Abstract
Chaperonin 60s are a ubiquitous class of proteins that promote folding and assembly of other cellular polypeptides in an ATP-dependent manner. The oligomeric state of chaperonin 60s has been shown to be crucial to their role as molecular chaperones. Chaperonin 60s are also known to be important stimulators of the immune system. Mycobacterium tuberculosis possesses a duplicate set of chaperonin 60s, both of which have been shown to be potent cytokine stimulators. The M. tuberculosis chaperonin 60s are present in the extracellular milieu at concentrations that are extremely low for the formation of an oligomer. Here we present the crystal structure of one of the chaperonin 60s of M. tuberculosis, also called Hsp65 or chaperonin 60.2, at 3.2-Å resolution. We were able to crystallize the protein in its dimeric state. The unusual dimerization of the protein leads to exposure of certain hydrophobic patches on the surface of the protein, and we hypothesize that this might have relevance in binding to immunogenic peptides, as it does in the eukaryotic homologs.
Chaperonin 60 (Cpn60), also commonly referred to as heat shock protein 60 (Hsp60), is one of the major molecular chaperones that are present ubiquitously in all forms of life. These molecular chaperones are known to assist the folding, assembly, and transport of several cellular proteins (16). Cpn60s have been shown to be overexpressed under a variety of unnatural conditions, such as thermal stress, hypoxia, nutrient deprivation, phagocytosis, etc. Invasion of a host is an apparent form of a stress, and induction of Cpn60s has also been observed in pathogenic organisms. The overexpressed pathogen-derived Cpn60s act as major antigens that result in strong immune responses from the host (43).
In Escherichia coli, the chaperonin GroEL has been shown to be essential for growth at all temperatures (13). The E. coli chaperonin has provided a paradigm for understanding protein folding mechanisms mediated by the chaperonins (41). GroEL promotes de novo folding of ∼10 to 15% of all proteins in the bacterial cytosol in coordination with the heptameric cochaperonin GroES (12).
X-ray studies combined with electron microscopy studies have provided valuable insights into the functional cycle of the GroEL chaperonin (41, 4, 8). The crystal structures of unliganded GroEL and the GroEL-GroES complex revealed a cylindrical complex with subunits of GroEL assembled into two heptameric rings stacked back to back to form the native 14-mer. The two rings enclose a large central channel that facilitates proper protein folding in an ATP-dependent manner (41, 4, 3). The crystal structure of Cpn60 from Paracoccus denitrificans also revealed a similar arrangement of Cpn60 subunits as a tetradecamer (15).
GroEL is assisted in its function by a 10-kDa cochaperonin, GroES. The cochaperonin exists as a heptamer and adopts a dome-like structure that can bind to either GroEL ring to enclose the central cavity (17, 21). GroES acts as a lid to seal off the folding chamber and helps displace bound substrate protein into the cavity, where the protein can undergo productive folding. Binding of GroES to GroEL is dependent on adenine nucleotide. ATP-dependent conformational changes in GroEL have been shown to be necessary for proper chaperonin function in vivo (34).
In most prokaryotic organisms, a single copy of the cpn60 gene usually occurs on the chromosome and is found in the groESL operon along with cpn10. Some of the actinomycete species, however, are known to express multiple cpn60 genes. For example, Mycobacterium tuberculosis contains two copies of the cpn60 gene (19). One of these, cpn60.1, is in an operon with cpn10, while the second, cpn60.2, occurs separately on the chromosome. Study of the regulation of heat shock proteins in M. tuberculosis has shown that there is overexpression of the two Cpn60s upon thermal shock (36), as well as upon phagocytosis by macrophages (25). It is therefore reasonable to believe that the Cpn60s of M. tuberculosis contribute to its defense response against external stress conditions. Moreover, both Cpn60.1 and Cpn60.2 of M. tuberculosis have been shown to be highly antigenic and potent cytokine inducers (23, 42). The Cpn60s therefore might represent important components of M. tuberculosis that have roles as immunomodulators and perhaps also are required for proper protein folding and transport.
Although the biochemical and functional properties of Cpn60s encoded on the groESL operon have been studied extensively, the necessity of a duplicate gene in certain bacteria is not known. The functional role of the second copy of Cpn60 as a chaperonin is poorly understood. A recent study suggested a proteolytic role for the second Cpn60 in Mycobacterium leprae (31). Thus, the second copy of Cpn60 might have different roles in the physiology of bacteria that possess multiple cpn60 genes. Structural studies of the second Cpn60 could therefore provide valuable insights into its functional properties. In this paper we present the crystal structure of M. tuberculosis Cpn60.2, also commonly referred to as Hsp65, and describe its relevance to the functional properties.
MATERIALS AND METHODS
Cloning, expression, and purification.
The full-length gene coding for the M. tuberculosis GroEL homolog, cpn60.2 (Rv0440), was PCR amplified from the M. tuberculosis H37Rv cosmid library kindly provided by Stewart Cole (9). The cpn60.2 fragment was cloned into E. coli expression vector pET28a (Novagen) through an intermediate subcloning step in pBluescript SK(+) (Stratagene). Primers used for amplification carried a His6 tag, which was incorporated into the protein at the C terminus. The gene was overexpressed in E. coli BL21(DE3) and was purified by Ni-nitrilotriacetic acid affinity chromatography. The protein used for crystallization was dialyzed extensively against 10 mM Tris-Cl (pH 8.0) and concentrated to 14 mg/ml.
