignant tumors are frequently hyper- or hypomethylated. Depending on the locus, methylation can result in gene transcription or repression [10]. Methylation analysis has already changed clinical practice for glioblastoma treatment, where promoter methylation of MGMT is predictive of response to the drug temozolomide [11]. So far, this is the only clinically implemented DNA methylation-based predictive biomarker test. DNA methylation can be quantified reliably and reproducibly via microarray, sequencing, or real-time PCR after bisulfite conversion of DNA [12–14]. Moreover, due to the stability of DNA methylation, the methylation status can be easily obtained even from degraded material like formalin-fixed paraffin-embedded (FFPE) tissue [15], which represents the most common clinically relevant sample material. Thus, studying epigenetic alterations is a promising approach for the development of biomarkers. Several epigenetic biomarkers have been tested in HNSCC, sometimes with conflicting results [16]. Hypermethylation of TIMP3 and CCNA1, for instance, have been associated with an increased risk of developing second primary tumors [17]. Hypermethylation of a gene panel in salivary samples prior to treatment was shown to be prognostic of local recurrence and overall survival [18]. Consequently, epigenetic markers are emerging as prognostic tools in patients with HNSCC.
The paired-like homeodomain transcription factor 3 (PITX3), located on chromosome 10q24 [19], plays an important role in the midbrain dopamine system development, and single-nucleotide polymorphisms within the PITX3 gene have been associated with Parkinson’s disease [20]. While the role of PITX3 in the brain development has been well characterized, its contribution to tumorigenesis remains more elusive. It has been found to be hypermethylated in breast cancer tumors [21]. Recently, Holmes et al. reported that PITX3 promoter methylation is strongly associated with biochemical recurrence-free survival in prostate cancer patients [22].
PITX3 shares a sequence similarity and at least partly redundant function with the paired-like homeodomain 2 (PITX2) [21, 23]. PITX2 promoter methylation is a well studied prognostic biomarker in various cancers, i.e. prostate, breast, and biliary tract cancers [24–31]. Recent studies investigating the methylation status of PITX2 in HNSCC and lung cancer cohorts found that hypermethylation was associated with improved survival [32, 33]. An in vitro diagnostic (IVD) test for anthracycline sensitivity in breast cancer patients based on PITX2 methylation is currently being developed for commercial distribution by the company Therawis GmbH (Munich, Germany). Similar to PITX2, the closely related transcription factor PITX1 plays a role in the embryonal development [34]. PITX1 has been found to be predictive of chemosensitivity in a small HNSCC cohort [35].
Given the need for new prognostic biomarkers for patients with curatively treated HNSCC and the promising results regarding PITX2 methylation as a biomarker, the methylation status of PITX3 was investigated in well-annotated HNSCC cohorts.
Methods
Patients and ethics
The training cohort comprised 326 retrospectively enrolled HNSCC patients treated at the University Hospital Bonn. Clinical data were obtained for the majority of patients. The study was approved by the Institutional Review Board of the University Hospital Bonn, which waived the need for written informed consent. The validation cohort consisted of 528 HNSCC patients analyzed by The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov/). All experiments were conducted in accordance with the Helsinki Declaration of 1975.
Sample preparation, DNA extraction, and bisulfite conversion
Freshly cut sections from FFPE blocks were stained with hematoxylin and eosin. The tumor area was annotated and macrodissected from unstained slides for subsequent methylation analysis. The process of DNA extraction and bisulfite conversion was performed using the innuCONVERT Bisulfite All-In-One Kit (Analytik Jena, Germany) as previously reported [36]. In order to assess HPV status, the surrogate marker p16 was evaluated by immunohistochemistry.
DNA and messenger RNA (mRNA) from samples from the validation cohort were prepared as described by the TCGA Research Network (www.cancergenome.nih.gov).
PITX3 quantitative methylation analysis
For the training cohort, quantitative PITX3 methylation analysis was performed by means of quantitative real-time PCR (qPCR) with primers and probes as previously described [22]. The analytical performance of the qPCR assay has been characterized earlier [22].
For the validation cohort, DNA methylation data were generated by the TCGA Research Network using the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, CA, USA). HumanMethylation450 data of level 2 were downloaded directly from the TCGA webpage. The data files included background-corrected methylated (bead_M) and unmethylated (bead_U) bead pair summary intensities as extracted by the R package ‘methylumi’. The bead pair (cg20023259) in proximity to the locus of the qPCR assay was selected. Methylation values for the bead pair were calculated by the formula 100% × bead_M/(bead_M + bead_U). The position of the qPCR assay and the bead pair cg20023259 within the PITX3 gene is shown in Fig. 1.
Fig. 1.