The untagged gene was similarly cloned in expression vector pET23d, and the protein was purified by conventional chromatography. All the protein purifications were carried out at 4°C. Site-directed mutagenesis was performed by using a Quik Change site-directed mutagenesis kit (Stratagene).
Size exclusion chromatography.
Size exclusion chromatography was performed at room temperature by using a fast protein liquid chromatography system (Pharmacia Amersham) equipped with a Superdex-200 HR 10/30 column. The column was equilibrated with at least 3 bed volumes of 50 mM Tris-Cl (pH 8.0) supplemented with 150 mM NaCl prior to each run. A typical flow rate of 0.35 ml/min was maintained. Absorbance at 280 nm was measured to monitor elution of the protein from the column.
Crystallization and data collection.
Crystals were grown at room temperature by the hanging drop vapor diffusion method by using a reservoir solution containing 100 mM HEPES (pH 7.5), 25% (wt/vol) polyethylene glycol 3350, and 10% (vol/vol) n-propanol. These conditions were similar to those reported previously by Adir et al. (1). Crystals were frozen after cryoprotection by addition of 10% (vol/vol) glycerol to the mother liquor. X-ray diffraction data for Cpn60.2 were recorded for two crystals under standard cryogenic conditions at Beamline BL1 at the Protein Structure Factory, BESSY. The data were processed, scaled, and merged with DENZO and SCALEPACK (30).
Structure determination and refinement.
The structure of Cpn60.2 was solved by the molecular replacement method by using the program AMoRe (28) with the crystal structure of E. coli (GroEL-K+-Mg2+-ATP)14 (Protein Data Bank code 1KP8) as the search model (40). The search model was a monomer of E. coli GroEL. The rotation and translation parameters of the second molecule were obtained by fixing those of the first molecule. Refinement was performed by using REFMAC5 (26) and CNS (6). The initial refinement consisted of rigid body, positional, and B-factor refinement. Models were rebuilt in O (18) by using σA-weighted 2Fo-Fc and Fo-Fc electron density maps. The progress of refinement was monitored by using crystallographic R factor, and parameterization at each stage was monitored throughout the study by using Rfree from a set of randomly selected 5% reflections. The quality of the structure was monitored by using PROCHECK (22). Refined coordinates and structure factors have been deposited in the Protein Data Bank under accession code 1SJP.
RESULTS
Purification and determination of quaternary structure.
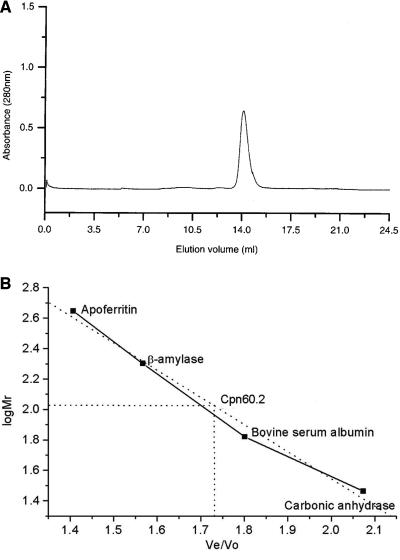
Cpn60.2 was expressed in large quantities and had a protomer molecular mass of ∼60 kDa. Intriguingly, during size exclusion chromatography, the purified Cpn60.2 eluted at a molecular mass of 110 kDa, unlike the tetradecameric E. coli GroEL (Fig. 1). This finding was surprising since Cpn60.2 exhibits significant sequence homology with other chaperonins that exist as higher oligomers. Moreover, sequence comparison with other Cpn60 homologs did not reveal any key alterations at the interface residues. Thus, the loss of the tetradecameric state of M. tuberculosis Cpn60.2 was perplexing. Native polyacrylamide gel electrophoresis showed that the protein migrated as a lower oligomer (32). Moreover, attempts to reconstitute native-like tetradecamers in vitro or by using different purification procedures did not yield the tetradecameric protein (32). In order to confirm that the His6 tag at the C terminus did not interfere with oligomerization, the untagged protein was purified by conventional chromatography, and it did not form the canonical tetradecamer (32).
FIG. 1.
Size exclusion chromatogram of Cpn60.2. (Top panel) Elution profile of purified Cpn60.2 on a Superdex-200 HR 10/30 column, showing the homogeneity of the purified protein. (Bottom panel) Elution of standard molecular weight marker proteins, shown as a function of log Mr. Ve and Vo are the elution and void volumes, respectively. A comparison with standard molecular mass markers showed that Cpn60.2 elutes as a 110-kDa species.
Crystallization, structure determination, and refinement.
Cpn60.2 was crystallized in space group P21. The Matthews number suggested the presence of a dimer in the crystallographic asymmetric unit. This corroborated our gel filtration chromatography results, although the concentration of protein used for crystallization was sufficiently high (200 μM) to be able to form an oligomer. Useful diffraction data could be collected with crystals soaked in glycerol for a short period and frozen in liquid N2. The protocol for the cryo conditions was arrived at after several trials of soaking the crystals in other cryoprotection solutions for different times. Soaking for a long time in glycerol or other cryoprotectants decreased the diffraction quality significantly. Data from two crystals could be merged well, with an overall completeness of more than 99% (Table 1).
TABLE 1.