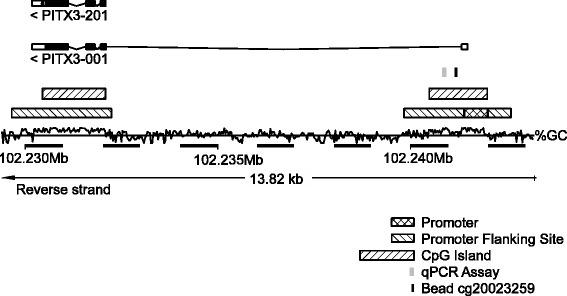
Genomic organization and assay position. Genomic organization of the PITX3 gene and locations of the PITX3 qPCR assay [22] and the Infinium HumanMethylation450 BeadChip bead cg20023259. The information was obtained from Ensembl Homo sapiens version GRCh38.p7
PITX3 mRNA expression analysis
The PITX3 mRNA data obtained from the TCGA Research Network were generated by means of the Illumina HiSeq 2000 RNA Sequencing version 2 analysis (Illumina, Inc., San Diego, CA, USA) as described by the TCGA Research Network (www.cancergenome.nih.gov). PITX3 mRNA expression (normalized counts) data of level 3 were downloaded from the TCGA webpage.
Statistical analysis
Statistical analyses were performed using SPSS, version 24 (SPSS Inc., Chicago, IL, USA). Bivariate correlations were tested using the Spearman rank correlation coefficient (ρ). Overall survival analyses were conducted by Kaplan-Meier and Cox proportional hazards regression analyses. The Mann-Whitney U test and one-way analysis of variance (one-way ANOVA) were employed for the comparison of groups. p values <0.05 were considered significant.
Results
PITX3 DNA methylation and mRNA expression in HNSCC tissue
The results from the validation cohort are entirely based upon data generated by the TCGA Research Network. The PITX3 locus was highly methylated in tumor tissues from the training and the validation cohort. Histograms of PITX3 methylation frequencies in both cohorts are shown in Fig. 2a, c. In the training cohort, methylation data were available for 466 patient samples including 140 normal and 326 tumor tissue samples. The comparison of PITX3 methylation in tumor and normal adjacent tissue (NAT) revealed a significant difference between tumor tissue (mean 69.0%, median 71.8%) and NAT (mean 32.1%, median 32.3%; p < 0.001; Fig. 2b). Methylation ranged from 0.79 to 100% in tumor tissue and from 0 to 82.1% in normal tissue. This finding was confirmed in the validation cohort, in which methylation data were available for 578 patient samples including 50 normal and 528 tumor tissue samples. Methylation was significantly different between tissue types (p < 0.001; Fig. 2d) and ranged from 6.9 to 76.3% in tumor tissue (mean 36.5%, median 35.9%) and from 11.7 to 26.1% in normal tissue (mean 17.1%, median 16.9%). The methylation differences observed between the two analyzed cohorts might be caused by the application of two different technologies: qPCR and Infinium HumanMethylation450K BeadChip.
Fig. 2.
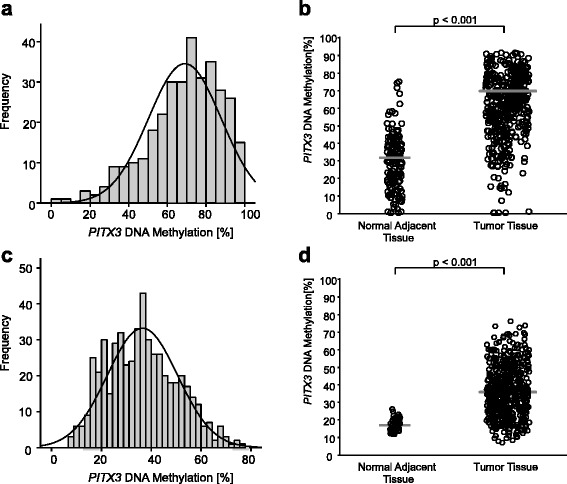
PITX3 DNA methylation in HNSCC specimens. Frequency of PITX3 DNA methylation in the training cohort (a) comprising 326 HNSCC patients and validation cohort (b) including 528 HNSCC patients. Shown are the methylation levels in tumor tissue only. PITX3 DNA methylation in tumor tissue compared to normal adjacent tissue from the training (c) and the validation (d) cohort, respectively. Tumor tissue was methylated significantly higher compared to corresponding normal adjacent tissue. p values refer to Wilcoxon-Mann-Whitney test
Furthermore, DNA methylation was shown to be inversely correlated with PITX3 mRNA (p = 0.036) in the validation cohort. Interestingly, mRNA levels were also significantly different in tumor (mean 6.55 normalized counts) compared to normal (mean 15.37 normalized counts) tissue (p < 0.001).