Data collection and refinement statistics
| Type of data | Parametera | Valueb |
|---|---|---|
| Data collection statisticsc | a (Å) | 58.7 |
| b (Å) | 113.8 | |
| c (Å) | 79.5 | |
| β (°) | 94.6 | |
| Matthews no. | 2.4 | |
| No. of unique reflections | 17,489 | |
| Redundancy | 5 (3) | |
| Completeness (%) | 99.5 (97) | |
| Average I/σ(I) | 20.2 (2.6) | |
| Rmerge (%) | 10.0 (41.5) | |
| Wavelength (Å) | 0.91840 | |
| Refinement statisticsd | No. of protein atoms | 6,595 |
| Resolution range (Å) | 50-3.2 | |
| Rwork (%) | 24.1 | |
| Rfree (%) | 28.5 | |
| Rmsd bond distances (Å) | 0.008 | |
| Rmsd bond angles (°) | 1.003 | |
| Average B factor (Å2) | 82 | |
| Main chain (Å2) | 80 | |
| Side chain (Å2) | 84 | |
| Ramachandran plot most favored (%) | 87.2 |
Rmerge = Σ|Ihkl − 〈Ihkl〉|/Σ|Ihkl|. Rwork = Σ|Fobs − Fcalc|/Σ|Fobs|. Rfree is the R factor calculated from a subset of reflections excluded from refinement.
The values in parentheses are data for the highest-resolution shell.
The space group is P21.
The Protein Data Bank code is 1SJP.
Molecular replacement calculations with E. coli GroEL monomers as the search models were attempted. E. coli GroEL is known to exist in two distinct conformational states, one corresponding to the cis form (the GroES-bound state) and one corresponding to the trans form (the uncomplexed form). Several crystal structures of GroEL are available in the Protein Data Bank. However, none of these yielded a convincing molecular replacement solution. The top rotation solutions were at best 4 σ levels above the background and did not stand out in comparisons with other solutions. Similarly, the translation searches also did not yield any significantly distinct solution. The difficult molecular replacement problem was solved by comparing rotation and translation functions of the different models.
Comparison of rotation functions yielded one consistent orientation as one of the top solutions in all the models. However, translation searches with this orientation and with different models did not yield a satisfactory solution. The best solution for the translation function was obtained by using the E. coli (GroEL-K+-Mg2+-ATP)14 complex structure (Protein Data Bank code 1KP8) as the model. The 1KP8 model was oriented according to the common orientation at the start of translation searches. The best translation peak showed a correlation coefficient and R factor of 0.65 and 0.56, respectively. This solution was only marginally better than the other possible solutions. This solution, however, did not yield satisfactory placement of the second molecule. In the final analysis, the next best translation solution of the first molecule yielded its correct placement. The correlation coefficient and R factor improved to 0.68 and 0.53, respectively, when the second molecule was also placed appropriately in the unit cell.
The first round of rigid body refinement showed a major conformational change from the starting coordinates. The apical and intermediate domains, as defined for the E. coli GroEL structure, showed a dramatic conformational change with respect to the equatorial domain. The movement was observed in both the molecules present in the asymmetric unit. After several iterative rounds of refinement and manual rebuilding in O (18), the R and Rfree converged to 24.1 and 28.5%, respectively. The final structure showed an excellent geometry, with 87.2% of the residues within the most favored regions of the Ramachandran plot (Table 1).
Overall structure.
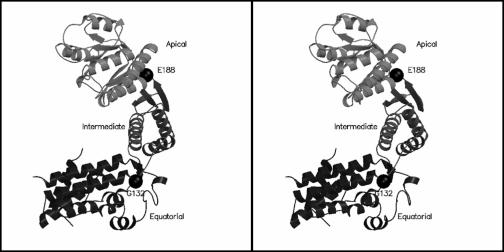
The final refined structure consists of residues 60 to 514 in chain A and residues 62 to 514 in chain B. No interpretable density could be obtained for the first 59 residues in chain A, the first 61 residues in chain B, and residues 80 to 87 and 515 to 539 in both chains. Hence, these regions were not modeled. Therefore, the final model comprises 447 amino acid residues in chain A and 445 residues in chain B. The overall molecular architecture of the protein, as shown in Fig. 2, was similar to that of E. coli GroEL with the protein comprising three distinct domains (equatorial, intermediate, and apical) and with the domain nomenclature like that for E. coli GroEL (4).
FIG. 2.
Stereo view of the overall molecular architecture of Cpn60.2. Cpn60.2 has a fold similar to that of E. coli GroEL, and the protein is divided into three domains (equatorial, intermediate, and apical). The hinge points resulting in movement of the domains are indicated by spheres. Figures 2, 3b, and 5a were generated by using MOLSCRIPT (20) and Raster3D (24).
Previously, size exclusion chromatography suggested that the molecule is a homogenous dimer (Fig. 1). The buried accessible area between two molecules in an asymmetric unit was estimated to be 1,384 Å2. This value, although not very high for the formation of a strong dimer, appears to be comparable to the typical values obtained for protein oligomers. Dimerization is mediated by several main chain and side chain charge-charge interactions across the two subunits. Several van der Waals, hydrophobic, and hydrogen-bonding interactions also occur.