Association of PITX3 DNA methylation with clinico-pathological parameters
Both in the training and validation cohort, PITX3 DNA methylation was significantly associated with lymph node status (p = 0.006 and p < 0.001, respectively). Moreover, a significant methylation difference was found in regard to HPV status in the training cohort. Using p16 as surrogate marker, p16-negative tumors were significantly lower methylated than p16-positive tumors (66.2 vs. 73.7%, p = 0.018). This was confirmed in the validation cohort: HPV-associated (p16-positive) tumors were significantly highly methylated than p16-negative tumors (40.0 vs. 33.1%, p = 0.015). In addition, a significant difference in methylation was found when analyzing tumor site in both cohorts (training cohort: p = 0.023, validation cohort: p = 0.017). A detailed analysis of both cohorts can be found in Table 1.
Table 1.
Association of clinicopathological parameters with PITX3 methylation in HNSCC patients of the training cohort (n = 326) and the validation cohort (n = 528)
| Training cohort | Validation cohort (TCGA) | |||||
|---|---|---|---|---|---|---|
| Variable | No. (%) of patients | Mean PITX3 methylation (%) | p value | No. (%) of patients | Mean PITX3 methylation (%) | p value |
| All patients | 326 (100) | 69.0 | 528 (100) | 36.5 | ||
| Sex | 0.58 | 0.42 | ||||
| Female | 74 (22.7) | 68.0 | 142 (26.9) | 35.6 | ||
| Male | 252 (77.3) | 69.3 | 386 (73.1) | 36.8 | ||
| Age (years) | 0.16 | 0.32 | ||||
| Mean | 62.2 | 60.9 | ||||
| Median | 62 | 0.55 | 61 | 0.48 | ||
| n < Median | 132 (40.5) | 70.3 | 282 (53.4) | 36.1 | ||
| n > Median | 119 (36.5) | 71.7 | 245 (46.4) | 36.9 | ||
| Unknown | 75 (23.0) | 1 (0.2) | ||||
| Smoking status | 0.91 | 0.92 | ||||
| Non-smoker | 25 (7.7) | 69.8 | 122 (23.1) | 36.7 | ||
| Smoker | 178 (54.6) | 70.2 | 393 (74.4) | 36.6 | ||
| Unknown | 123 (37.7) | 13 (2.5) | ||||
| Pack years | 0.17 | 0.60 | ||||
| <40 | 92 (28.2) | 72.0 | 168 (31.8) | 36.5 | ||
| >40 | 46 (14.1) | 76.0 | 130 (24.6) | 37.3 | ||
| Unknown | 188 (57.7) | 230 (43.6) | ||||
| Alcohol consumption | 0.10 | 0.37 | ||||
| Yes | 81 (24.8) | 74.8 | 352 (66.7) | 37.0 | ||
| No | 47 (14.4) | 69.8 | 165 (31.3) | 35.9 | ||
| Unknown | 198 (60.7) | 11 (2.1) | ||||
| Tumor site | 0.023a | 0.017a | ||||
| Oral cavity | 50 (15.3) | 69.0 | 320 (60.6) | 36.6 | ||
| Oropharynx | 139 (42.6) | 72.0 | 81 (15.3) | 40.2 | ||
| Hypopharynx | 22 (6.7) | 72.8 | 10 (1.9) | 33.6 | ||
| Larynx | 95 (29.1) | 67.0 | 117 (22.2) | 33.9 | ||
| Unknown | 20 (6.1) | |||||
| pT | 0.80 | 0.22 | ||||
| T1/T2 | 160 (49.1) | 69.8 | 190 (36.0) | 34.9 | ||
| T3/T4 | 119 (36.5) | 69.2 | 276 (52.3) | 36.5 | ||
| Unknown | 47 (14.4) | 62 (11.7) | ||||
| pN | 0.006a | <0.001a | ||||
| N0 | 128 (39.3) | 66.3 | 180 (34.1) | 32.8 | ||
| N1/2/3 | 173 (53.1) | 72.0 | 240 (45.5) | 37.7 | ||
| Unknown | 25 (7.7) | 108 (20.5) | ||||
| p16 | 0.013a | 0.015a | ||||
| Negative | 200 (61.3) | 66.2 | 74 (14) | 33.1 | ||
| Positive | 51 (15.6) | 73.7 | 41 (7.8) | 40.0 | ||
| Unknown | 75 (23.0) | 413 (78.2) | ||||
| Grade | 0.40 | 0.33 | ||||
| 1 | 7 (1.8) | 62.3 | 63 (11.9) | 34.7 | ||
| 2 | 199 (49.9) | 70.1 | 311 (58.9) | 36.3 | ||
| 3 | 107 (26.8) | 68.8 | 125 (23.7) | 37.5 | ||
| 4 | 0 (0) | 7 (1.3) | 43.5 | |||
| Unknown | 86 (21.6) | 22 (4.2) | ||||
| Surgical margin | 0.18 | 0.40 | ||||
| Negative | 225 (69.0) | 69.7 | 407 (77.1) | 36.0 | ||
| Positive | 41 (12.6) | 65.6 | 60 (11.4) | 37.6 | ||
| Unknown | 60 (18.4) | 61 (11.6) | ||||
| Second tumor | 0.070 | NA | ||||
| Yes | 38 (11.7) | 72.7 | ||||
| No | 140 (42.9) | 77.9 | ||||