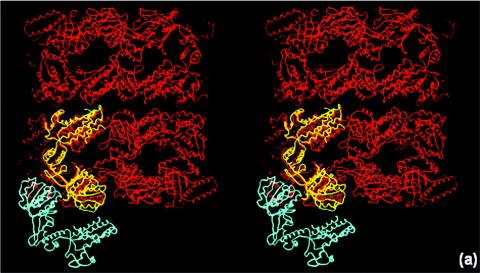
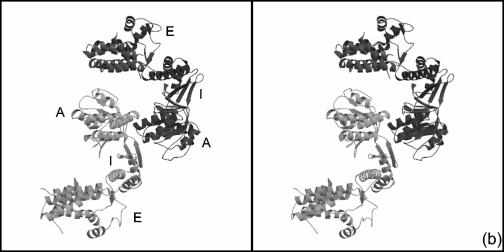
There are two types of monomer-monomer interactions in a GroEL tetradecamer; one type is within a heptameric ring, while the other is between subunits across the two heptameric rings. In both these types of associations, the principal protein-protein interactions are formed through the equatorial domains. The Cpn60.2 dimerization does not resemble any of the two possible GroEL intersubunit associations (Fig. 3a). Moreover, the two molecules in the asymmetric unit of Cpn60.2 interact with each other principally through their apical domains (Fig. 3b). Thus, the mode of dimerization of the Cpn60.2 molecules is much different from that observed in E. coli GroEL.
FIG. 3.
Unusual mode of dimerization in Cpn60.2 in stereo. (a) Superposition of Cpn60.2 (cyan and yellow) and E. coli GroEL (red), showing the difference in the monomer-monomer association between the two proteins. The Cpn60.2 dimer resembles neither of the two modes of GroEL dimerization (see text for details). (b) Dimerization of Cpn60.2, showing the intermonomer association mediated via the apical domains, unlike in GroEL. The two monomers are indicated by different shades of gray. A, I, and E represent the apical, intermediate, and equatorial domains, respectively.
Comparison of the two Cpn60.2 molecules.
Overall superposition of the two molecules in the dimer showed a somewhat high root mean square deviation (rms) deviation of 1.71 Å for 439 equivalent Cα atoms. The high rms value resulted from differences in the relative orientations of different domains with respect to one another, with the largest deviations occurring in the apical domain. Of the regions that deviate the most, residues 349 to 359 are involved in crystal packing. Crystal packing requirements thus seem to enforce local conformational changes in the two molecules within the asymmetric unit. Individual domain-wise superposition resulted in lower rms deviations. The rms deviation for the equatorial domain was 0.74 Å for 170 atoms, the rms deviation for the intermediate domain was 0.62 Å for 92 atoms, and the rms deviation for the apical domain was 0.73 Å for 183 atoms. The individual domains of Cpn60.2 thus seem to exhibit a high degree of similarity to each other.
Comparison of Cpn60.2 with E. coli GroEL.
Several crystal structures of E. coli GroEL, both in the uncomplexed state and as complexes with GroES, ADP, and ATP-γ-S, are available for comparison. Among the available crystal structures, the (GroEL-K+-Mg2+-ATP)14 complex showed the best match with Cpn60.2. The rms deviation between the two structures was 1.81 Å for 339 equivalent Cα atoms. A comparison of individual domains of the two proteins showed better superposition, with rms deviations of 0.86 Å for the equatorial domain (146 atoms), 1.28 Å for the intermediate domain (88 atoms), and 0.78 Å for the apical domain (181 atoms). The largest deviation was observed in helix-spanning residues 60 to 79. The orientation of this helix was dramatically changed compared to E. coli GroEL. The change was evident in the early stages of refinement, and there was a clear indication from the electron density maps (Fig. 4).
FIG. 4.
Conformational change in the 60-79 helix of Cpn60.2. A stereo view of the ‖Fo|-|Fc‖ map contoured at 1σ level clearly shows the different orientation of this helix. The thin line indicates the Cα trace of GroEL, while Cpn60.2 is indicated by the thick line. This figure was generated by using Bobscript (11).
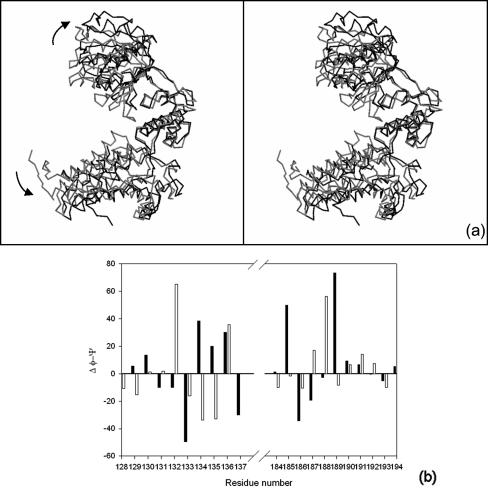
Cpn60.2 had a more open tertiary conformation than E. coli GroEL (Fig. 5a). This open conformation resulted from a large en bloc movement of the apical domain away from the equatorial domain and a concomitant inward movement of the intermediate domain. These movements were considered rigid body movements across two distinct hinge points, one at the interface of the apical and intermediate domains and the other at the interface of the intermediate and equatorial domains. The hinge points were identified by inspection of difference dihedral angle plots. At the interface of the intermediate and equatorial domains, the plot showed maximal deviation at the ψ and φ of G132 and A133, respectively (Fig. 5b). Changes in the dihedral angles at residues G132 and A133 may be considered a rigid body rotation across the virtual Cα-Cα bond between these residues. These alterations in the dihedral angles resulted in a 14° rotation of the intermediate domain toward the equatorial domain. A concomitant 19° swing of the apical domain relative to the intermediate domain resulted in an outward movement of the apical domain at residues E188 and G189. These differences once again may be considered as a rotation across the virtual Cα-Cα bond between E188 and G189. Corresponding residues in the E. coli GroEL-GroES complex bring about similar movements across the three domains. The variation in the relative arrangement of different domains in Cpn60.2, compared with GroEL, might have arisen due to the different quaternary association of subunits.