| Unknown | 148 (45.4) | 528 (100) | ||||
| Vascular invasion | 0.083 | 0.072 | ||||
| Yes | 22 (6.7) | 63.0 | 124 (23.5) | 38.4 | ||
| No | 170 (52.1) | 70.1 | 232 (43.9) | 35.6 | ||
| Unknown | 134 (41.1) | 172 (32.6) | ||||
Mann-Whitney U test for sex, age (dichotomized), smoking status, pT, p16, surgical margin, second tumor, lymphovascular invasion, and pack years; One-way ANOVA for alcohol consumption, lymph node, grade, and tumor site; Spearman’s rank correlation for age. Results from the validation cohort are entirely based upon the data generated by the TCGA Research Network: http://cancergenome.nih.gov/
NA data not available
asignificance
Association of PITX3 DNA methylation with survival
Higher PITX3 DNA methylation analyzed as continuous variable showed a strong trend towards poorer overall survival in the training cohort (Hazard ratio (HR) = 1.01, [95% confidence interval (95%CI) 1.00–1.02], p = 0.061). PITX3 DNA methylation levels were subsequently dichotomized using an optimized cut-off (72.5%). PITX3 DNA hypermethylation was significantly associated with an increased risk of disease-related death in univariate Cox proportional hazards (HR = 1.80, [95%CI 1.20–2.69], p = 0.005; Table 2) and Kaplan-Meier (p = 0.004; Fig. 3a) analyses. In addition, p16 expression and N category were significantly associated with survival (Table 2). Furthermore, vascular invasion (V) was significantly associated with poor overall survival (HR = 3.51, [95%CI 1.90–6.47], p < 0.001).
Table 2.
Univariate and multivariate Cox proportional hazard analysis of overall survival of HNSCC patients in the training cohort (n = 307)
| Univariate | Multivariate | |||
|---|---|---|---|---|
| Variable | HR (95%CI) | p value | HR (95%CI) | p value |
| PITX3 methylation (dichotomized, optimized cut-off 72.5%) | 1.80 (1.20–2.69) | 0.005a | 2.28 (1.29–4.00) | 0.004a |
| p16 expression (neg. reference) | 0.40 (0.20–0.81) | 0.010a | 0.27 (0.11–0.67) | 0.005a |
| N category | 1.28 (1.07–1.53) | 0.008a | 1.27 (0.97–1.67) | 0.82 |
| T category | 1.14 (0.96–1.36) | 0.14 | 1.33 (1.03–1.72) | 0.029a |
Nineteen patients from the 326 patients of the cohort were omitted from survival analysis due to missing follow-up data
NA not analyzed
asignificant feature
Fig. 3.
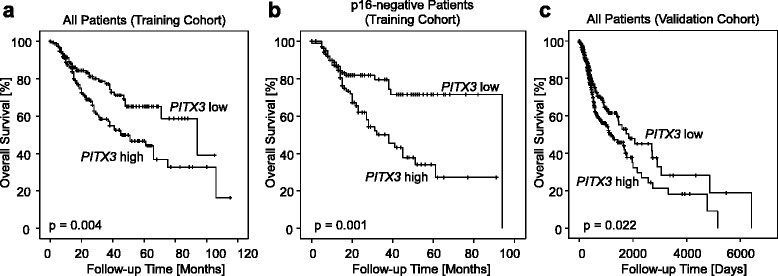
Overall survival in patients stratified according to PITX3 DNA methylation. Kaplan-Meier analysis of overall survival in HNSCC patients stratified according to PITX3 DNA methylation levels in tumor tissue. a Training cohort (all patients). b Training cohort (p16-negative patients). c Validation cohort (all patients). High PITX3 DNA methylation was associated with an adverse prognosis in both cohorts, in particular in p16-negative patients from the training cohort. A subgroup analysis of p16-negative patients from the validation cohort was not performed due to largely missing data
Interestingly, when performing a subgroup analysis among patients with p16-negative tumors, patients with PITX3 hypermethylated tumors revealed an HR of 2.44 ([95%CI 1.44–4.14], p = 0.001) compared to patients with hypomethylated cancers. This finding was confirmed in Kaplan-Meier analysis (p = 0.001; Fig. 3b). No significant survival differences were seen in PITX3-stratified patients with HPV-positive tumors. In a multivariate analysis, PITX3 methylation status added independent prognostic information (HR = 2.28, [95%CI 1.29–4.00], p = 0.004; Table 2) about the risk of dying. Vascular invasion status was excluded from multivariate analysis due to largely missing data from both cohorts.