FIG. 5.
Domain movement in Cpn60.2 as detected by superposition and difference φ-ψ plot. (a) Cα traces of Cpn60.2 and GroEL superposed with respect to the intermediate domain show significant domain movement. The comparison revealed the outward movement of the apical and equatorial domains in Cpn60.2 (black) with respect to GroEL (gray). The outward movement is more prominent for the apical domain. (b) Difference in φ-ψ angles of Cpn60.2 and GroEL identifies the putative hinge points leading to domain movement. The solid bars indicate Δφ, while the open bars indicate Δψ. The maximal deviations occur at residues 132 and 133 between the equatorial and intermediate domains and at residues 188 and 189 between the intermediate and apical domains.
The interfaces between the domains are remarkably populated with charged residues. Sequence comparison of the known Cpn60s indicated a high level of conservation of these charged residues, suggesting the importance of the charged interfaces (5). In the Cpn60.2 structure, these residues exhibit extensive interaction networks. R318 of the apical domain is involved in interdomain salt bridges with E185 and E175 of the intermediate domain. The orientations of the guanidino group of R318 and carboxylates of glutamates are determined by their interactions with E331 and K377, respectively. The other region where extensive charge-charge interactions are observed involves residues E394 and K390 of the intermediate domain and residues R207 and E209 of the apical domain. Although K390 does not interact directly with E209, these residues juxtapose the charge groups of E394 and R207 in an appropriate orientation for strong interactions. The equivalent residues in E. coli, however, do not show similar interactions. The loss of these interactions in E. coli seems to have arisen due to the closed tertiary conformation of the protein. Interestingly, the charge network in Cpn60.2 continues across the other monomer with the involvement of R281 and D358 from the other subunit. The extensive charge-charge interaction networks therefore might stabilize the open tertiary conformation of Cpn60.2.
Surface properties of Cpn60.2.
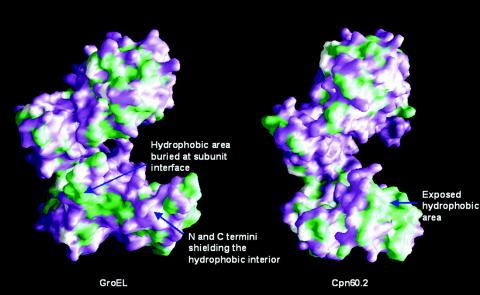
The crystal structure of Cpn60.2 reveals that large hydrophobic regions are exposed on the surface. At least two exposed hydrophobic patches are present on the surface of the protein (Fig. 6). Intriguingly, however, the exposed hydrophobic patches do not lead to self-aggregation of Cpn60.2. One of the two exposed hydrophobic regions lies in the equatorial domain and may arise due to the uninterpretable electron density of N-terminal residues. An elegant conformational change involving reorientation of the N-terminal helix (residues 60 to 79) (Fig. 4) in both the molecules in the asymmetric unit shields hydrophobic residues of the equatorial domain. In addition, this conformational change appears to promote the packing of the helices spanning residues 87 to 107 and 60 to 79. However, some amount of hydrophobic surface still remains exposed. Another hydrophobic patch occurs in the apical domain. The apical domain in GroEL has been shown to bind substrate proteins. Thus, the two hydrophobic patches in each of the Cpn60.2 monomers might have a role in binding unfolded polypeptides, thereby preventing their misfolding and aggregation.
FIG. 6.
Comparison of surface properties of the Cpn60.2 monomer with those of the GroEL monomer. Hydrophobic regions exposed on the surface of GroEL and Cpn60.2 are green. While in GroEL the hydrophobic surfaces of a monomer are buried upon oligomeric assembly of the protein, in Cpn60.2 these regions remain exposed even upon dimerization. Thus, the regions might bind nonnative polypeptides, preventing their aggregation. This figure was generated by using GRASP (29).
Consequences of disordered N-terminal residues in the ATP-binding pocket.
The equatorial domain spans the nucleotide-binding pocket in E. coli GroEL (3). Several charged and small hydrophobic residues in the region from residue 480 to residue 500 (E. coli residue numbers) contribute to the ATP-binding site in GroEL. In addition, residues at the N terminus of the protein also contribute to this site. For example, G31 is crucial for interaction with the α-phosphate of ATP, while P33 is involved in hydrophobic interactions with the adenosine moiety. Moreover, residues D87 and T91 in GroEL are involved in hydrogen bonding with Mg2+ and β-phosphate of ATP. The uninterpretable density of N-terminal residues leads to the loss of the nucleotide-binding pocket in Cpn60.2. Furthermore, the electron density of the loop (residues 83 to 90) involved in β-phosphate binding was also not interpretable. These structural changes suggest that Cpn60.2 might have altered ATP-binding properties. Indeed, when examined, Cpn60.2 behaved like a very weak ATPase (32). Thus, despite sequence conservation of ATP-binding residues, due to quaternary structural alterations Cpn60.2 appears to have lost ATPase activity.
The unexpected quaternary structure of the protein that we observed in our crystal structure, the loss of ATPase activity, and the unusual exposure of a hydrophobic surface area strongly indicate that Cpn60.2 has unique structural properties. The gene encoding this protein is not present in the groESL operon, unlike the genes in other bacteria, and this leads to speculation that this protein may not be a canonical chaperonin. The unusual structure might therefore suggest novel functional properties for Cpn60.2.
Structural comparison with HslVU protease.
M. leprae Cpn60.2 has recently been shown to possess HslVU-like proteolytic activity (31). Considering the high sequence identity (95%) between the M. tuberculosis and M. leprae Cpn60.2 molecules, the crystal structure of M. tuberculosis Cpn60.2 presents us with an opportunity to probe the possible role of Cpn60.2 in proteolysis. Portaro et al. (31), on the basis of 20% sequence similarity with HslVU protease, suggested two catalytic triads, T136-K168-Y264 and T375-K409-S502 (corresponding to residues T88-K120-Y216 and T327-K361-S454 in M. tuberculosis). The Cpn60.2 crystal structure, however, revealed that the residues suggested to be part of the triad were very far apart in space. In the first putative triad, T88 and K120 are located in the equatorial domain, 23 Å from one another, while Y216 is present in the apical domain and is 44 and 54 Å from residues T88 and K120, respectively. The structure does not seem to suggest any possible conformational movement to bring these three residues together for activity. In the second suggested triad, while residues T327 and K361 are present in the apical domain, 18 Å from one another, residue S454, in the equatorial domain, is 42 and 50 Å from T327 and K361, respectively. Thus, although the suggested triads might not be responsible for the proteolytic role of Cpn60.2, some other residues might contribute to this activity. We explored the likelihood that residues S177, T179, and K377 in Cpn60.2 might constitute the catalytic triad due to their spatial proximity. Structural superposition of the triads of Cpn60.2 and HslVU (2), however, did support this possibility.
Instability of the N-terminal region of Cpn60.2.
Matrix-assisted laser desorption ionization—time of flight analysis of the crystals indicated a molecular mass of ∼53 kDa, which was smaller than the expected size (data not shown). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the purified Cpn60.2 showed that the protein migrated as a doublet, with the high-molecular-mass band corresponding to ∼60 kDa. N-terminal sequencing revealed that the high-molecular-mass band sequence is AKTIAYD, while the sequence of the low-molecular-mass band is WGAPTITNTG. The former sequence corresponds to the intact protein, while the latter sequence corresponds to Cpn60.2 truncated at K41. In an attempt to prevent degradation of Cpn60.2, several combinations of protease inhibitors, such as leupeptin, aprotinin, soybean trypsin inhibitor, phenylmethylsulfonyl fluoride, and EDTA, were included during purification. However, none of the inhibitors, which were used separately or in a cocktail, prevented proteolytic cleavage. Degradation of the protein occurred at a tandem KK repeat, suggesting that there was proteolysis by a trypsin-like protease, and therefore the K41 residue was changed to serine or alanine. However, neither of the two mutants showed enhanced stability. Interestingly, the homologous Hsp65 molecule of M. leprae has been shown to cleave at a consensus site of tandem basic residues (31). We therefore hypothesize that the degradation of M. tuberculosis Cpn60.2 might be due to autoproteolysis.
DISCUSSION
E. coli GroEL has been one of the best-characterized ATP-dependent molecular chaperones, and its chaperoning ability is intricately related to its oligomeric state. M. tuberculosis possesses two copies of GroEL homologs, which are designated Cpn60.1 and Cpn60.2. Levels of sequence identity with GroEL of more than 50% for both the proteins suggest that these proteins might also function as molecular chaperones in M. tuberculosis. Interestingly, the mycobacterial chaperonins have previously been shown to be secreted into the extracellular environment, although their role as molecular chaperones is limited to the cytosol (10, 14). The existence of chaperonins in the extracellular environment thus suggests a possible alternate functional role. For example, Cpn10 has been hypothesized to bind divalent cations in the extracellular milieu (37, 38). Both the Cpn60s of M. tuberculosis are known to be strong antigens responsible for inducing humoral and cellular immune responses (43). The two chaperonins are also known to be involved in cell signaling at very low concentrations (33). It is thus possible that both the chaperonins have multiple functions.
Cpn60.2, also known as Hsp65 or the 65-kDa antigen, can elicit a strong delayed-type hypersensitivity reaction in experimental animals infected with M. tuberculosis (35). This protein is one of the major immunoreactive proteins of M. tuberculosis, and its immunodominant epitopes have recently been identified. It has also been explored for possible inclusion in a recombinant vaccine (39). Moreover, Cpn60.2 has been shown to enhance cross presentation of exogenous proteins by dendritic cells to the CD8+ T cells (7). Thus, the crystal structure of Cpn60.2 presents an opportunity to correlate the interesting structure-function properties of this protein.
When mapped onto the three-dimensional structure, the recently identified T-cell epitopes of M. leprae and M. tuberculosis Hsp65s spanned the equatorial and intermediate domains (27). These epitopes are the most exposed regions in the dimeric Cpn60.2 structure. Interestingly, this region is buried in the monomer-monomer interface of the oligomeric GroEL molecule. The apical domain, which is also the least accessible of the three domains due to the peculiar oligomerization mode, however, possessed only one immunodominant epitope. The occurrence of the immunodominant peptides in the exposed equatorial domain and their absence from the buried apical domain are intriguing. This is because the T-cell epitopes are not required to be surface exposed since their recognition involves processing of polypeptides prior to their loading onto the major histocompatibility complex molecules. Further studies of this important antigen could highlight the correlation between the immunodominance of peptides and their structural disposition.