The prognostic biomarker performance was validated in the TCGA cohort. Data were dichotomized by means of an optimized cut-off (35.1%). Univariate analysis showed a significant risk of death for patients with hypermethylated tumors (HR = 1.43, [95%CI 1.05–1.95], p = 0.022). In a multivariate Cox proportional hazard analysis, PITX3 methylation added significant prognostic information to T and N category for PITX3 methylation as dichotomized variable (HR = 1.49, [95%CI 1.02–2.16], p = 0.038; Table 3). However, mRNA expression analyzed as a continuous variable did not reveal any prognostic significance (HR = 1.00, [95%CI 0.98–1.02]; p = 0.74). The introduction of an optimized cut-off (normalized counts: 13.52) classified 58 patients as PITX3 mRNA expression-positive and 462 patients as negative. Only a trend towards a better prognosis of expression-positive patients was found (HR = 0.59, [95%CI 0.34–1.03], p = 0.062).
Table 3.
Univariate and multivariate Cox proportional hazard analysis of overall survival of HNSCC patients in the validation cohort from The Cancer Genome Atlas (n = 528)
| Univariate | Multivariate | |||
|---|---|---|---|---|
| Variable | HR (95%CI) | p value | HR (95%CI) | p value |
| PITX3 methylation (dichotomized, optimized cut-off 35.1%) | 1.43 (1.05–1.95) | 0.022a | 1.49 (1.02–2.16) | 0.038a |
| N category | 1.39 (1.14–1.70) | 0.001a | 1.25 (1.02–1.54) | 0.035a |
| T category | 1.31 (1.12–1.54) | 0.001a | 1.28 (1.06–1.56) | 0.013a |
p16 expression was omitted from analysis due to largely missing data
NA not analyzed
asignificant feature
Discussion and conclusions
Squamous cell cancer of the head and neck region poses a substantial global health burden and affects patients with a history of nicotine and alcohol abuse as well as patients harboring oral infections with high-risk HPV. The disease poses a large health burden worldwide. Surgery and radiotherapy are the mainstay of therapy with curative intent. Recent advantages in reconstructive techniques may help to reduce morbidity after extensive surgery of radiation-induced damage [37–39]. For patients undergoing curative therapy, biomarkers are urgently needed to identify individuals who are likely to experience tumor recurrence or metastasis and subsequent disease-related death. These patients could benefit from intensified surveillance or adjuvant therapy. Moreover, new immunotherapeutic drugs are emerging from which high-risk patients might benefit.
Epigenetic alterations in form of hypo- or hypermethylated DNA are frequent events in cancer. Investigating the DNA methylation status in patients with malignant diseases is a promising approach to establish new biomarkers. In the present study, PITX3 DNA methylation was analyzed in a well-annotated HNSCC cohort from the University Hospital Bonn. In multivariate analysis, PITX3 DNA hypermethylation was significantly associated with the risk of death. These results were validated in a HNSCC cohort from the TCGA Research Network. Furthermore, hypermethylation correlated inversely with PITX3 mRNA expression in the TCGA cohort. Although these findings lead to the conclusion that hypermethylation is associated with silencing of the PITX3 gene, the transcriptional relevance remains unclear. Hypomethylation may result in transcription of the gene; however, the role of PITX3 in tumorigenesis has not been described. One possible mechanism could be the co-transcription of other genes on chromosome 10q such as suppressor of fused homolog (SUFU), which acts as a negative regulator of the hedgehog signaling pathway. Combined loss of SUFU and p53 results in the development of medulloblastomas and rhabdomyosarcoma in mice [40]. Hedgehog signaling may also play a role in HNSCC tumorigenesis. Hence, the transcription of a negative regulator could result in a reduced tumor growth [41]. It has previously been shown that HPV infection may result in alteration of the DNA methylome. Therefore, HPV-mediated epigenetic alterations may account for the hypermethylation in HPV-positive tumors described in the present study [42, 43]. However, SUFU and PITX3 are not located on the immediately neighboring regions of 10q24. It is therefore unclear whether methylation of PITX3 could have any influence on SUFU transcription.
In a rat model for prostate cancer development following exposure to endocrine-disrupting chemicals, PITX3 was found to be one of several hypomethylated and therefore expressed genes [44]. While this model cannot serve as a model for HNSCC tumorigenesis, it is an interesting finding and possibly allows for the implication of PITX3 alteration being an early event in cancer development. This would be in line with the results of Holmes et al., who were able to show that PITX3 methylation is a strong prognostic marker for biochemical recurrence in localized prostate cancer [22].