In addition to their strong antigenicity, heat shock proteins have also been shown to be associated in vivo with a large repertoire of cellular peptides. The Hsp-bound peptides are presented to the immune system in a complex with the major histocompatibility complex class I molecules. This interesting role of heat shock proteins as antigen presenters and carriers has been demonstrated for the eukaryotic heat shock proteins. The exposure of hydrophobic surfaces on Cpn60.2 leads us to hypothesize that M. tuberculosis Cpn60.2 might also play such a role. This might be achieved by binding endogenous peptides by nonspecific interactions with the exposed hydrophobic regions. Thus, Cpn60.2 might serve a dual role, as a potent immunogenic molecule on its own and as a carrier of immunogenic peptides.
In conclusion, we obtained a glimpse into the structure of a lower oligomeric form of chaperonin 60. While many previous studies have shown that Cpn60.2 exhibits high antigenicity and cytokine stimulation abilities, the structural basis of these abilities was unknown. The presence of these proteins in the extracellular milieu and the very low concentrations at which they exist in the extracellular medium suggest that the proteins can exist in a lower oligomeric form. The lower oligomeric state might therefore represent the functionally relevant form for cytokine stimulation. Moreover, since the cpn60.2 gene does not form part of the canonical groESL operon, the lower oligomer might be the natural state of the protein. Since it would be difficult to crystallize proteins at low concentrations, the dimeric form that we obtained fortuitously serves as a good model to understand the cell signaling properties of Cpn60.2. The crystal structure of Cpn60.2 in its dimeric state thus is important for understanding the immunological and cell signaling properties of this protein.
Acknowledgments
We thank Anthony Coates and Douglas Young for useful discussions. The cosmid DNA library of M. tuberculosis H37Rv was a generous gift from Stewart Cole. We acknowledge the overall support of S. E. Hasnain and the support of the beam line staff at BESSY, Berlin, Germany, for diffraction and data collection.
Financial support for this work was provided by the Department of Biotechnology. R.Q. is a Council of Scientific and Industrial Research Senior Research Fellow, and S.C.M. is a Wellcome Trust International Senior Research Fellow.
REFERENCES
- 1.Adir, N., E. Dobrovetsky, I. Shafat, C. Cohen, and Y. Kashi. 2002. Isolation, purification and preliminary X-ray characterization of Cpn60-2 (65 kDa heat-shock protein) from Mycobacterium tuberculosis. Acta Crystallogr. Sect. D 58:1474-1475. [DOI] [PubMed] [Google Scholar]
- 2.Bochtler, M., C. Hartmann, H. K. Song, G. P. Bourenkov, H. D. Bartunik, and R. Huber. 2000. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403:800-805. [DOI] [PubMed] [Google Scholar]
- 3.Boisvert, D. C., J. Wang, Z. Otwinowski, A. L. Horwich, and P. B. Sigler. 1996. The 2.4Å crystal structure of the bacterial chaperonin GroEL complex with ATP-γ-S. Nat. Struct. Biol. 3:170-177. [DOI] [PubMed] [Google Scholar]
- 4.Braig, K., Z. Otwinowski, R. Hegde, D. C. Boisvert, A. Joachimiak, A. L. Horwich, and P. B. Sigler. 1994. The crystal structure of the bacterial chaperonin GroEL at 2.8Å. Nature 371:578-586. [DOI] [PubMed] [Google Scholar]
- 5.Brocchieri, L., and S. Karlin. 2000. Conservation among HSP60 sequences in relation to structure, function and evolution. Protein Sci. 9:476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Chen, K. J., J. Lu, L. Wang, and Y. H. Gan. 2004. Mycobacterial heat shock protein 65 enhances antigen cross-presentation in dendritic cells independent of Toll-like receptor 4 signaling. J. Leukoc. Biol. 75:260-266. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S., A. M. Roseman, A. S. Hunter, S. P. Wood, S. G. Burston, N. A. Ranson, A. R. Clarke, and H. R. Saibil. 1994. Location of a folding protein and shape changes in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature 371:261-264. [DOI] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.De Bruyn, J., K. Soetaert, P. Buyssens, D. I. Calonne, J. L. Coene, X. Gallet, R. Brasseur, R. Wattiez, P. Falmagne, H. Montrozier, M. A. Lanéelle, and M. Daffé. 2000. Evidence for specific and non-covalent binding of lipids to natural and recombinant Mycobacterium bovis BCG Hsp60 proteins, and to the Escherichia coli homologue GroEL. Microbiology 146:1513-1524. [DOI] [PubMed] [Google Scholar]
- 11.Esnouf, R. M. 1999. Further additions to Molscript version 1.4 including reading and contouring of electron density maps. Acta Crystallogr. Sect. D 55:938-940. [DOI] [PubMed] [Google Scholar]
- 12.Ewalt, K. L., J. P. Hendrick, W. A. Houry, and F. U. Hartl. 1997. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell 90:491-500. [DOI] [PubMed] [Google Scholar]