HNSCC display a high genomic instability with a mean of 141 copy number alterations per tumor, including amplification of 10q24 [45, 46]. Since amplification has been associated with hypermethylation, this might provide an explanation for the observed methylation differences [47]. Even though the biological function remains unclear for now, PITX3 DNA methylation status emerges as a promising biomarker particularly for patients with HPV-negative tumors.
In brief, this is the first study to show that the methylation status of PITX3 is an independent prognostic factor for overall survival in patients with HNSCC. These findings warrant further investigation in a prospective study setting.
Acknowledgements
Not applicable.
Funding
This work was funded by the University Hospital Bonn, Germany.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article or are available from the corresponding author on reasonable request.
Authors’ contributions
VS and DD analyzed the data, drafted the manuscript, and conceived and coordinated the study. FB, GK, DP, and AS participated in the design of the study and its supervision and revised the manuscript. HG, DG, FD, and AF provided and characterized the sample material. HG revised the manuscript. JD processed the TCGA data. EEH performed the real-time PCR and the DNA preparations and participated in the data analysis. All authors read and approved the final version of the manuscript.
Competing interests
Dimo Dietrich is a consultant for AJ Innuscreen GmbH (Berlin, Germany) and receives royalties from product sales. Dimo Dietrich is a consultant and receives or received compensation from Therawis GmbH (Munich, Germany). Therawis GmbH aims to commercialize the methylation biomarker PITX2. The other authors report to have no conflict of interest regarding the topic of the article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board (IRB) at the University Hospital of Bonn. The review board waived the need for written informed consent. All experiments were conducted in accordance with the Helsinki Declaration of 1975.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. [DOI] [PMC free article] [PubMed]
- 3.Dok R, Nuyts S. HPV positive head and neck cancers: molecular pathogenesis and evolving treatment strategies. Cancers. 2016;8(4):41. [DOI] [PMC free article] [PubMed]
- 4.Sacco AG, Cohen EE. Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J Clin Oncol. 2015;33(29):3305–13. doi: 10.1200/JCO.2015.62.0963. [DOI] [PubMed] [Google Scholar]
- 5.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116–27. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 6.Chow LQ, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, et al. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol. 2016;34(32):3838–45. [DOI] [PMC free article] [PubMed]
- 7.Echarri MJ, Lopez-Martin A, Hitt R. Targeted therapy in locally advanced and recurrent/metastatic head and neck squamous cell carcinoma (LA-R/M HNSCC). Cancers. 2016;8(3):27. [DOI] [PMC free article] [PubMed]
- 8.Murphy CT, Galloway TJ, Handorf EA, Egleston BL, Wang LS, Mehra R, et al. Survival impact of increasing time to treatment initiation for patients with head and neck cancer in the United States. J Clin Oncol. 2016;34(2):169–78. doi: 10.1200/JCO.2015.61.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chau NG, Li YY, Jo VY, Rabinowits G, Lorch JH, Tishler RB, et al.Incorporation of next-generation sequencing into routine clinical care to direct treatment of head and neck squamous cell carcinoma. Clin Cancer Res. 2016;22(12):2939–49. [DOI] [PubMed]
- 10.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 11.Seystahl K, Wick W, Weller M. Therapeutic options in recurrent glioblastoma—An update. Crit Rev Oncol Hematol. 2016;99:389–408. doi: 10.1016/j.critrevonc.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich D, Hasinger O, Banez LL, Sun L, van Leenders GJ, Wheeler TM, et al. Development and clinical validation of a real-time PCR assay for PITX2 DNA methylation to predict prostate-specific antigen recurrence in prostate cancer patients following radical prostatectomy. J Mol Diagn. 2013;15(2):270–9. doi: 10.1016/j.jmoldx.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schatz P, Dietrich D, Koenig T, Burger M, Lukas A, Fuhrmann I, et al. Development of a diagnostic microarray assay to assess the risk of recurrence of prostate cancer based on PITX2 DNA methylation. J Mol Diagn. 2010;12(3):345–53. doi: 10.2353/jmoldx.2010.090088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietrich D. Direct quantitative bisulfite sequencing using tag-modified primers and internal normalization. Anticancer Res. 2016;36(12):6343–6. [DOI] [PubMed]