- 13.Fayet, O., T. Ziegelhoffer, and C. Georgopoulos. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures, J. Bacteriol. 171:1379-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fossati, G., G. Izzo, E. Rizzi, E. Gancia, D. Modena, M. L. Moras, N. Niccolai, E. Giannozzi, O. Spiga, L. Bono, P. Marone, E. Leone, F. Mangili, S. Harding, N. Errington, C. Walters, B. Henderson, M. M. Roberts, A. R. Coates, B. Casetta, and P. Mascagni. 2003. Mycobacterium tuberculosis chaperonin 10 is secreted in the macrophage phagosome: is secretion due to dissociation and adoption of a partially helical structure at the membrane? J. Bacteriol. 185:4256-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukami, T. A., M. Yohda, H. Taguchi, M. Yoshida, and K. Miki. 2001. Crystal structure of chaperonin-60 from Paracoccus denitrificans. J. Mol. Biol. 312:501-509. [DOI] [PubMed] [Google Scholar]
- 16.Hartl, F. U. 1996. Molecular chaperones in cellular protein folding. Nature 381:571-580. [DOI] [PubMed] [Google Scholar]
- 17.Hunt, J. F., A. J. Weaver, S. Landry, L. Gierasch, and J. Deisenhofer. 1996. The crystal structure of the GroES co-chaperonin at 2.8Å resolution. Nature 379:37-45. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved method for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 19.Kong, T. H., A. R. M. Coates, P. D. Butcher, C. J. Hickman, and T. M. Shinnick. 1993. Mycobacterium tuberculosis expresses two chaperonin-60 homologs. Proc. Natl. Acad. Sci. USA 90:2608-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 21.Langer, T., G. Pfeifer, J. Martin, W. Baumeister, and F. U. Hartl. 1992. Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein within its central cavity. EMBO J. 11:4757-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 23.Lewthwaite, J. C., A. R. M. Coates, P. Tormay, M. Singh, P. Mascagni, S. Poole, M. Roberts, L. Sharp, and B. Henderson. 2001. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a CD14-binding domain. Infect. Immun. 69:7349-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merritt, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 25.Monahan, I., J. Betts, D. Banerjee, and P. Butcher. 2001. Differential expression of mycobacterial proteins following phagocytosis by macrophages. Microbiology 147:459-471. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr. Sect. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 27.Mustafa, A. S., K. E. Lundin, R. H. Meloen, T. M. Shinnick, and F. Oftung. 1999. Identification of promiscuous epitopes from the mycobacterial 65-kilodalton heat shock protein recognized by human CD4+ T cells of the Mycobacterium leprae memory repertoire. Infect. Immun. 67:5683-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. Sect. A 50:157-163. [Google Scholar]
- 29.Nicholls, A., K. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11:281-296. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 31.Portaro, F. C. V., M. A. F. Hayashi, L. J. de Arauz, M. S. Palma, M. T. Assakura, C. L. Silve, and A. C. M. de Camargo. 2002. The Mycobacterium leprae hsp65 displays proteolytic activity. Mutagenesis studies indicate that the M. leprae hsp65 proteolytic activity is catalytically related to the HslVU protease. Biochemistry 41:7400-7406. [DOI] [PubMed] [Google Scholar]
- 32.Qamra, R., V. Srinivas, and S. C. Mande. 2004. Mycobacterium tuberculosis GroEL homologues unusually exist as lower oligomers and retain the ability to suppress aggregation of substrate proteins. J. Mol. Biol. 342:605-617. [DOI] [PubMed] [Google Scholar]
- 33.Ranford, J. C., A. R. M. Coates, and B. Henderson. 2000. Chaperonins are cell-signaling proteins: the unfolding biology of molecular chaperones. Exp. Rev. Mol. Med. 15(September):1-17. [DOI] [PubMed] [Google Scholar]
- 34.Rye, H. S., S. G. Burston, W. A. Fenton, J. M. Beechem, Z. Xu, P. B. Sigler, and A. L. Horwich. 1997. Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature 388:792-798. [DOI] [PubMed] [Google Scholar]
- 35.Silva, C. L., R. L. Pietro, A. Januario, V. L. Bonato, V. M. Lima, M. F. da Silva, and D. B. Lowrie. 1995. Protection against tuberculosis by bone marrow cells expressing mycobacterial hsp65. Immunology 86:519-524. [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 37.Taneja, B., and S. C. Mande. 2001. Metal ions modulate the plastic nature of Mycobacterium tuberculosis chaperonin-10. Protein Eng. 14:391-395. [DOI] [PubMed] [Google Scholar]
- 38.Taneja, B., and S. C. Mande. 2002. Structure of Mycobacterium tuberculosis chaperonin-10 at 3.5Å resolution. Acta Crystallogr. Sect. D 58:260-266. [DOI] [PubMed] [Google Scholar]
- 39.Tascon, R. E., M. J. Colston, S. Ragno, E. Stavropoulos, D. Gregory, and D. B. Lowrie. 1996. Vaccination against tuberculosis by DNA injection. Nat. Med. 2:888-892. [DOI] [PubMed] [Google Scholar]
- 40.Wang, J., and D. C. Boisvert. 2003. Structural basis for Groel-assisted protein folding from the crystal structure of (Groel-KMgATP)14 at 2.0Å resolution. J. Mol. Biol. 327:843-855. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Z., A. L. Horwich, and P. B. Sigler. 1997. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388:741-750. [DOI] [PubMed] [Google Scholar]
- 42.Young, D. B., and T. R. Garbe. 1991. Heat shock proteins and antigens of Mycobacterium tuberculosis. Infect. Immun. 59:3086-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zügel, U., and S. H. E. Kaufmann. 1999. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 12:19-39. [DOI] [PMC free article] [PubMed] [Google Scholar]