- 15.Jung M, Uhl B, Kristiansen G, Dietrich D. Bisulfite conversion of DNA from tissues, cell lines, buffy coat, FFPE tissues, microdissected cells, swabs,sputum, aspirates, lavages, effusions, plasma, serum, and urine. Methods Mol Biol. 2015. [Epub ahead of print]. [DOI] [PubMed]
- 16.Juodzbalys G, Kasradze D, Cicciu M, Sudeikis A, Banys L, Galindo-Moreno P, et al. Modern molecular biomarkers of head and neck cancer. Part I. epigenetic diagnostics and prognostics: systematic review. Cancer Biomark. 2016;17(4):487–502. [DOI] [PubMed]
- 17.Rettori MM, de Carvalho AC, Longo AL, de Oliveira CZ, Kowalski LP, Carvalho AL, et al. TIMP3 and CCNA1 hypermethylation in HNSCC is associated with an increased incidence of second primary tumors. J Transl Med. 2013;11:316. doi: 10.1186/1479-5876-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho AL, Henrique R, Jeronimo C, Nayak CS, Reddy AN, Hoque MO, et al. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clin Cancer Res. 2011;17(14):4782–9. doi: 10.1158/1078-0432.CCR-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Song Z, Yuan L, Xiong W, Yang Z, Gong L, et al. Genetic analysis of PITX3 variants in patients with essential tremor. Acta Neurol Scand. 2016. [Epub ahead of print]. [DOI] [PubMed]
- 20.Jimenez-Jimenez FJ, Garcia-Martin E, Alonso-Navarro H, Agundez JA. PITX3 and risk for Parkinson’s disease: a systematic review and meta-analysis. Eur Neurol. 2014;71(1-2):49–56. doi: 10.1159/000353981. [DOI] [PubMed] [Google Scholar]
- 21.Dietrich D, Lesche R, Tetzner R, Krispin M, Dietrich J, Haedicke W, et al. Analysis of DNA methylation of multiple genes in microdissected cells from formalin-fixed and paraffin-embedded tissues. J Histochem Cytochem. 2009;57(5):477–89. doi: 10.1369/jhc.2009.953026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes EE, Goltz D, Sailer V, Jung M, Meller S, Uhl B, et al. PITX3 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients after radical prostatectomy. Clin Epigenetics. 2016;8:104. doi: 10.1186/s13148-016-0270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L'Honore A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P, et al. Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol. 2007;307(2):421–33. doi: 10.1016/j.ydbio.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Uhl B, Gevensleben H, Tolkach Y, Sailer V, Majores M, Jung M, et al. PITX2 DNA Methylation as Biomarker for Individualized Risk Assessment of Prostate Cancer in Core Biopsies. J Mol Diagn. 2017;19(1):107–14. [DOI] [PubMed]
- 25.Uhl B, Dietrich D, Branchi V, Semaan A, Schaefer P, Gevensleben H, et al. DNA Methylation of PITX2 and PANCR Is Prognostic for Overall Survival in Patients with Resected Adenocarcinomas of the Biliary Tract. PLoS One. 2016;11(10):e0165769. [DOI] [PMC free article] [PubMed]
- 26.Vinarskaja A, Schulz WA, Ingenwerth M, Hader C, Arsov C. Association of PITX2 mRNA down-regulation in prostate cancer with promoter hypermethylation and poor prognosis. Urol Oncol. 2013;31(5):622–7. [DOI] [PubMed]
- 27.Bañez LL, Sun L, van Leenders GJ, Wheeler TM, Bangma CH, Freedland SJ, et al. Multicenter Clinical Validation of PITX2 Methylation as a Prostate Specific Antigen Recurrence Predictor in Patients With Post-Radical Prostatectomy Prostate Cancer. J Urol. 2010;184(1):149–56. [DOI] [PubMed]
- 28.Weiss G, Cottrell S, Distler J, Schatz P, Kristiansen G, Ittmann M, et al. DNA Methylation of the PITX2 Gene Promoter Region is a Strong Independent Prognostic Marker of Biochemical Recurrence in Patients With Prostate Cancer After Radical Prostatectomy. J Urol. 2009;181(4):1678–85. [DOI] [PubMed]
- 29.Hartmann O, Spyratos F, Harbeck N, Dietrich D, Fassbender A, Schmitt M, et al. DNA Methylation Markers Predict Outcome in Node-Positive, Estrogen Receptor-Positive Breast Cancer with Adjuvant Anthracycline-Based Chemotherapy. Clin Cancer Res. 2009;15(1):315–23. [DOI] [PubMed]
- 30.Harbeck N, Nimmrich I, Hartmann A, Ross JS, Cufer T, Grützmann R, et al. Multicenter Study Using Paraffin-Embedded Tumor Tissue Testing PITX2 DNA Methylation As a Marker for Outcome Prediction in Tamoxifen-Treated, Node-Negative Breast Cancer Patients. J Clin Oncol. 2008;26(31):5036–42. [DOI] [PubMed]
- 31.Nimmrich I, Sieuwerts AM, Meijer-van Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, et al. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Res Treat. 2008;111(3):429–37. [DOI] [PubMed]
- 32.Sailer V, Holmes EE, Gevensleben H, Goltz D, Droge F, de Vos L, et al. PITX2 and PANCR DNA methylation predicts overall survival in patients with head and neck squamous cell carcinoma. Oncotarget. 2016;7(46):75827-38. [DOI] [PMC free article] [PubMed]
- 33.Holmes EE, Jung M, Meller S, Leisse A, Sailer V, Zech J, et al. Performance Evaluation of Kits for Bisulfite-Conversion of DNA from Tissues, Cell Lines, FFPE Tissues, Aspirates, Lavages, Effusions, Plasma, Serum, and Urine. PLoS One. 2014;9(4):e93933. [DOI] [PMC free article] [PubMed]
- 34.Marcil A, Dumontier E, Chamberland M, Camper SA, Drouin J. Pitx1 and Pitx2 are required for development of hindlimb buds. Development. 2003;130(1):45–55. doi: 10.1242/dev.00192. [DOI] [PubMed] [Google Scholar]
- 35.Takenobu M, Osaki M, Fujiwara K, Fukuhara T, Kitano H, Kugoh H, et al. PITX1 is a novel predictor of the response to chemotherapy in head and neck squamous cell carcinoma. Mol Clin Oncol. 2016;5(1):89–94. doi: 10.3892/mco.2016.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dietrich D, Hasinger O, Liebenberg V, Field JK, Kristiansen G, Soltermann A. DNA methylation of the homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell lung cancer patients. Diagn Mol Pathol. 2012;21(2):93–104. doi: 10.1097/PDM.0b013e318240503b. [DOI] [PubMed] [Google Scholar]
- 37.Laino L, Iezzi G, Piattelli A, Lo Muzio L, Cicciu M. Vertical ridge augmentation of the atrophic posterior mandible with sandwich technique: bone block from the chin area versus corticocancellous bone block allograft—clinical and histological prospective randomized controlled study. Biomed Res Int. 2014;2014:982104. doi: 10.1155/2014/982104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cicciu M, Herford AS, Cicciu D, Tandon R, Maiorana C. Recombinant human bone morphogenetic protein-2 promote and stabilize hard and soft tissue healing for large mandibular new bone reconstruction defects. J Craniofac Surg. 2014;25(3):860–2. doi: 10.1097/SCS.0000000000000830. [DOI] [PubMed] [Google Scholar]
- 39.Herford AS, Tandon R, Stevens TW, Stoffella E, Cicciu M. Immediate distraction osteogenesis: the sandwich technique in combination with rhBMP-2 for anterior maxillary and mandibular defects. J Craniofac Surg. 2013;24(4):1383–7. doi: 10.1097/SCS.0b013e318292c2ce. [DOI] [PubMed] [Google Scholar]
- 40.Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26(44):6442–7. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 41.Dimitrova K, Stoehr M, Dehghani F, Dietz A, Wichmann G, Bertolini J, et al. Overexpression of the Hedgehog signalling pathway in head and neck squamous cell carcinoma. Onkologie. 2013;36(5):279–86. doi: 10.1159/000350322. [DOI] [PubMed] [Google Scholar]
- 42.Zhang HZ, Shan CG, Huang AP, Wang JM. Characterization of gene methylation in human papillomavirus associated-head and neck squamous cell carcinoma. Genet Mol Res. 2016;15(3). https://www.ncbi.nlm.nih.gov/pubmed/27706614. [DOI] [PubMed]
- 43.Lechner M, Fenton TR. The genomics, epigenomics, and transcriptomics of HPV-associated oropharyngeal cancer—understanding the basis of a rapidly evolving disease. Adv Genet. 2016;93:1–56. doi: 10.1016/bs.adgen.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Cheong A, Zhang X, Cheung YY, Tang WY, Chen J, Ye SH, et al. DNA methylome changes by estradiol benzoate and bisphenol A links early-life environmental exposures to prostate cancer risk. Epigenetics. 2016;11(9):674–89. [DOI] [PMC free article] [PubMed]
- 45.Marucci G, Fabbri PV, Morandi L, De Biase D, Di Oto E, Tallini G, et al. Pathological spectrum in recurrences of glioblastoma multiforme. Pathologica. 2015;107(1):1–8. [PubMed] [Google Scholar]
- 46.Lin M, Smith LT, Smiraglia DJ, Kazhiyur-Mannar R, Lang JC, Schuller DE, et al. DNA copy number gains in head and neck squamous cell carcinoma. Oncogene. 2006;25(9):1424–33. doi: 10.1038/sj.onc.1209166. [DOI] [PubMed] [Google Scholar]
- 47.Schneider KU, Dietrich D, Fleischhacker M, Leschber G, Merk J, Schaper F, et al. Correlation of SHOX2 gene amplification and DNA methylation in lung cancer tumors. BMC Cancer. 2011;11:102. doi: 10.1186/1471-2407-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article or are available from the corresponding author on reasonable request.


